Significance
The Bateson–Dobzhansky–Muller (BDM) model describes negative epistatic interactions that occur between genes with a different evolutionary history to account for hybrid incompatibility and is a central theory explaining genetic mechanisms underlying speciation. Since the early 1900 s when the BDM model was forwarded examples of BDM incompatibility have been described in only a few nonvertebrate cases. This study reports the only vertebrate system, with clearly defined interacting loci, that supports the BDM model. In addition, this study also poses that tumorigenesis serves as a novel mechanism that accounts for postzygotic isolation.
Keywords: hybrid incompatibility, evolution, genetics, Bateson–Dobzhansky–Muller model, Xiphophorus
Abstract
Mixing genomes of different species by hybridization can disrupt species-specific genetic interactions that were adapted and fixed within each species population. Such disruption can predispose the hybrids to abnormalities and disease that decrease the overall fitness of the hybrids and is therefore named as hybrid incompatibility. Interspecies hybridization between southern platyfish and green swordtails leads to lethal melanocyte tumorigenesis. This occurs in hybrids with tumor incidence following progeny ratio that is consistent with two-locus interaction, suggesting melanoma development is a result of negative epistasis. Such observations make Xiphophorus one of the only two vertebrate hybrid incompatibility examples in which interacting genes have been identified. One of the two interacting loci has been characterized as a mutant epidermal growth factor receptor. However, the other locus has not been identified despite over five decades of active research. Here we report the localization of the melanoma regulatory locus to a single gene, rab3d, which shows all expected features of the long-sought oncogene interacting locus. Our findings provide insights into the role of egfr regulation in regard to cancer etiology. Finally, they provide a molecular explainable example of hybrid incompatibility.
In the late 1920s, three investigators, Myron Gordon, Georg Haeussler, and Kurt Kosswig, independently found that hybrids between two distant Xiphophorus fish species, Xiphophorus maculatus (southern platyfish), and Xiphophorus hellerii (green swordtail), develop spontaneous and lethal pigment cell tumors that were later determined to be melanoma (1–3). Since its establishment, this model system has been intensively studied to assess the underlying genetic contributions to tumor etiology. The development of hybridization-induced tumor has been viewed as a representation of the genome incompatibility hypothesis known as the Bateson–Dobzhansky–Muller (BDM) model (4–8). The BDM model states that negative epistatic interactions in hybrids serve as the molecular genetic mechanisms underlying genome incompatibility and is associated with problems in hybrid fitness. Although BDM incompatibility was identified in a few model organisms (9), Xiphophorus and mice represent the only vertebrate systems in which the incompatible loci have been identified (10, 11).
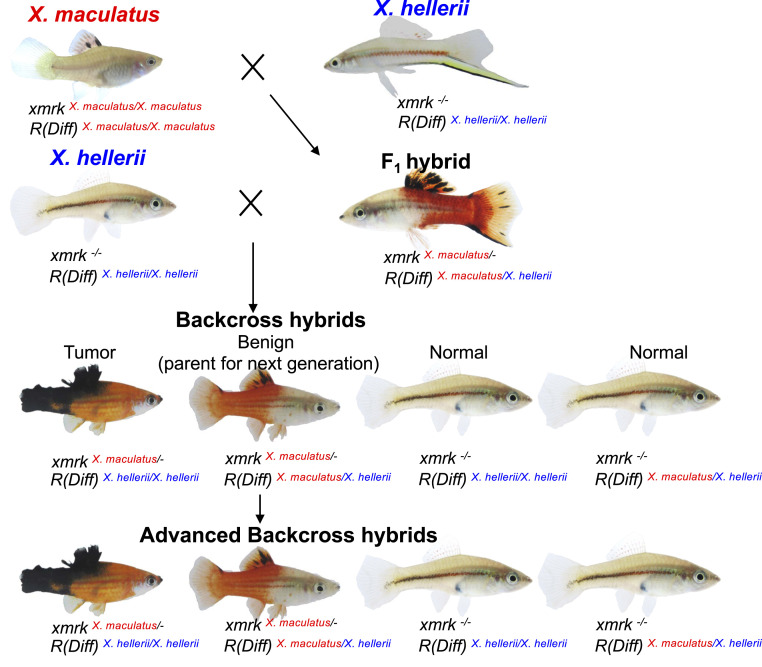
This Xiphophorus interspecies hybrid system, now termed the “Gordon–Kosswig–Anders (GKA) model,” was first described by Gordon and Kosswig in 1920s. This model employs crossing of X. maculatus and X. hellerii to produce F1 interspecies hybrids. X. maculatus exhibits a nevus-like pigmentation pattern in its dorsal fin (spotted dorsal, Sd), while X. hellerii does not exhibit this trait. In the F1 hybrid, the Sd pigmentation pattern becomes expanded, with melanin pigmentation covering the entire dorsal fin due to melanocyte hyperplasia (12, 13). Backcrossing the F1 hybrid to the X. hellerii parent leads to three distinct phenotypes among the backcross (BC) progeny that follow Mendelian distributions: 25% of hybrids exhibit hyperplasia of pigmentation pattern as observed in the F1 hybrid, 25% exhibit lethal and invasive nodular exophytic melanoma, and the remaining 50% of progeny do not display a black pigmentation pattern (14) (Fig. 1). The hybridization-induced disease observed in Xiphophorus interspecies hybrid represents a type of genetic incompatibility. In 1950s, Anders argued this spontaneous tumorigenesis is due to segregation of two loci from X. maculatus; one was named Tu for “tumor” and another locus was named R for “repressor” or Diff for “differentiation” [hereafter referred to as R(Diff)]. These concepts led to what we now know as oncogenes and tumor suppressors (15, 16). In the late 1980s, it was shown the Tu gene encodes a mutant duplicate copy of egfr, and this gene was named as Xiphophorus melanoma regulatory kinase (xmrk) (17, 18). The xmrk oncogene is tightly linked to or part of the Sd locus and controls melanocyte proliferation.
Fig. 1.
GKA model crossing scheme. The crossing scheme shows the Xiphophorus species used to produce F1 and BC1 interspecies hybrids. X. maculatus Jp163 A and X. hellerii are used to produce F1 hybrids artificially. The F1 hybrids are subsequently backcrossed to X. hellerii to produce BC hybrid progeny. BC hybrids exhibiting melanocyte hyperplasia and heterozygous R(Diff) are used as parents for next-generation BC.
In addition, the GKA model offers us a natural two-hit melanoma model wherein the oncogenic effect of xmrk can be fully eliminated by a regulatory locus that must have coevolved with xmrk (19). The EGFR gene is one leading oncogene of many human cancers (20). It is preproliferative and is an upstream activator of BRAF and NRAS signaling, which are the driver oncogenes in over 50% of all human melanomas (21). This evidence promotes the characterization of R(Diff) as having significant implications in cancer etiology. Therefore, identifying the R(Diff) gene will highlight the genetic interactions underlying the tumor induction due to hybrid incompatibility and in addition may forward novel molecular target(s) in regulation of EGFR function for human disease control.
