Summary
The diaphragm is critical for respiration and separation of the thoracic and abdominal cavities, and defects in diaphragm development are the cause of congenital diaphragmatic hernias (CDH), a common and often lethal birth defect. The genetic etiology of CDH is complex. Single-nucleotide variants (SNVs), insertions/deletions (indels), and structural variants (SVs) in more than 150 genes have been associated with CDH, although few genes are recurrently mutated in multiple individuals and mutated genes are incompletely penetrant. This suggests that multiple genetic variants in combination, other not-yet-investigated classes of variants, and/or nongenetic factors contribute to CDH etiology. However, no studies have comprehensively investigated in affected individuals the contribution of all possible classes of variants throughout the genome to CDH etiology. In our study, we used a unique cohort of four individuals with isolated CDH with samples from blood, skin, and diaphragm connective tissue and parental blood and deep whole-genome sequencing to assess germline and somatic de novo and inherited SNVs, indels, and SVs. In each individual we found a different mutational landscape that included germline de novo and inherited SNVs and indels in multiple genes. We also found in two individuals a 343 bp deletion interrupting an annotated enhancer of the CDH-associated gene GATA4, and we hypothesize that this common SV (found in 1%–2% of the population) acts as a sensitizing allele for CDH. Overall, our comprehensive reconstruction of the genetic architecture of four CDH individuals demonstrates that the etiology of CDH is heterogeneous and multifactorial.
Keywords: congenital diaphragmatic hernia, CDH, structural birth defect, whole-genome sequencing, WGS, genetics
Introduction
The diaphragm is a mammalian-specific muscle critical for respiration and separation of the abdominal and thoracic cavities.1 Defects in diaphragm development lead to congenital diaphragmatic hernias (CDHs), a common structural birth defect (1 in 3,000–3,500 births2, 3, 4, 5) in which the barrier function of the diaphragm is compromised. In CDH, a weakness develops in the diaphragm, allowing the abdominal contents to herniate into the thoracic cavity and impede lung development. The resulting lung hypoplasia and pulmonary hypertension are important causes of the neonatal mortality and long-term morbidity associated with CDH.5, 6, 7 The phenotype of CDH is highly variable, and the clinical outcomes are diverse.7,8 Underlying this phenotypic diversity is a complex genetic etiology.9
Genetic variants in many chromosomal regions and over 150 genes have been implicated in CDH. Molecular cytogenetic studies of individuals with CDH have identified multiple aneuploidies, chromosomal rearrangements, and copy-number variants in different chromosomal regions.9,10 Chromosomal abnormalities are found in 3.5%–13% of CDH cases and are most frequently associated with complex cases in which hernias appear in conjunction with other comorbidities.9 In addition, many individual genes have been identified through analyses of chromosomal regions commonly associated with CDH,11 exome sequencing studies,12, 13, 14, 15 and analyses of mouse mutants.16,17 Variants in these individual genes can lead to either isolated or complex CDH.
Most genetic studies of the etiology of CDH have focused on the role of germline de novo variants. The preponderance of CDH cases that occur sporadically without a family history of CDH7 and the low sibling recurrence rate (0.7%18) have argued for the importance of this class of genetic variants. Indeed, trio studies of CDH-affected children and their unaffected parents that employed cytogenetic analyses or exome or genome sequencing have identified de novo chromosomal anomalies and gene variants.12, 13, 14, 15,19 However, to date most identified genes recur in none or only a few CDH cases.20 Furthermore, one of these exome sequencing studies estimated that only 15% of sporadic non-isolated CDH cases can be attributed to de novo gene-disrupting or deleterious missense variants.15 In addition, variants in particular CDH-associated chromosomal regions or genes are often incompletely penetrant for CDH or associated with subtle subclinical diaphragm defects.11,21 Thus, while de novo chromosomal anomalies and variants in individual genes undoubtedly are important, the genetic etiology of CDH is more complex and likely polygenic and multifactorial.
Another class of variants that may contribute to CDH etiology is somatic de novo variants. A potential role of somatic variants has been suggested by the discordant appearance of CDH in monozygotic twins18,22 and the finding of tissue-specific genetic mosaicism in CDH individuals.23,24 More recently, our functional studies using mouse conditional mutants found that development of localized muscle-less regions leads to CDH and suggest that in humans somatic variants in the diaphragm may cause muscle-less regions that ultimately herniate.17
Although less commonly investigated, inherited variants have been linked to CDH. Analyses of families with multiple members affected by CDH revealed that autosomal recessive alleles can cause CDH.25, 26, 27, 28 Other familial CDH cases exhibit an inheritance pattern of autosomal dominance with incomplete penetrance. For instance, two families have been reported with multiple CDH offspring who inherited either a large deletion or frameshift variant in ZFPM2 but with unaffected carrier parents.29 In another case, monoallelic missense variants in GATA4 were inherited in three generations of one family and associated with a range of diaphragm defects, but only one family member had symptomatic CDH.21 Thus, these familial cases demonstrate that inherited variants can contribute to CDH etiology, but these genetic variants often exhibit incomplete penetrance and variable expressivity.
While human genetics studies have been essential for identifying candidate CDH chromosomal regions and genes, experiments with rodents have been critical for determining mechanistically how the diaphragm and CDH develop and explicitly testing whether candidate genes cause CDH. Embryological and genetic lineage experiments17,30, 31, 32 have shown that the diaphragm develops primarily from two transient embryonic tissues: the somites and the pleuroperitoneal folds (PPFs). The somites are the source of the diaphragm’s muscle, as muscle progenitors migrate from cervical somites into the PPFs.32 The PPFs give rise to the diaphragm’s muscle connective tissue and central tendon.17 Importantly, the PPFs regulate the development of the diaphragm’s muscle and control overall diaphragm morphogenesis, which takes place between embryonic day (E) 9.5 and E16.5 in the mouse (corresponding to E30–E60 in humans).17,32 Engineered mutations in mice of candidate CDH genes have definitively established that these genes are functionally important in CDH.17 In addition, conditional mutagenesis experiments indicate that the PPFs are an important cellular source of CDH, as inactivation of Gata4, WT1, or β-catenin in the PPFs results in hernias,17,33,34 while Gata4 inactivation in somites does not affect diaphragm development.17 Furthermore, these experiments established that mutations in CDH genes initiate aberrations in the development of the PPFs by E12.5 in the mouse.17,33,34 In contrast, mutations in the diaphragm’s muscle lead to diaphragms that are muscle-less or with thin or aberrant muscle but so far have not been found to lead to CDH.17,35, 36, 37, 38, 39, 40, 41, 42, 43, 44 Altogether, these data indicate that the PPFs are critical for diaphragm morphogenesis and a cellular source of CDH, while a direct role in CDH for genes expressed in the diaphragm’s muscle is less clear. Given the importance of the PPFs in CDH, prioritization of genes expressed in the early mouse PPFs is likely to be an effective strategy for evaluating new candidate CDH genes derived from human genetic studies.
In this study, we take a novel approach to studying the etiology of CDH. Complementing recent studies using large cohorts of CDH individuals that focus on one class of possible variants—de novo germline variants12, 13, 14, 15—we comprehensively examine the genome of four CDH individuals with multiple tissue samples and their unaffected parents. Using deep whole-genome sequencing and a sophisticated bioinformatics toolkit, we determine the contribution of germline and somatic de novo and inherited variants to CDH etiology. Our analysis includes variants of different sizes—single nucleotide variants (SNVs), small insertions and deletions (indels), and larger structural variants (SVs)—in all genomic regions (exons, introns, UTRs, and intergenic). We prioritize implicated genes not only based on their frequency in the general population and predicted effect on gene function, but also on their expression in early PPF fibroblasts and diaphragm muscle progenitors, using a newly generated mouse RNA-sequencing (RNA-seq) dataset. Altogether, we reconstruct the diverse genetic architecture underlying isolated CDH in four individuals, revealing the heterogeneous and multifactorial genetic etiology of CDH.
Material and methods
Ranking of CDH-implicated genes
CDH genes reported in the literature were gathered from recent reviews,9,45 large human cohort studies,12, 13, 14, 15,46 and recent studies.46,47 Original publications implicating genes in CDH were checked for the level of evidence (i.e., variants likely impacting gene function or deleterious as described in original studies), and the following ranking system was used.
Mouse Data Ranking: 10 = CDH, >80% frequency; 9 = CDH, 40%–60% frequency; 8 = CDH, <40% frequency; 7 = muscle-less patches, muscle-less diaphragm, thin diaphragm, >80% frequency; 6 = muscle-less patches, muscle-less diaphragm, thin diaphragm, 40%–60% frequency; 5 = muscle-less patches, muscle-less diaphragm, thin diaphragm, <40% frequency.
Human Data Ranking: 9 = inherited compound heterozygous or homozygous deleterious variant (2 alleles in 1 gene) + de novo deleterious variant, >1 individual; 8 = inherited compound heterozygous or homozygous deleterious variant (2 alleles in 1 gene), >1 individual; 7 = de novo deleterious variant (1 allele in 1 gene), >1 individual; 6 = inherited compound heterozygous or homozygous deleterious variant (2 alleles in 1 gene), 1 individual; 5 = inherited deleterious variant (1 allele in 1 gene), >1 individual; 4 = unknown inheritance deleterious variant (1 allele in 1 gene), >1 individual; 3 = de novo deleterious variant (1 allele in 1 gene), 1 individual; 2 = inherited deleterious variant (1 allele in 1 gene), 1 individual; 1 = unknown inheritance deleterious variants (1 allele in 1 gene), 1 individual. The final ranking of each gene was determined as the sum of the mouse data and the human data ranking and constitutes the order of genes found in Table S1. CDH-implicated genes were queried for expression in the mouse E12.5 PPF RNA-seq dataset and expression plotted using the ggplot2 R package.48 Gene networks within the list of CDH-associated genes were visualized using STRING,49 visualizing high and medium levels of evidence to connect gene nodes, using all evidence (except the “text mining” option was not used).
Patient samples
Participants were enrolled as previously described.15 All four probands had isolated CDH and were enrolled in Columbia University institutional review board (IRB) protocol AAAB2063 and provided informed written consent for participation in this study. Probands 411, 809, and 967 had left hernias with <50% of chest wall devoid of diaphragmatic tissue, while proband 716 had a large (>50% of chest wall devoid of diaphragmatic tissue) right hernia. Reflecting the severity of the hernia in proband 716, her liver was found in the chest cavity at 21 weeks in utero; the liver was found to be in the abdomen of probands 769 and 809 in utero. None of the probands were placed on extracorporeal membrane oxygenation (ECMO) or suffered pulmonary hypertension, and all are currently alive. The self-reported ancestries are probands 411 and 809 are European (non-Hispanic), proband 716 is Asian, and proband 967 is African and Hispanic. Whole-genome analysis of blood from probands 411, 716, and 809 and their parents were included in a previous study.14
Whole-genome sequencing
DNA was prepared for sequencing using TruSeq DNA PCR-free libraries (Illumina) and run on the Illumina HiSeq X Ten System at a minimum of 60× median whole genome coverage. Whole genome sequencing and data analysis at the University of Utah was covered by IRB protocol 00085165.
