Abstract
Motivation
Comparative plasmid genome analyses require complex tools, the manipulation of large numbers of sequences and constitute a daunting task for the wet bench experimentalist. Dedicated plasmid databases are sparse, only comprise bacterial plasmids and provide exclusively access to sequence similarity searches.
Results
We have developed Web-Assisted Symbolic Plasmid Synteny (WASPS), a web service granting protein and DNA sequence similarity searches against a database comprising all completely sequenced natural plasmids from bacterial, archaeal and eukaryal origin. This database pre-calculates orthologous protein clustering and enables WASPS to generate fully resolved plasmid synteny maps in real time using internal and user-provided DNA sequences.
Availability and implementation
WASPS queries befit all current browsers such as Firefox, Edge or Safari while the best functionality is achieved with Chrome. Internet Explorer is not supported. WASPS is freely accessible at https://archaea.i2bc.paris-saclay.fr/wasps/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The Darwinian evolution of genomes from the three domains of life is fast-tracked by the acquisition of traits through horizontal gene transfer (Cordaux and Batzer, 2009; Cossu et al., 2017; Seth-Smith et al., 2012). This process is mediated by plasmids and viruses defined as mobile genetic elements. In contact with their hosts, replicating plasmids in particular have been shown as remarkably plastic (Cury et al., 2018). The comparative analysis of plasmid genomes constitutes a daunting task due to the lack of dedicated, plasmid-centric resources. Similarity searches in public databases of the National Centre for Biotechnology Information (NCBI) cannot be restricted exclusively to plasmid DNA or protein. Recently, plasmid assets have been developed proposing either a comprehensive manually curated bacterial plasmid list (Brooks et al., 2019) or a bacterial plasmid database which can be interrogated using sequence similarity programs (Galata et al., 2019). The Easyfig synteny tool allows the efficient comparison of genomes and plasmids but requires user-installation (Sullivan et al., 2011). Bioinformatics web tools providing access to a database of bacterial, archaeal and eukaryal plasmids and their synteny are still missing at this time. In this work, we present the database-backed WASPS web service which in addition to similarity searches allows the real-time generation of fully resolved plasmid synteny maps. The evolutionary relationships between user-provided sequences and databases sequences can be inferred using these synteny maps. The WASPS database is updated at regular intervals, comprises all completely sequenced natural plasmids from the three domains of life and provides full pre-calculated orthologous gene clustering.
2 Materials and methods
WASPS consists of three modules.
The WASPS database is a relational database containing all entries from the NCBI RefSeq plasmid repository encompassing all completely sequenced bacterial, archaeal and eukaryal plasmids of natural origin. At database creation, each protein-encoding gene is hierarchically linked to its plasmid of origin using GenBank keys. All corresponding protein sequences undergo orthologous clustering using UCLUST (Edgar, 2010) and obtain a centroid identifier. At this date, the WASPS database contains 18 915 bacterial, 240 archaeal and 36 eukaryal plasmids.
The WASPS Webtool provides a user interface for the remote interrogation of the database and proposes 5 distinct features:
Text-based search in plasmid and proteins definitions and accession numbers.
Protein and DNA-based similarity searches with user-provided sequences using BLAST (Altschul et al., 1990) and DIAMOND (Buchfink et al., 2015) algorithms.
The drawing of synteny maps using a user-provided annotated DNA file in GenBank format. It proceeds by the extraction and matching of the corresponding protein sequences against WASPS centroids using BLAST or DIAMOND. The query sequence and related plasmids from the database are drawn using a consistent symbolic coloring.
The drawing of synteny maps using an unannotated raw or Fasta DNA file. It requires the initial six-frame translation of the query sequence using ATG, GTG or TTG as start codons and TAG, TAA or TGA as stop codons. All frames and related plasmids are drawn as above.
The similar drawing of plasmid synteny maps from the database according to orthologous clustering using WASPS plasmid and protein accession numbers.
Dedicated hyperlinks connect text and similarity searches with synteny queries. Wide genomic areas can be explored by panning and zooming directly within the browser. All synteny map genes grant access to relevant additional information, to their protein sequences and to all their orthologs from the WASPS database using specific gestures in the web interface (Supplementary Material). Maps can be exported as PNG or SVG.
The WASPS updater is fully automated and operates a pipeline in the background to refresh the database at fixed intervals (Supplementary Material).
