Abstract
Motivation
During development, progenitor cells undergo multiple rounds of cellular divisions during which transcriptional programs must be faithfully propagated. Investigating the timing of transcriptional activation, which is a highly stochastic phenomenon, requires the analysis of large amounts of data. In order to perform automatic image analysis of transcriptional activation, we developed a software that segments and tracks both small and large objects, leading the user from raw data up to the results in their final form.
Results
MitoTrack is a user-friendly open-access integrated software that performs the specific dual task of reporting the precise timing of transcriptional activation while keeping lineage tree history for each nucleus of a living developing embryo. The software works automatically but provides the possibility to easily supervise, correct and validate each step.
Availability and implementation
MitoTrack is an open source Python software, embedded within a graphical user interface (download here).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Live imaging of developing organisms provides the unprecedented opportunity to access to nuclei/cellular genealogy. The combination of lineage information and transcriptional history permits to decipher the extent of transcriptional memory, i.e. the influence of the transcriptional status of mother nuclei on that of their daughters (Ferraro et al., 2016). Here we present MitoTrack, a user-friendly and open source software, able to automatically track nuclei and their transcriptional activity across multiple cell divisions. The tool has been primarily developed for images of living Drosophila embryos, in which labeling nascent transcription has become popular (Garcia et al., 2013; Lucas et al., 2013; Pichon et al., 2018). However, MitoTrack could also be used for genealogy and memory tracking in other organisms, such as zebrafish embryos, mouse early embryos or even in organoids.
2 Software description
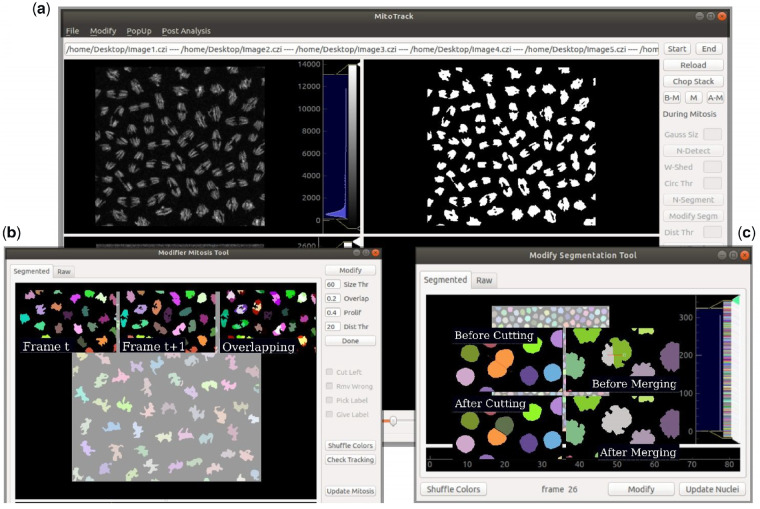
The typical data processed with MitoTrack are images from early Drosophila embryos where nuclei are labeled with a histone-RFP transgene and reporter mRNA detected upon binding of the MCP-GFP to MS2 containing nascent mRNAs (sample data can be downloaded following this link) (Fernandez and Lagha, 2019). Most of MitoTrack algorithms are not novel and several more sophisticated programs are similarly able to perform lineage tracking in living organisms (Amat et al., 2014; Tinevez et al., 2017; Tran et al., 2018; Wolff et al., 2018). However, MitoTrack was developed to fulfill the specific task of extracting lineage information and the timing of transcriptional activation directly from the raw data. All the developed algorithms work in a single package, embedded into a user-friendly graphical user interface (GUI) (Fig. 1a), released as open source (a tutorial with walk-through examples is provided as a Supplementary Material). The main objective of MitoTrack is to analyze transcriptional activation in a nucleus-by-nucleus basis, across multiple nuclear divisions to quantify the extent of transcriptional memory. The general tasks are (i) to segment and track nuclei across mitosis (keeping lineage information), (ii) to segment spots (transcription sites) and (iii) to merge these pieces of information and obtain the activation timing for each mother nucleus and its daughters. The analysis is organized into four steps: ‘before mitosis’, ‘during mitosis’, ‘after mitosis’ and ‘merging of partial analysis’.
Fig. 1.
MitoTrack GUI to track transcriptional activation and nuclei lineage. (a) Main window composed of four distinct panels, among which, two are shown here, with raw nuclei channel (upper left) and detected nuclei (upper right). MitoTrack typical tools and edit boxes are shown on the left. (b) ‘Modifier Mitosis Tool’ showing all its commands and parameters. Three typical images of nuclei at a time t, t + 1 and their overlap are shown to illustrate the basis of the algorithm for automatic nuclei tracking during mitosis. (c) Interface of the ‘Modify Segmentation Tool’ with its main commands visible. Exemplary results of manual corrections of nuclei segmentation
3 Features
Input data are 4D stack (.czi or .lsm) with two channels, one for nuclei and the other for transcription sites (spots). When loaded, data are automatically maximum intensity projected.