However, the R(Diff) gene has not been identified. Previous effort to define the R(Diff) locus forwarded a chromosomal region that includes cdkn2ab, an ortholog of the human tumor suppressor genes CDKN2A and B, which is mutant in 10% of high-melanoma-risk families (22–27). In this study, we found cdkn2ab is not the R(Diff) gene but is tightly linked to it. More importantly, we have identified the long-hypothesized R(Diff) locus to a strong candidate gene, rab3d.
Results
A Region on Chromosome 5 Determines Malignancy of Melanocytic Lesions.
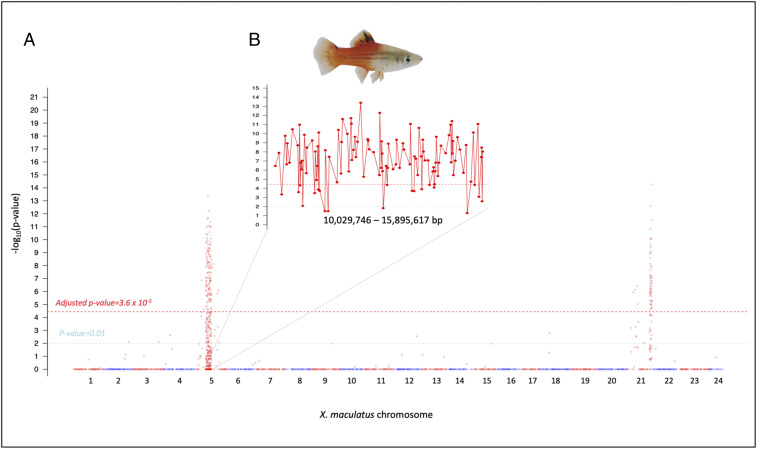
Hybrids between X. maculatus and X. hellerii exhibit enhanced pigmentation in their dorsal fin (i.e., melanocyte hyperplasia) due to hemizygosity for the xmrk oncogene and heterozygosity of the R(Diff) locus [i.e., xmrkX. mac/-; R(Diff)X. mac/X.hel]. This phenotype and genotype are also present in ∼25% of BC hybrid progeny (Fig. 1). Due to meiotic recombination, successive backcrossing of such animals to X. hellerii should stepwise reduce heterozygosity in the advanced BC progeny and finally result in a xmrkX. mac/-; R(Diff)X. mac/X.hel isogenic line (i.e., introgression). In this manner, advanced BC (BCn) fish were produced in order to validate the candidate gene cdkn2ab (chromosome 5, 15.8 Mbp) as a locus carrying R(Diff) function. The maternal parent (i.e., interspecies hybrid) for each successive BC generation was genotyped for inheritance of cdkn2abX. mac and only heterozygous hybrids (i.e., xmrkX. mac/-, cdkn2abX. mac/X.hel) exhibiting the enhanced dorsal fin pigmentation were selected for a next round of backcrossing. χ2 tests were performed on genotyping data of each variant site collected from 90 BCn hybrids that exhibited melanocyte hyperplasia to locate genes that displayed ancestral allele linkage disequilibrium and predominately showed a heterozygous inheritance pattern (SI Appendix, Figs. S4 and S5). Two genomic regions were found to correlate to the hyperplasia phenotype (Fig. 2A): one on chromosome 21 which encompasses xmrk, the melanoma driver oncogene that induces melanocyte proliferation, and a second region on chromosome 5 that corresponds to a previously mapped R(Diff) region of 5.8-Mbp region harboring the candidate cdkn2ab gene (Fig. 2B). An average loss of 50% heterozygous loci per BC generation is expected. Therefore, one expects to see a heterozygous region only accounting for an average of 5.5 and 2.7 Mbp of the 700-Mbp genome for BC7 and BC8 individuals, respectively. However, haploid maps produced from these advanced BC animals show the heterozygous content to be much higher than this expectation. In addition, heterozygous loci are predominantly surrounding the cdk2ab region (SI Appendix, Figs. S3 and S6), suggesting selection of individuals that exhibited melanocyte hyperplasia and a genotype of cdkn2abX. mac/X.hel for further backcrossing coselected an adjacent locus on chromosome 5, rendering higher-than-expected heterozygosity in BCn hybrids. These observations indicate that cdkn2ab itself is less likely to be R(Diff), while the coselected locus with cdkn2ab is the true R(Diff).
Fig. 2.
Genetic mapping of heterozygous loci in advanced BC hybrids. A total 90 BC hybrids (BC2 to BC8) that exhibited dorsal fin melanocyte benign hyperplasia and were produced by crossing cdkn2ab genotyped hybrid (cdkn2abX. mac/X. hel) with X. hellerii (cdkn2abX. hel/X. hel). (A) Manhattan plot showing –log10P value (χ2 test) across the genome. The y axis represents –log10P value and the x axis represents amplicon chromosomal coordinates, which are labeled as red or blue. Only the −log10 P value of loci that exhibited higher X. maculatus allele frequency is plotted due to introgression. The light blue dashed line represents a P value of 0.01 that is suggestive of statistical significance. χ2 test P values were corrected using Bonferroni method across the genome-wide data. The red dashed line represents adjusted P value of 0.05 corresponding to 3.6 × 10−5. (B) A zoom-in view of the chromosome 5 10,029,746- to 15,895,617-bp region. This region is highly correlated to the pigmentation phenotype observed in BC hybrids.
Genetic Mapping of a Mutant EGFR Regulator Locus.
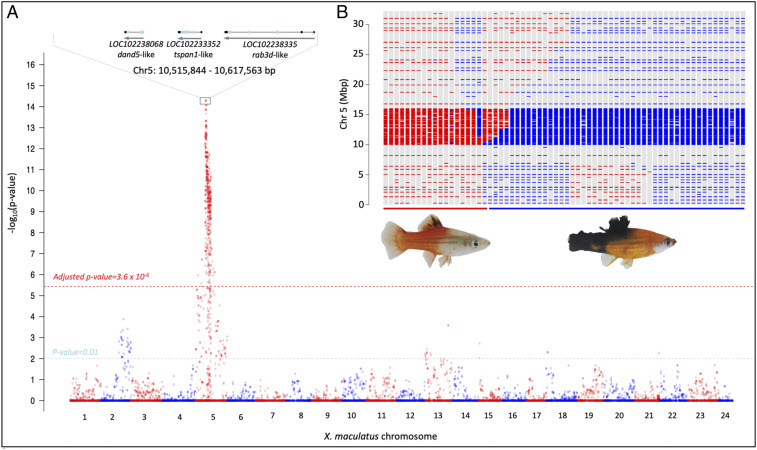
To locate the R(Diff) candidate gene(s) independent of artificial selection of genetic marker, we produced BC interspecies hybrid progeny of the GKA model, that is, X. hellerii (Rio Sarabia) × [X. maculatus Jp163 A × X. hellerii (Rio Sarabia)] and performed targeted genomic sequencing on BC progeny that developed two distinct melanocyte phenotypes: benign hyperplasia and melanoma tumor. Association analyses between parental allele inheritance and pigment cell phenotypes were performed on a total of 66 BC1 progeny (19 hyperplasia and 47 melanoma). Both melanocyte phenotypes are known to be xmrk-dependent; therefore, no association between xmrk and melanocyte phenotypes was observed, as expected (Fig. 3A). Genotypes of four linked polymorphic sites (10,582,852 10,582,855 10,582,868, and 10,582,870) on chromosome 5 are the most significantly correlated to melanocyte phenotypes, where individuals exhibiting benign pigment cell lesions inherited both parental alleles, while melanoma-bearing individuals only inherited the X. hellerii alleles (Fig. 3B).
