Abstract
Polycystic ovary syndrome (PCOS) is a gynecological endocrine disorder in women of reproductive age. There is adequate evidence that suggests several microRNAs (miRNAs) are of great importance for PCOS. It seems that dysregulated expression of miR-27a, miR-130b, and miR-301a are associated with PCOS. The aim of this study was to investigate whether plasma levels of these miRNAs are different between patients with PCOS and healthy controls. Fifty-three women with a definite diagnosis of PCOS, and 53 healthy controls were enrolled. MiRNAs expression levels in plasma were evaluated by real-time PCR. The diagnostic values of each miRNA were calculated by the receiver operating characteristic (ROC) curve and areas under the curves (AUC). The main clinical characteristics were not significantly different between the two groups. The circulating plasma expression levels of miR-27a and miR-301a had a significant increase (P = 0.0008 and P <0.0001, respectively) but miR-130b expression level decreased in the patient group (P <0.0001). The AUC for miR-27a, miR-130b, and miR-301a were 0.71, 0.77, and 0.66, respectively. A positive exponential was observed for miR-27a and miR-301a in multiple logistic regression. Changes in the plasma expressions of the studied miRNAs are likely to be associated with PCOS phenotypes. MiR-27a has a potential to serve as a diagnostic biomarker of PCOS.
Key Words: Polycystic ovary syndrome, plasma, miRNA-27a, miRNA-130b, miRNA-301a
Polycystic ovary syndrome (PCOS) is known as a heterogeneous disorder characterized by irregular menstrual cycles. About 6% of women of reproductive age suffer from PCOS (1). PCOS causes infertility, biochemical problems, hypera-ndrogenism, granulosa cell proliferation arrest, and insulin resistance. Dysregulated androgen metabo-lites and increased risk factors for type-2 diabetes mellitus have also been reported in the PCOS patients (2, 3). Familial aggregation can potentially exacerbate this disorder due to its genetic basis. The involvement of inheritance and dominant gene penetrance in PCOS incidence has been demons-trated (4).
MicroRNAs (miRNAs) are small non-coding, single-stranded RNA molecules with 18–24 nucleotides in length that regulate the gene expression by targeting 3'-untranslated region (3'UTR) of messenger RNAs (mRNAs) (5). These conserved molecules play a crucial role in various cell functions such as apoptosis, proliferation, and signaling. Extracellular/circulating miRNAs could be selectively secreted into the blood, plasma or serum, and can be considered a therapeutic target, or independent biomarker of different diseases (6). The pathogenic role of miRNAs in reproductive system diseases has drawn much attention (7). Previous research has reported alterations in the expression levels of certain miRNAs in PCOS women versus controls (8). The expression of miR-27a in granulosa cells has been reported to have an association with insulin resistance in women with PCOS compared to healthy women, and its target gene could be SMAD5 (9). Other studies showed that the loss of peroxisome proliferator- activated receptor gamma (PPAR-γ) in granulosa cells of PCOS patients could be due to the ability of miR-27a to down-regulate PPAR-γ gene expression (10,11). MiR-301a plays roles in glycemic control and appropriate beta-cell function (12). Based on various predictive bioinformatics algorithms, miR-301a is a potential PPAR-γ regulator, and therefore can affect PCOS pathways (13). Evidence suggests miR-130b is up-regulated in the follicular fluid of women with fertility problems compared to women without fertility problems (14). According to above evidence, the changes in the expression of these three miRNAs seem to be related to PCOS phenotypes. Besides, bioinformatics research has been aimed to select these miRNAs. So far, the plasma expression levels of miR-27a, miR-130b, and miR-301a in PCOS patients have not been investigated to examine their potential as a biomarker. Our aim was to investigate whether the plasma levels of these miRNAs could be used to detect women with PCOS.
Materials and methods
Bioinformatics and molecular pathways analysis
PCOS-related information was drawn from the MalaCards Database. The genes involved in predisposition to PCOS were assessed by DisGeNET, GS2D, DiSNOR, and particularly PCOSKB that has a PCOS-specific database. To identify target gene(s), sites were searched by TargetScan, DIANA-microT, PicTar, miRTarBase, miRDB, miRcodes, and miRWalk. Then, the relationships among the pathways, the expression of the candidate genes and the selected microRNAs were analyzed by the mimiRNA, miRmine, miRandola, and miRNA databases to determine the most effective and specific microRNAs according to their prediction scores. The miRNAs were identified in the molecular pathways that were involved in the progression of PCOS-related diseases such as diabetes. Bioinformatics analysis indicated miR-27a, miR- 130b, and miR-301a can potentially serve as efficient diagnostic biomarkers for PCOS.
