Significance
Nutrition and the body clock are deeply intertwined, both impinging on our physiological health. Food composition can dramatically rewire peripheral clock metabolism; however, whether food challenges can impact circadian metabolism of the master clock in the suprachiasmatic nucleus (SCN) or other brain areas has not been fully explored. Here we analyzed the complete diurnal metabolome of the SCN and medial prefrontal cortex (mPFC) in mice fed a balanced diet or a high-fat diet (HFD). Strikingly, our data reveal unexpected daily rhythmicity in both SCN and mPFC metabolites that is significantly impacted by HFD in a region-specific manner. Our findings unveil an unsuspected sensitivity of brain clocks to nutrition.
Keywords: circadian clock, suprachiasmatic nucleus, prefrontal cortex, high-fat diet, metabolome reorganization
Abstract
Food is a powerful entrainment cue for circadian clocks in peripheral tissues, and changes in the composition of nutrients have been demonstrated to metabolically reprogram peripheral clocks. However, how food challenges may influence circadian metabolism of the master clock in the suprachiasmatic nucleus (SCN) or in other brain areas is poorly understood. Using high-throughput metabolomics, we studied the circadian metabolome profiles of the SCN and medial prefrontal cortex (mPFC) in lean mice compared with mice challenged with a high-fat diet (HFD). Both the mPFC and the SCN displayed a robust cyclic metabolism, with a strikingly high sensitivity to HFD perturbation in an area-specific manner. The phase and amplitude of oscillations were drastically different between the SCN and mPFC, and the metabolic pathways impacted by HFD were remarkably region-dependent. Furthermore, HFD induced a significant increase in the number of cycling metabolites exclusively in the SCN, revealing an unsuspected susceptibility of the master clock to food stress.
Circadian cycles are controlled by an endogenous clock that contributes to the maintenance of tissue physiology and homeostasis (1). A variety of environmental cues, termed zeitgebers, entrain circadian clocks to the 24-h day–night cycle, allowing organisms to maximize fitness (2). Light is the principal zeitgeber for the suprachiasmatic nucleus (SCN) master clock, whereas entrainment of peripheral clocks is driven in large part by food (3), underscoring the intimate relationship among systemic metabolism, homeostasis, and circadian rhythms (4–9).
The time of feeding and composition of food deeply influence the cyclic functions of peripheral clocks (10–12). Modifications in dietary nutrients, such as in a high-fat diet (HFD), can reprogram the liver clock both transcriptionally and metabolically (13, 14), highlighting the intrinsic plasticity of the clock system. Although light is the dominant entrainment signal for the master clock, some reports suggest that specific feeding regimens could affect the SCN, leading to altered response to light and changes in circadian behavior (12, 15, 16).
The impact of diets on brain functions is intriguing, but its biochemical nature remains obscure (17–20). For example, a HFD influences brain health, leading to impaired cognitive function, anxiety, anhedonic behavior, and a higher incidence of emotional disorders, such as posttraumatic stress disorder (21–25). Furthermore, a ketogenic diet, which significantly rewires circadian rhythms (14), has beneficial effects on epilepsy, Alzheimer’s disease, and memory processes during aging (26–28). Despite these important discoveries, however, how a nutritional challenge affects metabolism in the SCN and in other brain regions has been virtually unexplored.
We sought to investigate the circadian metabolic profile in the brain of mice fed either an HFD or a balanced diet (normal chow [NC]) by high-throughput mass spectrometry (MS) metabolomics. This approach has previously led to the circadian metabolite profiling of nutritional challenges in the liver, serum, and a variety of other tissues to create what we call a circadian metabolome atlas (13, 29, 30).
In this study, we further analyzed the data of our circadian atlas (30) in depth and integrated the metabolite data directly with corresponding gene expression profiles, focusing on the central clock within the SCN (1, 31, 32) and the medial prefrontal cortex (mPFC), a brain region implicated in higher cognitive functions and neuropsychiatric disorders (33). Both the SCN and mPFC displayed remarkable oscillations in a variety of metabolites under NC. Importantly, HFD feeding had a profound influence on cyclic metabolic profiles of the SCN and mPFC, inducing de novo cycling metabolites and impacting their phase and amplitude of oscillation. The effect of HFD was brain region-specific, affecting different numbers and classes of metabolites and metabolic pathways in the SCN or mPFC, indicating that distinct brain areas have a diverse, intrinsic susceptibility to food stress. Furthermore, the circadian reorganization of metabolic pathways in the SCN was accompanied by corresponding changes in gene expression, suggesting a certain degree of coherence between transcriptome and metabolome oscillations in the master clock.
Our results unveil that nutritional challenges have a previously unsuspected impact on circadian brain metabolism, providing a framework for the interpretation of how food may influence neuronal function and ultimately, behavior.
Results
Reorganization of the Brain Metabolome on HFD Feeding.
To explore whether food stress may alter circadian metabolism in the brain, adult male C57BL/6J mice were fed an HFD for 10 wk. The metabolome profile was obtained by reverse phase ultra-high performance liquid chromatography-tandem mass spectrometry as described previously (13, 29, 30). The SCN and mPFC were isolated by punches from fresh brains every 4 h throughout the circadian cycle.
