Abstract
Post-translational modifications (PTMs) play pivotal roles in controlling the stability and activity of the tumor suppressor p53 in response to distinct stressors. Here we report an unexpected finding of a short chain fatty acid modification of p53 in human cells. Crotonic acid (CA) treatment induces p53 crotonylation, but surprisingly reduces its protein, but not mRNA level, leading to inhibition of p53 activity in a dose dependent fashion. Surpringly this crotonylation targets serine 46, instead of any predicted lysine residues, of p53, as detected in TCEP-probe labeled crotonylation and anti-crotonylated peptide antibody reaction assays. This is further confirmed by substitution of serine 46 with alanine, which abolishes p53 crotonylation in vitro and in cells. CA increases p53-dependent glycolytic activity, and augments cancer cell proliferation in response to metabolic or DNA damage stress. Since serine 46 is only found in human p53, our studies unveil an unconventional PTM unique for human p53, impairing its activity in response to CA. Because CA is likely produced by the gut microbiome, our results also predict that this type of PTM might play a role in early human colorectal neoplasia development by negating p53 activity without mutation of this tumor suppressor gene.
Keywords: p53, crotonylation, posttranslational modification (PTM), short chain fatty acids, cellular metabolism
INTRODUCTION
p53 is pivotally important for maintaining genomic stability and preventing tumor formation in response to various stressors, such as DNA damage, reactive oxygen species (ROS), ribosomal stress, nutrient depletion, hypoxia and oncogenic overloading [1]. Yet, when overly or abnormally activated, p53 can cause developmental defects and human diseases [2]. Thus, its protein level and activity are tightly regulated via multiple mechanisms. Cancers also evolve different strategies to control p53 activity in favoring their growth and survival [3], such as modifying its protein [4] in addition to mutating its gene, as it is the most frequently inactivated protein in all types of human cancers [5]. In fact, p53 can be modified via various forms of PTM, which either positively (acetylation or phosphorylation) or negatively (ubiquitination) regulate p53 stability and activity [6]. Recently, a type of unsaturated short chain fatty acid (SCFA) called Crotonic acid (CA), has been shown to modify histone proteins for epigenetic regulations [7]. However, this type of modifications has not been explored for p53 regulation.
MATERIALS AND METHODS
Plasmids and antibodies
The His-tagged p53 was generated into the pET30a vector, and His-p53-S46A was mutant based on His-p53. The no-tag plasmid pcDNA3.1-p53, pcDNA3.1-p53-S33A, pcDNA3.1-p53-S46A, pcDNA3.1-p53-T81A, pcDNA3.1-p53-4M were obtained from Giannino Del Sal. p53-K24R was mutant based on pcDNA3.1-p53. The Flag-pcDNA-p53, Flag-pcDNA-p53-1-300, Flag-pcDNA-p53-101-300, Flag-pcDNA-p53-101-393 were obtained from Mu-shui Dai. pcDNA-p53-8KR was obtained from Wei Gu. The following antibodies were commercially purchased: Anti-Flag (Sigma-Aldrich, St. Louis, MO, USA), anti-p53 (DO-1, FL-393 Santa Cruz Biotechnology), pan anti-crotonyl (ptm-501, PTM Biolabs), anti-GAPDH (#5174, Cell Signaling Technology), anti-β-actin (C4, Santa Cruz Biotechnology), anti-PFKP, anti-PKM2, anti-HK2, anti-LDHA (#8337, Glycolysis Antibody Sampler Kit, Cell signaling Technology).
Cell Culture
HCT116p53+/+, HCT116p53−/−, RKOp53+/+,, RKOp53−/−, H460, A549, cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Hyclone) supplemented with 10% fetal bovine serum (FBS) and penicillin and streptomycin, under 5% CO2. BC-1 and BCP-1 cells were obtained from Zhiqiang Qin and maintained in complete RPMI 1640 medium (ATCC) supplemented with 20% FBS. MCF7 was obtained from Heather Machado. MCF7, MDA-MB-468 were maintained in DMEM with 10% FBS.
Immunoblotting
Cells were harvested and lysed in lysis buffer consisting of 50 mM Tris/HCl (pH7.5), 0.5% Nonidet P-40 (NP-40), 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 10 μM pepstatin A and 1mM leupeptin. Equal amounts of clear cell lysate (20–80 μg) were used for immunoblotting (IB) analyses as described previously [4].