Fig. 3.
Genetic mapping of the R(Diff) locus. BC1 hybrids were produced by crossing F1 hybrid to X. hellerii. Pigmented hybrids were classified into two categories independent of any molecular marker. Sixty-six hybrids that include 19 exhibiting dorsal fin melanocyte hyperplasia and 47 displaying melanoma were genotyped. The number of animals of each genotype does not reflect the BC hybrid phenotypical distribution because the Xiphophorus Genetic Stock Center preferentially collects tumor-bearing fish for research purposes. (A) Manhattan plot showing –log10 P value of χ2 test across the genome. The y axis represents –log10 P value and the x axis represents amplicon order on each chromosome, which is labeled as red or blue. the light blue dashed line represents P value of 0.01 that is suggestive of statistical significance. χ2 test P values were corrected using Bonferroni method across the genome-wide data. the red dashed line represents adjusted P value of 0.05 corresponding to 3.6 × 10−6. Both melanocyte hyperplasia and melanoma are driven by xmrk, and therefore chromosome 21 is not related to separation of the two melanocyte phenotypes. A strong peak corresponding to linkage disequilibrium is found on chromosome 5, with polymorphisms located at 10,582,852, 10,582,855, 10,582,868, and 10,582,870 bp exhibited the top –log10 P value. These polymorphisms are adjacent to three gene models: dand5, tspan, and rab3d. (B) Chromosome 5 haploid maps of all BC hybrids. Blue and red bars represent genotypes in term of parental allele inheritance (red: heterozygous for both parental alleles; blue: homozygous for X. hellerii allele). Their locations on the bar graph correspond to amplicon physical location. Colored lines underneath the haploid maps represent phenotypes, with the red line corresponding to pigment cell lesion and the blue line corresponding to melanoma. A locus located between 10,515,844 and 10,617,563 bp is free of recombination in both groups of hybrids. Hybrids exhibiting melanocyte hyperplasia inherited both ancestral alleles in this locus, and tumor-bearing hybrids inherited only a recurrent parental (i.e., X. hellerii) allele.
Haploid maps produced from all BC individuals supported the result of association analyses and forwarded a region (i.e., 10,515,844 to 10,617,563) that is free of chromosomal cross-overs in both cohorts (Fig. 3B). The R(Diff) locus is assigned to this 101.7-kbp region (Fig. 3A). As expected, this locus maps to the vicinity (5.2 Mbp) upstream of cdkn2ab.
The rab3d Gene Is the Functional Carrier of the R(Diff) Function.
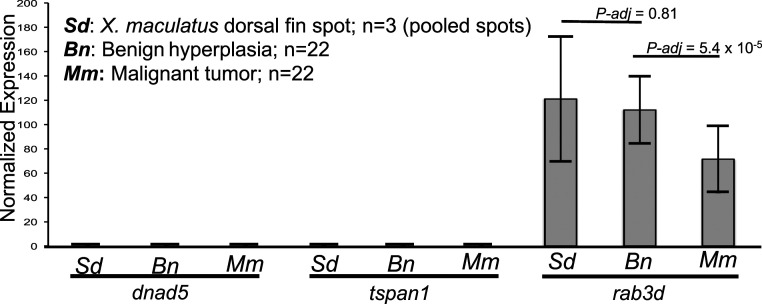
The 101.7-kbp R(Diff) locus encodes three gene models on the reverse strand: differential screening-selected gene aberrative in neuroblastoma (DAN) domain family member 5 (dnad5), tetraspanin-1 (tspan1), and ras-related protein Rab-3D (rab3d). Expression of a gene is the prerequisite for display of genetic function. To determine the gene expression pattern of these three candidate genes and to assess gene expression changes in parental nevus-like dorsal fin melanocyte spots, melanocyte hyperplasia, and melanoma tumor, transcription profiling was performed on these tissues. The rab3d gene is the only gene expressed among the three candidates in melanocyte spots of parental X. maculatus (three pools of dorsal fin spots), dorsal fin melanocyte hyperplasia (n = 22), and melanoma tumor (n = 22) of BC interspecies hybrids (Fig. 4). Therefore, rab3d serves as the only gene that can carry the R(Diff) function.
Fig. 4.
Gene expression profiling of the R(Diff) locus. Bar graph showing dnad5, tspan1, and rab3d expression in X. maculatus dorsal fin spots (Sd), benign hyperplasia (Bn), and malignant tumors (Mm) of BC hybrids. The y axis represents library size-normalized read counts. Error bars represent SD. Differential expression analyses between different genotypes were performed using DESeq2, and P values are corrected using FDR.
In addition, differential expression analysis of rab3d among the parental dorsal fin spots, interspecies hybrid melanocyte expansion, and in melanoma showed rab3d expressed at the same level between the melanocyte spots and hyperplastic pigmented cell lesions (adjusted P value = 0.81; Fig. 4), and expressed at a higher level in melanocyte hyperplasia (adjusted P value = 5.4 × 10−5) than in melanoma of BC fish.
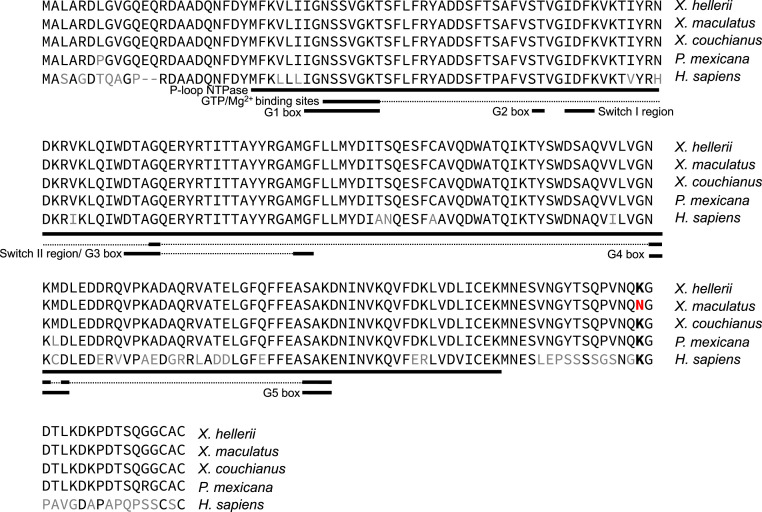
The two parental rab3d alleles differ by an Asn residue in X. maculatus and Lys in X. hellerii at the C-terminus tail downstream of the P-loop domain that harbors the guanosine triphosphatase (GTPase) activity (i.e., Asn/Lys-204; Fig. 5). Although this Asn/Lys site locates in the hypervariable C terminus of rab3d, comparative genomics showed that different from X. maculatus the Lys is conserved in human and 60% of all fish species analyzed (i.e., the next dominating allele is Arg) that include another xmrk-null Xiphophorus species, Xiphophorus couchianus, and other Poeciliidae fish, suggesting the X. maculatus allele is a genetic outlier (Fig. 5 and Dataset S1).