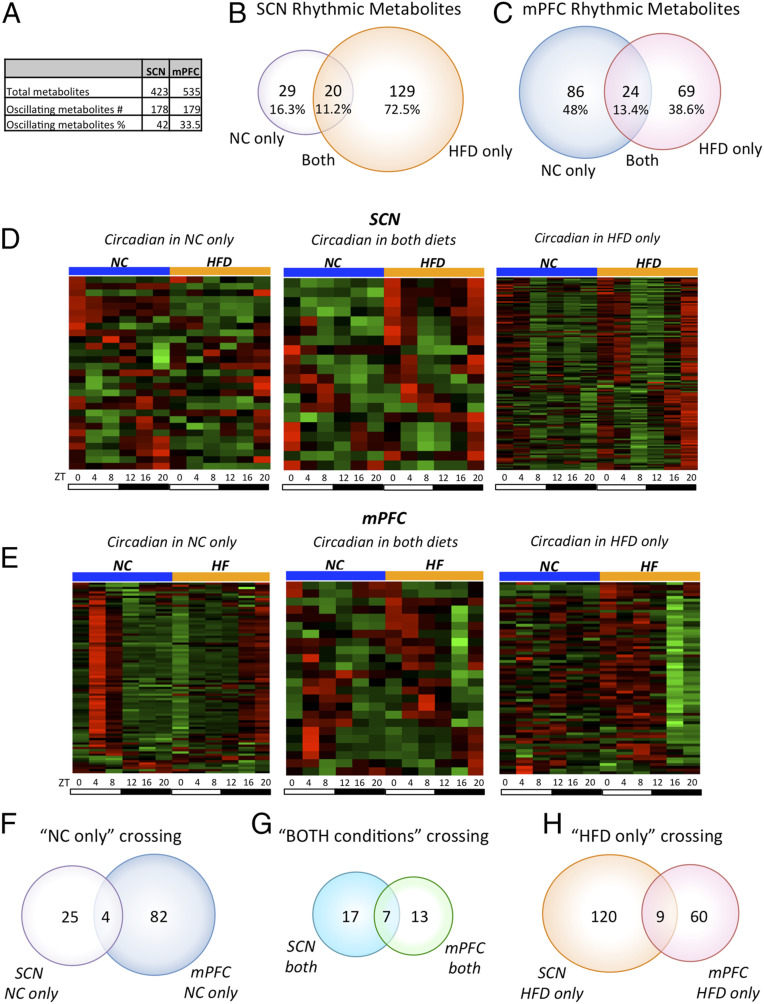
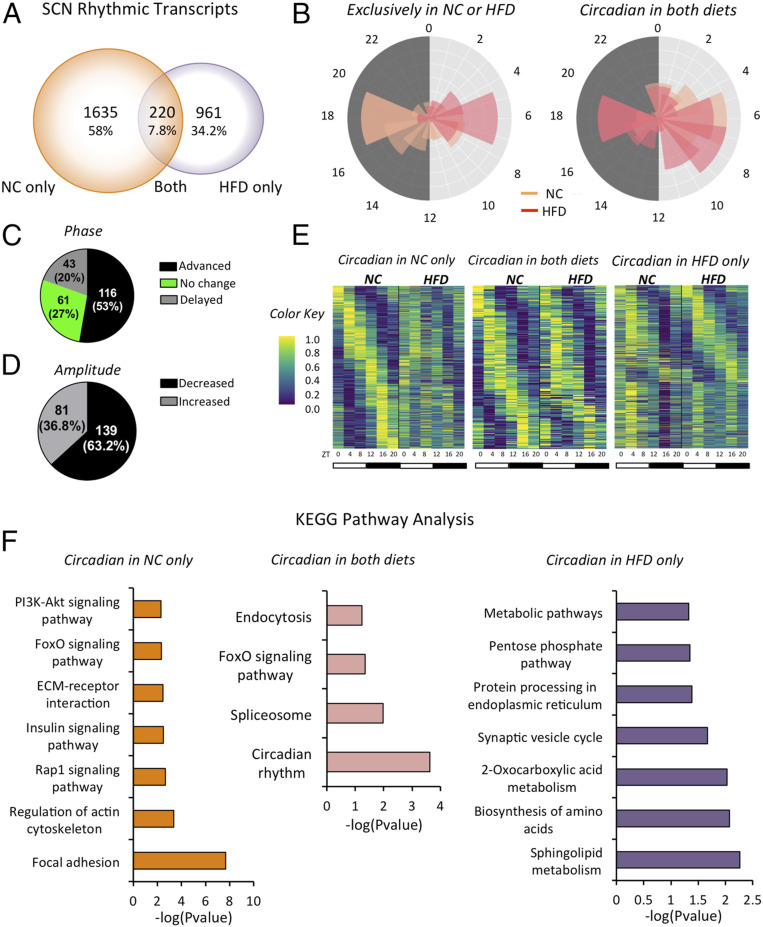
Two-way ANOVA showed that out of 423 total metabolites detected in the SCN, 224 (52.9%) displayed a diet main effect and 134 (31.7%) had a time point main effect (P < 0.05; Dataset S1). In the mPFC, out of 535 total metabolites detected, 175 (32.7%) displayed a diet main effect and 188 (35.1%) had a time point main effect (P < 0.05; Dataset S2). Analysis of rhythmic metabolites was performed using the nonparametric test JTK_CYCLE (34), incorporating a window of 20 to 28 h for the determination of circadian periodicity. A complete list of oscillating metabolites and JTK_CYCLE analysis is provided in Dataset S3 for the SCN and in Dataset S4 for the mPFC; see also circadiomics.ics.uci.edu (35). A high percentage of metabolites were rhythmic in both the SCN and mPFC—42% and 33.5%, respectively, of the total cyclic metabolites (P < 0.05, JTK_CYCLE; Fig. 1A).
Fig. 1.
An HFD remodels the circadian metabolome in the SCN and mPFC. (A) Total number of metabolites and number and percentage of oscillatory metabolites in the SCN and mPFC (JTK_CYCLE, P < 0.05). (B) Venn diagram showing the number and percentage of rhythmic metabolites in the SCN in NC-only, both diets, and HFD-only groups (JTK_CYCLE, P < 0.05). This Venn diagram is also shown in fig. 2 of ref. 30. (C) Venn diagram showing the number and percentage of rhythmic metabolites in the mPFC in the NC-only, both diets, and HFD-only groups (JTK_CYCLE, P < 0.05). This Venn diagram is also shown in fig. 2 of ref. 30. (B and C) Venn diagrams reprinted with permission from ref. 30. (D) Heat maps representing the circadian metabolites in the SCN in the NC-only, both diets, and HFD-only groups (n = 5 per time point per group; JTK_CYCLE, P < 0.05). (E) Heat maps representing the circadian metabolites in the mPFC in the NC-only, both conditions, and HFD-only groups (n = 5 per time point per group; JTK_CYCLE, P < 0.05). (F) Venn diagram for the NC-only group representing the circadian metabolites exclusively in the SCN (SCN NC only), in both the SCN and the mPFC, and exclusively in the mPFC (mPFC NC only). (G) Venn diagram for the both diets group showing the circadian metabolites exclusively in the SCN (SCN both), in both the SCN and the mPFC, and exclusively in the mPFC (mPFC both). (H) Venn diagram for the HFD-only group representing the circadian metabolites exclusively in the SCN (SCN HFD only), in both the SCN and mPFC, and exclusively in the mPFC (mPFC HFD only).
An unexpected finding was the unique impact of nutrition on the diurnal metabolic profiles of the SCN and mPFC. First, in the NC condition, only 29 out of 178 oscillating metabolites (16.3%) were circadian in the SCN (Fig. 1 B and D), while 86 metabolites out of 179 (48%) were cycling in the mPFC (Fig. 1 C and E). Notably, the HFD induced a large number of de novo rhythmic metabolites exclusively in the SCN (129 [72.5%]; Fig. 1 B and D), while driving only 69 de novo cycling metabolites in the mPFC (38.6%; Fig. 1 C and E). The number of circadian metabolites in both feeding conditions was 20 in the SCN and 24 in the mPFC (Fig. 1 B–E).
To gain further insight into how food stress reshapes the diurnal metabolome in a brain region-specific fashion, we crossed the metabolic profiles of the SCN and mPFC. Notably, only a small number of metabolites were commonly rhythmic in both the SCN and the mPFC for each condition analyzed (Fig. 1 F–H and SI Appendix, Tables S1–S3), underscoring the unique daily metabolic signature driven by the nutritional status in distinct brain areas.
HFD Affects Phase and Amplitude of Circadian Metabolites in a Brain Region-Specific Manner.