In Vitro Crotonylation of Serine (Ser) Assays
Recombinant human p300 proteins were purchased from Active Motif (Carlsbad, CA, catalog number 31124) For each reaction, 20 μg of HEK293T cell lysis complex or 200 ng of p300 protein, 10 μM of Cro-CoA were added in the reaction buffer (50 mM Tris-CI, pH 8.0, 10% glycerol, 100 nM TSA, 5 mM Nicotinamide, 0.1 mM EDTA, 1 mM DTT and 1x proteinase inhibitor cocktail). The reaction mixtures were incubated at 30°C for 1 hour. Crotonoyl-coenzyme A (Cro-CoA) was purchased from Sigma (St. Louis, MO, 28007), Substrates included 100 ng of His-p53 and 100 ng of His-p53-S46A [8], and Crotonylation assays were also done using unlabeled Cro-CoA (1 mM) followed by SDS–PAGE, and then Crotonylation S46 was detected by WB using the pan anti-Cro antibody[9].
In vitro Crotonylation Assay with Antibody and TCEP
Briefly WT or P53 beads were incubated with crotonylation buffer (50 mM TRIS pH 8.0, 10% glycerol and 0.1 mM EDTA, 1 mM DTT, 100 nM Trichostatin A, 5 mM nicotinamide, and 1x protease inhibitor cocktail) in the presence and absence of p300 and HEK293 lysate at 37°C for 1 hr. Subsequently, beads were washed and crotonylation was detected using SDS-PAGE and immunoblotting using the PTM501 Pan crotonyllysine antibody. In a separate trial, the crotonylation reaction was performed as mentioned above without use of TSA and NAD. Following the crotonylation reaction a TCEP-Biotin chemical probe was added to the beads at a final concentration of 2 mM and incubated for 16 hrs at 37°C. Subsequently, beads were washed and crotonylation was detected using SDS-PAGE and immunoblotting using a a high sensitivity HRP-Streptavidin antibody. In both trials, DO1 was used to detect the p53 loading control [10].
Reverse transcription and quantitative PCR analyses
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Total RNAs of 0.5 to 1μg were used as templates for reverse transcription using poly-(T)20 primers and M-MLV reverse transcriptase (Promega, Madison, WI, USA). Quantitative PCR (Q-PCR) was conducted using SYBR Green Mix according to the manufacturer’s protocol (BioRad, Hercules, CA, USA). The primers for human p53, p21, Actin were used as previously described [11].
Colony formation assay
Cells were trypsinized and seeded with the same amount on 6-well plates following siRNA transfection for 12 to 18 h. The medium was changed every 3 days until the colonies were visible. t. Cells were then fixed by methanol and stained by crystal violet solution at RT for 30 min. ImageJ was used for quantification of colonies.
RESULTS
Crotonic acid negatively regulates p53 level and activity
SCFAs, including acetic acid (AA), propionic acid (PA), butyric acid (BA), and CA whose synonym is but-2-enoic acid (Fig. 1A), are common products of the dietary fiber fermentation by microflora in the human gut. Since these SCFAs are essential for molecule functions and human health, we first tested whether they would affect p53 level and activity by treating human wild type (wt) p53-containing colon cancer RKO or lung cancer H460 cells with each of the acids individually. Interestingly, PA or BA induced the protein level of p53 and the expression of its target genes p21 and MDM2 (Figs. 1B and 1C) as expected [12], whereas CA reduced the protein level of p53 and p21 (Fig. 1B) or MDM2 (Fig. 1C) in a dose dependent fashion as detected by immunoblot (IB) analysis. Remarkably, the reduction of p53 level and activity by CA was observed in a variety of wt p53-containing cancer cells, including lymphoma BC-1 (Fig. 1D). However, the reduction of p53 level by CA was not observed in mutant (mt) p53-harboring cancer cells, such as lymphoma BCP-1 (Fig. 1D). Also, the reduction of p53 was not observed at its RNA level (Fig. S1). These results suggest that CA can negatively regulate p53 level and activity in human or cancer cells potentially at a post-translational step.
Fig. 1. Crotonic acids decrease p53 protein level.