Patients and study design
The participants of this case-control study were 53 women with PCOS (aged 22–35 years) and 53 matched healthy women with regular menstruation. Sampling was with informed consent and completing the questionnaire was confidential. From May to September 2017, all participants attended Kashani hospital, Shahrekord, Southwest Iran. Data including age, recent drug consumption, stress, and parity were collected by interview and from medical files.
The identification of PCOS phenotype was based on at least two of the diagnostic criteria of the revised Rotterdam European Society of Human Reproduction and Embryology/American Society for Reproductive Medicine as follows: (I) chronic oligo and/or anovulation (cycle length < 26 days or >35 days); (II) clinical hyperandrogenism (presence of hirsutism evaluated by a Ferriman-Gallwey score >8,severe acne) and biochemical hyperandrogenism (total testosterone concentration>0.5 ng/mL); and (III) polycystic ovaries on ultrasound (defined as the presence of 12 or more ovarian cysts with 2–10 mm diameter per ovary and/or ovarian volume ≥ 10 cm3) (15). None of the patients had a history of drug use that interferes with glucose and lipid metabolism, and none of the controls had clinical or biochemical hyperandrogenemia. Control women had no clinical or biochemical sign of androgen excess and no background of menstrual irregularities. The study of the protocol was approved by the Ethics Committee of Shahrekord University of Medical Sciences (IR.SKUMS. REC.1395.307). The informed consent form to participate in the study was also signed by all participants.
Plasma preparation and quantitative Real-Time PCR
Up to 3 mL of whole blood from each participant was collected in K2EDTA plasma tubes (Sunphoria Co., Ltd VP3082, Taiwan). Plasma was isolated by centrifugation (Hettich, MIKRO200R, Germany) of whole blood at 2700 rpm for 7 min at room temperature within 2 h after blood collection. Plasma supernatant was recovered, divided into aliquots, and immediately stored at -70 °C. RNA isolation was done using TRIzol® reagent (Gene all/RiboEx LS, catalog number. 15596026, Korea) according to the manufacturer’s instructions. The concentration of RNA was determined by Nano-Drop 2000 (Thermo Scientific™, USA), and then the purified RNA was stored at −80 °C. For all samples, RNA was incubated with 1 µl of DNase I and cDNA synthesis was performed using the Revert Aid first-strand cDNA Poly A Polymerase (PAP) synthesis kit (Stem Cell Technology, BN-0011.17, Iran) in a thermal cycler (25 °C for 10 min, 42 °C for 15 min, and 48 °C for 15 min). The real-time PCR was performed by a Rotor-Gene RG-300 (Corbett Research, Sydney, AU) and the SYBR Green real-time PCR Master Mix Kit (Stem cell technology, catalog number; BN-0011.17, Iran) in a thermal cycler (95 °C for 10 s, 58-60 °C for 20 s, and 72 °C for 15 s). The forward primers for miR-27a, miR-130b, miR-301a, and SNORD47 were designed by Gene Runner Software, version 4.0 (Table 1). The universal primer was used as reverse primer in real-time PCR (Stem cell technology, catalog number; BN-0011.17, Iran). SNORD47 level was also analyzed as an internal control in the real-time PCR. The specificity of primers was confirmed by melting curves analysis. Gene expression was normalized to internal controls, and the fold changes of differentially expressed miRNA levels were calculated by 2−ΔΔCt.
Table 1.