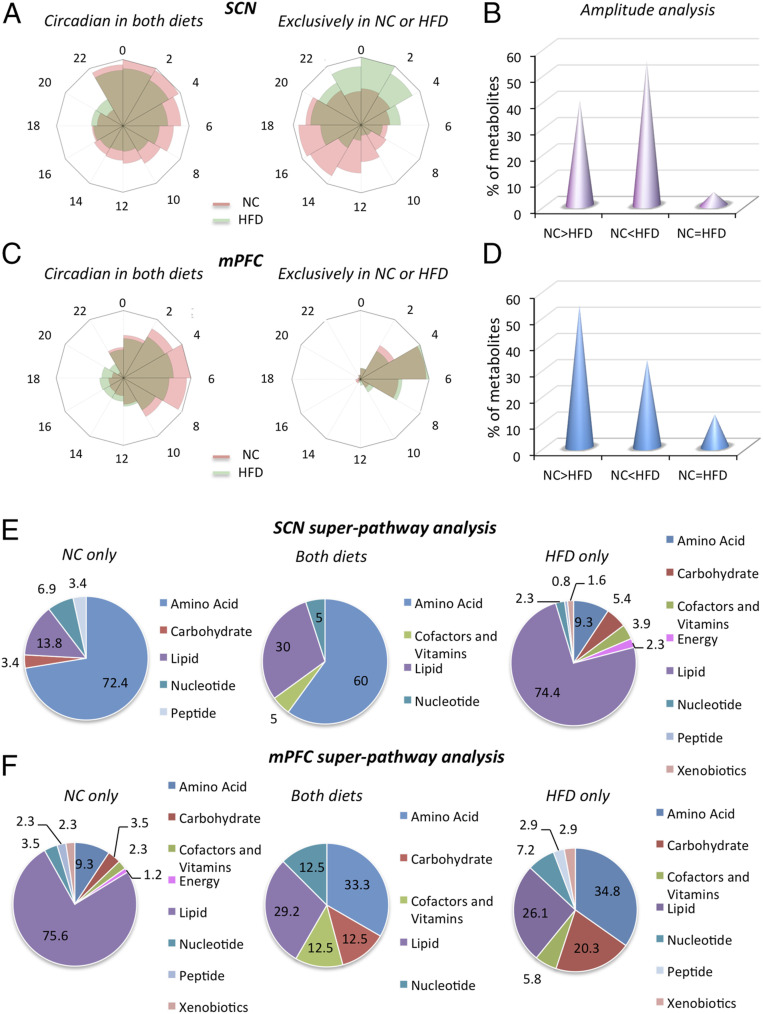
We next analyzed the phase and amplitude of the oscillations of brain metabolites. Rhythmic metabolites in the SCN under both NC and HFD regimens did not show profound differences in the phase of oscillation (Fig. 2A); however, metabolites circadian only with the NC or the HFD displayed a unique partitioning in their phase distribution throughout the daily cycle (Fig. 2A). The HFD-only metabolites tended to peak late during the night or early during the day, while the phase distribution of NC-only metabolites was mainly between zeitgeber time 12 (ZT12) and ZT20 (Fig. 2A). In contrast, there was only a marginal effect of HFD on the phase of mPFC rhythmic metabolites in the both diets group, the NC-only group, and the HFD-only groups (Fig. 2C). Notably, the amplitude of oscillation of rhythmic metabolites in the both diets group was uniquely affected by the HFD in the two brain regions. Of all common SCN oscillating metabolites, 55% exhibited an increase in the amplitude on the HFD, whereas 40% showed a decrease (Fig. 2B). In contrast, the HFD induced an overall decrease in the amplitude of the common cyclic metabolites in the mPFC, with only 33% displaying an increase and 54% showing a decrease (Fig. 2D).
Fig. 2.
The HFD influences the phase, amplitude, and class of oscillatory metabolites in a brain region-specific manner. (A) Radar plots representing the phase distribution of metabolites circadian in both NC and HFD (first plot) and exclusively circadian in NC and HFD (second plot) in the SCN (exclusive plot: Anderson–Darling test, phase distribution significance P < 0.0001). (B) Amplitude analysis of metabolites circadian in both diets in the SCN showing the percentages of metabolites with amplitude lower, higher, or equal with respect to NC condition. (C) Radar plots representing the phase of metabolites circadian in both NC and HFD (first plot) and exclusively circadian in the NC and HFD (second plot) in the mPFC. (D) Amplitude analysis of metabolites circadian in both diets in the mPFC, showing the percentages of metabolites with amplitude lower, equal, or higher with respect to the NC diet. (E) Superpathway analysis of oscillatory metabolites in the NC-only, both diets, and HFD-only groups in the SCN, showing the percentages of the different classes out of the total number of circadian metabolites for each specific condition. (F) Superpathway analysis of oscillatory metabolites in the mPFC in the NC-only, both diets, and HFD-only groups. The pie chart represents the % of the different classes out of the total number of circadian metabolites for each specific condition.
There are important differences in the specific classes of metabolites oscillating differentially in the SCN and mPFC. In the NC-only group, the most abundant class of metabolites rhythmic in the SCN was amino acids, accounting for 72.4% of the oscillators, followed by lipids (13.8%) and nucleotides (6.9%) (Fig. 2E). Strikingly, the HFD extensively modified the SCN circadian metabolome profile. Indeed, the lipid class became predominant (74.4%), while amino acids were reduced to only 9.3% of the cycling metabolites. Moreover, new classes of metabolites became oscillatory, including energy, xenobiotics, and cofactors and vitamins (Fig. 2E).
Remarkably, the mPFC metabolome profile was significantly different from that of the SCN. Indeed, in the NC-only condition, lipids were the prominent oscillators (75.6%; Fig. 2F), and the HFD shifted the composition toward amino acids (34.8%) and carbohydrates (20.3%) (Fig. 2F). Interestingly, not only the composition, but also the phase of oscillations displayed a unique circadian signature in the mPFC and SCN (SI Appendix, Fig. S1). Indeed, the peaks of the various classes were remarkably different between the NC and the HFD and also between the SNC and the mPFC (SI Appendix, Fig. S1 A and B), further emphasizing the profound effect of nutrition on the rhythmicity of unique metabolic pathways in these two distinct brain areas.
The metabolites cycling in the both diets group in the SCN and mPFC were more similar in composition, with high percentages of amino acids and lipids (Fig. 2 E and F); however, carbohydrates were present only in the mPFC (Fig. 2F), again emphasizing the area-specific diurnal signature driven by the nutritional status in the central nervous system.
HFD Impacts Unique Metabolic Pathways in the SCN and mPFC.