The ‘before mitosis’ and ‘after mitosis’ steps of the analysis work similarly: nuclei are segmented and tracked while transcriptional sites are detected and each of them is associated to the closest -if not overlapping- nucleus. Nuclei are pre-smoothed and then thresholded with the classical Otsu thresholding algorithm (Otsu, 1979); the black and white resulting images are labeled and segmented using a watershed segmentation algorithm (van der Walt et al., 2014) and finally tracked in consecutive frames following a minimum distance criterion of their centroids positions. Spots are segmented with a blob detection algorithm, namely a Laplacian of Gaussian. The resulting images are filtered and each spot is then associated to the closest nucleus. Potential segmentation errors can be corrected with a pop-up tool that allows the user to easily modify nuclei segmentation by merging or splitting nuclei (Fig. 1c). The ‘during mitosis’ step consists in a similar nuclei segmentation but differs in the tracking procedure, where mitotic duplication of chromosomes must be accounted for. MitoTrack follows nuclei across mitosis under an overlapping criterion, by projecting the mask of each nucleus at frame t onto the following frame. In this way, two nuclei (daughters) can be associated to a single nucleus (mother), thereby creating a lineage genealogy. A pop-up tool (Fig. 1b) allows the user to easily correct both segmentation and tracking during this particular step.
The last part of MitoTrack consists in merging the three aforementioned sections in order to update nuclei labels for the entirety of the analyzed movie encompassing mitosis. Moreover, in this step, the software attributes sub-labels to properly distinguish the two daughters from each other. In addition, MitoTrack allows for a spatial analysis and hence records the timing of transcriptional activation within user-defined domains. All the extracted quantitative data are organized into an .xls file.
4 Results and perspectives
The described program represents a robust analysis platform to quantify the timing of transcriptional activation prior and after mitosis while keeping the genealogy information. Using MitoTrack software, we can now quantify the extent of transcriptional memory in hundreds of nuclei from various genotypes, as recently performed in (Dufourt et al., 2018). One of the main advantages of MitoTrack resides in the various tools it provides for manual correction of nuclei tracking during mitosis (Fig. 1c). However, mainly depending on nuclei crowding and temporal resolution, MitoTrack is able to automatically track all nuclei during mitosis with near 100% accuracy (for example in mitosis from nc12 to nc13 with a temporal resolution of 20 s/frame, or in mitosis from nc13 to nc14 with images every 4 s, as the sample data accompanying the provided tutorial).
By comparing the timing of activation of neighboring nuclei, we can now directly evaluate the extent of temporal coordination [referred to as synchrony (Lagha et al., 2013)]. Thanks to MitoTrack spatial analysis tool, it is possible to measure the timing of activation in distinct spatial domains and thus quantify the impact of gradients of morphogens (for example, the Dorsal activator) on transcriptional dynamics.
Funding
We are grateful to CNRS and FRM for supporting this work. A.T was sponsored by the SyncDev ERC grant. This work was supported by the ERC_SynDev Grant # 679792 to M.L. and HFSP-CDA grant to M.L.
Conflict of Interest: none declared.
Supplementary Material
References
- Amat F. et al. (2014) Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods, 11, 951–958. [DOI] [PubMed] [Google Scholar]
- Dufourt J. et al. (2018) Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun., 9, 5194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C., Lagha M. (2019) Lightening up gene activation in living Drosophila embryos. Methods Mol. Biol., 2038, 63–74. [DOI] [PubMed] [Google Scholar]
- Ferraro T. et al. (2016) Transcriptional memory in the Drosophila embryo. Curr. Biol., 26, 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H.G. et al. (2013) Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol., 23, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M. et al. (2013) Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell, 153, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas T. et al. (2013) Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr. Biol., 23, 2135–2139. [DOI] [PubMed] [Google Scholar]
- Otsu N. (1979) A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern., 9, 62–66. [Google Scholar]
- Pichon X. et al. (2018) A growing toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol. Cell, 71, 468–480. [DOI] [PubMed] [Google Scholar]
- Tinevez J.Y. et al. (2017) TrackMate: an open and extensible platform for single-particle tracking. Methods, 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tran H. et al. (2018) LiveFly: a toolbox for the analysis of transcription dynamics in live Drosophila embryos. Methods Mol. Biol., 1863, 183–195. [DOI] [PubMed] [Google Scholar]
- van der Walt S. et al. (2014) scikit-image: image processing in Python. PeerJ, 2, e453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C. et al. (2018) Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb. Elife, 7, e34410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.