Primer sequences used for real-time PCR quantifications
| Forward primer | Sequences (5' to 3') |
|---|---|
| miR-27a | TTCACAGTGGCTAAGTTCC |
| miR-130b | CAATGATGAAAGGGCAT |
| miR-301a | CAGAGGAATAGTATTGTCAAAG |
| SNORD47 | ATCACTGTAAAACCGTT |
Statistical analysis
SPSS version 19 (SPSS Inc., Chicago, USA) and GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, CA, USA) were used to perform all data analyzes. The gene expression results have been investigated using the One-way ANOVA test. Statistically, significant differences in variables between groups were analyzed using the Mann-Whitney, and chi-squared tests. A receiver operating characteristic (ROC) curve was plotted to investigate the differential effects of gene expression in the patients and controls. 95% confidence interval (CI), along with the area under the ROC curve (AUC), is basically dependent on the sensitivity and specificity of the biomarker Multiple logistic regression analysis was used to investigate the diagnostic value of the studied miRNAs for PCOS by combining their expression levels. Significance level (P) was considered ≤0.05. All data were shown as mean±SD.
Results
Characteristics of participants
There were no significant differences between PCOS patients and control group with regard to age and BMI (P> 0.05). The mean (±SD) age of the participants was 26.81 (±4.58) years, and their mean BMI 22.73 (±1.78) Kg/m2. Participants’ BMI was in the normal range (approximately 18.5 (±29.4 kg/m2)) (Table 2). The prevalence of phenotypic characteristics is shown in Table 3. Main clinical characteristics were not significantly different between the two groups.
Table 2.
Demographic characteristics of women with PCOS and control subjects
| Variables | Controls (n=54) | Cases (n=54) | P value | |
|---|---|---|---|---|
| Age (years) | 26.50 ( 24-31) | 25 (22-30.25) | 0.143 | |
| Body mass index (Kg/m 2 ) | 23.40 (20.33-23.70) | 24.97 (21.40-24.70) | 0.798 | |
| Marital | Single | 20 (37.04) | 16(29.63 ) | 0.541 |
| status | Married | 34 (62.97 ) | 38(70.37 ) | |
Age and BMI were reported in median (interquartile ranges). Mann-Whitney U test was utilized to compare Age and BMI difference between two groups. Other data are expressed as percentage (number) Chi-Square test was used to compare the two groups with regards. No significant relationships were observed between groups. Significance level (P) was considered ≤ 0.05.
Table 3.
Prevalence of the component and composite phenotypes in PCOS
| P henotypes | Percent |
|---|---|
| Polycystic ovaries | 100 |
| Depression | 75.9 |
| Hirsutism | 72.2 |
| Acne | 70.4 |
| Alopecia | 63 |
| Stress | 55.6 |
| Oligomenorrhea/amenorrhea | 44 |
| Sleep problems | 37 |
| Family evidence | 32 |
| Skin darkness | 29.6 |
The most common apparent phenotypes are expressed in percentages in the patients group.
Analyzes of miRNAs pattern expression
First, the gene expression levels of miR-27a, miR-130b, and miR-301a in patients and controls were investigated with suitable primers (Table 1). A noteworthy increase in miRNA-27a expression was observed using real-time PCR in patients in comparison with controls. Similarly, the expression of miRNA-301a increased significantly in patients (fold changes: 1.50 and 1.77 in miR-27a and miR-301a, respectively; P=0.0008 and P<0.0001, respectively). In contrast, the circulating levels of miR-130b had a noticeable decrease (fold change: 0.41; P <0.0001) (Figure 1). According to the multivariate logistic regression, miR-27a and miR-301a were significantly associated with the risk of developing PCOS. In addition, the regression model showed that miR-130b had an inverse correlation with the risk of developing PCOS (95% CI, 0.174-0.492, P<0.0001). MiR-301a was significantly associated with the risk of developing PCOS. In addition, the regression model showed that miR-130b had an inverse correlation with the riskof developing PCOS (95% CI, 0.174-0.492, P<0.0001).
Fig. 1.
