Significance
Recruitment of the immune system to suppress malignant tumors can involve different mechanisms, including inhibition of suppressive immune checkpoints (the enemy of my enemy is my friend), generation of specific CAR-T cells that recognize tumor cells, and immunization against the patient’s tumor antigens. Here we describe a mechanism, where excess of the p50 subunit of the NF-ĸB transcription factor that when pairs with another subunit—p65—is oncogenic, but when generated in excess over p65 and as a result homodimerizes, it suppresses tumors. It appears to act via several mechanisms: it stimulates expression of other cancer suppressors, it inhibits generation of the immune inhibitor PD-L1 (thus unleashes the immune system), and it initiates secretion of chemokines, which are chemical attractants of immune cells.
Keywords: ubiquitin ligase KPC1, PD-L1, NF-ĸB p50, tumor suppression, chemokines
Abstract
Nuclear factor–ĸB (NF-ĸB) transcription factor is a family of essential regulators of the immune response and cell proliferation and transformation. A typical factor is a heterodimer made of either p50 or p52, which are limited processing products of either p105 or p100, respectively, and a member of the Rel family of proteins, typically p65. The transcriptional program of NF-ĸB is tightly regulated by the composition of the dimers. In our previous work, we demonstrated that the ubiquitin ligase KPC1 is involved in ubiquitination and proteasomal processing of p105 to generate p50. Its overexpression and the resulting high level of p50 stimulates transcription of a broad array of tumor suppressors. Here we demonstrate that additional mechanisms are involved in the p50-mediated tumor-suppressive effect. p50 down-regulates expression of a major immune checkpoint inhibitor, the programmed cell death-ligand 1 (PD-L1), both in cells and in tumors. Importantly, the suppression is abrogated by overexpression of p65. This highlights the importance of the cellular quantities of the two different subunits of NF-ĸB which determine the composition of the dimer. While the putative p50 homodimer is tumor-suppressive, the “canonical” p50p65 heterodimer is oncogenic. We found that an additional mechanism is involved in the tumor-suppressive phenomenon: p50 up-regulates expression of the proinflammatory chemokines CCL3, CCL4, and CCL5, which in turn recruit into the tumors active natural killer (NK) cells and macrophages. Overall, p50 acts as a strong tumor suppressor via multiple mechanisms, including overexpression of tumor suppressors and modulation of the tumor microenvironment by recruiting active immune cells.
Nuclear factor–ĸB (NF-ĸB), discovered in 1986 by David Baltimore’s group (1), is a major transcriptional regulator for the cell response to external signals. It orchestrates a broad range of cellular processes, among them cell division and differentiation and cell death and survival. Importantly, NF-ĸB controls the immune and inflammatory response. Dysregulated activity of NF-ĸB has been reported to be involved in a broad array of immune system-related disorders (2) and malignant transformation (3, 4).
NF-ĸB is typically a heterodimer that can be made of either p50 or p52 and RelA, RelB, or cRel. p50 and p52 homodimers were described as well (5). p50 and p52 are the products of ubiquitination and proteasome-dependent limited processing of the long precursors—p105 and p100, respectively (6–8). p50 and p52 NF-ĸB subunits lack a transactivation domain, which is present in the Rel proteins. As a result, homodimers based on p50 or p52 promote transcription only in case of complex formation with additional transcriptional activators, such as Bcl3, HDAC3, or IĸBZ (9). In resting cells, the dimers are trapped in the cytoplasm by specific inhibitors known as IĸBs (e.g., IĸBα, p100, p105, and Bcl3) (10). In response to a broad array of signals (e.g., oxidative stress, viral and bacterial infections, proinflammatory cytokines, and DNA damage), certain IĸBs are phosphorylated on specific serine residues by the IĸB kinase (IKK) complex (11) and, consequently, are ubiquitinated and degraded by the proteasome. This releases the dimers that are translocated to the nucleus to initiate the specific transcriptional program (12).
The crucial role of NF-ĸB in tumorigenesis is well established (13). In many solid malignances, the activity of NF-κB is up-regulated, modulating tumor initiation, promotion, and metastases (14). In most cases, the tumor-promoting activity of NF-κB is related to the p65p50 heterodimeric complex. Thus, in ovarian cancer, strong expression of both subunits was found compared to borderline and benign ovarian tumors (15). Enhanced p65 staining was found in human prostate adenocarcinoma, correlating with increased tumor grade. The study revealed that the DNA-binding complex is mainly made of NF-ĸB p50p65 heterodimers (16).
Although in most cases, NF-ĸB appears to be oncogenic, in some studies it has been demonstrated as a tumor suppressor (17), which is due in particular to its p50 subunit (18). For example, cells lacking NF-ĸB1, the p50-coding gene, accumulate alkylating agents-induced mutations, and NF-ĸB1−/− mice are more susceptible to lymphomas in response to alkylating agents-induced DNA damage (19). In another study, it was shown that in pancreatic cancer cells, IKKα regulates G1 to S-phase transition by exchange of the tumor promoter p52RelB with the tumor-suppressive p50RelB NF-ĸB dimers on the promoter of the F-box protein S-phase kinase-associated protein 2 (SKP2), inhibiting its expression (20). SKP2 is the ubiquitin ligase that negatively regulates the abundance of the cyclin-dependent kinase inhibitor p27KIP, thus promoting malignant transformation by increased transition to S phase (21). In another example, one of the well-defined p50-mediated tumor-suppressive mechanisms was described in human glioblastoma and breast cancer cells and xenografts, and in tumors derived from patients: it was shown that excessive generation of p50 by the ubiquitin ligase KPC1 triggers up-regulation of transcripts of numerous tumor suppressors, suggesting that p50p50 homodimers control their transcription (rather than the “canonical” tumorigenic p50p65) (22).
NF-ĸB–dependent modulation of the tumor microenvironment is another essential mechanism that regulates cancer dynamics—progression or suppression. It can combine two layers of activity: 1) it can affect immune cells in the adjacent tumor surroundings, ensuring their reactivity (23); and/or 2) it affects tumor cells by stimulating secretion of chemokines or generation of membrane-bound immune checkpoint molecules (24).
An example of a tumor suppression mechanism based on modulation of the tumor microenvironment is the induction of mitochondrial outer-membrane permeabilization that causes activation of NF-ĸB in tumor cells with subsequent anticancer immune response, including macrophage activation and T cell infiltration (25). In a mouse lung cancer model, activation of NF-ĸB up-regulates the T cell-recruiting chemokines CCL2 and CCL5, resulting in tumor rejection. In lung cancer patient specimen, NF-ĸB activity was linked to high infiltration of T cells to the tumor (26).
Chemokines play an important role in cancer progression not only through the direct autocrine effect on the tumor cells, but also through recruitment of specific immune cells (27). Genomic expression signature of 12 chemokines (CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21, CXCL9, CXCL10, CXCL11, and CXCL13) was identified in genomic arrays of colorectal carcinoma (28) and of ∼15,000 distinct solid tumors. It was correlated with the presence of tertiary lymph node-like structures and was associated with better survival of a subset of melanoma patients (29).