Our analysis of specific metabolic pathways led us to examine how an HFD influences the oscillatory metabolome in the SCN and mPFC. Our subpathway analysis revealed unique features of the two brain subregions under both NC and HFD. The most prominent pathways in the SCN were leucine, isoleucine, and valine metabolism; methionine, cysteine, S-adenosylmethionine, and taurine metabolism; and glycine, serine, and threonine metabolism in mice fed NC (SI Appendix, Fig. S2A), as predicted by the high enrichment in the amino acid class (Fig. 2E). The subpathways enriched in the SCN in the HFD-only group were mainly lipid-related: sphingolipid metabolism, phospholipid metabolism, diacylglycerol, and fatty acid metabolism (acyl carnitine) (SI Appendix, Fig. S2C). Notably, the mPFC exhibited almost an inverted trend with respect to the SCN. Indeed, phospholipid, sphingolipid and fatty acid metabolism, diacylglycerol, and lysolipid were highly enriched in the NC-only group (SI Appendix, Fig. S3A). On the other hand, amino acid pathways were mainly oscillating in the HFD-only group, specifically lysine metabolism; glycine, serine, and threonine metabolism; and histidine metabolism (SI Appendix, Fig. S3C). Moreover, as shown by the increase in carbohydrate diurnal fluctuation in the mPFC of HFD-fed mice (Fig. 2F), the nucleotide sugar; fructose, mannose, and galactose metabolism; and glycolysis, gluconeogenesis, and pyruvate metabolism pathways gained oscillation (SI Appendix, Fig. S3C). In the circadian in both diets group, a variety of pathways belonging mainly to amino acid and lipid classes were enriched in the SCN (SI Appendix, Fig. S2B), while the metabolites oscillating in the mPFC were clustered mainly in the amino acid, carbohydrate, and lipid categories (SI Appendix, Fig. S3B).
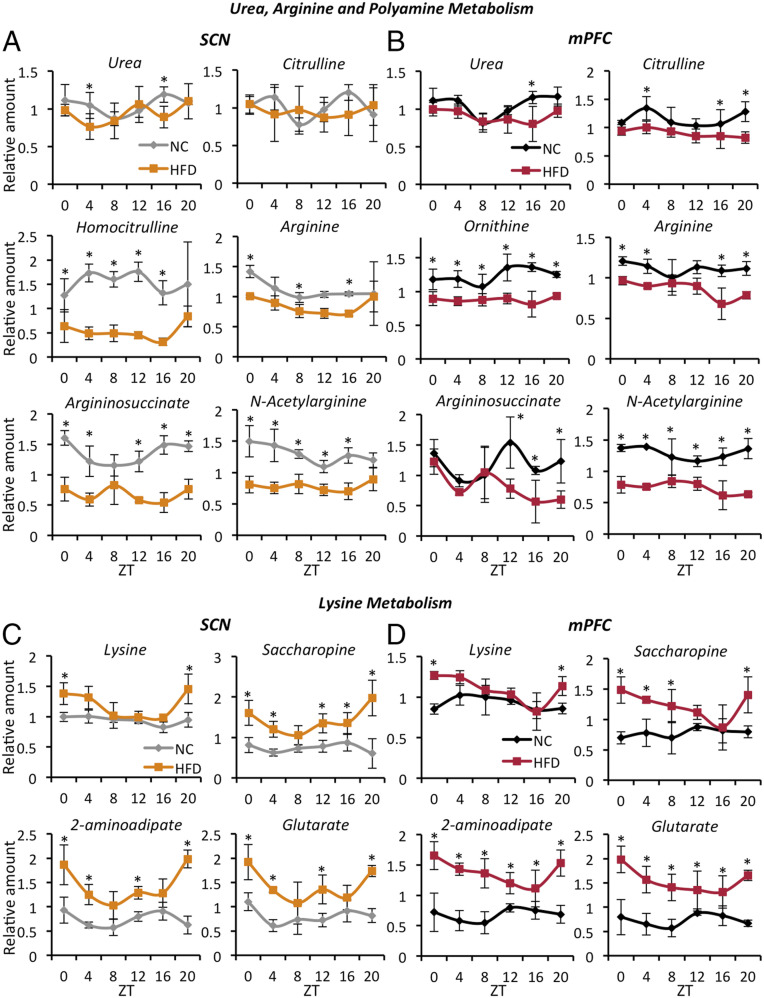
We next sought to identify the metabolites whose diurnal fluctuations and levels are impacted by an HFD in both the SCN and mPFC or exclusively in the SCN or the mPFC. To identify these cyclic metabolites, we crossed the SCN and mPFC circadian metabolome datasets (Fig. 1 F–H). In the NC-only crossing, among the common metabolites (SI Appendix, Table S1), urea caught our attention because of the importance of the urea cycle for the disposal of excess nitrogen and ammonia detoxification. Excessive ammonia levels result in neurologic disorders caused mainly by dysfunction in glutamate neurotransmission (36). Urea lost circadian rhythmicity and was decreased in both the SCN and the mPFC of HFD fed mice (Fig. 3 A and B). Interestingly, many metabolites belonging to the urea cycle and arginine metabolism pathway followed the same pattern. In particular, in the mPFC, citrulline, ornithine, arginine, argininosuccinate, and N-acetylarginine were all down-regulated under the HFD (Fig. 3B). Similar behavior was observed for the metabolites of the same pathway in the SCN (Fig. 3A). Ornithine was not detected in the SCN metabolome, although other related metabolites, such as homocitrulline (Fig. 3B), were significantly decreased. Similar data were obtained for the mPFC (SI Appendix, Fig. S4A). Related to the urea cycle, polyamine metabolism was also altered by the HFD with a tendency toward reduced levels in both the mPFC and the SCN (SI Appendix, Fig. S4A). The biological relevance of these losses of diurnal oscillation and down-regulation of urea and arginine metabolism in the brain with an HFD remains unclear. As the urea cycle takes place principally in the liver, where all of the relevant enzymes are located (37), we speculate that urea is reaching the brain through the circulation. Interestingly, and in keeping with our hypothesis, the urea diurnal rhythm was disrupted in the liver (13), as well as in the serum (29), of the HFD-fed mice, becoming low and flat. Moreover, changes in arginine metabolism may be important, considering that arginine is the precursor of synthesis of nitric oxide, a neurotransmitter and/or neuromodulator in the central nervous system (38) involved in neurodevelopment, synaptic plasticity, ischemic brain injury, and neurodegenerative diseases (39). Thus, alterations in this metabolic pathway might contribute to changes in the unique circuits of neurophysiology and behavior as a consequence of HFD feeding.
Fig. 3.
Diurnal metabolic pathways impacted by HFD in both the SCN and the mPFC. (A) SCN: representative oscillatory metabolites from the urea and arginine metabolism pathways. (B) mPFC: representative oscillatory metabolites from the urea and arginine metabolism pathways. Metabolites belonging to the same pathways and intersecting with the urea cycle are also represented (n = 3 to 5 per time point per group; CyberT test, *P < 0.05). (C) SCN: representative oscillatory metabolites from the lysine catabolic pathway. (D) mPFC: representative oscillatory metabolites from the lysine catabolic pathway. Metabolites belonging to the same pathway are also represented (n = 3 to 5 per time point per group; CyberT test, *P < 0.05). Error bars represent SD.