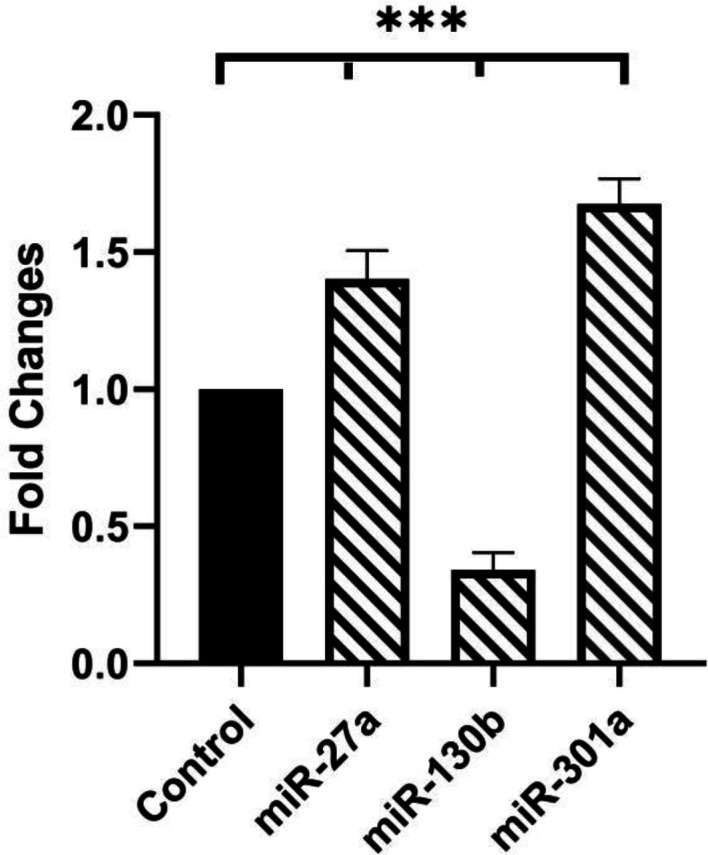
Investigation of miRNAs expressions. The data from the real-time PCR was normalized versus SNORD47. MiR-27a and miR-301a expression levels significantly increased (fold changes: 1.50 and 1.77; P = 0.0008 and P < 0.0001, respectively) in patients in comparison with controls, miR-130b expression had a noticeable decrease (fold change: 0.41; P < 0.0001). The One-way ANOVA was used to determine the normality of data distribution. MiRNAs expression was examined in triplicate, and the results are presented as mean ± SD. (***= P < 0.001). Significance level (P) was considered ≤ 0.05
Sensitivity and Specificity of miRNAs for PCOS
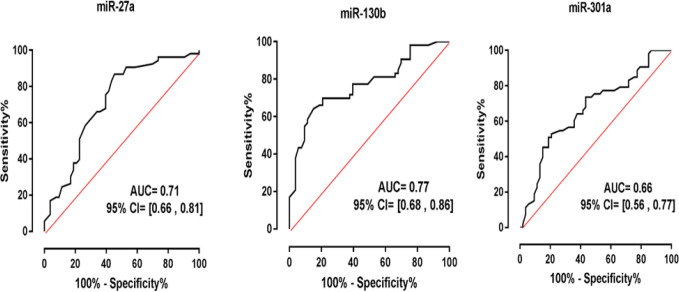
The diagnostic value of miRNA-27a, miRNA-130b and miRNA-301a was investigated by the ROC curve and AUC. The cut-off values for sensitivity and specificity of miR-27a, miR-130b, and miR-301a were 66.04 and 66.4, 69.81 and 79.25, and 66.04 and 58.49, respectively (Table 4). Based on the ROC analyzes, the areas under the curves (AUC) for miR-27a, miR-130b, and miR-301a were 0.71, 0.77 and 0.66, respectively (Figure 2). The highest sensitivity and specificity were considered to be the optimal diagnostic point, according to which miR-27a had more efficiency for the diagnosis of PCOS.
Table 4.
The cut-off values as an optimal diagnostic point for miRNAs characteristics
| MiRNA | Cut off | Sensitivity% | 95% CI | Specificity% | 95% CI |
|---|---|---|---|---|---|
| miR-27a | < 0.525 | 66.04 | 51.73% to 78.48% |
66.04 | 51.73% to 78.48% |
| miR-130b | > 1.55 | 69.81 | 55.66% to | 79.25 | 65.89% to |
| 81.66% | 89.16% | ||||
| miR-301a | < 3.325 | 66.04 | 51.73% to 78.48% |
58.49 | 44.13% to 71.86% |
Fig. 2.