An additional significant feature of immune cells invasion of different cancers is the surface expression of inhibitory ligands, programmed cell death-ligand 1 (PD-L1), for example (30). It is highly accepted that the PD-1/PD-L1 axis is one of the main mechanisms that inhibits the anticancer activity of T cells and macrophages (31). Recently, it was shown that PD-1 and PD-L1 blockade elicited a strong natural killer (NK) cell antitumoral activity (32). NF-κB can be involved in the regulation of immune checkpoints on the surface of tumor cells (24). For instance, p65-containing NF-ĸB dimers were shown to up-regulate CSN5—a deubiquitinating enzyme—which resulted in stabilization of PD-L1, thus bypassing immune suppression of the cancer cells (33).
The current study expands our previous findings that the ubiquitin ligase KPC1 acts as a tumor suppressor via its excessive activity on the p105 NF-ĸB precursor, generating excess of the p50 subunit. Our initial results demonstrated that p50 stimulates transcription of a broad array of tumor suppressors (22). Here we show that excess of p50 down-regulates the surface expression of PD-L1. In addition, it also up-regulates expression of the proinflammatory cytokines CCL3, CCL4, and CCL5, that recruit NK cells and macrophages to the tumor. These yet unreported findings of p50 reveal multilayered mechanisms that are mostly driven by a change in the cellular equilibrium between the different NF-ĸB subunits which determine the Dr. Jekyll and Mr. Hyde character of this transcription factor—moving on the broad range between a tumor promoter and suppressor.
Results
Suppression of Tumor Growth via Generation of Stoichiometric Excess of p50 over the p65 Subunit.
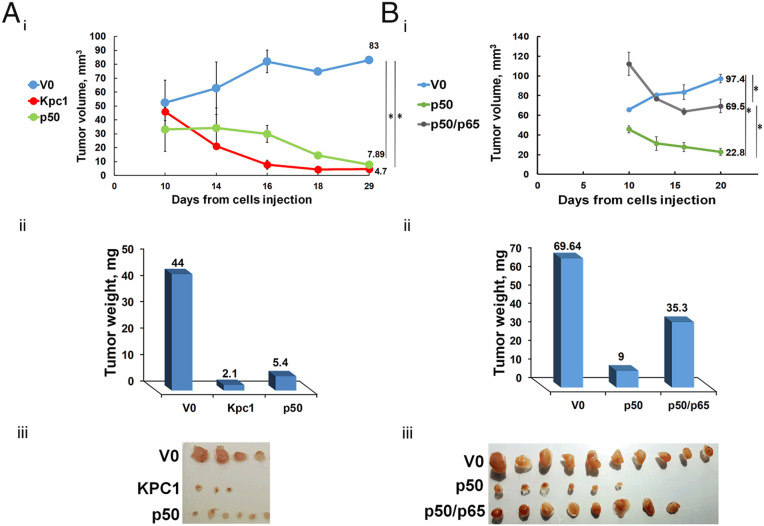
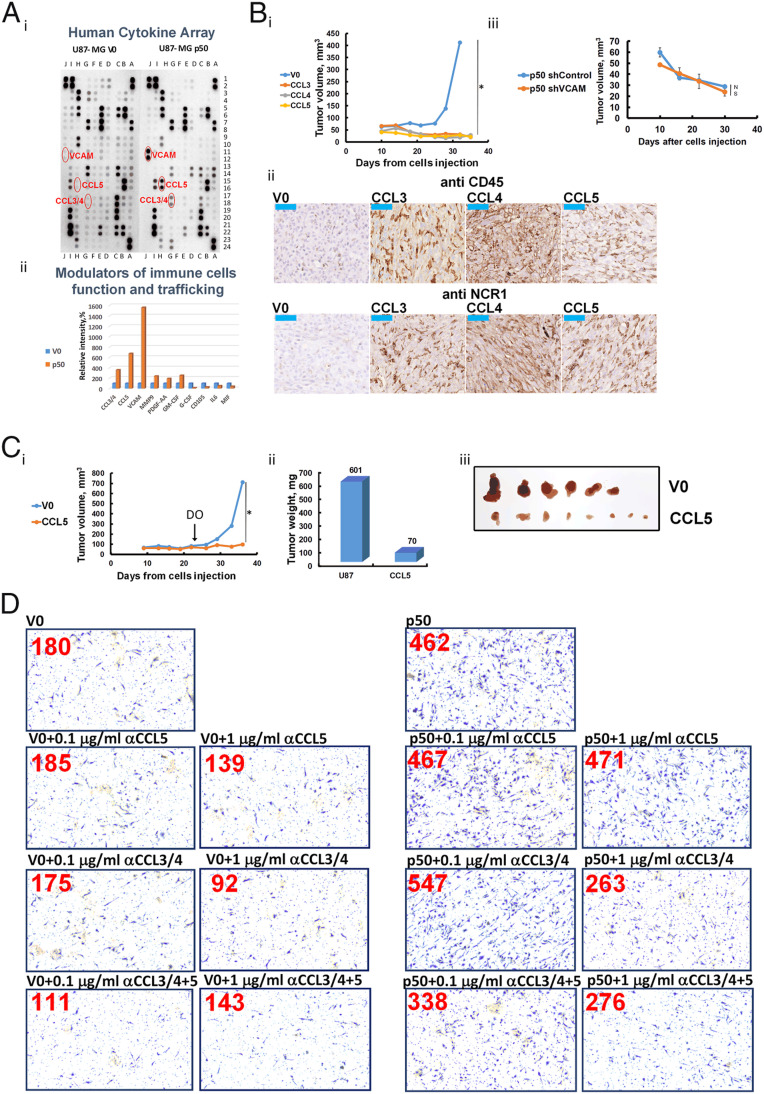
The different transcriptional programs of NF-ĸB are dependent on different compositions of the transcription factor dimers. In our previous work, we demonstrated that the tumor-suppressive effect of KPC1 is mediated via generation of excess of the p50 subunit, that probably homodimerizes. This in turn resulted in significant up-regulation of tumor suppressors in U87-MG glioblastoma and MDA–MB-231 breast (22) cancer cells-derived tumor. In order to further investigate the contribution of excess of p50 to the tumor-suppressive phenomenon, we overexpressed in the glioblastoma cells p65, the canonical oncogenic partner of p50. As can be seen in Fig. 1, the suppressive effect of p50 (Fig. 1A) was partially abrogated by a concomitant expression of p65: tumors in which we expressed both proteins were significantly larger compared to tumors that express p50 alone (compare Fig. 1A, iii to Fig. 1B, iii).
Fig. 1.
p50-dependent and KPC1-mediated tumor suppression is attenuated by p65. (A) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells that overexpress V0, KPC1, or FLAG-p50. (iii) The corresponding expressing tumors 4 wk after inoculation. (B) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells that overexpress V0, FLAG-p50, or FLAG-p50 and p65. (iii) The corresponding expressing tumors 3 wk after inoculation. * represents P value <0.05.
Excess of the p50 Subunits Suppresses Expression of PD-L1.