Fig. 5.
Sequence alignment of rab3d genes. Protein sequence comparison between X. hellerii rab3d and X. maculatus, X. couchianus, and Poeciliid fish Poecilia mexicana. Different amino acids between X. hellerii and X. maculatus and between X. hellerii and P. mexicana are labeled in red and black, respectively. Functional domains are labeled with black lines underneath protein sequences, with thinner lines linked functional domain in tertiary structure. The only amino acid change between X. maculatus to other Xiphophorus is Lys-204 > Asn-204 mutation. The Lys is conserved, and only X. maculatus encodes an Asn.
Discussion
The finding that rab3d is the only expressed gene located in the tumorigenesis-determining locus on chromosome 5 in Xiphophorus genome, forwarded by both genetic mapping and transcriptomics, concludes a search for the hypothetical locus R(Diff) that has been ongoing for over five decades (22, 25, 28–31). As a viviparous fish where embryonic development occurs within the female, the technical hurdles involved in transgenesis have not yet been overcome for Xiphophorus. Therefore, genetic manipulation of rab3d cannot be performed in the Xiphophorus system until our current development of Xiphophorus transgenesis is proven successful and efficient. Although a readily available system for such a test is currently not available, the negative epistasis between xmrk and rab3d hallmarks cancer, in addition to hybrid lethality (9) and sterility (32), an innovative mechanism for decreasing hybrid fitness, and reinforcing speciation.
Epitasis underlying human disease can be elucidated by investigating mechanisms that “evolutionary mutant models” developed to cope with similar mutations as in human disease and/or produce adaptive phenotypes that are similar to human disease (19, 33). X. maculatus is one such species where oncogenicity of a mutant EGFR (i.e., xmrk) is compromised by a regulatory allele R(Diff). EGFR is one of the most prevalent oncogenes exhibiting mutation and/or dysregulation in many varied human cancers (34–53). Despite over 40 y of effort in attempting to inhibit EGFR by blocking its kinase activity, and development of four generations of small molecules, or monoclonal antibodies, success in disease control is very limited to three cancer subtypes, that is, nonsmall cell lung cancers with kinase-activating mutations in EGFR (54–56), ∼10% of metastatic colorectal cancers (57, 58), and a subcategory of advanced head and neck cancers (59, 60). Current methodologies to inhibit EGFR attempt to directly block the adenosine 5′-triphosphate (ATP) binding pocket or ligand binding pocket or by targeting acquired mutations that lead to acquired resistance have all turned out to be inefficient in providing promising therapeutic benefit. Therefore, it is of utmost importance to revolutionarily reattack the question of EGFR-associated cancer etiology and identify reliable next-generation treatment strategies that enable disease control with higher response rates and lower resistance. The xmrk, originated from a gene duplication event (61), encodes a mutant EGFR that autodimerizes and activates downstream proproliferative pathways in a ligand-independent manner (62). The xmrk gene is a bona-fide oncogene because its ectopic expression leads to transformation and tumorigenesis of melanocytes in murine cells and medaka fish (63, 64). However, X. maculatus does not exhibit tumorigenesis, suggesting X. maculatus harbors a mechanism [i.e., R(Diff)] in suppressing the driver oncogene. Therefore, the identification of rab3d as gene exhibiting R(Diff) regulatory function delineate mechanisms of how X. maculatus counteracts the deleterious EGFR mutant in its genome. The molecular mechanism underlying rab3d suppression of xmrk can lead to innovative strategies in developing next-generation EGFR inhibitors.
The rab3d genes showed both codon mutation between the two parental alleles and transcriptional differences between normal dorsal fin pigment cells, pigment cell hyperplasia, and melanoma tumor. These results suggest both structural and expression divergences between the two parental alleles of rab3d are essential to elucidate molecular interactions we can learn in order to advance our knowledge in control of EGFR and associated disease. There are two questions to answer regarding rab3d regulation of xmrk. First, what is the molecular nature of this xmrk–rab3d interaction? RAB3D is a ras-related small G protein with GTPase activity controlling exocytosis (65–67) and has been shown to regulate secretion of a broad range of molecules in different cell types (68–71). For example, RAB3D-dependent secretion of matrix metalloproteinase in macrophages is a prerequisite for macrophage recruitment to tumor cells (68), while secretion of the same molecule by tumor cells is a signal for tissue invasion and metastasis (72). We hypothesize that the X. maculatus allele of rab3d regulates cancer cell invasion by either mediating immune cell recruitment to tumor microenvironment or by hampering tumor cell secretion of molecules that facilitate metastasis. However, RAB3D function in cancer has not been clearly characterized despite efforts made investigating its function in cancer cell proliferation and metastasis in vitro (73–83). Currently there is no understanding of the cell population within the Xiphophorus tumor microenvironment (e.g., tumor cells, endothelial cells, cancer-associated fibroblasts, and immune cells) where expression of different parental rab3d alleles may change the fundamental outcome of xmrk expression (84, 85). RAB3D is also involved in cell membrane–associated protein dynamics. It has been shown that human RAB3D interacts with GOLM1 and selectively assist cytoplasmic EGFR recycling to cell membrane (83). Therefore, it is our second hypothesis that rab3d regulates xmrk function by directly controlling xmrk protein turnover and cellular localization where it displays its full activity. Second, what functionality change is associated with the amino acid difference between the two parental alleles? The change of Asn204 in X. maculatus to Lys204 in X. hellerii is located in the C terminus of RAB3D, where proper modifications (i.e., methylation and geranylgeranylation) are required for subcellular localization (86, 87). The Lys (X. hellerii) and Asn (X. maculatus) exhibit both physical differences (i.e., Lys is charged and Asn is uncharged) and varied posttranslational modifications. This amino acid change may alter protein hydrophobicity, affect RAB3D subcellular localization, and hinder efficient transportation of secretion granules and eventually affect on-site and dynamics of the above mechanisms. In summary, the conclusion of epistasis underlying the interspecific hybridization-induced tumorigenesis provides insights into a strategy in counteracting detrimental conditions.
Overall, vertebrate organisms that support the molecular mechanism proposed by the BDM model is only limited to mice (32) and now Xiphophorus fishes. The discovery of xmrk–rab3d genetic interaction underlying spontaneous tumorigenesis in interspecies hybrids poses an example showing that hybrid-induced disease can act as a mechanism that reduces hybrid fitness. Characterizing mechanism of RAB3D functional regulation of EGFR can lead to development of innovative EGFR regulation strategy.
Materials and Methods
Animal Model.