ROC curve analysis of miR-27a, miR-130b, and miR-301a to discriminate women with PCOS from healthy controls. Based on the ROC analyzes, the areas under the curves (AUC) for miR-27a, miR-130b, and miR-301a were 0.71, 0.77 and 0.66, respectively. The highest sensitivity and specificity were considered to be the optimal diagnostic point, according to which miR-27a had far more efficiency for the diagnosis of PCOS
Discussion
The development of PCOS symptoms is associated with metabolic disorders and infertility. The presence of circulating miRNAs in serum, plasma, and the follicular fluid can serve as potential biomarkers for diagnosis of PCOS (16). Given the stable structure of miRNAs in the plasma, they can serve as highly potential biomarkers for the diagnosis of PCOS (17). With regards to the numerous genes and miRNAs that may contribute to developing PCOS, bioinformatics analysis can be used as a new prediction tool (7). In this study, the relationship of miR-27a, miR-130b, and miR-301a with the presence of PCOS was investigated using bioinformatics analysis to identify potential diagnostic biomarkers. To this end, three circulating miRNAs were studied in order to obtain more information on their potential role in PCOS. The elevated expression levels of miR-27a and miR-301a were detected in the patient group. In addition, the logistic regression analysis revealed that both miRNAs were correlated with PCOS development in patients. The sensitivity and specificity results demonstrated the potential of biomarkers to confirm PCOS diagnosis; however, miR-27a seems to be more efficient due to better specificity. A study has also shown miR-27a is comparatively over-expressed in the ovaries of PCOS patients (9). It has seemed that miR-27a-3p in the granulosa cells of PCOS patients plays a significant role in ovarian follicular development, and miR-27a-3p in the granulosa cells of the patients is dysregulated (18). In other words, miR-27a is highly associated with premature ovarian failure and granulosa cell apoptosis in human (19). In a rat model, in granulosa cells treated with steroid hormones, miR-27a expression was modulated (20). miR-27 a is involved in TLR2/4-activated macrophages and down-regulates interleukin 10, which is an essential regulatory cytokine in ovarian function (21). Besides, miR-27a is able to lead to inflammatory cytokines such as TNF-α and IL-6 production, leading to recruiting immune cells by suppressing the PPAR-γ gene (22). The estrogen receptor presence is essentially dependent on miR-27a, and elevated estrogen receptor level in PCOS patients is likely the sign of miR-27a function in PCOS (23, 24). In addition to miR-27a, miR-301a generally affects inflammatory cytokines through NF-κB and AP-1 pathways, resulting in obesity-related inflammation (25, 26). The marked difference in miR-130b between controls and the patient group showed that this circulating miRNA level was lower in the patients in comparison with controls. In contrast to miR-130b in ovarian cancer, the down-regulation of this miRNA was detected in human ovarian tissue (27). In addition, diabetes, which is common among PCOS patients, leads to a reduction in miR-130b expression (28). A lower expression of miR-130b in adipocyte differentiated cell line indicates that the metabolic process is likely to play a role in miR-130b expression (29). In the study of Sørensen et al., miR130b was seen only in non- PCOS women (30), and it is therefore logical that in our study, miR130b was decreased in PCOS patients in comparison with controls. In contradiction with our results, evidence suggests that in the follicular fluid of women with fertility problems, miR-130b is up-regulated in comparison with women without fertility problems. Therefore, over-expression of miR-130b seems to play a role in ovarian dysfunction (14). This contradiction may be due to the different mechanisms of infertility in PCOS patients and other patients investigated in other studies. Similar to the result of the current study about PCOS, up-regulation of miR-301a has been demonstrated in diabetes. MiR-301a may contribute greatly to controlling the Kv4.2 voltage-gated potassium channel gene expression in diabetes, and therefore may have therapeutic potential (31). One limitation of our study was the small sample size. A larger study including more phenotypes is needed for validation and comparison with the NIH and Androgen Excess and PCOS Society criteria. The lack of addressing clinical indices such as blood fasting glucose, insulin, and homeostasis may be the most noticeable limitation of our study. Besides that, the calculating levels of miR-130b and miR-301a might confirm our results because circulating plasma miRNAs may not fully represent miRNAs profile expression in the granulosa and follicular cells. Indeed, research with a larger cohort including different Rotterdam criteria is needed. Another limitation of this study was the lack of studying of the target gene(s). The study of granulosa cells (in the IVF processes) and functional studies in this field are necessary. However, the target gene(s) in this study were obtained only by means of the applicable software. Our study showed a common target (PPAR-γ) for all miRs. However, PPAR-γ has been reported to be related to PCOS (32, 33); similarly, the studied miRs in our study were observed to be related to PPAR-γ (34-36), but changes in the expression levels showed that PPAR-γ cannot be proposed as the target gene for all the three miRNAs in PCOS. Therefore, additional studies are needed in order to explain the biomarker role of these miRNAs and the target gene(s). In conclusion, the plasma gene expression of miR-27a and miR-301a were higher in the PCOS patients than in healthy controls, but miR-130b expression decreased in patients. It seems that miR-27a has the potential to serve as an efficient diagnostic biomarker for PCOS.