The comparisons of HFD-only rhythmic metabolites between the SCN and mPFC revealed a specific feature linked to lysine metabolism (SI Appendix, Table S3). Indeed, lysine, saccharopine, aminoadipate, and glutarate were increased, and all started to oscillate with a specific phase on an HFD (Fig. 3 C and D). This effect was particularly evident in the mPFC, in which all of the metabolites displayed a prominent nadir at ZT16 (Fig. 3D). Lysine catabolism can follow two different pathways: the saccharopine pathway, predominant in extracerebral tissues and in the developing brain, and the pipecolate pathway, previously thought to be more common in the adult brain. Both pathways converge in the synthesis of 2-aminoadipate semialdehyde (40) (SI Appendix, Fig. S4B). However, recent findings have challenged this notion, showing that the saccharopine pathway is highly active in the adult brain as well (41). Indeed, the HFD mainly modulated saccharopine concentration and rhythmicity in both the SCN and the mPFC. Pipecolate, a neuroactive compound that becomes neurotoxic at high levels (42), was detected only in the mPFC metabolome and did not show significant changes related to the NC diet (SI Appendix, Fig. S4B).
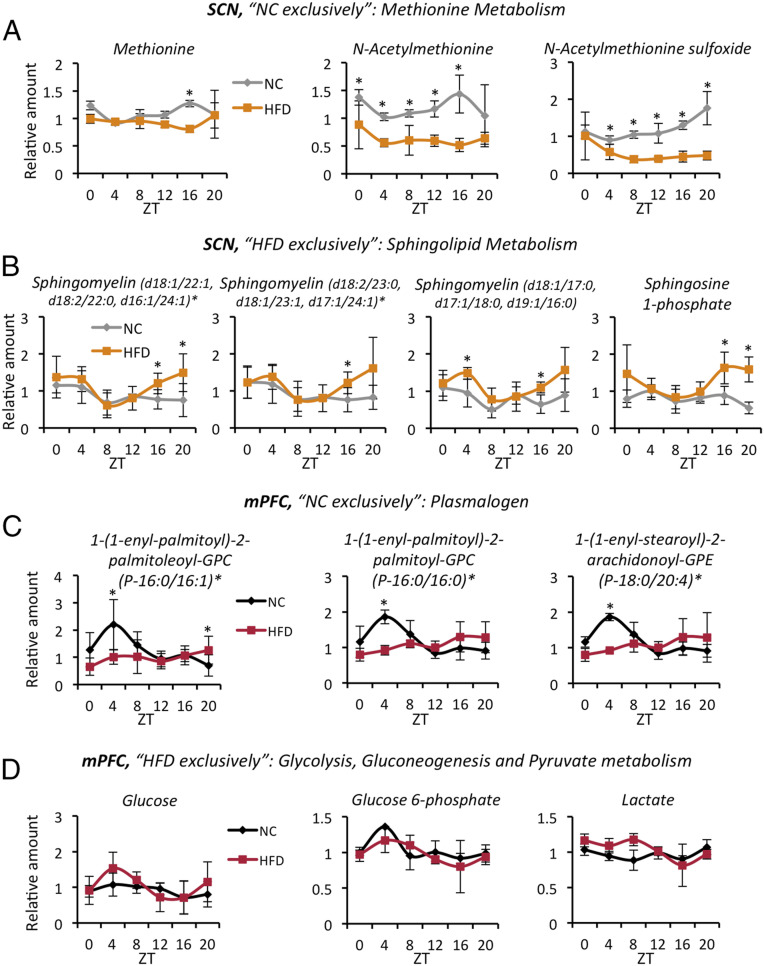
In addition to these pathways whose modulation by an HFD is similar in both the SCN and the mPFC, we identified specific pathways impacted uniquely in each of the brain areas analyzed. Methionine metabolism was exclusively diurnal under NC in the SCN (Fig. 4A). Methionine, N-acetylmethionine, and N-acetylmethionine sulfoxide showed significant dampening of their rhythmic behavior in the SCN with the HFD, an effect significantly less prominent in the mPFC (SI Appendix, Fig. S5A). The methionine cycle, together with the folate cycle and one-carbon metabolism, integrate the cellular metabolic status by cycling carbon units from amino acid donors into different cellular processes. These processes include biosynthesis of macronutrients, regulation of the redox state, regulation of the nucleotide pools, and ultimately control of the epigenetic status of the cell through methylation of histones and nucleic acids (43–46). Other components of the methionine cycle or connected metabolites did not show circadian oscillation, although some (e.g., SAH, cysteine) were down-regulated by the HFD in the SCN; see circadiomics.ics.uci.edu (35).
Fig. 4.
Circadian metabolites affected by HFD feeding exclusively in the SCN or the mPFC. (A) Metabolites exclusively circadian in the SCN with the NC diet (n = 5 per time point per group; CyberT test, *P < 0.05). (B) Metabolites exclusively circadian in the SCN with the HFD (n = 5 per time point per group; CyberT test, *P < 0.05). (C) Metabolites exclusively circadian in the mPFC with the NC diet (n = 3 to 5 per time point per group; CyberT test, *P < 0.05). (D) Metabolites exclusively circadian in the mPFC with the HFD (n = 3 to 5 per time point per group; CyberT test, *P < 0.05). Error bars represent SD.
Among the de novo rhythmic metabolites on the HFD, sphingolipid metabolism was uniquely present in the SCN (Fig. 4B). Sphingolipids are highly enriched in the central nervous system, where in addition to contributing structural roles in membranes forming the so-called “lipid rafts” (47), they also may operate as second messengers in response to various stimuli (48). They are also involved in memory formation (49), extinction (50), and a variety of neurologic disorders, including depression and Alzheimer’s disease (51, 52). Notably, sphingomyelin and sphingosine 1-phosphate started to oscillate in the SCN with the HFD, all following the same profile, with a nadir at ZT8 and a zenith at ZT20 (Fig. 4B). In contrast, these metabolites did not gain diurnal oscillation in the mPFC on HFD feeding (SI Appendix, Fig. S5B).
Plasmalogen is a specific class of membrane glycerophospholipids that has essential roles in brain functioning, including regulation of ion transport, fusion of synaptic vesicles for neurotransmitter release, and serving as a reservoir for second messengers (53). They are also important antioxidant factors (54), working as scavengers for free radical species, thereby protecting neuronal cells. Our analysis revealed plasmalogen as exclusively circadian in the mPFC in the NC condition (Fig. 4C). All of these oscillatory metabolites showed a specific peak at ZT4 with the NC diet that was completely lost with the HFD. The flat profile for plasmalogen in the mPFC induced by the HFD suggests a greater susceptibility of the mPFC to oxidative stress at specific times of the day, in line with recent findings (55). This drastic difference between the NC diet and the HFD was not seen in the SCN (SI Appendix, Fig. S5C), although some plasmalogens acquired a diurnal expression on the HFD (SI Appendix, Fig. S5C; second and third graphs).
Finally, glucose, glucose 6-phosphate, and lactate gained daily rhythmicity only in the mPFC (Fig. 4D), and not in the SCN (SI Appendix, Fig. S5D), of HFD-fed animals. This finding is of particular interest in the context of the so-called “astrocyte-neuron lactate shuttle” and the metabolic coupling between these two different cell types (56). Specifically, the astrocytes produce lactate through glycolysis for the neurons to use as an energy source (57). Indeed, we speculate that HFD may induce circadian rhythmicity in the glycolytic pathway, which is active predominantly in astrocytes (57), and in the production of lactate, which is subsequently shuttled to neurons and used as a fuel. Why this occurs exclusively in the mPFC and not in the SCN is not clear, and the underlying biological mechanisms and significance of this HFD-mediated effect remain to be clarified. Nevertheless, this finding reinforces our hypothesis of an area-specific rewiring of the circadian metabolome in the brain.