Whole-genome alignment, variant calling, and quality checks
Genomic sequencing reads were aligned with BWA-MEM50 against human GRCh37 reference genome (including decoy sequences from the GATK resource bundle). Aligned BAM files were de-duplicated using samblaster.51 Base quality score recalibration and realignment of small insertions and deletions was performed with the GATK package.52 Alignment quality was checked with samtools53 “stats” and “flagstats” functions. Variants (SNVs, indels) were called with GATK Haplotypecaller.52 Sample relatedness, sex, and reported ancestry were confirmed with Peddy.54
Germline and somatic de novo SNV and indel variant analysis
De novo SNVs and indels were called with RUFUS using standard parameters (25 length k-mers and 40 threads) and realigning k-mers to the human GRCh37 reference genome. Each proband tissue sample was run against both parents as control samples to call germline de novo variants, and all possible combinations of one proband tissue against the other two were run to call tissue-specific variants. Variants flagged “DeNovo” by RUFUS were retained, and any variants found in all three proband tissues were called germline de novo. Variants were further filtered, and only variants were retained with genotype quality (GQ) scores > 20, read depths > 15 at the variant site, and the variant was found in ≥ 20% of reads in the sample using annotations from the GATK haplotypecaller52 via a python script written with the cyvcf2 package55 and annotations in Integrative Genomics Viewer (IGV).56 Variant quality, alignment, and sample specificity were confirmed visually in IGV.56 5–20 bp indels located at the start or end of single-nucleotide repeats were filtered out, as these are potential false positives due to alignment error. Genetic locations were annotated with the University of California Santa Cruz (UCSC) Known Gene Annotation.57 Noncoding variants were intersected against a bed file of enhancers from the VISTA Enhancer Database58 using bedtools59 to determine whether variants were within annotated enhancers. Coding variant predicted damage was determined by both Protein Variation Effect Analyzer (PROVEAN)60 and combined annotation-dependent depletion (CADD) scores,61 and allele frequency within a large, healthy population was determined by gnomAD.62 Genes containing coding variants were annotated as intolerant for loss of function with ExAC Pli scores.63
Inherited SNV and indel variant analysis
The VAAST3 pipeline was used to call predicted damaging inherited recessive variants.64,65 The pipeline includes the following steps. First, GATK haplotypecaller52-identified variants were decomposed and normalized using VT,66 and variants with a GQ score < 30 or with > 25% of the samples in the variant call file (VCF) file not genotyped (“no-call”) were filtered out using vcftools.67 Variant effect predicted annotations were added to the filtered VCF file using variant effect predictor (VEP)68 with version 83 of the hg19 vep-cache. As a background population, a VCF file with variants from 1000 Genomes phase 369 were run through the same workflow. A VAAST variant prioritizer (VVP) background database was made from the 1000 Genomes filtered variants using the build background function of VVP.70 This background database and the filtered VCF file of variants discovered in this cohort were used as inputs to VVP to prioritize cohort-discovered variants, which were then passed to VAAST3 to be scored and ranked. Blood samples from parents and the proband were used in VAAST3’s trio recessive inherited model. Genes with a p value > 0.05 from VAAST3 were filtered out. The remaining genes were annotated with the predicted effect from VEP, the location of the variant from VVP, and the parental origin of the allele from VVP. Ensembl IDs given in the VAAST3 output were converted to gene names using GeneCard.71 Genes were then filtered out if (1) the genes were expressed at <10 transcripts per million (TPM) in E12.5 mouse PPFs (human gene names were converted to mouse orthologs using OrthoRetriever), (2) they were a pseudogene annotated by GENCODE,72 (3) they belonged to a highly mutable gene family, (4) the allele called is found in >0.01 of individuals in gnomAD, or (5) there are multiple alleles at the site in gnomAD. Variant impact was predicted by PROVEAN,60 CADD,61 and ExAC pli scores,63 and then genes were ranked based on the number and severity of damaging alleles.
Inherited and de novo SV analysis
SVs were called with the Lumpy Smooth pipeline.73 Regions with possible copy number variants based on read depth were called using CNVkit,74 CNVnator,75 and CN.MOPS76 with sample BAM files. Outputs from the three tools were converted into BEDPE files and merged together as one “deletions” file (evidence of a decrease in copy number) and one “duplications” (evidence of an increase in copy number) per sample. The copy number call BEDPE files and aligned BAM files were used as input to Lumpy73 and called with the lumpy_smooth script (in the LUMPY scripts directory). Lumpy output variants were used as inputs to SVTYPER77 to annotate each variant as an insertion, deletion, translocation or inversion within a VCF file. De novo and inherited SVs were identified with GQT78 using the LUMPY-created VCF file and a PED file describing family relations. We kept only variants with either spilt or discordant supporting read counts above 8 and but below 400 (to exclude noisy regions with high read mapping). SVs were confirmed in IGV,56 keeping variants with visible evidence in read coverage change and discordant reads. De novo and inherited SVs were queried in the complete VCF file from Abel et al79 to determine allele frequency in the general population, and we filtered out SVs with an allele frequency >10%. Variants located in intergenic regions, overlapping annotated repetitive element or elements,80 or homozygous in parental DNA were excluded. The discovered 343 bp deletion was confirmed with PCR using primers: forward, 5′-TTCCTCTACCATTGGGCGTTT-3′ and reverse, 5′-AGGTAGTACGGCTGACTTGC-3′.
E12.5 PPF RNA sequencing
PPFs were isolated from wild-type E12.5 mouse embryos by cutting embryos just above the hindlimbs and below the forelimbs and removing the heart and lungs cranially, leaving the trunk with attached nascent diaphragm. The PPFs were manually dissected and stored in RNALater (Thermo Fisher, #AM7020) at −80°C. RNA was isolated using a Rneasy Micro Kit (QIAGEN, #74004), RNA quality confirmed with RNA TapeStation ScreenTape Assay (Agilent, # 5067-5576), and sequenced using TruSeq Stranded mRNA Library Preparation Kit with polyA selection (Illumina) with HiSeq 50 Cycle Single-Read Sequencing v4 (Illumina) through the High-Throughput Genomics and Bioinformatic Analysis Shared Resource at Huntsman Cancer Institute at the University of Utah. Two biological replicates were sequenced (two pooled PPF pairs per replicate) and analyzed. Sequencing reads were aligned to the mouse genome (mm9) with Spliced Transcripts Alignment to a Reference (STAR),81 using standard parameters. featureCounts82 was used to count reads per gene and then normalized by TPM using R. Mouse data were gathered under the purview of the University of Utah Institutional Animal Care and Use Committee protocol 19-05009 to G.K.
Figure creation and statistics
Figures 1A, 3B, 4B, and S1 were created with R package ggplot2.48 Figure S2 was created with Prism 7 (Graphpad). Figures 2, 3A, 4A, and 6 were created with Adobe Illustrator. Figure 5 was created with Samplot and Adobe Photoshop. Genome tracks were plotted using the Gviz R package,83 with UCSC Known Gene,57 genomic evolutionary rate profiling (GERP) conservation scores,84 ENCODE Dnase 1 hypersensitivity clusters, H3K27 acetylation in normal human lung fibroblasts (NHLF) cells,85 and VISTA enhancer element tracks.58 Except for the VISTA elements, all data were downloaded from the UCSC Genome Table Browser. The 343 bp deletion sequence and the orthologous sequence in mouse (mm9 reference genome) were downloaded from the UCSC Genome Browser and aligned with Geneious.86 Statistics for Figure S1 were generated with Prism 7, using a one-way ANOVA with multiple comparisons to test differences between somatic or germline de novo variants found across probands or tissues and unpaired t tests to compare somatic and germline de novo variants within probands or tissues.
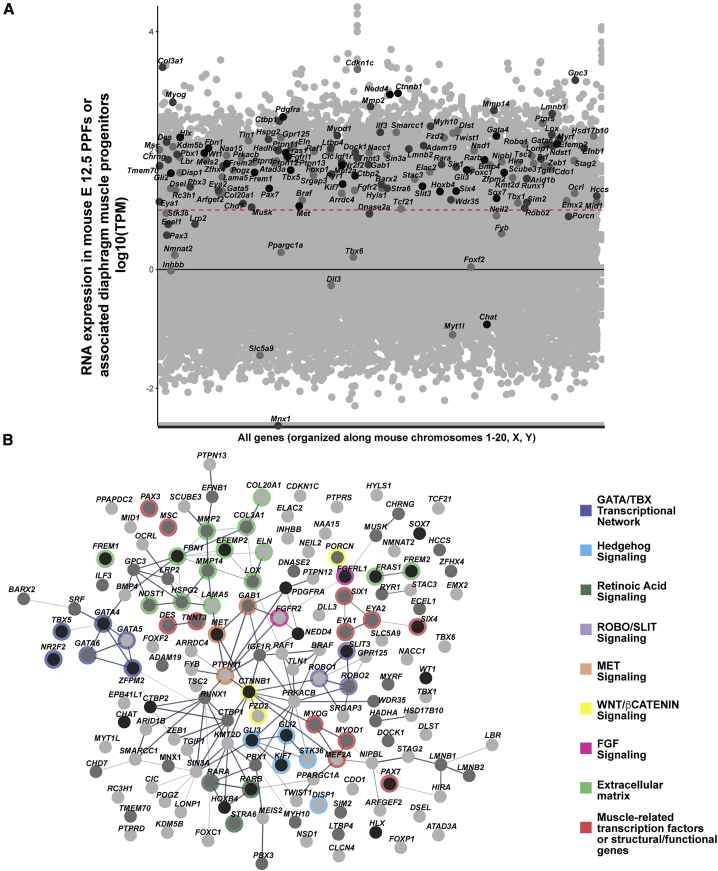
Figure 1.
Genes associated with CDH in the literature were collected and ranked based on the amount of human CDH individual and mouse model functional evidence (see Table S1 for rankings)
(A) CDH-associated genes overlaid on all genes expressed in mouse PPFs at E12.5. RNA-seq reads normalized using TPM. Black line denotes TPM = 0, and red dashed line denotes TPM = 10 (Log1). Black dots, CDH genes highly supported by human and mouse data; dark gray dots, genes with moderate data support; light gray dots, CDH genes with modest data support.
(B) STRING protein network of CDH-associated genes. Within the STRING tool all active interaction sources except text-mining were used, with a minimum required interaction score of 0.4. Edges are based on the strength of data support. Nodes without edges are genes that do not interact with any other listed genes. Thick edges, interaction score > 0.9; thinner edges, 0.9–0.7 interaction score; thinnest edges, interaction score < 0.4. Prominent GATA/TBX transcriptional network, extracellular matrix, and muscle-related genes are highlighted as well as Hedgehog, Retinoic Acid, ROBO/SLIT, MET, WNT/β-CATENIN, and FGF signaling pathways. In both RNA-seq expression and protein network genes ranked 1–27 (with a total score of ≥ 10) are shown in black, genes ranked 28–88 (with a total score 5–9) are shown in dark gray, and genes ranked 89–153 (with a total score < 5) are shown in light gray. Details on rankings are described in Table S1 and Material and methods.
Figure 3.
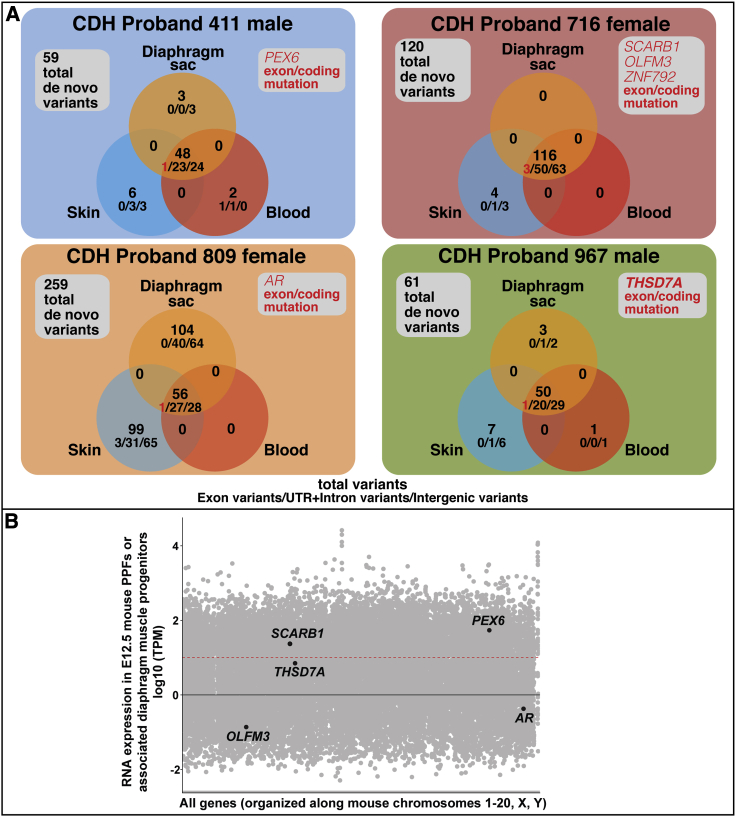
Germline and somatic de novo variants, identified via RUFUS, in CDH probands
(A) De novo germline and somatic variants unique to a single tissue and shared between two tissues (coding, gene-associated non-coding, or non-gene-associated variants) were found in the 4 CDH probands. 5 genes (PEX6, SCARB1, OLFM3, ZNF792, AR, and THSD7A) contained germline coding variants. Genes with coding variants are highlighted in red. #/#/# = exon (coding) variants/UTR + intron variants/intergenic variants.
(B) Five genes with germline de novo coding variants overlaid on all genes expressed in mouse PPFs at E12.5 (ZNF792 has no mouse ortholog). RNA-seq reads normalized using TPM. Black line denotes TPM = 0, and red dashed line denotes TPM = 10. Only SCARB1, PEX6, and THSD7A are expressed in the PPFs at approximately ≥10 TPM.
Figure 4.