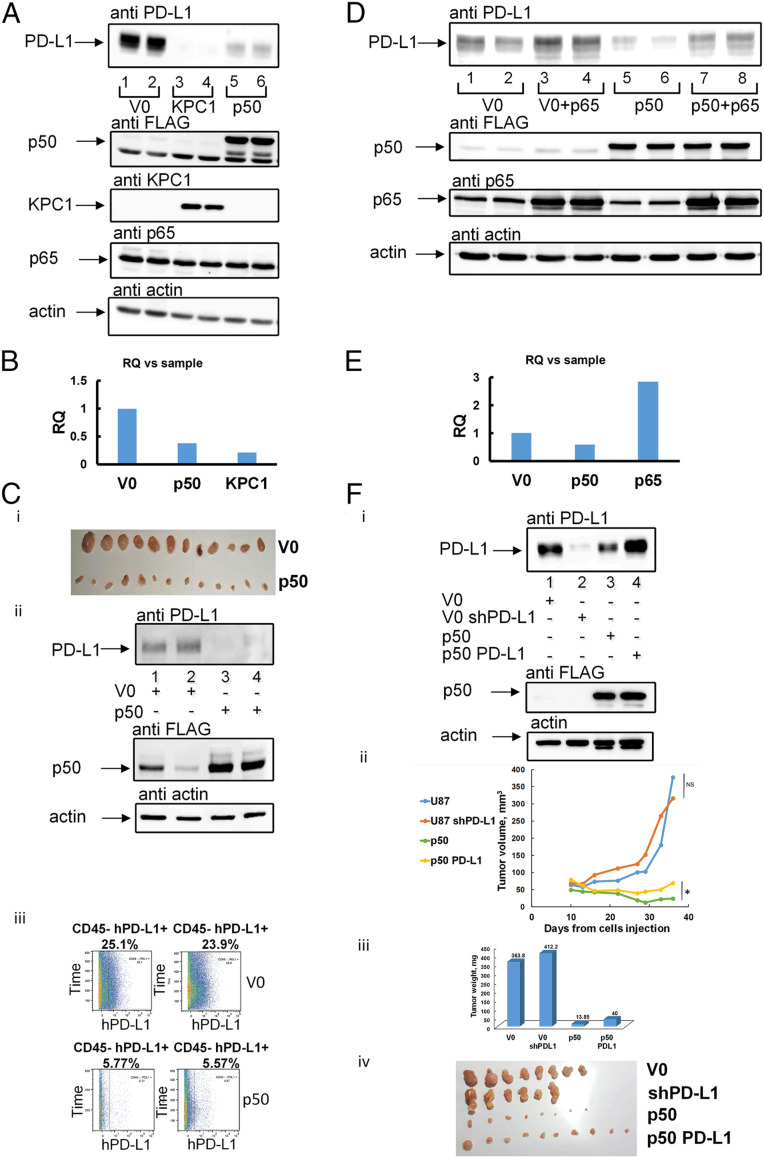
When investigating tumor suppression mechanisms, immune checkpoints are certainly important targets. Therefore, we decided to study the effect of p50 on PD-L1 expression in tumor cells and found that overexpression of either KPC1 or FLAG-p50 down-regulates its expression (Fig. 2A). In order to check if the reduction in the PD-L1 level is due to the transcriptional activity of p50, we compared the relative levels of PD-L1 messenger RNA (mRNA) isolated from U87-MG cells expressing V0, KPC1, or FLAG-p50 using quantitative real-time PCR (qRT-PCR). As can be seen in Fig. 2B, the level of PD-L1 mRNA is decreased significantly following expression of either KPC1 or FLAG-p50, which explains the corollary decrease in PD-L1 protein level. The finding in cells was further corroborated in tumors. To show this, we generated in nude mice U87-MG–derived xenografts stably expressing V0 or FLAG-p50. After 4 wk, the tumors were removed (Fig. 2C, i), and PD-L1 was monitored in the cell lysates (Fig. 2C, ii). Again, we found that p50 overexpression results in a significant decrease in the expression of PD-L1. In addition, we used mass cytometry by time-of-flight (CyTOF) of single-cell suspensions generated from the tumors to monitor the presence of the PD-L1 ligand on the surface of the human tumor cells. Since the bulk of the invading mouse immune cells is significant, we used anti-mouse CD45 to remove them from the analysis. We found that only ∼5% of p50-expressing tumor cells also expressed a high level of PD-L1 compared to ∼25% of cells expressing V0 (Fig. 2C, iii).
Fig. 2.
PD-L1 is down-regulated in KPC1- and p50-, but not in p65-overexpressing cells. (A) Overexpression of KPC1 or FLAG-p50 down-regulates PD-L1 in cells. U87-MG cells were stably transfected with an empty vector (V0) (lanes 1 and 2), myc-KPC1 (lanes 3 and 4), or FLAG-p50 (lanes 5 and 6). Cells were grown to confluence of ∼80%, lysed, and resolved via SDS/PAGE. Proteins were visualized using anti–PD-L1, anti-KPC1, anti-FLAG (for detection of p50), anti-p65, or anti-actin antibodies, as indicated. (B) Relative level of PD-L1 transcripts isolated from U87-MG cells expressing V0, Myc-KPC1, or FLAG-p50. Expression of the transcripts was analyzed using qRT-PCR as described under Materials and Methods. (C) Overexpression of FLAG-p50 down-regulates PD-L1 in U87-MG–derived tumors. (i) Shown are tumors expressing V0 or FLAG-p50 3 wk after inoculation. (ii) Tumors were lysed, and proteins were resolved via SDS/PAGE, blotted onto nitrocellulose membrane, and detected using the appropriate antibodies. (iii) Single-cell suspensions were generated from tumors expressing FLAG-p50 or V0, and cells were stained with human anti–PD-L1 and mouse anti-CD45 (to remove mouse immune cells in the CyTOF analysis) antibodies, followed by CyTOF analysis. Numbers represent percentage of human PD-L1–positive cells from the CD45-negative population. (D) Overexpression of p65 increases the level of PD-L1 in U87-MG glioblastoma cells. U87-MG cells were stably transfected with V0 (lanes 1 and 2) or FLAG-p50 (lanes 5 and 6) and then transiently transfected with p65 (lanes 3, 4, 7, and 8). Cells were grown to confluence of ∼80%, lysed, and resolved via SDS/PAGE. Proteins were visualized using anti–PD-L1, anti-FLAG (for detection of p50), anti-p65, or anti-actin antibodies, as indicated. (E) Relative level of PD-L1 transcripts isolated from U87-MG cells expressing V0, FLAG-p50, or p65. Expression of the transcripts was analyzed using qRT-PCR as described under Materials and Methods. (F) Overexpression of PD-L1 in U87-MG cells that also express p50 attenuates the tumor-suppressive effect of p50. (i) Silencing of PD-L1 (lane 2) or overexpression of FLAG-p50 (lane 3) or FALG-p50 and PD-L1 (lane 4) in U87-MG cells. Cells were lysed, and proteins were resolved via SDS/PAGE, blotted onto nitrocellulose membrane, and detected using the appropriate antibodies. Growth rates (ii) and weights (iii) of tumors derived from the U87-MG cells described under F, i. * represents P value < 0.05. NS represents nonsignificant. (iv) Shown are the corresponding tumors 5 wk after inoculation. RQ, relative quantification.
To confirm the hypothesis that tumorigenesis in this model of glioblastoma relies, among other factors, on the difference between the transcriptional programs set by different pairs of NF-ĸB factors made of either putative dimers of p50 or heterodimers of p50 and p65, we checked the PD-L1 level in p65 overexpressing cells. As can be seen in Fig. 2D, we found that expression of p65 in either U87-MG cells expressing V0 or p50 up-regulates PD-L1 protein level. The level of PD-L1 mRNA was similarly increased (Fig. 2E). Moreover, we found that overexpression of PD-L1 in the presence of overexpressed p50 weakens the suppressive effect of the later in tumors, giving rise to tumors which are larger than those expressing only p50 (Fig. 2F). This finding suggests that recruiting the immune system might be one of the mechanisms that p50 utilizes to suppress tumors. Silencing of PD-L1 in U87 cells does not reduce the tumors’ size (Fig. 2F). This is probably due to the fact that several factors are involved in the suppressive effect (including p50-induced expression of a broad array of tumor suppressors (22) and that PD-L1 expression is not involved in recruiting immune cells, but rather in inhibiting their activity once recruited (see also under Discussion).