Acknowledgements
We thank Dr. Akram Alizadeh for her help. The authors also are grateful to the staffs of Students Research Committee, Cellular & Molecular Research Center, and Basic Health Sciences Institute of Shahrekord University of Medical Sciences, Shahrekord, Iran.This study was financially supported by the Research and Technology Deputy of Shahrekord University of Medical Sciences (grant no. 1241).
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Azziz R, Carmina E, Chen Z, et al. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16057. doi: 10.1038/nrdp.2016.57. [DOI] [PubMed] [Google Scholar]
- 2.Al-Jefout M, Alnawaiseh N, Al-Qtaitat A. Insulin resistance and obesity among infertile women with different polycystic ovary syndrome phenotypes. Sci Rep. 2017;7:5339. doi: 10.1038/s41598-017-05717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu M, Feng R, Sun X, et al. Polymorphisms of pentanucleotide repeats (tttta)n in the promoter of CYP11A1 and their relationships to polycystic ovary syndrome (PCOS) risk: a meta-analysis. Mol Biol Rep. 2014;41:4435–45. doi: 10.1007/s11033-014-3314-3. [DOI] [PubMed] [Google Scholar]
- 4.Ranjzad F, Mahmoudi T, Irani Shemirani A, et al. A common variant in the adiponectin gene and polycystic ovary syndrome risk. Mol Biol Rep. 2012;39:2313–9. doi: 10.1007/s11033-011-0981-1. [DOI] [PubMed] [Google Scholar]
- 5.Naji M, Aleyasin A, Nekoonam S, et al. Differential Expression of miR-93 and miR-21 in Granulosa Cells and Follicular Fluid of Polycystic Ovary Syndrome Associating with Different Phenotypes. Sci Rep. 2017;7:14671. doi: 10.1038/s41598-017-13250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–65. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- 8.McCallie B, Schoolcraft WB, Katz - Jaffe MG. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril. 2010;93:2374–82. doi: 10.1016/j.fertnstert.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Sun J, Xu B, et al. Functional Characterization of MicroRNA-27a-3p Expression in Human Polycystic Ovary Syndrome. Endocrinology. 2018;159:297–309. doi: 10.1210/en.2017-00219. [DOI] [PubMed] [Google Scholar]
- 10.Tang ST, Wang CJ, Tang HQ, et al. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with polycystic ovary syndrome: a meta-analysis. Mol Biol Rep. 2012;39:9649–60. doi: 10.1007/s11033-012-1830-6. [DOI] [PubMed] [Google Scholar]
- 11.Kim SY, Kim AY, Lee HW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392:323–8. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Samandari N, Mirza AH, Nielsen LB, et al. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia. 2017;60:354–63. doi: 10.1007/s00125-016-4156-4. [DOI] [PubMed] [Google Scholar]
- 13.Banwait JK, Bastola DR. Contribution of bioinformatics prediction in microRNA-based cancer therapeutics. Adv Drug Deliv Rev. 2015;81:94–103. doi: 10.1016/j.addr.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez RM, Liang L, Racowsky C, et al. Extracellular microRNAs profile in human follicular fluid and IVF outcomes. Sci Rep. 2018;8:17036. doi: 10.1038/s41598-018-35379-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lujan ME, Chizen DR, Pierson RA. Diagnostic criteria for polycystic ovary syndrome: pitfalls and controversies. J Obstet Gynaecol Can. 2008;30:671–9. doi: 10.1016/S1701-2163(16)32915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorensen AE, Wissing ML, Salo S, et al. MicroRNAs Related to Polycystic Ovary Syndrome (PCOS) Genes (Basel) 2014;5:684–708. doi: 10.3390/genes5030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long W, Zhao C, Ji C, et al. Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers. Cell Physiol Biochem. 2014;33:1304–15. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Liu M, Sun J, et al. MicroRNA-27a-3p affects estradiol and androgen imbalance by targeting Creb1 in the granulosa cells in mouse polycytic ovary syndrome model. Reprod Biol. 2017;17:295–304. doi: 10.1016/j.repbio.