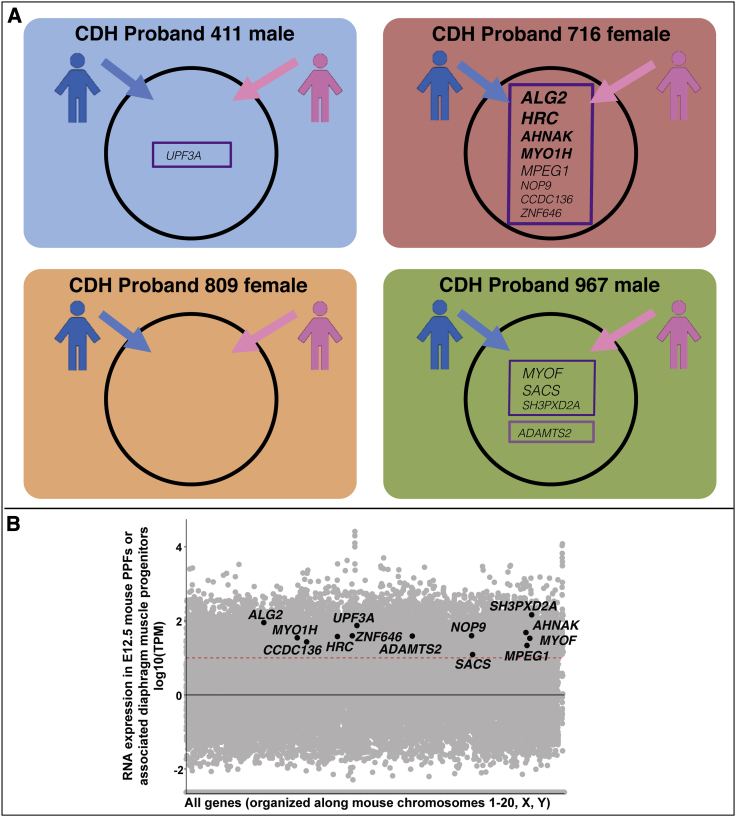
Inherited variants potentially contributing to etiology of CDH probands
(A) Compound heterozygous (boxed in dark purple) or homozygous (boxed in light purple) inherited variants highly ranked as damaging and rare by VAAST (with pseudogenes, highly mutable genes, and genes expressed at low levels in PPFs filtered out). Genes in bold and in larger font harbor variants that are ranked as more damaging (see Table S4 and its legend).
(B) Candidate genes with inherited variants overlaid on all genes expressed in mouse PPFs at E12.5. RNA-seq reads normalized using TPM. Black line denotes TPM = 0, and red dashed line denotes TPM = 10.
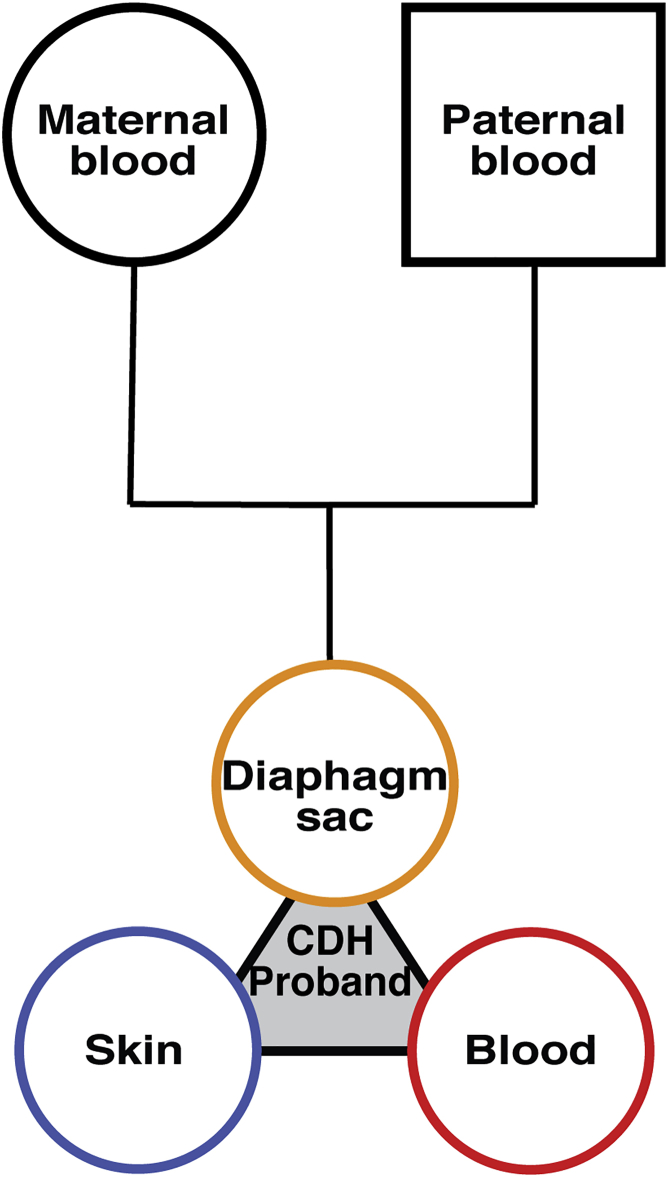
Figure 2.
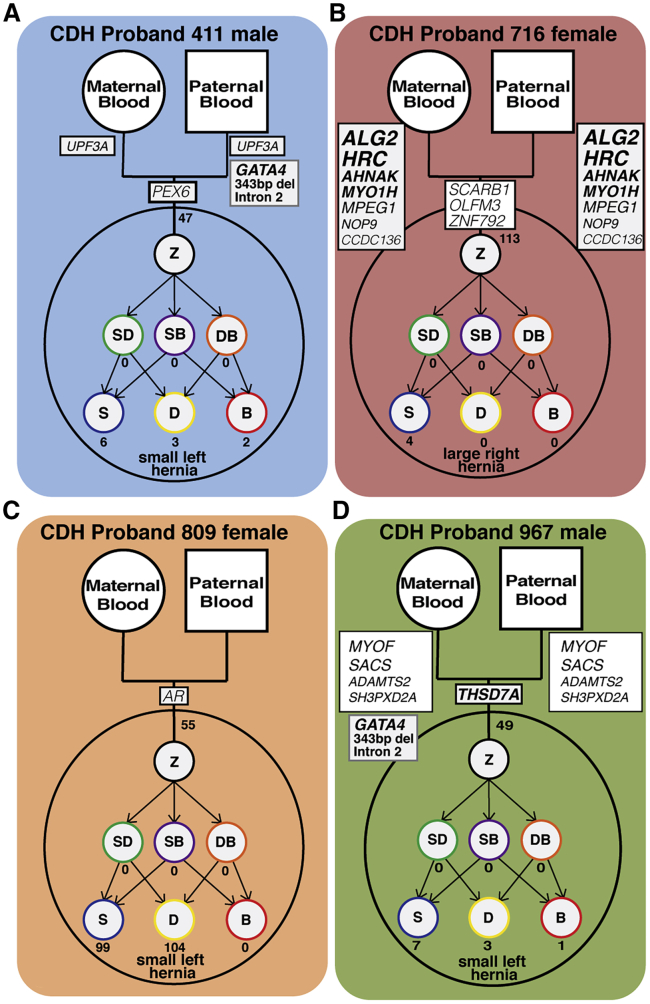
Sample cohorts analyzed in this study
Each cohort consists of DNA samples from diaphragm sac (orange), skin (blue), and blood (red) of the CDH proband and blood from the proband’s mother and father.
Figure 6.
Models of four CDH probands with inherited, germline, and somatic de novo variants, including coding, gene-associated (but non-coding), and non-gene-associated SNVs, indels, and SVs
Each model highlights the genetic complexity and heterogeneity underlying CDH. Z, zygote; SD, skin-diaphragm progenitor; SB, skin-blood progenitor; DB, diaphragm-blood progenitor; S, skin cell; D, diaphragm cell; B, blood cell.
Figure 5.
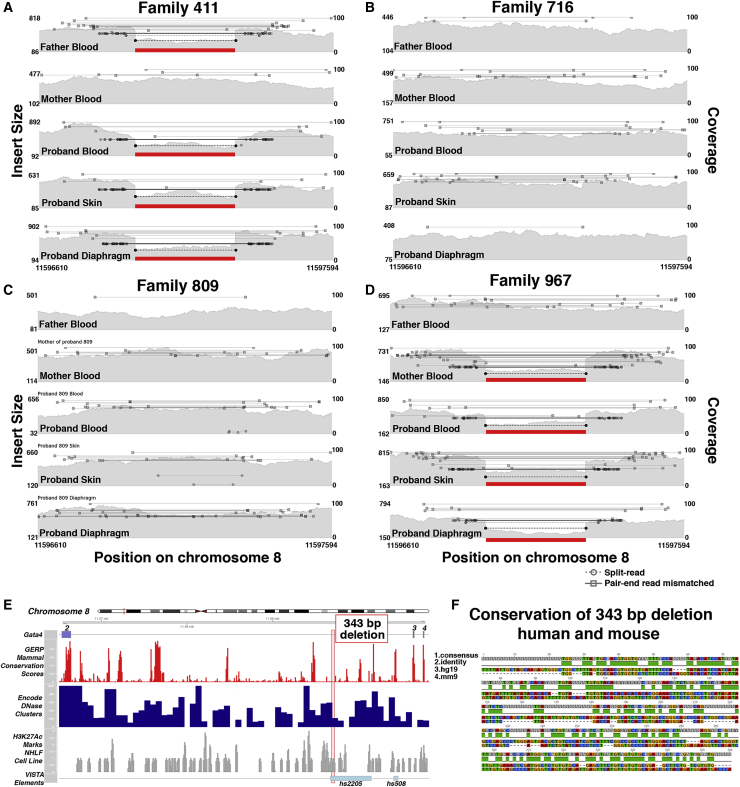
Inherited deletion in intron 2 of GATA4, in a region that overlaps an enhancer conserved in mouse and humans, found in CDH probands 411 and 967
(A–D) Samplot figures show coverage in gray (right y axis) and insert size between pair-end reads (left y axis), with split and discordant reads with mismatched insert size above 500 shown as bars. Paternal inheritance in proband 411 (A) and maternal inheritance in proband 967 (D) of intron 2 deletion.
(E) GATA4 intron 2 and 343 bp deleted region (red box) with tracks of gene location, conservation, DNase 1 hypersensitivity, and H3K27 acetylation in human lung fibroblasts. Bottom track shows enhancer element 2205 from the VISTA enhancer database, within which the deletion resides. Built on the hg19 human reference genome.
(F) Pairwise alignment of 343 bp region deletion in CDH families to the orthologous region in the mouse genome (mm9 reference genome). Track 1, consensus sequence; track 2, base pair identity; track 3, human (hg19) sequence; track 4, mouse (mm9) minus strand sequence.
Results
Strongly supported CDH genes are expressed at high levels in early mouse PPF fibroblasts or diaphragm muscle progenitors
In anticipation of needing a strategy to prioritize candidate genes discovered in our human genetic studies, we systematically and comprehensively analyzed CDH-implicated genes in the literature and then determined whether these genes were strongly expressed in early mouse PPF fibroblasts or diaphragm muscle progenitors.
We compiled a list of 153 CDH-implicated genes from three large cohort studies,13, 14, 15 previous literature reviews,9,45 and recently published studies46,47 (Table S1) and ranked the genes based on their level of support. We included genes that either had mouse functional data or human genetic data found in more than 1 CDH individual, associated with other developmental disorders or structural birth defects that co-occur in individuals with complex CDH, and/or implicated by Longoni et al.13 via their interaction with known CDH genes and expression in the developing diaphragm, as determined by Russell et al.45 We ranked each gene based on the nature of the variants, penetrance, and frequency reported in mouse and human data (for details, see Material and methods). For mouse data, genes in which variants resulted in herniation of abdominal contents into the thoracic cavity were ranked more highly than those that simply resulted in muscle-less regions or entirely muscle-less diaphragms. In addition, genes in which variants led to highly penetrant phenotypes were more highly ranked. For human data, genes in which inherited homozygous or biallelic putative deleterious (as described by original publication) variants or de novo deleterious variants were found in more than one CDH individual (either more than one individual in one study or across multiple studies) were ranked more highly than genes with putative deleterious variants of unknown inheritance or found in only one individual. The scores for mouse and human data were added to produce a final score and ranking. Twenty-seven genes had a score of 10–19 and were deemed highly likely to contribute to CDH etiology, 51 genes had a score of 5–9, and 75 genes had a score of <5.