Recruitment of the Host Immune Cells to the Microenvironment of KPC1- and p50-Overexpressing Glioblastoma Tumors.
Since expression of the inhibitory ligand PD-L1 on the surface of cancer cells is a major feature of tumors which is “used” by them to inhibit immune cells, we wanted to test if the reduction in PD-L1 level is associated, in parallel and independently, with infiltration of immune cells to the tumor’s niche.
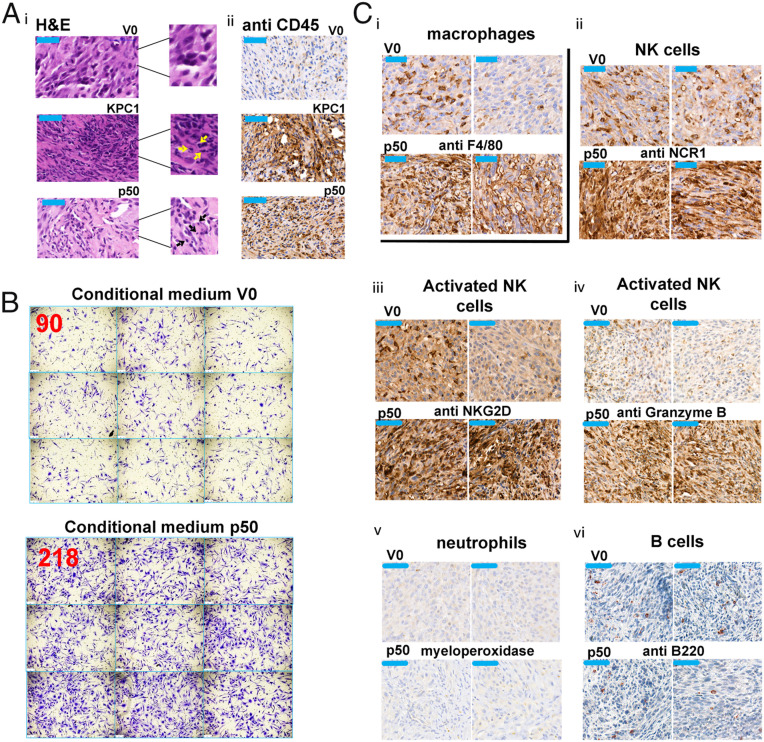
Hematoxylin eosin and immune staining for CD45 (pan-leukocyte marker) of xenografts derived from U87-MG cells expressing V0, KPC1, or p50 revealed that both KPC1- and p50-overexpressing tumors are highly infiltrated with leukocytes (Fig. 3A).
Fig. 3.
KPC1 and p50 recruit immune cells to the U87-MG–derived xenografts. (A) Hematoxylin eosin staining (i) and immunohistochemical staining for CD45 (ii) in xenografts of U87-MG cells stably expressing V0, Myc-KPC1, or FLAG-p50. All scale bars (in the upper left corner of each panel), 50 µm. Arrows indicate infiltrated immune cells. Tumors were grown in nude mice and stained as described under the Materials and Methods. (B) Conditional medium of U87-MG cells expressing FLAG-p50 recruits immune cells in a transwell migration assay. Representative microscopic images of macrophages that migrated through the chamber membrane to the lower compartment that contained the conditional medium. The numbers in red represent the average number of cells/field (out of at least three fields counted) migrated through the membrane. (C) Immunohistochemical staining of the macrophage marker F4/80 (i), the NK markers—NCR1 (ii), NKG2D (iii), and granzyme B (iv), the neutrophils marker myeloperoxidase (v), and the B cells marker B220 (vi) in U87-MG cells-derived tumors expressing either V0 or FLAG p50.
In addition, we used the conditional medium of U87-MG cells that overexpress V0 or p50 in order to test whether it attracts macrophages in a transwell migration assay. As can be seen in Fig. 3B, the medium that is derived from p50-overexpressing cells attracts a significantly higher (∼2.5-fold) number of macrophages compared to a medium derived from control cells.
Last, in order to identify the leukocytes attracted to the p50-overexpressing cells, we immunostained the tumors for specific markers of neutrophils (myeloperoxidase), B cells (B220), NK cells (NCR1), and macrophages (F4/80). We found that the p50-expressing tumors were infiltrated with macrophages (Fig. 3C, i) and NK cells (Fig. 3C, ii), but not with neutrophils (Fig. 3C, v) or B cells (Fig. 3C, vi). Notably, the NK cells infiltrated to the p50-expressing tumors were activated, as is evident from the positive staining for the NKG2D (Fig. 3C, iii) and Granzyme B (Fig. 3C, iv) markers. We suggest that activation of NK cells was possible due to the reduced expression of PD-L1 on the surface of p50-expressing U87-MG cells.
Involvement of Immune Cells in the p50-Mediated Tumor-Suppressive Mechanism.
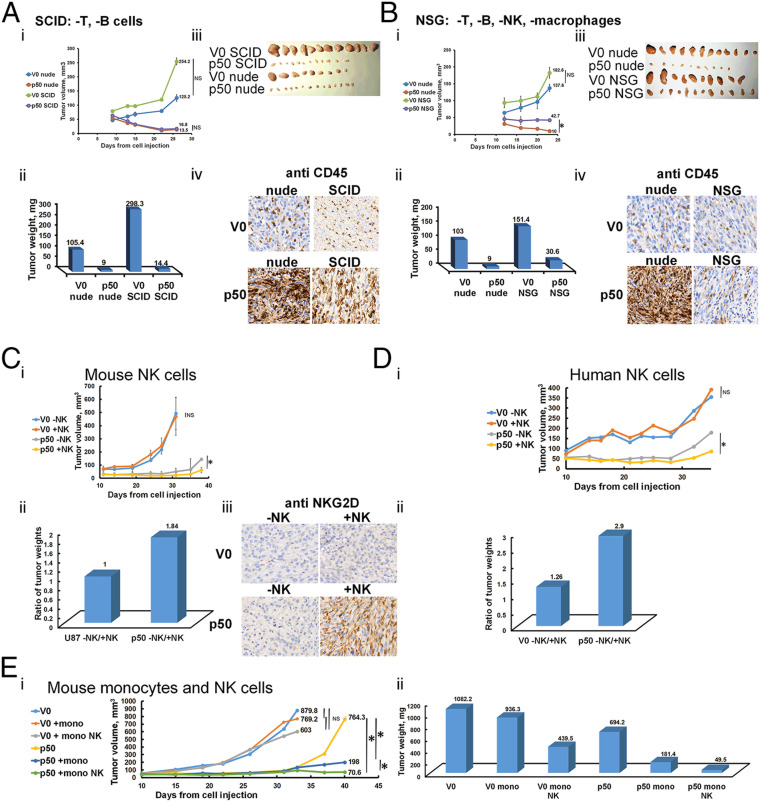
Since the p50-overexpressing tumors recruit the host immune cells, it was important to study whether the recruited cells are involved in tumor suppression. For this purpose, we compared growth of xenografts derived from U87-MG cells overexpressing V0 or FLAG-p50 cells in several types of immune-compromised mice: 1) nude mice that lack only T cells; 2) severe combined immunodeficiency (SCID) mice that lack T and B cells; and 3) Nod SCID gamma (NSG) mice that lack T, B, and NK cells, and macrophages. We found that in NSG mice, the tumor-suppressive effect of p50 was significantly smaller than that observed in SCID mice compared to nude mice (Fig. 4 A and B). This result strongly suggests the contribution of NK cells and/or macrophages to the suppressive mechanism of p50. To demonstrate directly the effect of NK cells on tumor growth, we generated in NSG mice xenografts of U87-MG cells expressing V0 or p50. We then replenished to NSG mice NK cells isolated from either mice or human peripheral blood mononuclear cells (PBMCs). Reappearance of NK cells from both origins in the host’s circulation significantly added to the suppressive effect of p50 (Fig. 4 C and D). Furthermore, we injected isolated mouse monocytes that delayed significantly the “escape” of p50-suppressed tumors from the suppressive effect. The effect of monocytes was further strengthened by injection of both monocytes and NK cells (Fig. 4E). It should be noted that the escape from suppression seen in tumors that overexpress p50 (Fig. 4E, i) is probably typical to NSG mice that lack almost all elements of the immune system. Similar tumors grown in nude mice completely regressed after several weeks.