X. maculatus Jp163 A, X. hellerii (Rio Sarabia), and first-generation BC (BC1) animals used in this study were supplied by the Xiphophorus Genetic Stock Center (https://www.xiphophorus.txstate.edu/). X. maculatus Jp163A strain female fish were artificially inseminated with sperm from male X. hellerii (Rio Sarabia strain) to produce F1 interspecies hybrids. F1 hybrid males were then backcrossed to X. hellerii females to generate the BC1 animals. At dissection, all fish were anesthetized in an ice bath and upon loss of gill movement were killed by cranial resection. Organs were dissected into RNAlater (Ambion Inc.) and kept at −80 °C until use. All BC1 fish were kept and samples taken in accordance with protocols approved by Texas State University Institutional Animal Care and Use Committee (IACUC 2015107711).
The advanced BC interspecies hybrids (i.e., BC2 through BC8) were produced in an independent series of crosses in the Biocenter of the University of Wurzburg, Germany. F1 interspecies hybrids originated from the reciprocal cross: X. maculatus Jp163A males were mated to X. hellerii (Rio Lancetilla strain) females. The F1 hybrid females (cdkn2abX. mac/X. hel genotype) were then successively backcrossed to X. hellerii males to produce the advanced-generation of BC hybrids. For each generation of backcrossing, only the interspecies hybrid BC fish that exhibited benign pigment cell hyperplasia and had the cdkn2abX. mac/X. hel genotype were used to produce the next generation of BC progeny. Fin clips were collected from all advanced BC fish and stored in ethanol at 4 °C. All advanced BC fish were kept and samples taken in accordance with the applicable European Union and national German legislation governing animal experimentation. When needed, fish were killed by overanesthetization with MS222. These experiments were performed under authorization (568/300-1870/13) of the Veterinary Office of the District Government of Lower Franconia, Germany, in accordance with the German Animal Protection Law (TierSchG).
DNA and RNA Isolation.
Fin clip, or muscular tissue, was digested by Protease K at room temperature for 1 h. The lysate was then transferred to 2.0-mL collection tubes. DNA isolation was performed by a QIAcube HT (Qiagen) automated biosample isolation system, with reagent contained in the QIAamp 96 DNA QIAcube HT Kit. The isolation system is equipped with a robotic arm with eight pipettes. Each pipette is able to pick and eject pipette tips, self-clean, and transfer liquids between wells/columns, or between master reservoirs and wells/columns in standard 96-well plate formats. Each sample was independently maintained throughout the isolation process. Concentrations of DNA samples were measured using Qubit 2.0 fluorometer (Life Technologies) and adjusted for sequencing library preparation.
Dorsal fin spots, dorsal fin exhibiting benign hyperplasia, and melanoma tumors were excised from X. maculatus Jp163A, and BC interspecies hybrid. Tissue samples were homogenized in TRI-reagent (Sigma Inc.) followed by addition of 200 μL/mL chloroform, vigorously shaken, and subjected to centrifugation at 12,000 × g for 5 min at 4 °C. Total RNA was further purified using an RNeasy mini RNA isolation kit (Qiagen). Column DNase digestion at 25 °C for 15 min removed residual DNA. Total RNA concentration was determined using a Qubit 2.0 fluorometer (Life Technologies). RNA quality was verified on an Agilent 2100 Bioanalyzer (Agilent Technologies) to confirm that RNA integrity number scores were above 8.0 prior to subsequent gene expression profiling.
Genetic Variants Identification and Annotation.
To identify interspecies polymorphisms between the X. maculatus and X. hellerii, genomic DNAs of 4 X. maculatus and 4 X. hellerii were isolated. DNA samples were forwarded for genome shotgun sequencing library preparation using Illumina Nextera sequencing Library Prep Kit, followed by sequencing on HiSEq. 2000 (Illumina, Inc.) using 150-bp paired-end sequencing strategy. Raw sequencing reads were trimmed and filtered using a custom Perl script, and adapter sequences were removed from the sequencing reads. The reads were truncated based on similarity to library adaptor sequences using custom Perl scripts. Then, low-scoring sections of each read were removed, preserving the longest remaining sequencing read fragment (88). Filtered genome sequencing reads were mapped to the reference X. maculatus genome (GenBank assembly accession no. GCA_002775205.2) using Bowtie2 “head-to-head” mode to show mismatches between sequenced animals and reference genome (89). Alignment files were sorted using Samtools (90). Subsequently, pileup files were generated for each X. maculatus, and X. hellerii sample, and variant calling was processed by both BCFtools and VarScan for polymorphisms detection, with minimum variant locus coverage of 2 and a P value for variant detection of 0.05 for VarScan and variant genotyping call Phred score of 0 and alternative genotyping Phred score ≥ 20 for BCFtools (90–92). Only the variants that were identified by both pipelines were forwarded for further analyses.
To localize fixed variants between the X. maculatus and X. hellerii, homozygous loci of X. maculatus were compared to those of X. hellerii. Such loci were identified if all X. maculatus were homozygous for one allele and all X. hellerii homozygous for the alternative allele.
These fixed species-specific genetic variants were functionally annotated using snpEff (93). A genome database was created using the X. maculatus genome sequence and annotation files (GenBank assembly accession no. GCA_002775205.2). Each variant was queried to the genome database to determine if it was located in a genetic or intergenic region, and to determine what effect each variant may have on the peptide sequence structure.
Amplicon Sequencing, Data Filter, and Genotyping.
Variants between X. maculatus and X. hellerii were used as references to design specific capture probes for targeted genomic sequencing. Variants with very high sequencing depth were removed due to the possibility of locating them in repetitive sequences. Sequencing probes were designed to amplify regions surrounding genetic variants. To genetically map candidate R(Diff) loci in a region (chromosome 5: 10,000,000 to 16,000,000) identified in a previous study (22), 406 sets of probe were designed to reach a resolution of 14.8 kbp within the 6-Mbp locus; for sex-determining regions (chromosome 21: 23,750,000 to 26,250,000; SI Appendix, Fig. S2 and Dataset S2), 101 sets or capture probes were designed for 24.8-kbp definition genetic mapping; for the rest of the genome, 1,510 sets of probes were designed for genotyping and establish individual BC progeny haploid map at definition of 459 kbp (SI Appendix, Fig. S1 and Dataset S2). Therefore, a total of 2,017 probe sets were produced for amplicon sequencing. Amplicons were custom-made using Illumina Genotype Ne library preparation kit, with i7 and i5 indices incorporated into adaptor sequences added to each end of PCR products amplified by capture probes. Pooled sequencing libraries were sequenced on Illumina MiSeq platform employing a 75-bp paired-end sequencing strategy (Illumina).