Previous mouse studies of the development of CDH demonstrated that the PPFs are a critical cellular source of CDH, and many CDH-implicated genes are expressed and required in early PPF fibroblasts.17,33,34,87 To test whether the CDH genes identified in the literature are expressed in the PPF fibroblasts or associated diaphragm muscle progenitors, we micro-dissected and isolated whole E12.5 PPFs with their associated fibroblasts and diaphragm muscle progenitors from wild-type mice and performed RNA sequencing. We found that nearly all (26/27, 96%) highly ranked genes are expressed at levels of at least 10 TPM reads (which includes 29% of total transcripts), while 45/51 (88%) of moderately ranked genes and 63/75 (84%) of lowly ranked genes are expressed at this level (Figure 1A). These data suggest that genes involved in CDH are expressed at levels of at least 10 TPM in E12.5 PPFs or diaphragm muscle progenitors. In evaluating the significance of newly identified putative CDH genes, a finding that such genes are expressed at ≥10 TPM increases confidence that such genes indeed are important to CDH etiology.
Analysis of the highly ranked CDH genes reveals gene families and pathways that likely lead to CDH. To discover protein networks, we inputted the 153 genes into STRING49 (using all active interaction sources except text mining and requiring a minimum interaction score of 0.4) to generate a protein network (Figure 1B). The two highest-scoring genes, GATA4 and ZFPM2 (FOG2), encode a transcription factor and a co-factor that directly interact with each other.88 In addition, in the heart GATA4 and ZFPM2 interact with the protein encoded by the highly ranked gene NR2F2,89 and GATA4 interacts with the protein encoded by the highly ranked gene TBX5.90,91 Thus, not only are variants in GATA4, GATA6, ZFPM2, NR2F2, and TBX5 highly implicated in CDH, but GATA4 (GATA6), ZFPM2, NR2F2, and TBX5 proteins may function together in a complex to regulate diaphragm development. Another class of highly ranked genes is those involved in the extracellular matrix (EFEMP2, FREM1, FREM2, FRAS1, FBN1, HSPG2, COL3A1, COL20A1, LAMA5, and ELN) as well as genes that modify matrix components (MMP2, MMP14, NDST1, and LOX). Also prominent are genes involved in several critical developmental pathways: ROBO/SLIT signaling (SLIT3, ROBO1, and ROBO2), Retinoic Acid signaling (RARB, RARA, and STRA6), SHH signaling (GLI2, GLI3, KIF7, DISP1, and STK36), WNT signaling (CTNNB1, FZD2, and PORCN), MET signaling (MET, GAB1, and PTPN11), and FGF signaling (FGFRL1and FGFR2). The high degree of connectivity between members of the WNT and SHH signaling pathways and other CDH-implicated genes, coupled with strong functional studies in mouse, suggest these two pathways may be particularly important.
Present in the list of CDH-implicated genes are those expressed in myogenic cells and critical for myogenesis (SIX4, SIX1, EYA1, EYA2, MEF2A, MSC, PAX7, PAX3, MYOD1, MYOG, DES, and TNNT3). However, while these genes are important for myogenesis, and variants lead to muscle-less regions or muscle-less diaphragms, it is not clear that they lead to diaphragmatic hernias. For instance, mutations in Pax3 in mouse lead to completely muscle-less diaphragms, and while the diaphragms are highly domed, they do not allow abdominal contents to herniate into the thoracic cavity.17 Thus, while multiple reviews have included these genes as potentially implicated in CDH (e.g., Kardon et al.,9 Longoni et al.,13 and Russell et al.45) their true role in CDH is less clear.
Cohort of 4 CDH probands and their parents with multiple proband tissue samples enable unique genetic insights into CDH etiology
To gain greater insight into the genetic etiology of CDH, we analyzed four probands with CDH and their parents with a unique array of tissue samples (Figure 2). Our cohort consists of four unrelated individuals (male probands 411 and 967 and female probands 716 and 809) who had an isolated CDH in which the diaphragmatic hernias were encased by a connective tissue sac. Reflecting the increased prevalence of left versus right CDH,7 three probands (411, 809, and 967) had left CDH and one proband (716) had right CDH. Three of the probands (411, 809, and 967) had hernias with <50% of the chest wall devoid of diaphragmatic tissue, and one (716) had a large hernia (>50% of the chest wall devoid of diaphragmatic tissue). From each CDH proband, the sac was surgically removed during corrective surgery and saved, and skin biopsies and blood draws were taken. In addition, blood samples were taken from each parent. Altogether, five samples (sac, skin, and blood from CDH proband and blood from 2 parents) from each family were paired-end, whole-genome sequenced to an average coverage > 50 reads across each genome. Using Peddy,54 for each pentad of samples we confirmed sample quality, sex, relatedness, and reported ancestry (Table S2).
Analysis of germline de novo variants identifies THSD7A as a novel CDH candidate gene
To discover germline de novo variants, we analyzed whole genome sequences of the pentad of samples using the variant calling tool RUFUS. RUFUS subdivides each genome into a series of k-mers and aligns k-mers from samples of interest to control samples to identify unique SNVs and INDELs in the sample of interest. CDH proband genomes were compared with parental genomes, and variants present in ≥20% of reads (with GQ scores > 20 and read depths > 15) of proband diaphragm sac, skin, and blood genomes but not in parental genomes were designated as germline de novo variants and confirmed by visual inspection in IGV.56 It should be noted that our experimental design differs from all previous studies.12, 13, 14, 15 These studies used blood samples from CDH probands and their parents, inferred that variants present in the proband and not the parents arose in the egg or sperm giving rising to the child, and designated the discovered variants as germline de novo variants. However, blood samples may also harbor somatic variants that arose later in development (see results below), and such somatic variants could be erroneously designated as a germline variant. Because we have samples from three different tissues of each CDH proband as well as the parents, variants present in the proband diaphragm sac, skin, and blood but not present in the parents are definitively identified as germline (or at least early embryonic) variants. In addition, the identification of these variants in three separate samples independently confirms that the variants are true variants.
Germline de novo SNVs and INDELs were found in all four CDH probands (shown in intersection of diaphragm sac, skin, and blood of 4 CDH probands in Figure 3A and detailed in Table S3). CDH probands have 48–116 germline de novo variants, falling close to the 70 de novo SNVs expected in each newborn in an average population.92,93 As expected, the largest number of de novo germline variants were in intergenic regions (24–63 variants), and fewer were in UTRs or intronic regions (20–50 variants). In the 4 probands, germline de novo non-synonymous coding (exon) variants in six genes (PEX6, SCARB1, OLFM3, ZNF792, AR, and THSD7A) were discovered. De novo coding variants impacting these genes have not been found in other CDH individuals, and these genes are not located within CDH-associated chromosomal regions. All of these coding variants are rare (gnomAD frequency < 0.0001). The PEX6 variant is a damaging frameshift variant, while the THSD7A variant is a damaging nonsense variant (stop gain, p.Trp260Ter). SCARB1, OLFM3, ZNF792, and AR are all missense variants, but only the OLFM3 and ZNF792 variants are predicted to be damaging by PROVEAN (which takes into account conservation of orthologous sequences60). THSD7A is the only one of these four genes (PEX6, THSD7A, OLFM3, and ZNF792) with a damaging variant predicted by ExAC Pli63 to be haploinsufficient (intolerant of loss of one allele) and also substantially expressed in mouse PPFs or diaphragm muscle progenitors (TPM of 7.0; Figure 3B). Thus THSD7A is a promising new candidate gene in which variants lead to CDH. THSD7A (Thrombospondin Type I Domain Containing 7A) is a protein containing 10 thrombospondin type I repeats and through its co-localization with αvβ3 integrin and paxillin has been shown to promote endothelial cell migration during development.94, 95, 96 In diaphragm development, THSD7A may similarly regulate diaphragm vascularization or it may regulate PPF fibroblast migration, which is essential for diaphragm morphogenesis.17
Somatic de novo variants are present in diaphragm, skin, and blood, but diaphragm somatic variants are unlikely to contribute to CDH in the probands analyzed
Our previous mouse genetic functional study of CDH17 suggested the hypothesis that somatic variants in the PPF-derived muscle connective tissue fibroblasts contribute to CDH etiology. Our cohort of CDH proband samples that include the diaphragm sac, which is composed of PPF-derived connective tissue, uniquely allow us to test this hypothesis. In addition, because the samples were collected within a few weeks of birth, we are able to determine the background frequency of somatic variants in the skin and blood prior to exposure of any potential environmental mutagens.
To identify somatic de novo variants, diaphragm sac, skin, or blood CDH proband genomes were compared via RUFUS with the other 2 genomes derived from the same CDH proband, and variants present in ≥20% of reads (with GQ scores > 20 and read depths > 15) in only 1 or 2 tissues of each proband genome and not present in parental genomes were designated as somatic de novo variants. Variants were only included in which all three tissue samples had at least 15× coverage and Phred genotype quality score97,98 above 20. Variants were designated as tissue-specific if ≥20% alternate variant reads were present in one or two tissue samples and no alternate reads were present in the other samples.
Somatic de novo variants were found in all four CDH probands (Figure 3; Table S3). Across both individuals and tissues, the alternative allele read depth of somatic variants was significantly lower than germline de novo variants (Figure S1).99, 100, 101 This indicates that the somatic de novo variants are present in a subset of the sampled cells; in the diaphragm, this reflects either that not all PPF-derived connective fibroblasts harbor the variant and/or that the sampled tissue includes several cells types (e.g., connective tissue fibroblasts and endothelial cells), of which only one cell type (e.g., fibroblasts) harbors the variant. An analysis of the mutational spectrum (Figure S2A) reveals that the spectrum was generally similar across all probands (although probands 411 and 967 harbor a larger number of deletions) and, as expected, transitions (C/T or A/G changes) are more frequent than transversions (C/G, C/A, A/T, A/C changes). The mutational spectrum also did not vary widely among diaphragm, skin, and blood (Figure S2B).
Diaphragm sac, skin, and blood all harbored private variants, but no variants were shared between two tissues (Figure 3A; Table S3). Intergenic variants were the most common class of somatic variants, with variants in UTRs and introns as the next most common. Somatic coding variant were only found in the blood of proband 411 (missense variant in MIR4717) and in the skin of proband 809 (missense variants in DHX57 and TNFAIP8L3 and a synonymous variant in Cxorf57). In all four probands, the diaphragm contained somatic variants, but none of these were in coding regions, and variants in UTRs and intron regions did not overlap any annotated enhancers. Thus, while somatic variants were found in the diaphragm, none of the variants are likely to contribute to the etiology of CDH in these children.
One striking feature of our analysis was the extremely high number of somatic variants in the diaphragm sac and skin, but not blood, of proband 809. This high number of variants was not an artifact of technical issues, as the samples passed all quality control filters (see Material and methods). Furthermore, not only does this child harbor more somatic variants (Figure 3A), but the alternate allele frequency is significantly higher than that found in the somatic variants in the other probands (Figure S1). This child also harbors a missense germline de novo in the androgen receptor gene AR. AR has been well characterized as a tumor suppressor gene102 and shown to be critical in DNA repair through activation of transcriptional targets.103,104 However, arguing against a causative role of AR is that the particular AR missense variant in proband 809 is not predicted by PROVEAN to be damaging.
Our analysis of somatic variants also provides insights into how representative variants in the blood are of germline variants. Blood is the most commonly sampled human tissue, and variants in the blood are typically designated as “germline” variants. However, our analysis shows that blood harbors somatic variants that would typically be erroneously tallied as germline variants. In the four children analyzed, an average of 1.5% (range of 1%–4%) of variants in the blood is unique to blood. Thus, our analysis suggests that, in general, 1.5% of variants found in the blood are not germline variants but instead are somatic variants.
Inherited variants in genes regulating muscle structure and function potentially contribute to the etiology of CDH in one proband
Damaging variants inherited from parents may also be a genetic source of CDH. As the parents of the four probands investigated do not have CDH, it is unlikely that an inherited variant of one allele would directly cause CDH, while homozygous or biallelic variants, in which each parent contributes a gene damaging allele, are more likely to contribute to CDH. Given this, recessive homozygous and biallelic inherited variants were identified and prioritized using the GATK variant calling pipeline and the variant prioritizing tool VAAST364,105 (Table S4). VAAST identifies and ranks genes based on whether they are predicted to be damaging based on protein impact and rare compared against a mixed control population from 1000 Genomes phase 3.69 Pseudogenes,72 highly mutable genes,106,107 and genes with multiple alleles in the variant region reported in gnomAD62 were removed and genes expressed at higher than 10 TPM in E12.5 mouse PPF fibroblasts or diaphragm muscle progenitors prioritized (Figure 4A and 4B; Table S4).