Fig. 4.
Infiltrated immune cells contribute to the tumor-suppressive effect of p50. (A and B) The suppressive effect mediated by p50 is partially abrogated in tumors grown in NSG mice lacking active macrophages and NK cells. Growth rates (i) and weights (ii) of tumors derived from U87-MG cells expressing V0 or FLAG-p50 in SCID and nude (A) or in NSG and nude (B) mice. (iii) The corresponding tumors 4 wk after inoculation in either SCID (A, iii), NSG (B, iii), or nude (A, iii and B, iii) mice. (iv) Immunohistochemical staining for CD45 of U87-MG cells overexpressing V0 or FLAG-p50 in nude and SCID (A, iv), or in nude and NSG (B, iv) mice. (C) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells expressing V0 or FLAG-p50 in NSG mice in the presence (reconstituted) or absence of mouse NK cells. (iii) Immunohistochemical staining for NKG2D in tumors derived from U87-MG cells expressing V0 or FLAG-p50 in the presence (reconstituted) or absence of mouse NK cells. (D) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells expressing V0 or FLAG-p50 in NSG mice in the presence (reconstituted) or absence of human NK cells. (E) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells expressing V0 or FLAG-p50 in NSG mice in the presence (reconstituted) or absence of mouse monocytes or monocytes and NK cells. * in the relevant panels represents P value < 0.05. NS represents nonsignificant.
p50 Regulates Expression of the Chemokines CCL3, CCL4, and CCL5 that Suppress Tumor Growth via Recruitment of Immune Cells.
The finding that host’s immune cells populate p50-expressing tumors prompted us to study the mechanism of their attraction. For that, we monitored the level of different modulators of the immune system that might have been released to the medium of p50-expressing cells. We found that CCL3, CCL4, CCL5, and vascular cell adhesion molecule (VCAM) are significantly up-regulated in p50-overexpressing cells (Fig. 5A). Therefore, it was important to examine whether the increase in their level is involved in the recruitment of the immune cells and the subsequent tumor suppression. For this purpose, we generated glioblastoma xenografts that overexpress the chemokines under control of the doxycycline (DO) promoter. As can be seen in Fig. 5B, i, expression of CCL3, CCL4, and CCL5 suppressed completely tumor growth. Moreover, the expression of the chemokines recruited NK cells to the tumors (Fig. 5B, ii). This result is in line with our observation that p50-coding tumors express a high level of these chemokines (Fig. 5A) and as a result, are infiltrated with immune cells (Fig. 3). In contrast, silencing of VCAM in U87-MG p50-overexpressing cells had no effect on the tumor-suppressive effect of p50 (Fig. 5B, iii).
Fig. 5.
p50 up-regulates cytokines expression that have a tumor-suppressive effect mediated via recruitment of immune cells. (A) Analysis of cytokines secreted by U87-MG cells expressing V0 or FLAG-p50. Representative images (i) and quantification (ii) of cytokines analyzed in a human cytokine array. (B) (i) Growth rates of tumors derived from U87-MG cells overexpressing V0, CCL3, CCL4, or CCL5, induced with DO from day 0 after inoculation of the cells in nude mice. (ii) Immunohistochemical staining for CD45 and NCR1 of V0-, CCL3-, CCL4-, or CCL5-expressing tumors. (iii) Growth rate of tumors derived from U87-MG cells overexpressing FLAG-p50 and in which VCAM was silenced. (C) Growth rates (i) and weights (ii) of tumors derived from U87-MG cells expressing V0 or CCL5 induced with DO on day 23 after inoculation of the cells in nude mice. (iii) Shown are tumors derived from cells expressing V0 or CCL5 5 wk after inoculation. Induction of CCL5 was done by addition of DO to the drinking water of the mice at day 23. (D) Representative microscopic images of macrophages that migrated through the chamber membrane to the lower compartment that contained conditional medium of U87-MG cells that express V0 or p50, in the presence or absence of anti-CCL3/4, and/or anti-CCL5, as indicated. The numbers in red represent the average number of cells/field migrated through the membrane. At least three fields were counted. * represents P value < 0.05. NS represents nonsignificant.
To bring this finding closer to the clinical reality, where tumors are diagnosed when they are already developed, it was interesting to check if expression of the chemokines can suppress the growth after the tumor has already been established (rather than inducing their expression on the day of inoculation). We found that CCL5, that its expression initiated 23 d after inoculating the cells in the mouse, has nevertheless exerted its suppressive effect on tumor growth (Fig. 5C).
In order to explore further the mechanism of recruitment of immune cells to the p50-expressing tumor, we examined the contribution of each chemokine. For that, we attracted macrophages to the conditional medium of U87-MG cells expressing either V0 or p50 and added to the medium neutralizing antibodies to CCL3 and CCL4 (a single antibody) or CCL5. We demonstrated that the antibody to CCL3 and 4 inhibited macrophages attraction, and the addition of the antibody to CCL5 strengthened this effect even further (Fig. 5D). Surprisingly, the antibody to CCL5 by itself did not alter the attractive effect of the conditional medium (Fig. 5D), although overexpression of the chemokine did attract NK cells (Fig. 5B, ii; although its effect was weaker than that of CCL3 and 4) and suppressed tumor growth (Fig. 5 B, i and C, i). The difference may be due to a dose effect.
Discussion
The discovery of involvement of the host’s immune system in attenuating tumor growth and the resulting development of different modalities of immune therapy have revolutionized the landscape of cancer treatment in recent years. The immune system can be recruited to combat tumors in several different ways, among them inhibition of immune checkpoints, generation of specific CAR-Ts, immunization against specific tumor antigens, and administration of cytokines that recruit the cellular immunity machineries (34).
