Abstract
Summary
Retroviruses and their vector derivatives integrate semi-randomly in the genome of host cells and are inherited by their progeny as stable genetic marks. The retrieval and mapping of the sequences flanking the virus-host DNA junctions allows the identification of insertion sites in gene therapy or virally infected patients, essential for monitoring the evolution of genetically modified cells in vivo. However, since ∼30% of insertions land in low complexity or repetitive regions of the host cell genome, they cannot be correctly assigned and are currently discarded, limiting the accuracy and predictive power of clonal tracking studies. Here, we present γ-TRIS, a new graph-based genome-free alignment tool for identifying insertion sites even if embedded in low complexity regions. By using γ-TRIS to reanalyze clinical studies, we observed improvements in clonal quantification and tracking.
Availability and implementation
Source code at https://bitbucket.org/bereste/g-tris.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Viruses and retroviral vectors (RV) stably insert DNA sequences in the genome of the host cells in integration sites (IS) and provide a permanent and unique genetic mark for each independently transduced cell clone and its progeny. Several Gene Therapy (GT) applications exploit RV to add the therapeutic transgene in diseased target cells. Molecular studies using IS assessed the safety and long-term efficacy of hematopoietic stem and progenitor cell based GT applications using gamma-retroviral and lentiviral vectors (LV) by identifying and tracking thousands of IS across different hematopoietic lineages over time. Results from IS studies allowed to monitor clonal evolution in vivo (Naldini, 2015), including clonal expansions caused by insertional mutagenesis (Afzal et al., 2017; Berry et al., 2017; Sherman et al., 2017; Spinozzi et al., 2017). PCR-based or targeted enrichment techniques, combined with Next-Generation Sequencing and bioinformatics analyses, allow the retrieval and mapping of the cellular genomic sequences flanking the vector ISs present in the DNA extracted from vector marked cells. Most of the existing bioinformatics tools for IS identification return IS from reads aligned to unique genomic positions while reads with multiple hits are discarded or handled separately to generate basic information of ISs landing within repetitive elements (Afzal et al., 2017; Berry et al., 2017; Sherman et al., 2017; Spinozzi et al., 2017). The amount of IS that cannot be univocally mapped averages 30% of the entire dataset (Calabria et al., 2014), therefore corresponds to a significant portion of patients’ IS. This could lead to miss potentially malignant clones caused by IS landing in repeats within or near oncogenes or to mis-quantify the abundance of each clone accounted as relative percentage of the IS on the overall set of IS. To identify the whole repertoire of IS in from sequence datasets of PCR products containing vector genome junctions, we developed γ-TRIS (Graph-based algorithm for Tracking Retroviral Insertion Sites).
2 Design and implementation
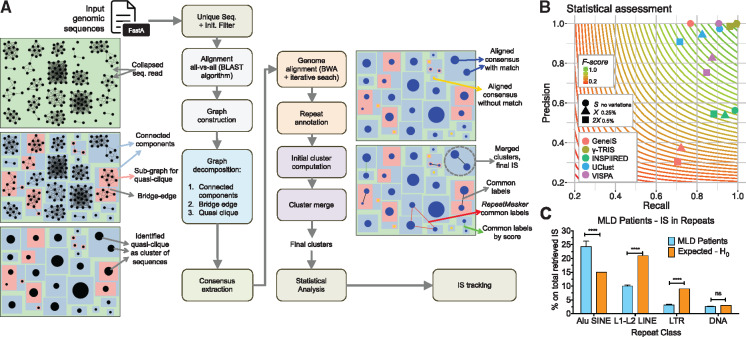
The basic idea of γ-TRIS is to identify IS from clusters of highly similar sequences as result of all-versus-all reads alignment, rather than a direct alignment against an indexed genome and then using a consensus sequence from each cluster as IS sequence to be mapped to the reference genome. γ-TRIS starts by aligning each unique sequence of the dataset to each other, identifying clusters of sequences containing vector-host genome junctions originating from the same IS represented by a graph structure (Fig. 1A). To reduce the number of false IS, the graph is iteratively decomposed in sub-graphs of highly connected sequences and from each atomic (highly-connected) cluster a consensus sequence is computed and mapped against the reference genome. These consensus sequences are also searched for known genomic repetitive elements to annotate each cluster with all the available genomic information. All the alignment results and repeat annotations of the consensus sequences are used as labels of the corresponding predicted clusters. Then, γ-TRIS compares the annotation labels by rolling-back the decomposition graph structure and tries to merge clusters that likely represent the same IS. The reliability of each putative IS of being an independent cluster is statistically assessed through a heuristic permutation test, therefore allowing to further merge similar clusters and to associate an upper-bound P-value to each cluster (P < 0.05). See Supplementary Material, Section S1 for development details.
Fig. 1.