Using these criteria, we identified 13 genes with biallelic or homozygous variants in the three CDH probands (Figure 4A and Table S4 with VAAST, CADD, PROVEAN impact, and ExAC Pli scores). None of these genes has been identified in previous CDH studies. Of these 13 genes, 3 genes (MPEG1, MYOF, and SACS) harbor 1 deleterious frameshift allele and 1 missense allele predicted by PROVEAN to be neutral, 5 genes (UPF3A, CCDC136, NOP9, ZNF646, and SH3PXD2A) harbor 1 missense allele predicted to be deleterious and 1 missense allele predicted to be neutral by PROVEAN, and 1 gene (ADAMTS2) is homozygous for an inherited in-frame insertion, predicted by PROVEAN to be neutral, although predicted by ExAC to be intolerant of loss-of-function (Pli score of 0.99). Because each of these genes has at least one predicted functional allele, these genes are unlikely to directly cause CDH but may act as sensitizing alleles that act in combination with other genetic variants produce CDH.
Proband 716, with a large right hernia, harbored four genes—ALG2, HRC, AHNAK, and MYO1H—in which both inherited maternal and paternal alleles were frameshift null or missense predicted damaging alleles and therefore more likely to contribute to CDH etiology. All four genes contain variants that are rare compared to the background population (1000 Genomes phase 3), based on the probabilistic framework underlying VAAST, and all variants are rare, allele frequency (AF) < 0.01, in gnomAD (Table S4). Interestingly, three of these genes are involved in skeletal muscle function. ALG2 encodes an α1,3-mannosyltransferase that catalyzes early steps of asparagine-linked glycosylation and is expressed at neuromuscular junctions.108 Human ALG2 variants have been found to affect the function of the neuromuscular junction and mitochondria organization in myofibers, leading to congenital myasthenic syndrome (fatigable muscle weakness) and mitochondrial myopathy.108, 109, 110 HRC encodes a histidine-rich calcium-binding protein that is expressed in the sarcoplasmic reticulum of skeletal, cardiac, and smooth muscle111 and in the heart has been shown to regulate calcium cycling.112,113 AHNAK encodes a large nucleoprotein that acts as a structural scaffold in multi-protein complexes.114 In particular, AHNAK interacts with dysferlin, which is a transmembrane protein critical for skeletal muscle membrane repair, and loss of dysferlin causes several types of muscular dystrophy.115,116 AHNAK is proposed to play a role in dysferlin-mediated membrane repair.115,116 The finding of deleterious biallelic variants in these three genes suggests that aberrations in the diaphragm muscle’s neuromuscular junctions, mitochondria, calcium handling, or membrane integrity contributes to the development of CDH in this child. In addition, proband 716 has one predicted null and one missense deleterious allele in MYO1H. MYO1H is a motor protein involved in intracellular transport and vesicle trafficking, expressed in retrotrapezoid neurons critical for sensing CO2 and regulating respiration, and variants in MYO1H cause a recessive form of central hypoventilation.117 While unlikely to directly contribute to CDH, the MYO1H variants’ potentially deleterious effect on neuronal regulation of respiration would have a detrimental impact on a CDH child.
An inherited deletion in intron 2 of Gata4 in two probands is a candidate common sensitizing allele for CDH
Another important potential source of genetic variants underlying CDH are SVs46 and include ≥50 bp insertions, deletions, inversions, and translocations. To identify SVs that could contribute to the etiology of the four CDH probands, we used the Lumpy smooth pipeline,73 which uses three copy number variant callers: cn.MOPS,76 CNVkit,74 and CNVnator.75 We then determined whether identified SVs were de novo or inherited in the CDH probands using the tool GQT78 and confirmed SVs visually using IGV.56 Though de novo SVs were discovered in multiple probands (Table S5), all were common (allele frequency > 0.1) in a large SV dataset of 14,623 ancestrally diverse individuals79 or located in an intergenic region. Thus, no discovered de novo SVs are likely to contribute to CDH etiology.
To discover inherited SVs potentially contributing to CDH, we analyzed variants within chromosomal regions highly associated with CDH.10 Multiple candidate inherited SVs were identified (with allele frequencies < 0.1 and not found in intergenic or repetitive regions; Table S5), but in no case were the probands homozygous for the SV or biallelic with any of the identified SNVs. However, one SV, a 343 bp deletion within the second intron of the highly ranked CDH-associated transcription factor, GATA4 (Table S1),16,17,21 was discovered in two probands (Figure 5) and subsequently confirmed by PCR. In proband 411 the deletion is paternally inherited (Figure 5A), while in proband 967 the deletion is maternally inherited (Figure 5D).
Comparison of the 343 bp deleted region in human with the orthologous region in mouse reveals that this region lies within an enhancer (element 2205) annotated in the VISTA enhancer database (Figure 5E).118 This enhancer drives reporter expression at E10.5 in the developing heart in a domain similar to the GATA4 expression domain119,120 and demonstrates that this region is a bona fide GATA4 enhancer. Although yet to be tested, this enhancer may also be important for GATA4 expression in the PPF cells that are critical for diaphragm development. As a parent of proband 411 and 967 possesses the deletion and does not have any reported CDH, this deletion alone is unlikely to cause CDH in the two probands. However, we hypothesize that this deletion reduces expression levels of GATA4 (a notably dosage-sensitive gene121) and thus sensitizes the two probands to develop CDH.
Given that two of the CDH individuals in this cohort harbor the 343 bp deletion, this deletion within the GATA4 intron 2 enhancer may be a sensitizing variant enriched in CDH individuals. To test this, we used PCR to identify the presence of the 343 bp deletion on a small cohort of 141 CDH individuals (including the 4 CDH individuals from this study) for which DNA was available. In addition, we compared the frequency of this deletion in the CDH cohort with that of a larger control population (which includes an ancestrally diverse population but also includes individuals with common cardiovascular, neuropsychiatric, and immune- related diseases) with SVs discovered using a similar, Lumpy software-based pipeline.79 We found that CDH individuals had an allele frequency of 0.98% (4 heterozygous individuals of 204 tested individuals, of which 141 had isolated CDH and 63 had complex CDH) and in the larger control population an allele frequency of 1.9%. These data suggest that this deletion has a roughly similar frequency of 1%–2% in CDH and control populations and is a common variant in the population. This relatively common 343 bp deletion may be a sensitizing variant that reduces GATA4 expression and, in conjunction with other variants, contributes to the genetic etiology of CDH.
Discussion
In this study, we comprehensively assessed the contribution of germline and somatic de novo and inherited SNVs, indels, and SVs to CDH etiology and reconstruct for the first time the genetic architecture of four individuals with isolated CDH. Our ability to perform such a comprehensive analysis of CDH, identifying genetic variants of different sizes (SNVs, indels, and SVs) in various genomic regions (exons, introns, UTRs, and intergenic) with different inheritance patterns (inherited and germline and somatic de novo) was enabled by three factors. First, the unique collection of diaphragm sac, skin, and blood samples from individuals with isolated CDH and blood from their parents allowed us to (1) confidently identify germline variants (present in all three proband samples and absent in parent samples); (2) discover de novo somatic variants (present in sac, but not in other samples) in the diaphragm’s connective tissue, which has previously been shown to be an important cellular source of CDH;9 and (3) determine which variants are inherited (present in all three proband samples and present in at least one parent). Second, whole-genome sequencing DNA samples to an average of >50× coverage was essential for identifying non-coding DNA regions that contribute to CDH etiology and positively calling somatic variants, which have relatively low numbers of reads. Finally, we employed a collection of computational tools that uses Illumina pair-end, whole genome sequences to discover de novo variants (RUFUS), identify and prioritize inherited variants (VAAST), and discover SVs (Lumpy pipeline). Altogether, the unique cohort of samples, high-depth whole genome sequences, and computational toolkit were essential to comprehensively interrogate the genetic architecture of the four CDH individuals. Our pipeline lays the groundwork for future, larger-scale studies investigating the genetic etiology of CDH. In addition, our methodology should be useful for investigating other birth defects with complex genetic etiologies, such as congenital heart defects.122
Our analysis of germline de novo variants revealed a potential pitfall of using blood samples to infer germline de novo variants and also identified a new candidate CDH-causative gene. Previous studies searching for de novo variants that cause CDH have used DNA from blood samples and then inferred that such variants are germline de novo variants.12, 13, 14, 15 However, while blood is the most readily available source of DNA, variants in its DNA may not have originally arisen in the germline but instead may have arisen later in somatic cells. Our cohort, with three proband-derived tissue samples, allows us to explicitly test this alternative hypothesis. We found that an average of 1.5% of the de novo variants in the probands’ blood were not germline in origin (not present in all three proband tissues) and is a similar rate as found in other recent papers.93,123 In fact, in proband 411, a blood-specific somatic variant in MIR4717 would have been classified as a germline variant. Thus, researchers should be cautious about inferring that all de novo variants in the blood are germline in origin. With DNA samples from three tissues from each CDH proband, we are able to confidently identify germline de novo variants because such variants will be present in all proband tissues but not in parental samples. We found an average of 68 germline de novo variants per proband, and this is similar to the 70 germline de novo variants found in the average population.93,124, 125, 126, 127 Of these, only a few are in gene-coding regions, and only one of these genes, THSD7A, harbors a deleterious variant and is predicted to be haploinsufficient and thus is a strong candidate CDH-causative allele.
The largely discordant appearance of CDH in monozygotic twins22,128 and our previous mouse genetic studies17 suggested that somatic variants in the diaphragm’s connective tissue may be a genetic feature of some CDH individuals. Previous studies of the role of somatic variants in other structural birth defects, such as congenital heart defects, have relied on blood, saliva, or skin samples and inferred that low frequency (<30%) alternate alleles represent somatic variants that may potentially contribute to birth defect etiology (e.g., Manheimer et al.129). In our study, we have DNA directly derived from proband tissue, the PPF-derived diaphragm connective sac, hypothesized to harbor somatic variants. Because we also have DNA derived from skin and blood proband samples, we were able to positively identify any alternate allele, regardless of its frequency, present in the sac, but not in skin or blood (or parental blood), as a somatic variant. Using similar logic, we were able to identify somatic variants in blood and skin. To confidently identify somatic variants, we conservatively included alleles present in at least 20% of the reads (and not present in the other tissues). Using this strategy, we identified in three of the probands 1–7 private somatic variants in the sac, skin, and blood. Proband 809 has an aberrantly high number of somatic variants, but we currently have no mechanistic explanation (e.g., variants in DNA repair genes) for this individual’s high somatic mutational load. In all probands, because no somatic variants were shared between two of the proband tissues, these somatic variants must have arisen after the developmental divergence of diaphragm connective tissue, skin, and blood. Importantly, no variants are shared between blood and diaphragm. Thus, blood samples are unlikely to be informative about somatic variants in the diaphragm. Furthermore, the presence of private somatic mutations in blood suggests that some de novo mutations in blood (which would be called as germline de novo mutations) identified in CDH individuals are unlikely to contribute to CDH etiology, as these mutations would not be found in the developing diaphragm. Our analysis of the diaphragm revealed multiple somatic variants in the diaphragm’s connective tissue, but none in coding or annotated enhancers, and so these somatic variants are unlikely to be deleterious. A previous study130 also found no evidence of damaging somatic variants, although this study examined tissue sampled around the periphery of the herniated region and so did not specifically sample the connective tissue that mouse genetic studies predict to be a cellular source of CDH.17,33,34 While our study did not find potentially damaging somatic variants in the diaphragm’s connective tissue, we have established an effective discovery strategy. A more definitive test of the role of somatic variants in CDH etiology awaits a future larger study.
The role of inherited variants in the etiology of CDH has received relatively little attention. Using VAAST, we identified multiple compound heterozygous or homozygous inherited, presumably recessive, SNVs and indel variants in all four probands. However, only a small number of these variants were rare, predicted damaging, and were in genes expressed in mouse PPFs fibroblasts or associated diaphragm muscle progenitors. Of particular note is proband 716, who inherited multiple damaging variants, including maternal and paternal damaging variants in ALG2, HRC, AHNAK, and MYO1H. ALG2, HRC, and AHNAK are all involved in skeletal muscle structure and function, and potentially variants in these genes may weaken muscle and lead to CDH. This is a surprising finding, as mouse genetic studies have found that while variants in muscle-specific genes lead to muscle-less diaphragms or diaphragms with aberrant muscle, none cause CDH (see Table S1). However, the inherited damaging variants in three muscle-related genes in proband 716, with an unusually large hernia, suggest that genetic alterations in muscle may lead to CDH. Another interesting aspect of proband 716 is the maternal and paternal damaging variants in MYO1H. MYO1H regulates the function of neurons critical for sensing CO2 and respiration,117 and so loss of MYO1H function may further compound the respiratory issues introduced by CDH.