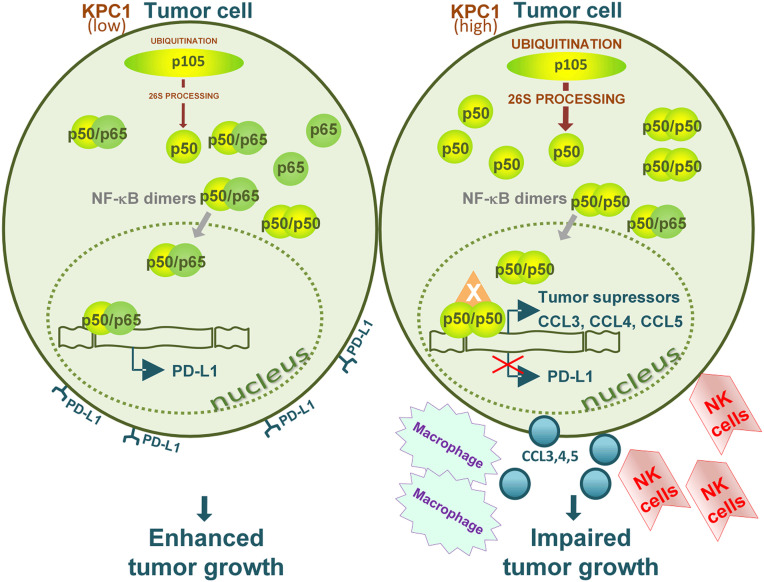
In a previous study (22) we demonstrated that processing of the long and inactive NF-ĸB p105 precursor to the active p50 subunit is mediated by the KPC1 ubiquitin ligase. The generation of stoichiometric excess of p50 over its canonical oncogenic p65 partner turned out to have a strong tumor-suppressive effect. Initial dissection of the mechanism of tumor suppression revealed that the putative p50 homodimers stimulate—under conditions when KPC1 is overexpressed—expression of a broad array of tumor suppressors (22). A further search for the mechanism(s) that underlie the effect of the tumor suppressors with the expectation that apoptosis might be involved unraveled a surprising finding—the shrinking tumors were infiltrated with the host’s (mouse) leukocytes, and the infiltration was dependent on the presence of either overexpressed KPC1 or p50 (Fig. 3). The immediate obvious questions raised are the nature of these white blood cells and the mechanism(s) that mediates their attraction to the tumor. Use of a variety of antibodies against different immune system’s cells and of different mice with different repertoires of immune cells revealed that the infiltrating cells are mostly macrophages and activated NK cells (Figs. 3 and 4). To test whether the infiltrating immune cells have an effect on tumor growth, or whether they are bystanders, we replenished NK (human and mouse) and monocytes (mouse; that are converted to macrophages in the tumor) to mice that lack functional immune cells. Replenishment of the cells resulted in tumor growth inhibition (Fig. 4). As for the mechanism of their attraction, measurement of cytokines secreted by the tumor cells showed that the p50-containing transcription factor stimulates secretion mostly of CCL3, 4, and 5 (Fig. 5). These cytokines led, in turn, to recruitment of the NK cells and macrophages (Fig. 5). In sum, we describe an aspect of involvement of the immune response against tumors where the KPC1 ubiquitin ligase ubiquitinates the long and inactive p105 precursor of NF-ĸB which is consequently processed by the 26S proteasome to the active p50 subunit. Excess of this subunit generates apparently a form of a transcription factor, which is probably a p50 homodimer, which is joined by an as yet unknown third subunit that has a transactivation domain. This factor initiates a transcription program that transcribes an array of cytokines which are secreted by the tumor cells and lead to recruitment of immune cells to the tumor which in turn help to suppress its growth (the sequence of events of this pathway is depicted in Fig. 6).
Fig. 6.
A graphical scheme describing the mechanism of tumor suppression by the ubiquitin ligase KPC1. Overexpressed KPC1 (Right) stimulates the limited processing of the NF-ĸB p105 inactive and long precursor to yield excess of the p50 subunit of the mature dimeric transcriptional regulator. In the absence of stoichiometrically available partner—typically p65—p50 probably homodimerizes. Along with an as yet unidentified additional subunit that contains a transactivation domain (X), it suppresses the expression of PD-L1 and stimulates the expression of a set of chemokines. This in turn attracts to the tumor activated NK cells and macrophages. Along with stimulation of expression of tumor suppressors (22), the overall effect of overexpressed KPC1 is strong tumor suppression.
Interestingly, we found an additional mechanism by which p50 mediates its tumor-suppressive effect. It down-regulates the expression of the immune checkpoint molecule PD-L1, which also results in tumor shrinkage (Fig. 2).
Thus, it appears that KPC1 and its downstream product p50 act on several layers to suppress tumor growth. They 1) activate transcription of tumor suppressors; 2) suppress expression of PD-L1; and 3) recruit immune cells via stimulation of secretion of an array of cytokines. It should be noted that our data concerning the recruitment of the immune system is an additional and independent decaying mechanism to the already strong effect of the p50-induced expression of tumor suppressors. Therefore, it is not an all or none effect, but rather a partial one. Furthermore, trying to isolate each immune effect independently from the other (e.g., the effect on tumor growth by overexpressing PD-L1, or the effect of replenishing the white blood cells) is even more partial. It should be noted, however, that all partial effects are nevertheless statistically significant.
The study also unraveled an interesting, yet still elusive aspect of the mode of action of the family of NF-ĸB transcription factors. It has been known that NF-ĸB can be both oncogenic and tumor-suppressive (35). It appears that these Dr. Jekyll and Mr. Hyde characteristics depend not only on tissue context and the tumor’s microenvironment, but also on the dimeric composition of the factor. Thus, while p50p65 is oncogenic, the suggested homodimeric p50 is tumor-suppressive. Indeed, reconstitution of the “normal” stoichiometric relationship between the two by overexpressing p65 in p50-overexpressing cells abrogated—although partially—the tumor-suppressive effect of p50 (Fig. 1) and resulted in reinitiation of expression of PD-L1 (Fig. 2). The partial effect may be due to the fact that even under conditions where p65 is overexpressed, there is a sufficient quantity of p50 dimers to still exert the suppressive effect. An interesting and obvious question that is related to the quantitative relationship between the two major subunits of NF-ĸB—p50 and p65—has to do with tumor initiation, development, and aggressiveness. One can imagine that the effect is continuous: with a very high proportion of p50, the overall effect is suppressive (a situation that may prevail in normal tissues). As the proportion gradually changes, the suppressive effect disappears and is gradually converted and becomes oncogenic. While it is difficult to titrate and monitor the proteins’ level in a transfection experiment, we can imagine that in different “real” tumors, the proportion varies and, accordingly, so does the aggressiveness of the tumors. Therefore, it will be interesting to monitor directly the levels of the transcripts of the two subunits in different tumors and healthy tissues (and, if possible, the level of the appropriate proteins) and to compare them to the development of the tumor and its aggressiveness—both histologically and clinically.
Materials and Methods
Materials.