Connection between the Circadian Metabolome and Transcriptome in the SCN.
To further investigate the HFD-driven rewiring of circadian physiology in brain clocks, we performed a diurnal transcriptome analysis of the SCN master clock via RNA sequencing (JTK_CYCLE details in Datasets S5 and S6). Out of 2,816 oscillating transcripts, 1,635 (58%) were diurnal on NC feeding, and only 220 (7.8%) were circadian in both diets. Notably, the HFD decreased the number of cycling genes to 961 out of 2,816 (34.2%) (Fig. 5A), highlighting a different effect of the HFD on the number of oscillators in the transcriptome (Fig. 5A) compared with the metabolome (Fig. 1B). Our phase analysis showed a specific partitioning of NC and HFD exclusively cycling transcripts, with HFD genes prevalently peaking in the daytime (around ZT6) and NC genes peaking at night (around ZT18) (Fig. 5 B and E). The oscillating transcripts in both diets did not exhibit a clear separation in their peaks between light and dark phases with the NC diet, while they still peaked preferentially in the light phase with the HFD (Fig. 5 B and E). Furthermore, HFD significantly phase-advanced (53%) the transcripts cycling in both dietary regimens, whereas only 20% were phase-delayed by HFD feeding in the SCN (Fig. 5C). Remarkably, the HFD resulted mainly in a decreased amplitude of rhythmic genes oscillating in both diets (Fig. 5D), in contrast to an increased amplitude of cyclic metabolites by the HFD in the SCN metabolome (Fig. 2B).
Fig. 5.
Impact of HFD feeding on the SCN circadian transcriptome. (A) Venn diagram showing the number and percentage of rhythmic transcripts in the SCN in the NC-only, both diets, and HFD-only groups (JTK_CYCLE, P < 0.05). (B) Radar plots representing the phase of transcripts exclusively circadian in NC and HFD (first plot) and in both NC and HFD (second plot) in the SCN. (C) Analysis of transcripts circadian in both conditions and phase advanced or delayed in HFD feeding with respect to NC feeding. (D) Amplitude analysis of genes circadian in both diets in the SCN, showing the percentage of genes with amplitude lower or higher with the HFD with respect to the NC diet. (E) Heat maps representing the transcripts circadian in the SCN in the NC-only, both diets, and HFD-only groups (n = 3 per time point per group; JTK_CYCLE, P < 0.05). (F) KEEG pathway analysis of rhythmic transcripts in the NC-only, both diets, and HFD-only groups. The x-axis reports the –log of the P value.
To study the classes of SCN circadian genes impinged on by a nutritional challenge, we performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Fig. 5F and Dataset S7). As expected, circadian rhythm was enriched in both diets together with endocytosis, FoxO signaling pathway, and spliceosome. NC-only rhythmic transcripts clustered in various categories, while HFD diurnal genes clustered in multiple pathways related to metabolism, including metabolic pathways, the pentose phosphate pathway, 2-oxocarboxylic acid metabolism, biosynthesis of amino acids, and sphingolipid metabolism. Notably, the most highly enriched pathway, sphingolipid metabolism, was consistent with the de novo rhythmic metabolites driven by the HFD in the SCN (Fig. 4B), indicating a certain degree of coherence between the SCN diurnal transcriptome and metabolome on an HFD-induced lipid overload.
Transcription factor binding site analysis demonstrated that distinct transcription factor binding motifs are enriched in the SCN rhythmic genes (SI Appendix, Fig. S6). Specifically, KID3 (ZNF354c) and FOXO1A motifs were enriched in genes cycling only in the NC diet, while the NMYC and ZFP206 motifs were overrepresented in genes oscillating only in the HFD (SI Appendix, Fig. S6). Interestingly, NFAT1 was highly enriched in genes oscillating in both diets, indicative of a possible important role for NFAT1 in circadian gene expression in SCN.
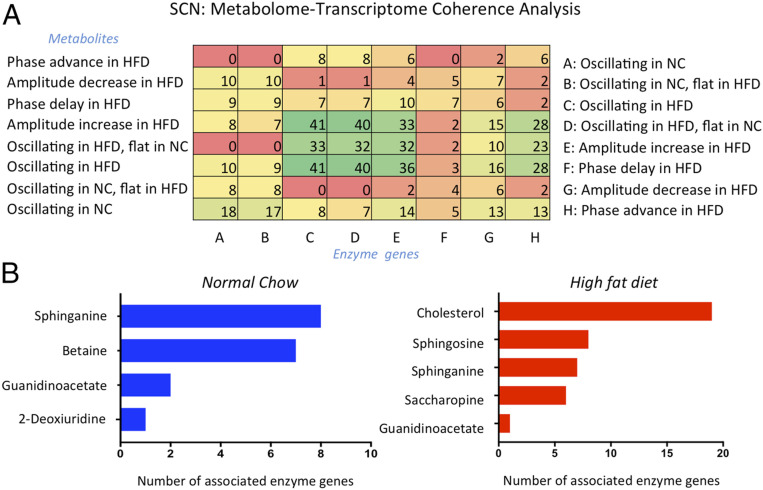
Recent evidence demonstrates coherence between the circadian transcriptome and metabolome in the liver with an HFD (13). To assess for the presence of similar cohesiveness in the SCN clock, we crossed our rhythmic transcriptome and metabolome data and then classified and grouped metabolite-enzyme gene edges based on the presence or absence of oscillation along with its amplitude and phase (Fig. 6A). The heat map resulting from this analysis showed that the most common edges were related to the oscillation of metabolites and related enzymes and to increases in the amplitude of the corresponding metabolites on HFD feeding. An additional observation was the phase-advancement of enzymes in the SCN on HFD feeding. All of the top edges belonged to the HFD condition, although consistency in oscillating metabolites and enzymes was also observed in the NC condition (Fig. 6A). Specifically, cholesterol and sphingosine were enriched in both the rhythmic metabolite levels and cyclic expression of genes encoding for enzymes involved in their metabolic pathway under HFD, while sphinganine and betaine were overrepresented under the NC diet (Fig. 6B). Our results highlight a clear link between the diurnal transcriptome and metabolome in the SCN driven by nutritional state.
Fig. 6.
Coherence between the circadian transcriptome and metabolome in the SCN. (A) Heat map representing the relationship between oscillating metabolites and corresponding enzymes (transcripts), obtained by crossing transcriptome and metabolome data in the SCN. (B) Graphs representing the number of oscillating enzymes for each reported circadian metabolite in the NC and HFD groups.