In our search for de novo or inherited SVs that could contribute to CDH etiology, we discovered in two probands a 343 bp deletion in intron 2 of GATA4, a highly ranked CDH-associated gene, that disrupts an annotated enhancer regulating GATA4 expression.58 We hypothesize that disruption of this enhancer leads to lower levels of GATA4 expression. GATA4 has notably dosage-sensitive effects on heart development121 and likely also on diaphragm development. Given that this deletion is inherited from unaffected parents and has an allele frequency of 1%–2% in the general population, we hypothesize that this deletion is a relatively common SV that acts as a sensitizing allele for CDH. We hypothesize that decreased expression of GATA4 expression resulting from the 343 bp deletion confers CDH susceptibility and in the context of other genetic variants (or environmental factors) leads to CDH. To test this hypothesis, future experiments in our lab will test in mice whether this 343 bp region regulates GATA4 expression in the PPFs and whether a deletion in this region sensitizes mice to develop CDH.
Our comprehensive analysis of the genomes of four individuals with isolated CDH allows us to reconstruct the diverse genetic architectures underlying CDH (Figure 6). Proband 809 is the most enigmatic of the four cases. She harbors no obvious candidate genetic variants leading to CDH. Yet, her genome is unusual in that it contains an abnormally high somatic mutational load in her skin and diaphragm connective tissue, but the variants in the diaphragm do not affect coding or annotated enhancer regions. The source of large number of somatic mutations is unclear, as she harbors no mutations in DNA repair genes. Proband 411 harbors the inherited 343 bp intron 2 GATA4 deletion that we hypothesize acts as a sensitizing CDH allele, but collaborating variants that drive CDH are unclear. Proband 716 differs from the other probands in that she has inherited multiple rare and damaging variants in myogenic genes that likely lead to CDH. Notably, while three of the probands in our cohort have small left hernias, she is the only proband who has a large (where >50% of the chest wall is devoid of diaphragm tissue) right hernia. Potentially, the origin of her atypical large right hernia is linked to the variants in myogenic genes as opposed to genes expressed in PPF fibroblasts. Proband 967 is the individual for which we have the strongest hypothesis about the genetic origin of CDH. This individual harbors the inherited 343 bp intron 2 GATA4 deletion that we postulate acts as a sensitizing CDH allele and a rare germline de novo damaging missense variant in the haploinsufficient-intolerant gene THSD7A. These two variants suggest the hypothesis that during the early development of proband 967, the PPFs of his nascent diaphragm were prone to apoptosis, were unable to proliferate sufficiently (as GATA4 promotes proliferation and survival17), and had defects in migration (due to low expression levels of THSD7A) that ultimately led to defects in diaphragm morphogenesis and CDH. Such a hypothesis could be tested by generating mice that contain the intron 2 Gata4 deletion as well as one Thsd7a damaging or null allele.
In summary, our comprehensive analysis demonstrates that the genetic etiology of every CDH individual is heterogeneous and likely multifactorial. A challenge for future studies will be to determine whether, despite a diverse array of initiating genetic variants, a small set of molecular pathways are consistently impacted in CDH. Identification of a few key molecular pathways common to all CDH individuals will be critical for designing potential in utero therapies to rescue or minimize the severity of herniation.
Data and code availability
Sequence data for the four probands and parents are accessible through the Kids First Data Resource Portal and/or dbGaP, accession phs001110. RNA-seq data from E12.5 PPFs are deposited at GEO GSE155840.
Acknowledgments
We would like to thank the individuals and their families for their generous contribution. We are grateful for the technical assistance provided by Patricia Lanzano, Jiangyuan Hu, Jiancheng Guo, and Liyong Deng. We thank A. Quinlan and R.M. Layer for help with LUMPY, D. Neklason at Utah Genome Project for coordinating sequencing, and C.Y. Chow, L.B. Jorde, A. Quinlan, E.M. Sefton, and B. Collins for critical reading of the manuscript. The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. E.L.B. was supported by the University of Utah Genetics Training grant (NIH T32 GM007464). Research was supported by NIH R01HD087360 to G.K.; March of Dimes 6FY15203 to G.K.; Utah Genome Project to G.K.; Wheeler Foundation to G.K.; NIH R01GM120609 to Y.S.; NIH R03HL138352 to Y.S.; NIH R01HD057036 to W.K.C.; NIH UL1 RR024156 to W.K.C.; NIH P01HD068250 to W.K.C.; and Wheeler Foundation to W.K.C. Additional funding support was provided by grants to W.K.C. from CHERUBS, CDHUK, and the National Greek Orthodox Ladies Philoptochos Society, and generous donations from the Williams Family, Wheeler Foundation, Vanech Family Foundation, Larsen Family, Wilke Family, and many other families.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xhgg.2020.100008.
Web resources
Samplot, https://github.com/ryanlayer/samplot
dbGaP phs001110, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001110.v2.p1
GEO GSE155840, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155840
Kids First Data Resource Portal, https://kidsfirstdrc.org
OrthoRetriever, http://lighthouse.ucsf.edu/orthoretriever/
VISTA Enhancer Browser, https://enhancer.lbl.gov/
Supplemental Information
Genes were collated from multiple studies, including family and single individual case studies, large human patient sequencing studies and functional studies using mouse genetics. A gene from human studies is included if multiple individuals harbor variants in the gene, the gene is associated with other implicated genes and expressed in the developing diaphragm, and/or the gene is associated with other developmental disorders or structural birth defects that co-occur in individuals with complex CDH. A gene from mouse studies is included if a variant in the gene leads to a diaphragm defect. Genes were then ranked based on the amount and type of evidence implicating their contribution to CDH (see Material and Methods). Literature cited.11–17,21,23,27,29,33–45,47 Genes involved in GATA/TBX transcriptional network are highlighted in blue; in Hedgehog signaling in cyan; in Retinoic Acid signaling in dark green; in ROBO/SLIT signaling in purple; in MET signaling in orange; in WNT/βCATENIN signaling in yellow; in FGF signaling in pink; in muscle-related transcription factors or structural/functional genes in red and ECM proteins are in light green.
Sheet 1 reports results of Peddy analysis of heterozygosity and indicates that sequences were of high quality and ancestry congruent with self-reported ancestry. Sheet 2 reports results of Peddy sex analysis and shows that sex of DNA samples is congruent with known sex of sample origins. Sheet 3 reports results of Peddy analysis of relatedness and shows that relatedness is as expected. Highlighted rows show 3 different samples (blood, skin, and diaphragm) from the same proband, and these samples will be tagged by Peddy as “duplicated” samples.
True positive variants were identified by RUFUS and then filtered based on read depth (> 15 reads total in all samples compared in the RUFUS call), call quality (> 20 GQ score) and alternate allele frequency (sample of interest > 20% alternate read frequency and other samples in comparison < 20% alternate read frequency). Sheet 1: summary of all de novo variants identified. Sheet 2: de novo coding variants identified. Sheets 3-6: de novo gene-associated variants identified in all samples (Sheet 3) or diaphragm (Sheet 4), skin (Sheet 5), and blood (Sheet 6). Sheets 7-10: de novo intergenic variants identified in all samples (Sheet 7) diaphragm (Sheet 8), skin (Sheet 9), or blood (Sheet 10). In all sheets darker cells highlight variants called with > 20% alternate reads.
VAAST3 recessive model comparing CDH individual families to 1000 Genomes phase 3 individuals was used to call predicted damaging, rare recessive variants. Genes harboring variants with a VAAST p value < 0.05 were saved as candidate genes potentially contributing to CDH (Sheet 2 shows all genes). Genes were then further filtered based on expression in PPFs (genes needed to be expressed at ≥ 10 TPM in PPFs), and pseudogenes and highly mutable genes were removed. Variants that pass these filters are highlighted in blue (proband 411), pink (proband 716), orange (proband 809), and green (proband 967). All variants which pass the filters are displayed on Sheet 1. In addition, genes on Sheet 1 were ranked 1 if both proband alleles were predicted deleterious (or null) and had a gnomAD frequency < 0.001, 2 if both proband alleles were predicted deleterious (or null) and only one allele had a gnomAD frequency < 0.001, 3 if one proband allele was predicted null and the other allele had a neutral variant, and 4 if one proband allele was predicted deleterious and the other allele had a neutral variant of if both alleles were predicted neutral, but gene was predicted intolerant of haploinsufficiency.
Structural variants were identified using the Lumpy Smooth workflow. Germline de novo variants were identified by querying variants unique to the CDH proband compared to the parents, filtered based on reads that Lumpy scored as supporting the variant (> 8 reads either split or discordant) and finally visually inspected in IGV to determine whether parents had evidence of the same variant. Inherited structural variants were filtered based on chromosomal regions highly associated with CDH10. Structural variants were visually inspected and filtered out if published alternate allele frequency > 0.1 for inherited SVs or > 0.01 for de novo SVs,79 if SV was located in an intergenic region, or overlapped a repetitive element.80 Sheet 1 shows inherited SVs that have published alternate allele frequency < 0.1. Sheet 2 shows all inherited SVs, with SVs passing filtering criteria highlighted in blue (proband 411), pink (proband 716), in orange (proband 809), or in green (proband 967). Sheet 3 shows all de novo SVs, none of which passed filtering criteria.