3 Results
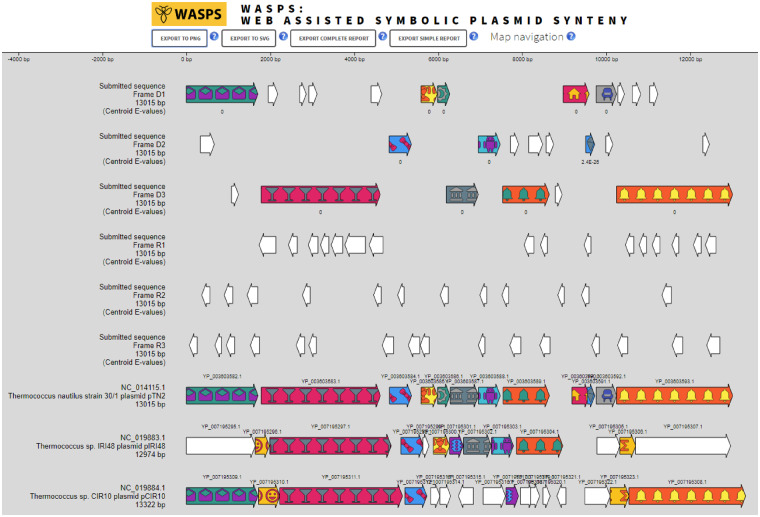
To illustrate WASPS capabilities, we submitted for synteny analysis the unannotated Fasta sequence of the 13 015 bp archaeal plasmid pTN2 (NC_014115.1) from Thermococcus nautili (Soler et al., 2010). Using the option ‘primary hits only’ and DIAMOND searching algorithm, three hits were detected: plasmid pTN2, already existing in the database, pIRI48 from Thermococcus sp. IRI18 and pCIR10 from Thermococcus sp. CIR10. All three plasmids were selected to generate a plasmid synteny map (Fig. 1; Supplementary Material). Interestingly, WASPS synteny results for pTN2 resembled a prior analysis where the same Thermococcus plasmids were aligned and drawn manually (Krupovic et al., 2013). In this representation, all plasmids shared orthologous uvrD genes whereas for the WASPS synteny pIRI48 gene YP_007195295.1 is not orthologous. To ascertain WASPS clustering quality, we assessed the orthology of the various uvrD genes using an alternative method. Protein sequence similarity among WASPS UvrD cluster members exceeded 26%, compatible with the proposed 30% orthology threshold (Lerat et al., 2003). Strikingly, similarity between each member of the UvrD cluster and the translated YP_007195295.1 gene never exceeded 15.2% (Supplementary Material). These results demonstrated the robustness of the WASPS synteny mapping.
Fig. 1.
Synteny map of plasmid pTN2 from T. nautili. The first six tracks correspond to genes predicted in the Fasta query sequence in the six reading frames with corresponding E-values. The following tracks refer to plasmids from the WASPS database sharing synteny with the submitted sequence. Gene orthology is indicated by consistent graphic symbolism throughout the map. Genes in white color are singletons, devoid of ortholog in the database. As indicated, plasmids maps are accurately drawn to scale
In a second experiment, the predictive capabilities of WASPS were tested on NCBI sequence contigs. We discovered that QMOB01000129.1, a 10 757 bp contig assigned to a Chloroflexi bacterium metagenome (Dombrowski et al., 2018) shared an excessive number of genes with archaeal plasmids (Supplementary Material). We surmised that this particular contig corresponded to a low level of archaeal DNA contamination in the metagenomic sample. WASPS was therefore able to provide an effortless characterization of metagenomes.
Funding
This work was funded by CNRS and the European Research Council under the European Union's Seventh Framework Program (FP/2007-2013)/Project EVOMOBIL - ERC Grant Agreement no. 340440 (P.F.). C.B. is supported by ‘Ecole Normale Supérieure de Lyon’.
Conflict of Interest: none declared.
Supplementary Material
References
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Brooks L. et al. (2019) A curated, comprehensive database of plasmid sequences. Microbiol. Resour. Announc., 8, e01325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B. et al. (2015) Fast and sensitive protein alignment using DIAMOND. Nat. Methods, 12, 59–60. [DOI] [PubMed] [Google Scholar]
- Cordaux R., Batzer M.A. (2009) The impact of retrotransposons on human genome evolution. Nat. Rev. Genet., 10, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu M. et al. (2017) Flipping chromosomes in deep-sea archaea. PLoS Genet., 13, e1006847.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cury J. et al. (2018) Host range and genetic plasticity explain the co-existence of integrative and extrachromosomal mobile genetic elements. Mol. Biol. Evol., 35, 2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N. et al. (2018) Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun., 9, 4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Galata V. et al. (2019) PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res., 47, D195–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M. et al. (2013) Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS One, 8, e49044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E. et al. (2003) From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol., 1, E19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H.M. et al. (2012) Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J. Bacteriol., 194, 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N. et al. (2010) Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res., 38, 5088–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.J. et al. (2011) Easyfig: a genome comparison visualizer. Bioinformatics, 27, 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.