To gain insight into HFD-dependent transcriptional changes in the mPFC, we investigated diurnal gene expression. mPFC core clock genes were minimally affected by the HFD (SI Appendix, Fig. S7), in agreement with observations in the liver (13, 58), and hypothalamus (12, 59). Since diets rich in fat have been linked to emotional disorders (22, 24), we analyzed the diurnal profile of transcripts involved in stress responses. Glucocorticoid receptor gained oscillation with a peak at nighttime on HFD feeding, indicating a possible stress response of mPFC neuronal circuits to the dietary change. Notably, mineralocorticoid receptor, which is also involved in the stress response (60), lost oscillation and showed increased expression during the active phase, similar to glucocorticoid receptor. However, genes implicated in signaling during brain stress (e.g., Fkbp1a, Sgk, Bdnf) were not significantly impacted in the mPFC by HFD feeding. Our analysis suggests a certain effect of HFD feeding on specific rhythmic gene expression in the mPFC. Further investigations are needed to provide a deeper understanding of how a diet rich in fat could affect transcription in the mPFC and ultimately behavior.
Discussion
Our brain consumes a high amount of energy despite representing only 2% of the total body mass. To maintain metabolic homeostasis, it is essential to ensure appropriate brain functions. Important advances have been made in understanding the metabolic coupling between neurons and astrocytes and how the lactate shuttle works (61); however, how brain metabolism is influenced by nutritional challenges remains somewhat obscure. We have demonstrated that a high fat nutritional stress uniquely impacts the circadian rhythmicity of distinct brain areas. A 10-wk HFD regimen deeply reshaped the diurnal metabolic profiles of the SCN and mPFC, influencing the number, phase, and amplitude of rhythmicity of oscillatory metabolites. Surprisingly, there was minimal overlap between the pathways rewired in both the SCN and the mPFC, suggesting high specificity in how a food stress is integrated by brain areas dedicated to different functions. Furthermore, diurnal transcriptional rewiring of the SCN transcriptome with the HFD was accompanied by a certain degree of temporal coherence between the oscillation of enzymes and their related metabolites. These data demonstrate a diet-dependent coordination between the daily transcriptome and metabolome in the SCN, indicating food as prominent regulator of the master clock, both metabolically and transcriptionally.
Finally, future experiments will shed light on the persistence of the effect of an HFD on the daily metabolome in the SCN and mPFC after the dietary regimen is changed to a balanced diet. These experiments may provide important insights into brain plasticity from a novel metabolic perspective, hopefully finding missing links between metabolism and neural function, which may lead to improved understanding of and treatment for neuropsychiatric disorders.
In conclusion, our study reveals an unexpected nutrition-driven plasticity of the brain circadian metabolome and a unique diurnal signature induced by the same food stress in specific areas. Our results open future avenues to explore how daily oscillations of specific metabolites may cause detrimental effects (e.g., cognitive, emotional) of diet-induced obesity on brain physiology and behavior.
Materials and Methods
Animals and Feeding.
Here 6-wk-old male C57BL/6J mice (JAX 00064; The Jackson Laboratory), maintained at 24 °C on a 12-h light/12-h dark cycle, were fed an HFD (60% kcal from fat; Research Diets, D12492) or an NC diet (Prolab, RMH 2500) ad libitum for 10 wk. Animals were sacrificed every 4 h throughout the circadian cycle. The mPFC- and SCN-enriched regions were dissected from the fresh brains and immediately snap-frozen in liquid nitrogen. The rest of the brain was frozen in dry ice, sliced, stained with cresyl-violet, and analyzed under a microscope, to verify proper dissection of the SCN and mPFC.
Animal care and use were in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of California Irvine.
Metabolomics Profile.
The metabolome analysis was performed by Metabolon. In brief, the samples were prepared using the automated MicroLab STAR system (Hamilton) and analyzed by reverse-phase ultra-high performance liquid chromatography-tandem mass spectrometry (SI Appendix). Normalized levels of metabolites were scanned for outliers among the replicates at each ZT/condition using Dixon’s test, with a percentile threshold of 95. At most, 2 out of 5 replicates were removed before downstream analysis. Data were further divided into two circadian groups (NC and HFD) per tissue, and pairwise analysis between the groups was performed using pipelines for CircadiOmics (35, 62).
All of the metabolomics data associated with this work are available at circadiomics.ics.uci.edu.
RNA-seq Analysis.
RNA-seq analysis was performed using the Tuxedo Suite tools Tophat and Cufflinks. Tophat was used to align the RNA-seq reads to the reference genome assembly mm10 and corresponding transcriptome, and Cufflinks was used to assemble the aligned reads and calculate gene expression levels. Expression levels output by Cufflinks are reported as fragments per kilobase per million mapped reads. The transcriptome data associated with this work are available at circadiomics.ics.uci.edu, and the RNA-seq data have been deposited in the Gene Expression Omnibus database.
Statistical Analysis.
The data are presented as mean ± SD. The analysis of circadian rhythmicity was performed using the nonparametric JTK_CYCLE test (34). Two-way ANOVA was performed to analyze the effect of diet or time point (ZT) on the levels of single metabolites. Differences among the different levels of metabolites between the two diets at specific time points were identified using the CyberT test (SI Appendix).
Supplementary Material
Acknowledgments
We thank all the members of the P.S.-C. laboratory for constructive comments and Jeff Bucktal and Jason Kinchen (Metabolon) for advice on the analysis of metabolic profiles. This work was supported by the NIH (Grant DA036408), INSERM, and Novo Nordisk (Foundation Challenge Grant, to P.S.-C.). K.K. was supported by a Japan Society for the Promotion of Science fellowship. The work of P.B. and Y.L. was supported in part by NIH Grant GM123558 and Defense Advanced Research Projects Agency Grant D17AP00002 (to P.B.). P.T. was supported by the Human Frontiers Science Program (LT 000576/2013) and the European Commission (H2020-MSCA-IF-2016 749697 GaMePLAY).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016589117/-/DCSupplemental.
Data Availability.
The metabolome data discussed in this study can be found either in the main text, SI Appendix, or Datasets S1–S4. RNA-seq data have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE157077), and can be also found in Datasets S5–S7.