References
- 1.Perry S.F., Similowski T., Klein W., Codd J.R. The evolutionary origin of the mammalian diaphragm. Respir. Physiol. Neurobiol. 2010;171:1–16. doi: 10.1016/j.resp.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Stege G., Fenton A., Jaffray B. Nihilism in the 1990s: the true mortality of congenital diaphragmatic hernia. Pediatrics. 2003;112:532–535. doi: 10.1542/peds.112.3.532. [DOI] [PubMed] [Google Scholar]
- 3.Torfs C.P., Curry C.J., Bateson T.F., Honoré L.H. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- 4.Parker S.E., Mai C.T., Canfield M.A., Rickard R., Wang Y., Meyer R.E., Anderson P., Mason C.A., Collins J.S., Kirby R.S., Correa A., National Birth Defects Prevention Network Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004-2006. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 5.Shanmugam H., Brunelli L., Botto L.D., Krikov S., Feldkamp M.L. Epidemiology and Prognosis of Congenital Diaphragmatic Hernia: A Population-Based Cohort Study in Utah. Birth Defects Res. 2017;109:1451–1459. doi: 10.1002/bdr2.1106. [DOI] [PubMed] [Google Scholar]
- 6.Lally K.P. Congenital diaphragmatic hernia - the past 25 (or so) years. J. Pediatr. Surg. 2016;51:695–698. doi: 10.1016/j.jpedsurg.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Pober B.R. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am. J. Med. Genet. C. Semin. Med. Genet. 2007;145C:158–171. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman K.G., Vargas S.O., Wilson J.A., Jennings R.W., Kozakewich H.P., Pober B.R. Congenital diaphragmatic defects: proposal for a new classification based on observations in 234 patients. Pediatr. Dev. Pathol. 2012;15:265–274. doi: 10.2350/11-05-1041-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kardon G., Ackerman K.G., McCulley D.J., Shen Y., Wynn J., Shang L., Bogenschutz E., Sun X., Chung W.K. Congenital diaphragmatic hernias: from genes to mechanisms to therapies. Dis. Model. Mech. 2017;10:955–970. doi: 10.1242/dmm.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holder A.M., Klaassens M., Tibboel D., de Klein A., Lee B., Scott D.A. Genetic factors in congenital diaphragmatic hernia. Am. J. Hum. Genet. 2007;80:825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longoni M., Lage K., Russell M.K., Loscertales M., Abdul-Rahman O.A., Baynam G., Bleyl S.B., Brady P.D., Breckpot J., Chen C.P., et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am. J. Med. Genet. A. 2012;158A:3148–3158. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longoni M., High F.A., Qi H., Joy M.P., Hila R., Coletti C.M., Wynn J., Loscertales M., Shan L., Bult C.J., et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum. Genet. 2017;136:679–691. doi: 10.1007/s00439-017-1774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longoni M., High F.A., Russell M.K., Kashani A., Tracy A.A., Coletti C.M., Hila R., Shamia A., Wells J., Ackerman K.G., et al. Molecular pathogenesis of congenital diaphragmatic hernia revealed by exome sequencing, developmental data, and bioinformatics. Proc. Natl. Acad. Sci. USA. 2014;111:12450–12455. doi: 10.1073/pnas.1412509111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi H., Yu L., Zhou X., Wynn J., Zhao H., Guo Y., Zhu N., Kitaygorodsky A., Hernan R., Aspelund G., et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L., Sawle A.D., Wynn J., Aspelund G., Stolar C.J., Arkovitz M.S., Potoka D., Azarow K.S., Mychaliska G.B., Shen Y., Chung W.K. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum. Mol. Genet. 2015;24:4764–4773. doi: 10.1093/hmg/ddv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jay P.Y., Bielinska M., Erlich J.M., Mannisto S., Pu W.T., Heikinheimo M., Wilson D.B. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev. Biol. 2007;301:602–614. doi: 10.1016/j.ydbio.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrell A.J., Ellis B.J., Fox Z.D., Lawson J.A., Weiss J.A., Kardon G. Muscle connective tissue controls development of the diaphragm and is a source of congenital diaphragmatic hernias. Nat. Genet. 2015;47:496–504. doi: 10.1038/ng.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pober B.R., Lin A., Russell M., Ackerman K.G., Chakravorty S., Strauss B., Westgate M.N., Wilson J., Donahoe P.K., Holmes L.B. Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am. J. Med. Genet. A. 2005;138A:81–88. doi: 10.1002/ajmg.a.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu L., Wynn J., Ma L., Guha S., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D., et al. De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet. 2012;49:650–659. doi: 10.1136/jmedgenet-2012-101135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L., Hernan R.R., Wynn J., Chung W.K. The influence of genetics in congenital diaphragmatic hernia. Semin. Perinatol. 2020;44:151169. doi: 10.1053/j.semperi.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L., Wynn J., Cheung Y.H., Shen Y., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D., et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum. Genet. 2013;132:285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veenma D., Brosens E., de Jong E., van de Ven C., Meeussen C., Cohen-Overbeek T., Boter M., Eussen H., Douben H., Tibboel D., de Klein A. Copy number detection in discordant monozygotic twins of Congenital Diaphragmatic Hernia (CDH) and Esophageal Atresia (EA) cohorts. Eur. J. Hum. Genet. 2012;20:298–304. doi: 10.1038/ejhg.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantarci S., Ackerman K.G., Russell M.K., Longoni M., Sougnez C., Noonan K.M., Hatchwell E., Zhang X., Pieretti Vanmarcke R., Anyane-Yeboa K., et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am. J. Med. Genet. A. 2010;152A:2493–2504. doi: 10.1002/ajmg.a.33618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veenma D., Beurskens N., Douben H., Eussen B., Noomen P., Govaerts L., Grijseels E., Lequin M., de Krijger R., Tibboel D., et al. Comparable low-level mosaicism in affected and non affected tissue of a complex CDH patient. PLoS ONE. 2010;5:e15348. doi: 10.1371/journal.pone.0015348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farag T.I., Bastaki L., Marafie M., al-Awadi S.A., Krsz J. Autosomal recessive congenital diaphragmatic defects in the Arabs. Am. J. Med. Genet. 1994;50:300–301. doi: 10.1002/ajmg.1320500316. [DOI] [PubMed] [Google Scholar]
- 26.Hitch D.C., Carson J.A., Smith E.I., Sarale D.C., Rennert O.M. Familial congenital diaphragmatic hernia is an autosomal recessive variant. J. Pediatr. Surg. 1989;24:860–864. doi: 10.1016/s0022-3468(89)80582-2. [DOI] [PubMed] [Google Scholar]
- 27.Kantarci S., Al-Gazali L., Hill R.S., Donnai D., Black G.C., Bieth E., Chassaing N., Lacombe D., Devriendt K., Teebi A., et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007;39:957–959. doi: 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell S.J., Cole T., Redford D.H. Congenital diaphragmatic hernia with probable autosomal recessive inheritance in an extended consanguineous Pakistani pedigree. J. Med. Genet. 1997;34:601–603. doi: 10.1136/jmg.34.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longoni M., Russell M.K., High F.A., Darvishi K., Maalouf F.I., Kashani A., Tracy A.A., Coletti C.M., Loscertales M., Lage K., et al. Prevalence and penetrance of ZFPM2 mutations and deletions causing congenital diaphragmatic hernia. Clin. Genet. 2015;87:362–367. doi: 10.1111/cge.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allan D.W., Greer J.J. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J. Comp. Neurol. 1997;382:459–468. doi: 10.1002/(sici)1096-9861(19970616)382:4<459::aid-cne3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Babiuk R.P., Zhang W., Clugston R., Allan D.W., Greer J.J. Embryological origins and development of the rat diaphragm. J. Comp. Neurol. 2003;455:477–487. doi: 10.1002/cne.10503. [DOI] [PubMed] [Google Scholar]
- 32.Sefton E.M., Gallardo M., Kardon G. Developmental origin and morphogenesis of the diaphragm, an essential mammalian muscle. Dev. Biol. 2018;440:64–73. doi: 10.1016/j.ydbio.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmona R., Cañete A., Cano E., Ariza L., Rojas A., Muñoz-Chápuli R. Conditional deletion of WT1 in the septum transversum mesenchyme causes congenital diaphragmatic hernia in mice. eLife. 2016;5:e16009. doi: 10.7554/eLife.16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paris N.D., Coles G.L., Ackerman K.G. Wt1 and β-catenin cooperatively regulate diaphragm development in the mouse. Dev. Biol. 2015;407:40–56. doi: 10.1016/j.ydbio.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P.X., Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 36.Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M.J., Hamard G., Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 37.Inanlou M.R., Dhillon G.S., Belliveau A.C., Reid G.A., Ying C., Rudnicki M.A., Kablar B. A significant reduction of the diaphragm in mdx:MyoD-/-(9th) embryos suggests a role for MyoD in the diaphragm development. Dev. Biol. 2003;261:324–336. doi: 10.1016/s0012-1606(03)00319-1. [DOI] [PubMed] [Google Scholar]
- 38.Ju Y., Li J., Xie C., Ritchlin C.T., Xing L., Hilton M.J., Schwarz E.M. Troponin T3 expression in skeletal and smooth muscle is required for growth and postnatal survival: characterization of Tnnt3(tm2a(KOMP)Wtsi) mice. Genesis. 2013;51:667–675. doi: 10.1002/dvg.22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laclef C., Hamard G., Demignon J., Souil E., Houbron C., Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- 40.Li J., Liu K.C., Jin F., Lu M.M., Epstein J.A. Transgenic rescue of congenital heart disease and spina bifida in Splotch mice. Development. 1999;126:2495–2503. doi: 10.1242/dev.126.11.2495. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Colucci-Guyon E., Pinçon-Raymond M., Mericskay M., Pournin S., Paulin D., Babinet C. Cardiovascular lesions and skeletal myopathy in mice lacking desmin. Dev. Biol. 1996;175:362–366. doi: 10.1006/dbio.1996.0122. [DOI] [PubMed] [Google Scholar]
- 42.Lu J.R., Bassel-Duby R., Hawkins A., Chang P., Valdez R., Wu H., Gan L., Shelton J.M., Richardson J.A., Olson E.N. Control of facial muscle development by MyoR and capsulin. Science. 2002;298:2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- 43.Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 44.Tseng B.S., Cavin S.T., Booth F.W., Olson E.N., Marin M.C., McDonnell T.J., Butler I.J. Pulmonary hypoplasia in the myogenin null mouse embryo. Am. J. Respir. Cell Mol. Biol. 2000;22:304–315. doi: 10.1165/ajrcmb.22.3.3708. [DOI] [PubMed] [Google Scholar]
- 45.Russell M.K., Longoni M., Wells J., Maalouf F.I., Tracy A.A., Loscertales M., Ackerman K.G., Pober B.R., Lage K., Bult C.J., Donahoe P.K. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc. Natl. Acad. Sci. USA. 2012;109:2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Q., High F.A., Zhang C., Cerveira E., Russell M.K., Longoni M., Joy M.P., Ryan M., Mil-Homens A., Bellfy L., et al. Systematic analysis of copy number variation associated with congenital diaphragmatic hernia. Proc. Natl. Acad. Sci. USA. 2018;115:5247–5252. doi: 10.1073/pnas.1714885115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan V.K., Beck T.F., Hernandez-Garcia A., Kundert P.N., Kim B.J., Jhangiani S.N., Gambin T., Starkovich M., Punetha J., Paine I.S., et al. The role of FREM2 and FRAS1 in the development of congenital diaphragmatic hernia. Hum. Mol. Genet. 2018;27:2064–2075. doi: 10.1093/hmg/ddy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham H., Sievert C. Springer International Publishing; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 49.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 https://arxiv.org/abs/1303.3997 1303.3997. [Google Scholar]
- 51.Faust G.G., Hall I.M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen B.S., Quinlan A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedersen B.S., Quinlan A.R. cyvcf2: fast, flexible variant analysis with Python. Bioinformatics. 2017;33:1867–1869. doi: 10.1093/bioinformatics/btx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 58.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210. 10.1101/531210. [Google Scholar]
- 63.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yandell M., Huff C., Hu H., Singleton M., Moore B., Xing J., Jorde L.B., Reese M.G. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu H., Huff C.D., Moore B., Flygare S., Reese M.G., Yandell M. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet. Epidemiol. 2013;37:622–634. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan A., Abecasis G.R., Kang H.M. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flygare S., Hernandez E.J., Phan L., Moore B., Li M., Fejes A., Hu H., Eilbeck K., Huff C., Jorde L., et al. The VAAST Variant Prioritizer (VVP): ultrafast, easy to use whole genome variant prioritization tool. BMC Bioinformatics. 2018;19:57. doi: 10.1186/s12859-018-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klambauer G., Schwarzbauer K., Mayr A., Clevert D.A., Mitterecker A., Bodenhofer U., Hochreiter S. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiang C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., Marth G.T., Quinlan A.R., Hall I.M. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Layer R.M., Kindlon N., Karczewski K.J., Quinlan A.R., Exome Aggregation Consortium Efficient genotype compression and analysis of large genetic-variation data sets. Nat. Methods. 2016;13:63–65. doi: 10.1038/nmeth.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abel H.J., Larson D.E., Chiang C., Das I., Kanchi K.L., Layer R.M., Neale B.M., Salerno W.J., Reeves C., Buyske S., et al. Mapping and characterization of structural variation in 17,795 deeply sequenced human genomes. bioRxiv. 2018:508515. doi: 10.1038/s41586-020-2371-0. 10.1101/508515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 81.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 83.Hahne F., Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol. Biol. 2016;1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- 84.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R., et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clugston R.D., Zhang W., Greer J.J. Early development of the primordial mammalian diaphragm and cellular mechanisms of nitrofen-induced congenital diaphragmatic hernia. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:15–24. doi: 10.1002/bdra.20613. [DOI] [PubMed] [Google Scholar]