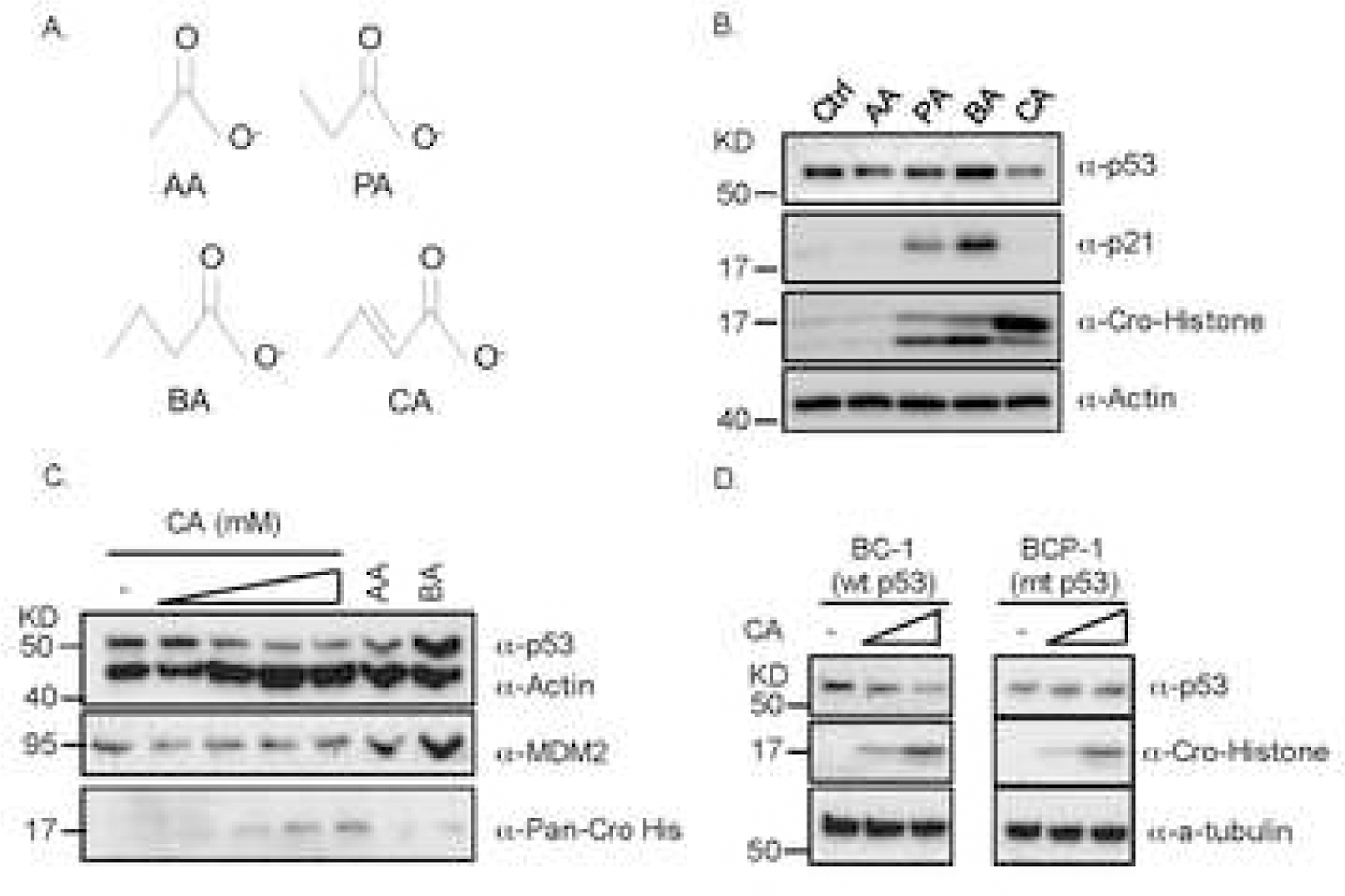
(A) The chemical structures of four short-chain fatty acids (SCFAs), such as acetic acid (AA), propionic acid (PA), butyric acid (BA), and crotonic acid (CA). (B) CA reduces the protein, but not mRNA, level of p53 as well as p21 level and induces histone crotonylation. RKO cells were treated with indicated SCFAs for 24h and harvested for IB with indicated antibodies (Ctrl: Control). Portions of cells were harvested for Q-PCR analysis of p53 mRNA levels (see Fig. S1E). (C) CA reduces p53 levels in a dose dependent fashion. H460 cells were treated with different doses of CA (2,5,10 and 20mM) for 24h and harvested for IB analysis with indicated antibodies. (D) CA reduces wt, but not mutant (mt), p53 in lymphoma cells. p53-containing BC-1 and mt p53-containing BCP-1 cells were treated with two indicated doses of CA (2,5mM) for 24h and harvested for IB with indicated antibodies.
Crotonylation of p53 at its N-terminus but not lysine residues.