Sequencing adaptor contamination was first removed from raw sequencing reads using fastx_toolkit, followed by trimming of low-quality sections of each sequencing read. Low-quality sequencing reads were further removed from sequencing result (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Processed sequencing reads were mapped to X. maculatus genome v5.0 (GenBank assembly accession no. GCA_002775205.2) using Bowtie2 (89). Mpileup files were made using samtools and genotyping was processed using both Bcftools and VarScan (90–92). Genetic variant call and genotype were required to be supported by both pipelines for further analyses (i.e., Bcftools: MAPQ ≥30, Phred score of genotype call = 0, with alternative genotype call Phred score ≥20; VarScan: MAPQ ≥ 30, P value <0.05, depth ≥20). Herein “genotype” refers to inheritance of ancestral alleles, with “heterozygous” meaning that a locus exhibited genetic material from both ancestors (i.e., X. maculatus and X. hellerii), and “homozygous” means that a locus exhibited genetic material from only the recurrent ancestor (i.e., X. hellerii). A haploid map was produced for each individual of BC progeny. To control the amplicon-sequencing-based genotyping result target specificity, only genotyping calls that locate less than 75 bp from the designed polymorphic sites and also supported by at least another variant genotyping call within a 75-bp range were kept. Qualified genotyping calls were subsequently ordered by chromosome name and their chromosomal location to produce the haploid map. For each BC hybrid individual, percentage of heterozygous loci is calculated as Heterozygous %= (number of heterozygous loci)/(total number of genotyped loci). Because BCn were selected for a marker on chromosome 5 (i.e., cdkn2ab) and chromosome 21 (i.e., Sd), percentage of heterozygous was calculated using all genotyped loci, or loci that are outside of chromosomes 5 and 21 (SI Appendix, Fig. S3).
Calculation of Allele Frequency.
Genotyped variants loci of all BC hybrids were combined together first to yield a data table with the rows as chromosomal coordinates and columns as hybrid individuals. Heterozygous, designated as “0/1,” refers to inheritance of both X. maculatus and X. hellerii parental alleles, while homozygous, designated as “1/1,” refers to inheritance of only X. hellerii alleles. The X. maculatus allele frequency for each locus is calculated as
Linkage Analyses.
For advanced BC samples exhibiting benign melanocyte hyperplasia their genetic backgrounds are predominantly represented by the recurrent parental genome (X. hellerii). An average X. hellerii genome component per BC generation follows a rule determined by (1 − 0.5n+1), where n equals the BC generation. Therefore, every locus is expected to exhibit dominance of X. hellerii allele and therefore disequilibrium (SI Appendix, Fig. S4). Since our previous studies had determined that the R(Diff) locus is heterozygous within the BC hybrids exhibiting melanocyte benign hyperplasia, we only plotted the −log10 P values of loci where X. maculatus allele frequency is higher and assigned −log10 P values of X. hellerii dominated loci arbitrarily to 0, in order to visualize dominantly heterozygous loci within the BC hybrids (SI Appendix, Fig. S5). Because there is only one pigment cell phenotype of the BCn hybrids, numbers of heterozygous and homozygous individuals per variant site were used to form a one-dimensional contingency table and tested using a goodness-of-fit χ2 test. χ2 test P values were adjusted using Bonferroni correction.
For BC1 progeny, numbers of heterozygous and homozygous individuals were counted for each pigmentation phenotype (i.e., melanoma and pigment cell hyperplasia), and numbers of each genotype per phenotype group (i.e., tumor or pigment cell hyperplasia) were used to form a contingency table and subsequently tested using a χ2 contingency table test, with the null hypothesis that both genotypes’ distribution follows random assortment. χ2 test P values were adjusted using Bonferroni correction.
Gene Expression Profiling.
RNA sequencing was performed upon sequencing libraries construction using the Illumina TruSeq messenger RNA (mRNA) library preparation kit (Illumina, Inc.). RNA libraries were sequenced as 125-bp paired-end fragments using an Illumina Hi-SEq. 2000 system (Illumina, Inc.). Short sequencing reads were filtered using an in-house data processing pipeline (88). RNA-sequencing reads produced from three sets of pooled dorsal fin spots collected from X. maculatus, dorsal fin exhibiting melanocyte hyperplasia of 22 BC hybrid, and melanoma of 22 BC hybrid were produced. Sequencing reads were mapped to X. maculatus reference genome (GenBank assembly accession no. GCA_002775205.2) using STAR (94). Gene expression was subsequently profiled by counting number of sequencing reads that mapped to gene models annotated by Ensembl using RSEM (95). For data visualization, gene expression read counts were normalized to library size and were plotted as bar graph using R (v3.5.1). Differentially expressed genes were identified using R package DESeq2, with P value adjusted using the false discovery rate (FDR) method (96). FDR <0.05 was used to determine differential expression.
Annotation of X. Hellerii Allele of rab3d.
The genome sequences of X. maculatus chromosome 5 (GenBank assembly accession no. GCA_002775205.2) and X. hellerii chromosome 5 (GenBank assembly accession no. GCA_003331165.2) were aligned to each other using lastz (http://www.bx.psu.edu/miller_lab/dist/). The R(Diff) region alignment was extracted using genomic coordinates of R(Diff) for data visualization. To localize the X. hellerii rab3d gene the coordinates of rab3d exons and coding sequences for X. maculatus were transferred to the X. hellerii allele using the lastz alignment. Sequence comparisons between mRNA and genomic DNA, protein sequence comparisons between X. hellerii to other species were processed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical Information.
Goodness-of-fit and contingency table χ2 tests were used to identify linkage disequilibrium for advanced BC (n = 90) and BC1 hybrids (n = 66), respectively. χ2 test P values were adjusted using Bonferroni correction.
Two-tailed t tests were used to test if sizes of BC7 (n = 23) and BC8 (n = 3) hybrid heterozygous loci are different from expected 5.5 and 2.7 Mbp, respectively.
Differential gene expression among parental dorsal fin melanocyte spots (n = 3 pooled samples), hybrid melanocyte hyperplasia samples (n = 22), and hybrid melanoma tumors (n = 22) were performed using the R package DESeq2 that implemented a modified Fisher’s exact test. Multiple test P values were corrected using the FDR method (96).
Supplementary Material
Acknowledgments
This work was supported by the NIH, National Cancer Institute grants R15-CA-223964 and R24 OD-011120 from the NIH Division of Comparative Medicine.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010133117/-/DCSupplemental.
Data Availability.
Sequencing data have been deposited in the NCBI Sequence Read Archive (BioProject number PRJNA610523, accession nos. SAMN14300088–SAMN14300232).