References
- 1.Mohawk J. A., Green C. B., Takahashi J. S., Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roenneberg T., Merrow M., Entrainment of the human circadian clock. Cold Spring Harb. Symp. Quant. Biol. 72, 293–299 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Asher G., Sassone-Corsi P., Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Marcheva B., et al. , Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turek F. W., et al. , Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass J., Takahashi J. S., Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckel-Mahan K., Sassone-Corsi P., Metabolism and the circadian clock converge. Physiol. Rev. 93, 107–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerhart-Hines Z., Lazar M. A., Circadian metabolism in the light of evolution. Endocr. Rev. 36, 289–304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tognini P., Murakami M., Sassone-Corsi P., Interplay between microbes and the circadian clock. Cold Spring Harb. Perspect. Biol. 10, a028365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang P., Turek F. W., Timing of meals: When is as critical as what and how much. Am. J. Physiol. Endocrinol. Metab. 312, E369–E380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatori M., et al. , Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohsaka A., et al. , High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Eckel-Mahan K. L., et al. , Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tognini P., et al. , Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metabol. 26, 523–538.e525 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Mendoza J., Pévet P., Challet E., High-fat feeding alters the clock synchronization to light. J. Physiol. 586, 5901–5910 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza J., Pévet P., Challet E., Circadian and photic regulation of clock and clock-controlled proteins in the suprachiasmatic nuclei of calorie-restricted mice. Eur. J. Neurosci. 25, 3691–3701 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Gómez-Pinilla F., Brain foods: The effects of nutrients on brain function. Nat. Rev. Neurosci. 9, 568–578 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenwood C. E., Winocur G., High-fat diets, insulin resistance and declining cognitive function. Neurobiol. Aging 26, 42–45 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Mattson M. P., Moehl K., Ghena N., Schmaedick M., Cheng A., Intermittent metabolic switching, neuroplasticity and brain health. Nat. Rev. Neurosci. 19, 63–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pizzorusso T., Tognini P., Interplay between metabolism, nutrition and epigenetics in shaping brain DNA methylation, neural function and behavior. Genes (Basel) 11, E742 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon J. B., et al. , Severely obese people with diabetes experience impaired emotional well-being associated with socioeconomic disadvantage: Results from diabetes MILES–Australia. Diabetes Res. Clin. Pract. 101, 131–140 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Dutheil S., Ota K. T., Wohleb E. S., Rasmussen K., Duman R. S., High-fat diet induced anxiety and anhedonia: Impact on brain homeostasis and inflammation. Neuropsychopharmacology 41, 1874–1887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis H., Stevenson R., The longer-term impacts of Western diet on human cognition and the brain. Appetite 63, 119–128 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Pagoto S. L., et al. , Association of post-traumatic stress disorder and obesity in a nationally representative sample. Obesity (Silver Spring) 20, 200–205 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Hassan A. M., et al. , High-fat diet induces depression-like behaviour in mice associated with changes in microbiome, neuropeptide Y, and brain metabolome. Nutr. Neurosci. 22, 877–893 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Elia M., Klepper J., Leiendecker B., Hartmann H., Ketogenic diets in the treatment of epilepsy. Curr. Pharm. Des. 23, 5691–5701 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Francis H. M., Stevenson R. J., Potential for diet to prevent and remediate cognitive deficits in neurological disorders. Nutr. Rev. 76, 204–217 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Newman J. C., et al. , Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metabol. 26, 547–557.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbondante S., Eckel-Mahan K. L., Ceglia N. J., Baldi P., Sassone-Corsi P., Comparative circadian metabolomics reveal differential effects of nutritional challenge in the serum and liver. J. Biol. Chem. 291, 2812–2828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyar K. A., et al. , Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T. J., Takasu N. N., Nakamura W., The suprachiasmatic nucleus: Age-related decline in biological rhythms. J. Physiol. Sci. 66, 367–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saper C. B., The central circadian timing system. Curr. Opin. Neurobiol. 23, 747–751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maren S., Phan K. L., Liberzon I., The contextual brain: Implications for fear conditioning, extinction and psychopathology. Nat. Rev. Neurosci. 14, 417–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes M. E., Hogenesch J. B., Kornacker K., JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel V. R., Eckel-Mahan K., Sassone-Corsi P., Baldi P., CircadiOmics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat. Methods 9, 772–773 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Gropman A. L., Summar M., Leonard J. V., Neurological implications of urea cycle disorders. J. Inherit. Metab. Dis. 30, 865–879 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris S. M., Jr, Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 22, 87–105 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Philippu A., Nitric oxide: A universal modulator of brain function. Curr. Med. Chem. 23, 2643–2652 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Picón-Pagès P., Garcia-Buendia J., Muñoz F. J., Functions and dysfunctions of nitric oxide in brain. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1949–1967 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Hallen A., Jamie J. F., Cooper A. J., Lysine metabolism in mammalian brain: An update on the importance of recent discoveries. Amino Acids 45, 1249–1272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pena I. A., et al. , Mouse lysine catabolism to aminoadipate occurs primarily through the saccharopine pathway; implications for pyridoxine dependent epilepsy (PDE). Biochim. Biophys. Acta Mol. Basis Dis. 1863, 121–128 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Hallen A., Cooper A. J., Reciprocal control of thyroid binding and the pipecolate pathway in the brain. Neurochem. Res. 42, 217–243 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Locasale J. W., Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaelin W. G. Jr, McKnight S. L., Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C., Thompson C. B., Metabolic regulation of epigenetics. Cell Metab. 16, 9–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katada S., Imhof A., Sassone-Corsi P., Connecting threads: Epigenetics and metabolism. Cell 148, 24–28 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Cremesti A. E., Goni F. M., Kolesnick R., Role of sphingomyelinase and ceramide in modulating rafts: Do biophysical properties determine biologic outcome? FEBS Lett. 531, 47–53 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Levade T., Jaffrézou J. P., Signalling sphingomyelinases: Which, where, how and why? Biochim. Biophys. Acta 1438, 1–17 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Delint-Ramírez I., Salcedo-Tello P., Bermudez-Rattoni F., Spatial memory formation induces recruitment of NMDA receptor and PSD-95 to synaptic lipid rafts. J. Neurochem. 106, 1658–1668 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Huston J. P., et al. , A sphingolipid mechanism for behavioral extinction. J. Neurochem. 137, 589–603 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Haughey N. J., Bandaru V. V., Bae M., Mattson M. P., Roles for dysfunctional sphingolipid metabolism in Alzheimer’s disease neuropathogenesis. Biochim. Biophys. Acta 1801, 878–886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olsen A. S. B., Færgeman N. J., Sphingolipids: Membrane microdomains in brain development, function and neurological diseases. Open Biol. 7, 170069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farooqui A. A., Horrocks L. A., Plasmalogens: Workhorse lipids of membranes in normal and injured neurons and glia. Neuroscientist 7, 232–245 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Kuczynski B., Reo N. V., Evidence that plasmalogen is protective against oxidative stress in the rat brain. Neurochem. Res. 31, 639–656 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Freeman L. R., Haley-Zitlin V., Rosenberger D. S., Granholm A. C., Damaging effects of a high-fat diet to the brain and cognition: A review of proposed mechanisms. Nutr. Neurosci. 17, 241–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bélanger M., Allaman I., Magistretti P. J., Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 14, 724–738 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Magistretti P. J., Allaman I., A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Murakami M., et al. , Gut microbiota directs PPARγ-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep. 17, 1292–1303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodie L. N., et al. , Western diet-induced obesity disrupts the diurnal rhythmicity of hippocampal core clock gene expression in a mouse model. Brain Behav. Immun. 88, 815–825 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Caudal D., Jay T. M., Godsil B. P., Behavioral stress induces regionally-distinct shifts of brain mineralocorticoid and glucocorticoid receptor levels. Front. Behav. Neurosci. 8, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allaman I., Bélanger M., Magistretti P. J., Astrocyte-neuron metabolic relationships: For better and for worse. Trends Neurosci. 34, 76–87 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Patel V. R., et al. , The pervasiveness and plasticity of circadian oscillations: The coupled circadian-oscillators framework. Bioinformatics 31, 3181–3188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolome data discussed in this study can be found either in the main text, SI Appendix, or Datasets S1–S4. RNA-seq data have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE157077), and can be also found in Datasets S5–S7.