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Nie M, Yu S, Peng S, et al. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol Reprod. 2015;93:98. doi: 10.1095/biolreprod.115.130690. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z, Shen WJ, Cortez Y, et al. Hormonal regulation of microRNA expression in steroid producing cells of the ovary, testis and adrenal gland. PLoS One. 2013;8:e78040. doi: 10.1371/journal.pone.0078040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie N, Cui H, Banerjee S, et al. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol. 2014;193:327–34. doi: 10.4049/jimmunol.1400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minge CE, Ryan NK, Van Der Hoek KH, et al. Troglitazone regulates peroxisome proliferator-activated receptors and inducible nitric oxide synthase in murine ovarian macrophages. Biol Reprod. 2006;74:153–60. doi: 10.1095/biolreprod.105.043729. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Mertens-Talcott SU, Zhang S, et al. MicroRNA-27a Indirectly Regulates Estrogen Receptor {alpha} Expression and Hormone Responsiveness in MCF-7 Breast Cancer Cells. Endocrinology. 2010;151:2462–73. doi: 10.1210/en.2009-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heikkila K, Ebrahim S, Lawlor DA. Systematic review of the association between circulating interleukin-6 (IL-6) and cancer. Eur J Cancer. 2008;44:937–45. doi: 10.1016/j.ejca.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Patel N, Tahara SM, Malik P, et al. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem J. 2011;434:473–82. doi: 10.1042/BJ20101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z, Li Y, Takwi A, et al. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Cai J, Wang Q, et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124:325–34. doi: 10.1016/j.ygyno.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Balducci S, Sacchetti M, Haxhi J, et al. Physical exercise as therapy for type 2 diabetes mellitus. Diabetes Metab Res Rev. 2014;30 Suppl 1:13–23. doi: 10.1002/dmrr.2514. [DOI] [PubMed] [Google Scholar]
- 29.Shi C, Huang F, Gu X, et al. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget. 2016;7:40830–45. doi: 10.18632/oncotarget.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen AE, Wissing ML, Englund AL, et al. MicroRNA Species in Follicular Fluid Associating With Polycystic Ovary Syndrome and Related Intermediary Phenotypes. J Clin Endocrinol Metab. 2016;101:1579–89. doi: 10.1210/jc.2015-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panguluri SK, Tur J, Chapalamadugu KC, et al. MicroRNA-301a mediated regulation of Kv4 2 in diabetes: identification of key modulators. PLoS One. 2013;8:e60545. doi: 10.1371/journal.pone.0060545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portius D, Sobolewski C, Foti M. MicroRNAs-Dependent Regulation of PPARs in Metabolic Diseases and Cancers. PPAR Res. 2017;2017:7058424. doi: 10.1155/2017/7058424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahimi Z, Chamaie-Nejad F, Saeidi S, et al. The Association of PPARgamma Pro12Ala and C161T Polymorphisms with Polycystic Ovary Syndrome and Their Influence on Lipid and Lipoprotein Profiles. Int J Fertil Steril. 2018;12:147–51. doi: 10.22074/ijfs.2018.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esmaili A, Baharvand H, Karbalaii K, et al. Intranuclear localization of EGFP-mouse PPARγ1 in bovine fibroblast cells. Yakhteh medical journal. 2010;12:97–104. [Google Scholar]
- 35.Pan S, Yang X, Jia Y, et al. Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression. J Cell Physiol. 2014;229:631–9. doi: 10.1002/jcp.24486. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Xue M, Xu J, et al. MiR-301a is involved in adipocyte dysfunction during obesity-related inflammation via suppression of PPARgamma. Pharmazie. 2016;71:84–8. [PubMed] [Google Scholar]