Tissue culture sera, media, and supplements for growing U87-MG American Type Culture Collection HTB 14 (ATCC HTB14), human embryonic kidney (HEK)293T, and bone marrow-derived macrophage (BMDM) were from Biological Industries. For growing primary human NK cells, we purchased cell culture media from CellGenix, sera from Sigma, supplements from Biological Industries, and human interleukin (IL)-2 from Peprothec. Collagenase I was from Sigma, and Dispase II was from Roche. Murine MCSF, granulocyte-macrophage colony-stimulating factor (GM-CSF), Interferon (INF)γ, and IL-4 were from Peprothec. Lipopolysaccharides (LPS) was from Sigma. Eosin 1% Aqueous and Haematoxylin (Harris) were from Pioneer Research Chemicals Ltd (PRC). Rabbit anti-CD45 (ab 10558), anti-F4/80 (ab 111101), anti-NCR1 (ab 199128), anti-NKG2D (ab 203353), anti-Granzyme B (ab 4059), and rat anti-CD45R/B220 (RA3-6B2; ab64100) antibodies for immunohistochemistry were from Abcam. Polyclonal antimyeloperoxidase was from Dako. iVIEW DAB Detection Kit for immunohistochemistry staining was from Ventana Medical Systems. Mouse anti-CCL5/RANTES (21445) and anti-CCL3/CCL4 (93321) antibodies for neutralization of the appropriate proteins were from R&D Systems. For Western blotting, mouse anti-FLAG (M2) antibody was from Sigma, rabbit anti–PD-L1 (E1L3N) antibody was from Cell Signaling Technology, and rabbit anti-p65 (C-20) antibody and mouse anti-actin were from Millipore. Peroxidase-conjugated (for Western blotting) secondary antibody was from Jackson ImmunoResearch Laboratories. Bradford reagent and materials for SDS/PAGE were from Bio-Rad. CalFectin Mammalian DNA Transfection Reagent was from Signagen Laboratories. Short hairpin RNAs (shRNA) were synthesized by Dharmacon. TaqMan Gene Expression Assay and TaqMan Fast Universal PCR Master Mix were from Applied Biosystems. Oligonucleotides were from Syntezza Bioscience. Restriction enzymes were from New England Biolabs. Falcon Permeable Support for 24-well plate with transparent polyethylene terephthalate (PET) membrane was from Corning. Doxycycline Hyclate was from Sigma. Proteome Profiler Human XL Cytokine Array Kit was from R&D Systems. EasySep Human NK Cell Isolation Kit, EasySep Mouse NK Cell Isolation Kit, and EasySep Mouse Monocyte Isolation Kit, were from STEMCELL technologies. Human PBMCs as a source for isolating human NK cells were from AllCells. Mouse anti-CD274 (PD-L1) (29E.2A3) and mouse anti-CD45 (HI30) for mass cytometry were from Bio Legend. BALB/cOlaHsd-Foxn1nu and C.B-17/IcrHsd-Prkdcscid mice were from Envigo, and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were from Jackson Laboratories (36) (JAX stock #005557). RNA purification kit was from Macherey-Nagel, and complementary DNA (cDNA) synthesis kit was from PCR Biosystem Ltd. Lenti-X Tet-One Inducible Expression System (pLVX) was from Invitrogene.
Methods.
Plasmid construction.
cDNAs coding for Myc-KPC1 and FLAG-p50 were described previously (22). cDNAs coding for p65 and PD-L1 were amplified and cloned into the NSPI-CMV MCS lentiviral expression vector (37) between BamHI and XhoI, or XhoI and AgeI, respectively. CCL3, CCL4, and CCL5 were amplified and cloned into the pLVX lentiviral expression vector between EcoRI and BamHI.
Cultured cells.
Dulbecco’s Modified Eagle Medium (DMEM) that contains penicillin–streptomycin and 10% fetal calf serum was used for growing U87-MG and HEK293T cells (at 37 °C and 5% CO2). Primary human NK cells were grown at 37 °C and 5% CO2 in Good Manufacturing Practice Stem Cell Growth Medium (GMP SCGM) medium supplemented with 10% human AB plasma, 1 mM sodium pyruvate, 1% nonessential amino acids, penicillin–streptomycin, 10 mM Hepes, and 300 Unit/mL IL-2 (38). Mouse BMDM were grown at 37 °C and 5% CO2 in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% fetal calf serum, penicillin–streptomycin, and 10 ng/mL macrophage colony-stimulating factor (MCSF) or 10 ng/mL MCSF and IL-4.
Stable transfection.
U87-MG cells were transfected with cDNAs coding for Myc-KPC1, FLAG-p50, p65, PD-L1, CCL3, CCL4, and CCL5, or with shRNA against PD-L1 (clones V2LHS_53668 and V3LHS_315809) using a lentiviral transduction system. Cells that stably expressed the proteins or shRNA were selected by adding puromycin (5 μg/mL) to the growth medium.
Tumorigenicity.
U87-MG cells that stably express Myc-KPC1, FLAG-p50, p65, PD-L1, CCL3, CCL4, and CCL5 were harvested at the exponential growth phase. Cells were dissociated with trypsin, washed with phosphate buffered saline (PBS), and diluted to 60 × 106 cells/mL. Cell suspension (6 × 106 cells; 0.1 mL) was injected s.c. at both flanks of 6- to 10-wk-old BALB/cOlaHsd-Foxn1nu, C.B-17/IcrHsd-Prkdcscid, or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Grown xenografts were measured, resected, and prepared for further analysis as described previously (22). Animal experiments were approved by the Animal Care Committee of the Technion in Haifa, Israel.
qRT-PCR.
RNAs were isolated (using an RNA purification kit) from U87-MG cells overexpressing Myc-KPC1, FLAG-p50, p65, or an empty vector. cDNAs were obtained from RNAs in a reverse transcription reaction using cDNA Synthesis Kit. The qRT-PCR was performed in triplicates with primers for PD-L1 or HPRT (control) genes using the TaqMan Gene Expression Assay.
Isolation of primary NK cells and monocytes.
Primary human NK cells were isolated from frozen PBMCs using the negative isolation-based EasySep Human NK Cell Isolation Kit according to the manufacturer’s protocols.
Primary mouse NK cells and Primary Mouse Monocytes were isolated from spleens or bone marrow, respectively, of C57BL/6JOlaHsd mice using the negative isolation-based EasySep Mouse NK Cell or EasySep mouse monocytes isolation kits, respectively, according to the manufacturer’s protocols.
Xenogeneic NK cells adoptive transfer.
NSG mice were infused i.v. (IV) with 4–5 × 106 human NK cells once a week following i.p. (IP) administration of 10 μg hu-IL-2 every other day along the duration of the experiment.
Allogeneic NK cells and monocytes adoptive transfer.
NSG mice were infused (IV) with 0.5–1 × 106 mice NK cells and/or with 1–2 × 106 monocytes once a week along the duration of the experiment.
BMDM differentiation and transmigration.
Bone marrow from the tibia and fibula of C57BL/6JOlaHsd mice was flushed out and prepared for culture as described above (39). BMDMs were grown in complete IMDM supplemented with 10 ng/mL MCSF. After 7 d, the growth medium was replaced with complete IMDM supplemented with 10 ng/mL MCSF and 10 ng/mL IL-4. On day 10, the growth medium was changed to a starvation medium that contained 1% fetal bovine serum (FBS). After 24 h of starvation, 2 × 105 BMDMs were seeded on a Falcon permeable chamber with 8.0 µm transparent PET membrane. To produce the conditional medium, 24 h prior to the transmigration assay, the growth medium of exponentially growing U87-MG cells that expressed either an empty vector (V0) or FLAG-p50 was changed to a starvation medium supplemented with 1% FBS. The media were then filtered using a 45-µm polyvinylidene fluoride (PVDF) filter and added to the lower part of the transmigration chamber. The BMDMs placed in the upper chamber were allowed to migrate (at 37 °C for 3 h) into the lower compartment that contains the conditional medium isolated from the U87-MG cells that express V0 or FLAG-p50. Antibodies that neutralize CCL5 and/or CCL3/4 were added to the conditional medium as indicated.