- 88.Chlon T.M., Crispino J.D. Combinatorial regulation of tissue specification by GATA and FOG factors. Development. 2012;139:3905–3916. doi: 10.1242/dev.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huggins G.S., Bacani C.J., Boltax J., Aikawa R., Leiden J.M. Friend of GATA 2 physically interacts with chicken ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and represses COUP-TF2-dependent activation of the atrial natriuretic factor promoter. J. Biol. Chem. 2001;276:28029–28036. doi: 10.1074/jbc.M103577200. [DOI] [PubMed] [Google Scholar]
- 90.Ang Y.S., Rivas R.N., Ribeiro A.J.S., Srivas R., Rivera J., Stone N.R., Pratt K., Mohamed T.M.A., Fu J.D., Spencer C.I., et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell. 2016;167:1734–1749.e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luna-Zurita L., Stirnimann C.U., Glatt S., Kaynak B.L., Thomas S., Baudin F., Samee M.A., He D., Small E.M., Mileikovsky M., et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Besenbacher S., Liu S., Izarzugaza J.M., Grove J., Belling K., Bork-Jensen J., Huang S., Als T.D., Li S., Yadav R., et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat. Commun. 2015;6:5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sasani T.A., Pedersen B.S., Gao Z., Baird L., Przeworski M., Jorde L.B., Quinlan A.R. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. eLife. 2019;8:e46922. doi: 10.7554/eLife.46922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuo M.W., Wang C.H., Wu H.C., Chang S.J., Chuang Y.J. Soluble THSD7A is an N-glycoprotein that promotes endothelial cell migration and tube formation in angiogenesis. PLoS ONE. 2011;6:e29000. doi: 10.1371/journal.pone.0029000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang C.H., Chen I.H., Kuo M.W., Su P.T., Lai Z.Y., Wang C.H., Huang W.C., Hoffman J., Kuo C.J., You M.S., Chuang Y.J. Zebrafish Thsd7a is a neural protein required for angiogenic patterning during development. Dev. Dyn. 2011;240:1412–1421. doi: 10.1002/dvdy.22641. [DOI] [PubMed] [Google Scholar]
- 96.Wang C.H., Su P.T., Du X.Y., Kuo M.W., Lin C.Y., Yang C.C., Chan H.S., Chang S.J., Kuo C., Seo K., et al. Thrombospondin type I domain containing 7A (THSD7A) mediates endothelial cell migration and tube formation. J. Cell. Physiol. 2010;222:685–694. doi: 10.1002/jcp.21990. [DOI] [PubMed] [Google Scholar]
- 97.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 98.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 99.Dong X., Zhang L., Milholland B., Lee M., Maslov A.Y., Wang T., Vijg J. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Behjati S., Huch M., van Boxtel R., Karthaus W., Wedge D.C., Tamuri A.U., Martincorena I., Petljak M., Alexandrov L.B., Gundem G., et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vijg J., Dong X., Zhang L. A high-fidelity method for genomic sequencing of single somatic cells reveals a very high mutational burden. Exp. Biol. Med. (Maywood) 2017;242:1318–1324. doi: 10.1177/1535370217717696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Culig Z., Santer F.R. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014;33:413–427. doi: 10.1007/s10555-013-9474-0. [DOI] [PubMed] [Google Scholar]
- 103.Polkinghorn W.R., Parker J.S., Lee M.X., Kass E.M., Spratt D.E., Iaquinta P.J., Arora V.K., Yen W.F., Cai L., Zheng D., et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu H., Roach J.C., Coon H., Guthery S.L., Voelkerding K.V., Margraf R.L., Durtschi J.D., Tavtigian S.V., Shankaracharya, Wu W., et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat. Biotechnol. 2014;32:663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lenz T.L., Spirin V., Jordan D.M., Sunyaev S.R. Excess of Deleterious Mutations around HLA Genes Reveals Evolutionary Cost of Balancing Selection. Mol. Biol. Evol. 2016;33:2555–2564. doi: 10.1093/molbev/msw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shyr C., Tarailo-Graovac M., Gottlieb M., Lee J.J., van Karnebeek C., Wasserman W.W. FLAGS, frequently mutated genes in public exomes. BMC Med. Genomics. 2014;7:64. doi: 10.1186/s12920-014-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cossins J., Belaya K., Hicks D., Salih M.A., Finlayson S., Carboni N., Liu W.W., Maxwell S., Zoltowska K., Farsani G.T., et al. WGS500 Consortium Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain. 2013;136:944–956. doi: 10.1093/brain/awt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monies D.M., Al-Hindi H.N., Al-Muhaizea M.A., Jaroudi D.J., Al-Younes B., Naim E.A., Wakil S.M., Meyer B.F., Bohlega S. Clinical and pathological heterogeneity of a congenital disorder of glycosylation manifesting as a myasthenic/myopathic syndrome. Neuromuscul. Disord. 2014;24:353–359. doi: 10.1016/j.nmd.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 110.Thiel C., Schwarz M., Peng J., Grzmil M., Hasilik M., Braulke T., Kohlschütter A., von Figura K., Lehle L., Körner C. A new type of congenital disorders of glycosylation (CDG-Ii) provides new insights into the early steps of dolichol-linked oligosaccharide biosynthesis. J. Biol. Chem. 2003;278:22498–22505. doi: 10.1074/jbc.M302850200. [DOI] [PubMed] [Google Scholar]
- 111.Pathak R.K., Anderson R.G., Hofmann S.L. Histidine-rich calcium binding protein, a sarcoplasmic reticulum protein of striated muscle, is also abundant in arteriolar smooth muscle cells. J. Muscle Res. Cell Motil. 1992;13:366–376. doi: 10.1007/BF01766464. [DOI] [PubMed] [Google Scholar]
- 112.Arvanitis D.A., Vafiadaki E., Fan G.C., Mitton B.A., Gregory K.N., Del Monte F., Kontrogianni-Konstantopoulos A., Sanoudou D., Kranias E.G. Histidine-rich Ca-binding protein interacts with sarcoplasmic reticulum Ca-ATPase. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1581–H1589. doi: 10.1152/ajpheart.00278.2007. [DOI] [PubMed] [Google Scholar]
- 113.Tzimas C., Johnson D.M., Santiago D.J., Vafiadaki E., Arvanitis D.A., Davos C.H., Varela A., Athanasiadis N.C., Dimitriou C., Katsimpoulas M., et al. Impaired calcium homeostasis is associated with sudden cardiac death and arrhythmias in a genetic equivalent mouse model of the human HRC-Ser96Ala variant. Cardiovasc. Res. 2017;113:1403–1417. doi: 10.1093/cvr/cvx113. [DOI] [PubMed] [Google Scholar]
- 114.Davis T.A., Loos B., Engelbrecht A.M. AHNAK: the giant jack of all trades. Cell. Signal. 2014;26:2683–2693. doi: 10.1016/j.cellsig.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 115.Huang Y., Laval S.H., van Remoortere A., Baudier J., Benaud C., Anderson L.V., Straub V., Deelder A., Frants R.R., den Dunnen J.T., et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 2007;21:732–742. doi: 10.1096/fj.06-6628com. [DOI] [PubMed] [Google Scholar]
- 116.Zacharias U., Purfürst B., Schöwel V., Morano I., Spuler S., Haase H. Ahnak1 abnormally localizes in muscular dystrophies and contributes to muscle vesicle release. J. Muscle Res. Cell Motil. 2011;32:271–280. doi: 10.1007/s10974-011-9271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spielmann M., Hernandez-Miranda L.R., Ceccherini I., Weese-Mayer D.E., Kragesteen B.K., Harabula I., Krawitz P., Birchmeier C., Leonard N., Mundlos S. Mutations in MYO1H cause a recessive form of central hypoventilation with autonomic dysfunction. J. Med. Genet. 2017;54:754–761. doi: 10.1136/jmedgenet-2017-104765. [DOI] [PubMed] [Google Scholar]
- 118.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 119.Watt A.J., Battle M.A., Li J., Duncan S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuo C.T., Morrisey E.E., Anandappa R., Sigrist K., Lu M.M., Parmacek M.S., Soudais C., Leiden J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 121.Pu W.T., Ishiwata T., Juraszek A.L., Ma Q., Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 122.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J., et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsieh A., Morton S.U., Willcox J.A.L., Gorham J.M., Tai A.C., Qi H., DePalma S., McKean D., Griffin E., Manheimer K.B., et al. Early post-zygotic mutations contribute to congenital heart disease. bioRxiv. 2019 doi: 10.1101/733105. [DOI] [Google Scholar]
- 124.Besenbacher S., Sulem P., Helgason A., Helgason H., Kristjansson H., Jonasdottir A., Jonasdottir A., Magnusson O.T., Thorsteinsdottir U., Masson G., et al. Multi-nucleotide de novo Mutations in Humans. PLoS Genet. 2016;12:e1006315. doi: 10.1371/journal.pgen.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jónsson H., Sulem P., Arnadottir G.A., Pálsson G., Eggertsson H.P., Kristmundsdottir S., Zink F., Kehr B., Hjorleifsson K.E., Jensson B.O., et al. Multiple transmissions of de novo mutations in families. Nat. Genet. 2018;50:1674–1680. doi: 10.1038/s41588-018-0259-9. [DOI] [PubMed] [Google Scholar]
- 126.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A., et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rahbari R., Wuster A., Lindsay S.J., Hardwick R.J., Alexandrov L.B., Turki S.A., Dominiczak A., Morris A., Porteous D., Smith B., et al. UK10K Consortium Timing, rates and spectra of human germline mutation. Nat. Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wat M.J., Shchelochkov O.A., Holder A.M., Breman A.M., Dagli A., Bacino C., Scaglia F., Zori R.T., Cheung S.W., Scott D.A., Kang S.H. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am. J. Med. Genet. A. 2009;149A:1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Manheimer K.B., Richter F., Edelmann L.J., D’Souza S.L., Shi L., Shen Y., Homsy J., Boskovski M.T., Tai A.C., Gorham J., et al. Robust identification of mosaic variants in congenital heart disease. Hum. Genet. 2018;137:183–193. doi: 10.1007/s00439-018-1871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsunami N., Shanmugam H., Baird L., Stevens J., Byrne J.L., Barnhart D.C., Rau C., Feldkamp M.L., Yoder B.A., Leppert M.F., et al. Germline but not somatic de novo mutations are common in human congenital diaphragmatic hernia. Birth Defects Res. 2018;110:610–617. doi: 10.1002/bdr2.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes were collated from multiple studies, including family and single individual case studies, large human patient sequencing studies and functional studies using mouse genetics. A gene from human studies is included if multiple individuals harbor variants in the gene, the gene is associated with other implicated genes and expressed in the developing diaphragm, and/or the gene is associated with other developmental disorders or structural birth defects that co-occur in individuals with complex CDH. A gene from mouse studies is included if a variant in the gene leads to a diaphragm defect. Genes were then ranked based on the amount and type of evidence implicating their contribution to CDH (see Material and Methods). Literature cited.11–17,21,23,27,29,33–45,47 Genes involved in GATA/TBX transcriptional network are highlighted in blue; in Hedgehog signaling in cyan; in Retinoic Acid signaling in dark green; in ROBO/SLIT signaling in purple; in MET signaling in orange; in WNT/βCATENIN signaling in yellow; in FGF signaling in pink; in muscle-related transcription factors or structural/functional genes in red and ECM proteins are in light green.
Sheet 1 reports results of Peddy analysis of heterozygosity and indicates that sequences were of high quality and ancestry congruent with self-reported ancestry. Sheet 2 reports results of Peddy sex analysis and shows that sex of DNA samples is congruent with known sex of sample origins. Sheet 3 reports results of Peddy analysis of relatedness and shows that relatedness is as expected. Highlighted rows show 3 different samples (blood, skin, and diaphragm) from the same proband, and these samples will be tagged by Peddy as “duplicated” samples.
True positive variants were identified by RUFUS and then filtered based on read depth (> 15 reads total in all samples compared in the RUFUS call), call quality (> 20 GQ score) and alternate allele frequency (sample of interest > 20% alternate read frequency and other samples in comparison < 20% alternate read frequency). Sheet 1: summary of all de novo variants identified. Sheet 2: de novo coding variants identified. Sheets 3-6: de novo gene-associated variants identified in all samples (Sheet 3) or diaphragm (Sheet 4), skin (Sheet 5), and blood (Sheet 6). Sheets 7-10: de novo intergenic variants identified in all samples (Sheet 7) diaphragm (Sheet 8), skin (Sheet 9), or blood (Sheet 10). In all sheets darker cells highlight variants called with > 20% alternate reads.
VAAST3 recessive model comparing CDH individual families to 1000 Genomes phase 3 individuals was used to call predicted damaging, rare recessive variants. Genes harboring variants with a VAAST p value < 0.05 were saved as candidate genes potentially contributing to CDH (Sheet 2 shows all genes). Genes were then further filtered based on expression in PPFs (genes needed to be expressed at ≥ 10 TPM in PPFs), and pseudogenes and highly mutable genes were removed. Variants that pass these filters are highlighted in blue (proband 411), pink (proband 716), orange (proband 809), and green (proband 967). All variants which pass the filters are displayed on Sheet 1. In addition, genes on Sheet 1 were ranked 1 if both proband alleles were predicted deleterious (or null) and had a gnomAD frequency < 0.001, 2 if both proband alleles were predicted deleterious (or null) and only one allele had a gnomAD frequency < 0.001, 3 if one proband allele was predicted null and the other allele had a neutral variant, and 4 if one proband allele was predicted deleterious and the other allele had a neutral variant of if both alleles were predicted neutral, but gene was predicted intolerant of haploinsufficiency.
Structural variants were identified using the Lumpy Smooth workflow. Germline de novo variants were identified by querying variants unique to the CDH proband compared to the parents, filtered based on reads that Lumpy scored as supporting the variant (> 8 reads either split or discordant) and finally visually inspected in IGV to determine whether parents had evidence of the same variant. Inherited structural variants were filtered based on chromosomal regions highly associated with CDH10. Structural variants were visually inspected and filtered out if published alternate allele frequency > 0.1 for inherited SVs or > 0.01 for de novo SVs,79 if SV was located in an intergenic region, or overlapped a repetitive element.80 Sheet 1 shows inherited SVs that have published alternate allele frequency < 0.1. Sheet 2 shows all inherited SVs, with SVs passing filtering criteria highlighted in blue (proband 411), pink (proband 716), in orange (proband 809), or in green (proband 967). Sheet 3 shows all de novo SVs, none of which passed filtering criteria.
Data Availability Statement
Sequence data for the four probands and parents are accessible through the Kids First Data Resource Portal and/or dbGaP, accession phs001110. RNA-seq data from E12.5 PPFs are deposited at GEO GSE155840.