Since CA treatment clearly led to histone crotonylation as detected in all of the above experiments (Figs. 1B–D), we tested if CA could also result in p53 crotonylation. To do so, we individually introduced three members of the human p53 family into HCT116p53−/− cells and performed co-immunoprecipitation (IP)-IB analysis. Surprisingly, only p53, but not p73 and p63, was crotonylated (Fig. 2A) as detected by a validated pan crotonylation antibody [13]. The endogenous p53 in HCT116 cells was also crotonylated (Fig. 2B). To identify which amino acid(s) of p53 can be crotonylated, we first introduced different p53 fragments into HCT116p53−/−cells and performed a co-IP with anti-Flag followed by IB analysis with anti-crotonylated peptide antibodies. Interestingly, crotonylation was detected only in the full length p53 and its N-terminus (aa 1–300), but not the central- and C-terminal (aa 100–393) fragments (Fig. 2C), suggesting that crotonylation might occur within the first 100 amino acids. Since crotonylation was reported to target lysine only [14], we replaced the only lysine residue (K24) in this region with arginine (Fig. S2A) and conducted the same co-IP-IB assay. To our surprise, p53-K24R was still crotonylated (Fig. 2D), suggesting that crotonylation might target a non-lysine residue in the N-terminus of p53. A previous study showed that aspirin can induce COX-1/2 acetylation at serine 529/516 [15, 16]. We thus suspected that crotonylation might target one of the serine or threonine residues within this region, and thus tested the crotonylation of p53 by comparing the wild type with its mutant p53–4M that has quadruple mutations of Ser33Ala, Ser46Ala, Thr81Ala and Thr315Ala and is available in our lab [4], using the same assay. To our further surprise, we did not detect crotonylation of p53–4M (Fig. 2E). Next, we used individual mutations at these amino acids and used them for the same co-IP-IB assay. As shown in Fig. 2F, crotonylation was detected in p53-S33A and p53-T81A, but not in p53-S46A and p53–4M, indicating that Ser46 might be the target residue for crotonylation.
Fig. 2. p53 is crotonylated at Serine 46.
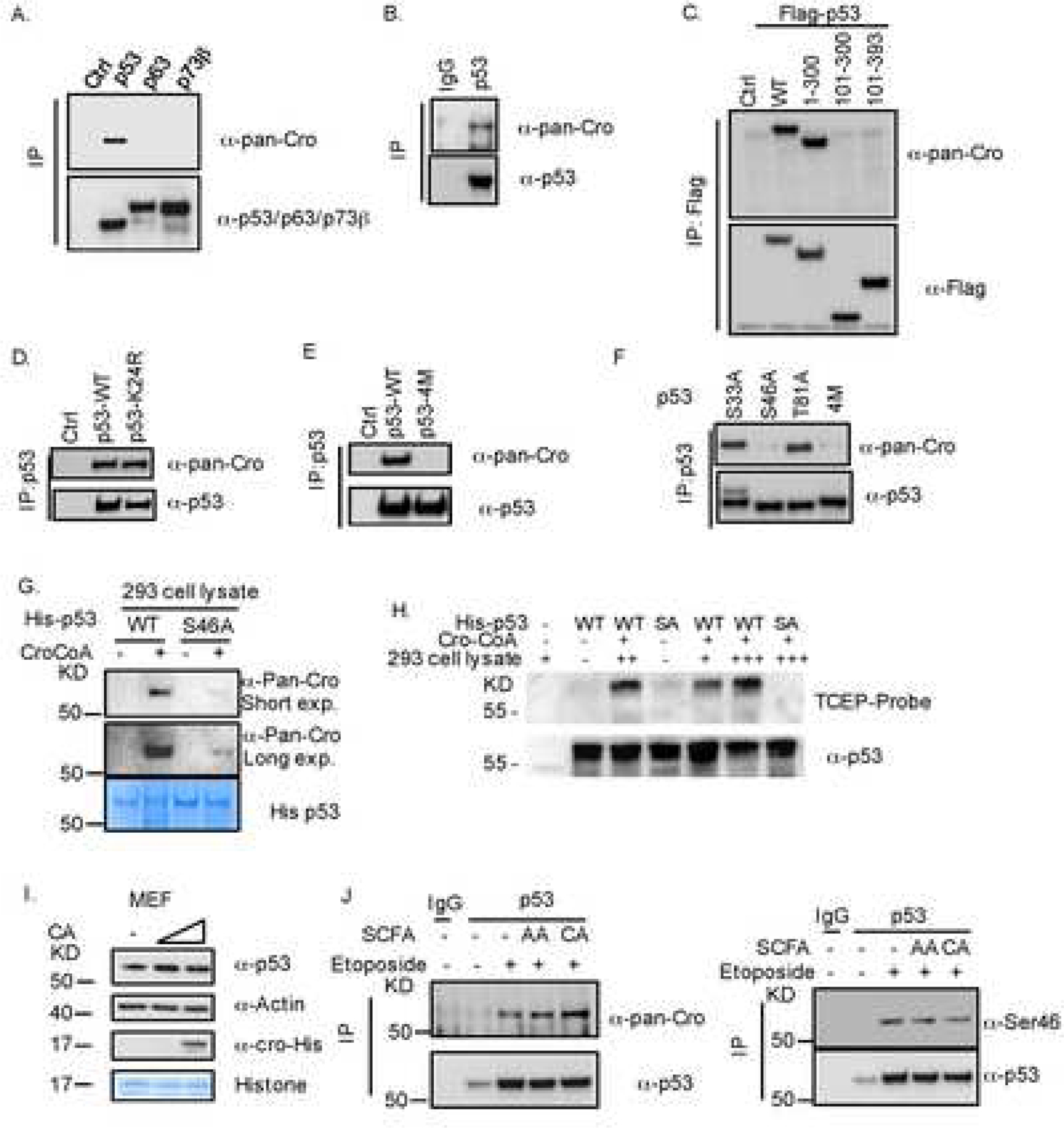
(A) Ectopic p53 is crotonylated. p53, p63 and p73β plasmids were introduced into HCT116p53−/− cells. Cells 30h after transfection (the same time for the following IP-IB experiments) were harvested for IP-IB or straight IB analysis with indicated antibodies. (B) Endogenous p53 is crotonylated. HCT116 cells were used for IP with the anti-p53 antibody followed by IB with indicated antibodies. (C) The N-terminus of p53 is crotonylated. Deletion