After the incubation, the membranes of the transmigration chamber were fixed with 4% Paraformaldehyde (PFA) for 10 min, stained with 0.4% Crystal Violet (70% ethanol solution) for 10–15 min, and extensively washed with H2O, followed by removal of cells’ debris from the upper side of the membrane with cotton swabs. Quantification (of the cells adhering to the lower side of the membrane) was carried out by Image-Pro Premier.
Cytokine profiling.
U87-MG cells that overexpress V0 or FLAG-p50 were grown in DMEM supplemented with 1% FBS. After 48 h, the conditional media were collected and incubated with The Proteome Profiler Human XL Cytokine Array membrane according to the manufacturer’s protocols.
Immunohistochemistry.
Immunohistochemical staining of 5-µm tissue sections of formalin-fixed, paraffin-embedded, U87-MG–derived tumors was performed using a Ventana BenchMark ULTRA IHC/ISH system. Sections were immunostained for CD45 (1:3,000), CD45R/B220 (1:200), myeloperoxidase (1:5,000), F4/80 (1:500), NCR1 (1:200), NKG2D (1:200), and Granzyme B (1:100). Visualization was carried out using the Ventana iVIEW DAB detection according to the manufacturer’s protocols.
Mass cytometry analysis of tumors.
Twenty-five days after injection of U87-MG cells that overexpress either FLAG-p50 or V0, xenografts were resected and prepared as single-cell suspensions as described previously (40). Cells were stained using different metal-tagged antibodies. The acquisition was performed using Fluidigm CyTOF mass cytometry. FlowJo v9.3 (TreeStar) software was used for the analysis of the FCS 3.1 files.
Statistical analysis.
Data in graphs represent the mean ± SD. Experiments were statistically analyzed by the two-tailed Student’s t test. A P value < 0.05 was considered as statistically significant.
Acknowledgments
A.C. and E.P. were supported by a grant from the Adelson Medical Research Foundation. A.C. was supported by grants from the Israel Science Foundation and by a grant from the Albert Sweet Foundation via the American Technion Society. A.C. is an Israel Cancer Research Fund Professor. We thank Dr. Bassem Fares for his technical help.
Footnotes
The authors declare no competing interest.
Data Availability.
All study data are included in the article.
References
- 1.Sen R., Baltimore D., Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Etzioni A., Ciechanover A., Pikarsky E., Immune defects caused by mutations in the ubiquitin system. J. Allergy Clin. Immunol. 139, 743–753 (2017). [DOI] [PubMed] [Google Scholar]
- 3.DiDonato J. A., Mercurio F., Karin M., NF-κB and the link between inflammation and cancer. Immunol. Rev. 246, 379–400 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi K., Karin M., NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Siggers T., et al. , Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 13, 95–102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts J. C., Nabel G. J., Differential regulation of NF-kappaB2(p100) processing and control by amino-terminal sequences. Mol. Cell. Biol. 16, 6363–6371 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan C.-M., Maniatis T., Generation of p50 subunit of NF-kappa B by processing of p105 through an ATP-dependent pathway. Nature 354, 395–398 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T., The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kB. Cell 78, 773–785 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Hayden M. S., Ghosh S., NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 26, 203–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napetschnig J., Wu H., Molecular basis of NF-κB signaling. Annu. Rev. Biophys. 42, 443–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman M. M., McFadden G., Modulation of NF-κB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagerlund R., Kinnunen L., Köhler M., Julkunen I., Melén K., NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 280, 15942–15951 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Ben-Neriah Y., Karin M., Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12, 715–723 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Xia Y., Shen S., Verma I. M., NF-κB, an active player in human cancers. Cancer Immunol. Res. 2, 823–830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giopanou I., et al. , Metadherin, p50, and p65 expression in epithelial ovarian neoplasms: An immunohistochemical study. BioMed Res. Int. 2014, 178410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla S., et al. , Nuclear factor-kappaB/p65 (Rel A) is constitutively activated in human prostate adenocarcinoma and correlates with disease progression. Neoplasia 6, 390–400 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkin S., Pikarsky E., NF-κB in Liver Cancer: The Plot Thickens (Springer, Berlin, 2010). [DOI] [PubMed] [Google Scholar]
- 18.Cartwright T., Perkins N. D., L Wilson C., NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 283, 1812–1822 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Voce D. J., et al. , Nfkb1 is a haploinsufficient DNA damage-specific tumor suppressor. Oncogene 34, 2807–2813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider G., et al. , IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 25, 3801–3812 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloom J., Pagano M., Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 13, 41–47 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Kravtsova-Ivantsiv Y., et al. , KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 p105 to p50 restricts tumor growth. Cell 161, 333–347 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Bao B., et al. , The immunological contribution of NF-κB within the tumor microenvironment: A potential protective role of zinc as an anti-tumor agent. Biochim. Biophys. Acta 1825, 160–172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betzler A. C., et al. , NF-κB and its role in checkpoint control. Int. J. Mol. Sci. 21, 3949 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giampazolias E., et al. , Mitochondrial permeabilization engages NF-κB-dependent anti-tumour activity under caspase deficiency. Nat. Cell Biol. 19, 1116–1129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopewell E. L., et al. , Lung tumor NF-κB signaling promotes T cell-mediated immune surveillance. J. Clin. Invest. 123, 2509–2522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilgelm A. E., Richmond A., Chemokins modulate immune surveillance in tumorignesis, metastatsis, and response to immunotherapy. Front. Immunol. 10, 333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coppola D., et al. , Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am. J. Pathol. 179, 37–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messina J. L., et al. , 12-Chemokine gene signature identifies lymph node-like structures in melanoma: Potential for patient selection for immunotherapy? Sci. Rep. 2, 765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong H., et al. , Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Ray A., et al. , Targeting PD1-PDL1 immune checkpoint in plasmacytoid dendritic cell interactions with T cells, natural killer cells and multiple myeloma cells. Leukemia 29, 1441–1444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu J., et al. , Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Invest. 128, 4654–4668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim S. O., et al. , Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell 30, 925–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei X., et al. , Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 470, 126–133 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Pikarsky E., Ben-Neriah Y., NF-kappaB inhibition: A double-edged sword in cancer? Eur. J. Cancer 42, 779–784 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Ohbo K., et al. , Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor γ chain. Blood 87, 956–967 (1996). [PubMed] [Google Scholar]
- 37.Akiri G., et al. , Wnt pathway aberrations including autocrine Wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene 28, 2163–2172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edri A., et al. , The Ebola-Glycoprotein modulates the function of Natural killer cells. Front. Immunol. 9, 1428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying W., Cheruku P. S., Bazer F. W., Safe S. H., Zhou B., Investigation of macrophage polarization using bone marrow derived macrophages. J. Vis. Exp. 76, 50323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timaner M., Beyar‐Katz O., Shaked Y., Analysis of the stromal cellular components of the solid tumor microenvironment using flow cytometry. Curr. Protoc. Cell Biol. 70, 19.18.1–19.18.12 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article.