γ-TRIS flowchart and comparative performances. (A) Schematic representation of the γ-TRIS pipeline and its graphical representation. (B) Scatter plots of average precision and recall for three tools on the simulated datasets. Background gradient color scale and contour lines are based on the F-score as indicated in the legend box. All tools have been tested with default parameters without any adjustments nor optimizations of the configurations for the specific simulated datasets, that could impact in the assessment of precision and recall. (C) MLD IS landing in repetitive elements within the main repeat classes SINE, LINE, LTR and DNA versus expected percentage under null hypothesis reflecting the proportion of repeats in the human genome
3 Results
We tested the reliability of γ-TRIS on simulated IS sequence datasets composed of 98 596 random coordinates as IS from the human genome (see Supplementary Material, Section S2). We compared the performances of γ-TRIS to those of the most recent IS mapping pipelines GENE-IS (Afzal et al., 2017), INSPIIRED (Sherman et al., 2017), VISPA (Spinozzi et al., 2017) and UClust (Edgar, 2010), a heuristic clustering method recently exploited for IS analysis in HIV-1 patients (Maldarelli et al., 2014; Wagner et al., 2014) (Supplementary Material, Sections S3 and S4). To avoid introducing any biases in final IS results, we did not change the default parameters in all tested pipelines (see Supplementary Material, Section S3 for details), whose optimal configuration has been developed and released by Authors. However, further parameter optimization and tuning on the simulated datasets might produce more favorable results for the other methods. Statistical assessment exploited positive predictive values (or precision) and sensitivity (or recall) by considering an IS as true positive (TP) if correctly assigned to the source locus, false positive (FP) if misplaced in any other genomic loci and false negative (FN) if missing or not returned in the result list. For the dataset without mutations, γ-TRIS returned 98 152 IS of the total 98 596 IS, 98 147 of which (>99.5%) were TPs, 5 FPs and 449 FNs, thus giving rise to an average recall of 0.994 and precision of 0.999, corresponding to the best F-score (>0.997) (Fig. 1B, Supplementary Table S1). In the artificially mutagenized datasets, precision and recall decreased at different rates depending on the tool used, but γ-TRIS maintained the best F-score (0.99 with 0.25% of errors, dataset X and 0.973 with 0.5% of errors, dataset 2X), without major unbalancing of precision and recall when compared to other tools, even when high levels of sequence variations were introduced. To evaluate γ-TRIS results, we also generated in vitro experiments from a purified human cell line with six known IS (see Supplementary Material, Section S5). UClust identified only five out of six IS while γ-TRIS and the other IS tools identified all six known IS (mapping >98% of the produced reads). We then re-analyzed the raw sequencing data from the published IS datasets of three GT patients of the LV-based clinical trial of Metachromatic Leukodystrophy (MLD), previously reported with VISPA to assess safety and efficacy of the treatment (Biffi et al., 2013). MLD results with γ-TRIS, compared to those published (Supplementary Fig. S1), showed improvements in the overall number of IS (average fold increase of 1.29 among the three patients), in the amount of sequencing reads (average fold increase of 1.59) and in the number of recaptured IS over time or lineages (average fold increase of 1.38) important readouts to evaluate the HSC marking and tracking of cell clones across lineages and time. Interestingly, the diversity index of the clonal population did not change using γ-TRIS. The number of additional IS found by γ-TRIS corresponded to the previous amount of IS discarded by VISPA (∼30% of the total sequencing reads). Although the vast majority of the new IS identified by γ-TRIS could not be precisely mapped on the host genome, about 5% of those lost by VISPA could be univocally mapped thanks to the improved quality of the consensus sequences. Moreover, MLD IS in repeated regions showed a marked preference to target SINE-Alu repetitive elements (Fig. 1C) as observed in a recent study of HIV-1 integration (Cohn et al., 2015).
Supplementary Material
Acknowledgements
The authors dedicate this work to the memory of all patients in San Raffaele Telethon Institute for Gene Therapy and their families, their hope is our strength. They are grateful to all the lab member of the Montini’s lab especially to Daniela Cesana. They finally thank CINECA for the ICT support.
Funding
This work was supported by Telethon Foundation TGT11D1, TGT16B01 and TGT16B03 to EM; ISCRA Grant HP10CEUWXF 2015 and Giovani Ricercatori GR-2016-02363681 by the Ministry of Health to AC; University and Research flagship initiative Interomics (PB05) to LM.
Conflict of Interest: none declared.
References
- Afzal S. et al. (2017) GENE-IS: time-efficient and accurate analysis of viral integration events in large-scale gene therapy data. Mol. Ther. Nucleic Acids, 6, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C.C. et al. (2017) INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol. Ther. Methods Clin. Dev., 4, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A. et al. (2013) Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science, 341, 1233158. [DOI] [PubMed] [Google Scholar]
- Calabria A. et al. (2014) VISPA: a computational pipeline for the identification and analysis of genomic vector integration sites. Genome Med., 6, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L.B. et al. (2015) HIV-1 integration landscape during latent and active infection. Cell, 160, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Maldarelli F. et al. (2014) HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science, 345, 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L. (2015) Gene therapy returns to centre stage. Nature, 526, 351–360. [DOI] [PubMed] [Google Scholar]
- Sherman E. et al. (2017) INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev., 4, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinozzi G. et al. (2017) VISPA2: a scalable pipeline for high-throughput identification and annotation of vector integration sites. BMC Bioinformatics, 18, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.A. et al. (2014) HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science, 345, 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.