References
- 1.Haiissler G., Über Melanombildungen bei Bastarden von Xiphophorus Helleri und Platypoecilus Maculatus var. Rubra. J. Mol. Med. 7, 1561–1562 (1928). [Google Scholar]
- 2.Gordon M., The genetics of a viviparous top‐Minnow platypoecilus; the inheritance of two kinds of melanophores. Genetics 12, 253–283 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosswig C., Über bastarde der teleostier platypoecilus und xiphophorus. Z. Indukt. Abstamm. Vererbungsl. 44, 253 (1928). [Google Scholar]
- 4.Bateson W., “Heredity and variation in modern lights” in Darwin and Modern Science, Ac S., Ed. (Cambridge University Press, 1909), pp. 85–101. [Google Scholar]
- 5.Dobzhansky T., Genetics and the Origin of Species (Columbia University Press, 1937). [Google Scholar]
- 6.Muller H., “Bearing of the Drosophila work on systematics” in The New Systematics, Huxley J., Ed. (Clarendon Press, 1940), pp 185–268. [Google Scholar]
- 7.Coyne J. A., Orr H. A., Speciation (Sinauer Associates, 2004). [Google Scholar]
- 8.Mack K. L., Nachman M. W., Gene regulation and speciation. Trends Genet. 33, 68–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maheshwari S., Barbash D. A., The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45, 331–355 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Pennisi E., Evolution. Two rapidly evolving genes spell trouble for hybrids. Science 314, 1238–1239 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Noor M. A., Evolutionary biology: Genes to make new species. Nature 423, 699–700 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Patton E. E., Mathers M. E., Schartl M., Generating and analyzing fish models of melanoma. Methods Cell Biol. 105, 339–366 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Walter R. B., Kazianis S., Xiphophorus interspecies hybrids as genetic models of induced neoplasia. ILAR J. 42, 299–321 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Schartl M., Walter R. B., Xiphophorus and medaka cancer models. Adv. Exp. Med. Biol. 916, 531–552 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Todaro G. J., Huebner R. J., N.A.S. Symposium: New evidence as the basis for increased efforts in cancer research. Proc. Natl. Acad. Sci. U.S.A. 69, 1009–1015 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knudson A. G., Jr, Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 68, 820–823 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adam D., Dimitrijevic N., Schartl M., Tumor suppression in Xiphophorus by an accidentally acquired promoter. Science 259, 816–819 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Wittbrodt J., et al. , Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341, 415–421 (1989). [DOI] [PubMed] [Google Scholar]
- 19.Schartl M., Beyond the zebrafish: Diverse fish species for modeling human disease. Dis. Model. Mech. 7, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas R., Weihua Z., Rethink of EGFR in cancer with its kinase independent function on board. Front. Oncol. 9, 800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutiérrez-Castañeda L. D., Nova J. A., Tovar-Parra J. D., Frequency of mutations in BRAF, NRAS, and KIT in different populations and histological subtypes of melanoma: A systemic review. Melanoma Res. 30, 62–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y., et al. , Molecular genetic analysis of the melanoma regulatory locus in Xiphophorus interspecies hybrids. Mol. Carcinog. 56, 1935–1944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazianis S., et al. , Comparative structure and characterization of a CDKN2 gene in a Xiphophorus fish melanoma model. Oncogene 18, 5088–5099 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Aitken J., et al. , CDKN2A variants in a population-based sample of Queensland families with melanoma. J. Natl. Cancer Inst. 91, 446–452 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Kazianis S., et al. , Localization of a CDKN2 gene in linkage group V of Xiphophorus fishes defines it as a candidate for the DIFF tumor suppressor. Genes Chromosomes Cancer 22, 210–220 (1998). [PubMed] [Google Scholar]
- 26.Nairn R. S., et al. , A CDKN2-like polymorphism in Xiphophorus LG V is associated with UV-B-induced melanoma formation in platyfish-swordtail hybrids. Proc. Natl. Acad. Sci. U.S.A. 93, 13042–13047 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nairn R. S., Morizot D. C., Kazianis S., Woodhead A. D., Setlow R. B., Nonmammalian models for sunlight carcinogenesis: Genetic analysis of melanoma formation in Xiphophorus hybrid fish. Photochem. Photobiol. 64, 440–448 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Förnzler D., Wittbrodt J., Schartl M., Analysis of an esterase linked to a locus involved in the regulation of the melanoma oncogene and isolation of polymorphic marker sequences in Xiphophorus. Biochem. Genet. 29, 509–524 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Ahuja M. R., Schwab M., Anders F., Linkage between a regulatory locus for melanoma cell differentiation and an esterase locus in Xiphophorus. J. Hered. 71, 403–407 (1980). [DOI] [PubMed] [Google Scholar]
- 30.Anders F., Tumour formation in platyfish-swordtail hybrids as a problem of gene regulation. Experientia 23, 1–10 (1967). [DOI] [PubMed] [Google Scholar]
- 31.Anders A., Anders F., Etiology of cancer as studied in the platyfish-swordtail system. Biochim. Biophys. Acta 516, 61–95 (1978). [DOI] [PubMed] [Google Scholar]
- 32.Flachs P., et al. , Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS Genet. 8, e1003044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albertson R. C., Cresko W., Detrich H. W. 3rd, Postlethwait J. H., Evolutionary mutant models for human disease. Trends Genet. 25, 74–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Conchas G. A., et al. , Epidermal growth factor receptor overexpression and outcomes in early breast cancer: A systematic review and a meta-analysis. Cancer Treat. Rev. 62, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Cossu-Rocca P., et al. , EGFR kinase-dependent and kinase-independent roles in clear cell renal cell carcinoma. Am. J. Cancer Res. 6, 71–83 (2015). [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsson J., Wester K., De La Torre M., Malmström P. U., Gårdmark T., EGFR-expression in primary urinary bladder cancer and corresponding metastases and the relation to HER2-expression. On the possibility to target these receptors with radionuclides. Radiol. Oncol. 49, 50–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohgaki H., Kleihues P.. The definition of primary and secondary glioblastoma. Clin. Cancer Res. 19, 764–772 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Boeck S., et al. , EGFR pathway biomarkers in erlotinib-treated patients with advanced pancreatic cancer: Translational results from the randomised, crossover phase 3 trial AIO-PK0104. Br. J. Cancer 108, 469–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M. A., et al. , EGFR in gastric carcinomas: Prognostic significance of protein overexpression and high gene copy number. Histopathology 52, 738–746 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Dancer J., Takei H., Ro J. Y., Lowery-Nordberg M., Coexpression of EGFR and HER-2 in pancreatic ductal adenocarcinoma: A comparative study using immunohistochemistry correlated with gene amplification by fluorescencent in situ hybridization. Oncol. Rep. 18, 151–155 (2007). [PubMed] [Google Scholar]
- 41.Galizia G., et al. , Epidermal growth factor receptor (EGFR) expression is associated with a worse prognosis in gastric cancer patients undergoing curative surgery. World J. Surg. 31, 1458–1468 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Nishio M., et al. , Gefitinib efficacy associated with multiple expression of HER family in non-small cell lung cancer. Anticancer Res. 26, 3761–3765 (2006). [PubMed] [Google Scholar]
- 43.Spindler K. L., et al. , Epidermal growth factor receptor analyses in colorectal cancer: A comparison of methods. Int. J. Oncol. 29, 1159–1165 (2006). [PubMed] [Google Scholar]
- 44.Bloomston M., Bhardwaj A., Ellison E. C., Frankel W. L., Epidermal growth factor receptor expression in pancreatic carcinoma using tissue microarray technique. Dig. Surg. 23, 74–79 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Stadlmann S., et al. , Epithelial growth factor receptor status in primary and recurrent ovarian cancer. Mod. Pathol. 19, 607–610 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Italiano A., et al. , Epidermal growth factor receptor (EGFR) status in primary colorectal tumors correlates with EGFR expression in related metastatic sites: Biological and clinical implications. Ann. Oncol. 16, 1503–1507 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Rogers S. J., Harrington K. J., Rhys-Evans P., O-Charoenrat P., Eccles S. A., Biological significance of c-erbB family oncogenes in head and neck cancer. Cancer Metastasis Rev. 24, 47–69 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Franklin W. A., Carbone D. P., Molecular staging and pharmacogenomics. Clinical implications: From lab to patients and back. Lung Cancer 41, S147–S154 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Hirsch F. R., Scagliotti G. V., Langer C. J., Varella-Garcia M., Franklin W. A., Epidermal growth factor family of receptors in preneoplasia and lung cancer: Perspectives for targeted therapies. Lung Cancer 41, S29–S42 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Daveau M., et al. , Hepatocyte growth factor, transforming growth factor alpha, and their receptors as combined markers of prognosis in hepatocellular carcinoma. Mol. Carcinog. 36, 130–141 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Di Lorenzo G., et al. , Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin. Cancer Res. 8, 3438–3444 (2002). [PubMed] [Google Scholar]
- 52.Suo Z., Nesland J. M., Type 1 protein tyrosine kinases in breast carcinoma: A review. Ultrastruct. Pathol. 26, 125–135 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Maihle N. J., et al. , EGF/ErbB receptor family in ovarian cancer. Cancer Treat. Res. 107, 247–258 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Arteaga C. L., Engelman J. A., ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25, 282–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreri A. J., Illerhaus G., Zucca E., Cavalli F.; International Extranodal Lymphoma Study G , Flows and flaws in primary central nervous system lymphoma. Nat. Rev. Clin. Oncol. 7, 1–2 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Sharma S. V., Bell D. W., Settleman J., Haber D. A., Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Van Cutsem E., et al. , Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J. Clin. Oncol. 25, 1658–1664 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Cunningham D., et al. , Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 351, 337–345 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Cohen R. B., Current challenges and clinical investigations of epidermal growth factor receptor (EGFR)- and ErbB family-targeted agents in the treatment of head and neck squamous cell carcinoma (HNSCC). Cancer Treat. Rev. 40, 567–577 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Bonner J. A., et al. , Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 11, 21–28 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Schartl A., Dimitrijevic N., Schartl M., Evolutionary origin and molecular biology of the melanoma-inducing oncogene of Xiphophorus. Pigment Cell Res. 7, 428–432 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Gómez A., Wellbrock C., Gutbrod H., Dimitrijevic N., Schartl M., Ligand-independent dimerization and activation of the oncogenic Xmrk receptor by two mutations in the extracellular domain. J. Biol. Chem. 276, 3333–3340 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Wellbrock C., Weisser C., Geissinger E., Troppmair J., Schartl M., Activation of p59(Fyn) leads to melanocyte dedifferentiation by influencing MKP-1-regulated mitogen-activated protein kinase signaling. J. Biol. Chem. 277, 6443–6454 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Schartl M., et al. , A mutated EGFR is sufficient to induce malignant melanoma with genetic background-dependent histopathologies. J. Invest. Dermatol. 130, 249–258 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Millar A. L., Pavios N. J., Xu J., Zheng M. H., Rab3D: A regulator of exocytosis in non-neuronal cells. Histol. Histopathol. 17, 929–936 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Martelli A. M., et al. , Rab3A and Rab3D control the total granule number and the fraction of granules docked at the plasma membrane in PC12 cells. Traffic 1, 976–986 (2000). [PubMed] [Google Scholar]
- 67.Pavlos N. J., et al. , Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol. Cell. Biol. 25, 5253–5269 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanania R., et al. , Classically activated macrophages use stable microtubules for matrix metalloproteinase-9 (MMP-9) secretion. J. Biol. Chem. 287, 8468–8483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim H., Han J. K., Rab3d is required for Xenopus anterior neurulation by regulating Noggin secretion. Dev. Dyn. 240, 1430–1439 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Williams J. A., Chen X., Sabbatini M. E., Small G proteins as key regulators of pancreatic digestive enzyme secretion. Am. J. Physiol. Endocrinol. Metab. 296, E405–E414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., et al. , Traffic of endogenous, transduced, and endocytosed prolactin in rabbit lacrimal acinar cells. Exp. Eye Res. 85, 749–761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mehner C., et al. , Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 5, 2736–2749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J., Kong R., Sun L., Silencing of Rab3D suppresses the proliferation and invasion of esophageal squamous cell carcinoma cells. Biomed. Pharmacother. 91, 402–407 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Luo Y., et al. , High expression of Rab3D predicts poor prognosis and associates with tumor progression in colorectal cancer. Int. J. Biochem. Cell Biol. 75, 53–62 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Yang J., et al. , High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget 6, 11125–11138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie J., Zheng Y., Xu X., Sun C., Lv M., Long noncoding RNA CAR10 contributes to melanoma progression by suppressing miR-125b-5p to induce RAB3D expression. OncoTargets Ther. 13, 6203–6211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kolankiewicz A. C. B., et al. , Patient safety culture from the perspective of all the workers of a general hospital. Rev. Gaúcha Enferm. 41, e20190177 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Jin T., et al. , Lcn2-derived circular RNA (hsa_circ_0088732) inhibits cell apoptosis and promotes EMT in glioma via the miR-661/RAB3D Axis. Front. Oncol. 10, 170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin H., Liu Y., Yiu S., Three dimensional culture of potential epithelial progenitor cells in human lacrimal gland. Transl. Vis. Sci. Technol. 8, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao K., et al. , The lncRNA HOXA11-AS regulates Rab3D expression by sponging miR-125a-5p promoting metastasis of osteosarcoma. Cancer Manag. Res. 11, 4505–4518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren Y., Shi G., Jiang P., Meng Q., MicroRNA-761 is downregulated in colorectal cancer and regulates tumor progression by targeting Rab3D. Exp. Ther. Med. 17, 1841–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiashi W., et al. , MicroRNA-506-3p inhibits osteosarcoma cell proliferation and metastasis by suppressing RAB3D expression. Aging (Albany NY) 10, 1294–1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye Q. H., et al. , GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell 30, 444–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Qian B. Z., Pollard J. W., Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qiu X., Valentijn J. A., Jamieson J. D., Carboxyl-methylation of Rab3D in the rat pancreatic acinar tumor cell line AR42J. Biochem. Biophys. Res. Commun. 285, 708–714 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Raffaniello R. D., Raufman J. P., Cytosolic RAB3D is associated with RAB escort protein (REP), not RAB-GDP dissociation inhibitor (GDI), in dispersed chief cells from Guinea pig stomach. J. Cell. Biochem. 72, 540–548 (1999). [PubMed] [Google Scholar]
- 88.Garcia T. I., et al. , Effects of short read quality and quantity on a de novo vertebrate transcriptome assembly. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 155, 95–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koboldt D. C., et al. , VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the NCBI Sequence Read Archive (BioProject number PRJNA610523, accession nos. SAMN14300088–SAMN14300232).