mutants of p53 were introduced into HCT116p53−/− cells. Cells were harvested for IP-IB with indicated antibodies. (D) p53-K24R is crotonylated. WT p53 and p53-K24R plasmids were introduced into HCT116p53−/− cells. Cells were harvested for IP-IB with indicated antibodies. (E) p53–4M is not crotonylated. WT p53 and p53–4M plasmids were introduced into HCT116p53−/− cells, which were harvested IP-IB with indicated antibodies. (F) p53-S46A is not crotonylated. The S33A, S46A, T81A or 4M mutants of p53 were introduced into HCT116p53−/− cells that were harvested for IP-IB with indicated antibodies. (G) p53 is crotonylated in vitro. His-p53WT or His-p53S46A was purified from E.coli for in vitro enzyme reactions using HEK293T cell lysate and Cro-CoA. Crotonylated proteins were detected by IB with the anti-pan cro antibody (G). (H) Crotonylation reactions were performed with WT and S46A(SA) His p53 beads in the presence and absence of HEK293 lysate. Crotonylation was detected using a TCEP-Biotin probe was used in the in vitro reaction and a high sensitivity HRP-Streptavidin antibody was used for detection of crotonylation. (I) CA does not alter murine p53 level. MEF cells were treated with different doses of CA (1,5mM) for 24h as indicated and harvested for IB with indicated antibodies. (J) CA induces crotonylation, but reduces Ser46 phosphorylation, of p53. HCT116 cells were treated with etoposide and AA or CA for 24h and harvested for IP-IB analysis with indicated antibodies.
Crotonylation of p53 at serine 46
To confirm the result above and also to determine if p53 can be crotonylated in an in vitro reconstituted system, we purified recombinant wt His-p53 or His-p53-S46A from E coli and used them for an in vitro crotonylation assay with crotonyl-CoA (Cro-CoA) as a co-factor. For this in vitro reaction, we used HEK293T cell lysates as the resources of yet unidentified crotonyl transferases. This choice was made because we detected a strong crotonylation reaction in this cell line, and also we found that acetyl-transferase p300 cannot catalyze p53 crotonylation in vitro (Fig. S2E). Only wt p53, but not p53-S46A, was crotonylated in a Cro-CoA dose-dependent fashion (data not shown). This result was further validated using the TCEP-probe label crotonylation assay (Fig. 2H). Because Ser46 is only found in human, but not mouse, p53 (Fig. S2F), we tested if CA can affect mouse p53 level or not by treating mouse embryonic fibroblasts (MEF) with CA. As shown in Fig. 2I, CA failed to reduce mouse p53 level, suggesting that CA-caused decrease of p53 level is Ser46-dependent. Because previous studies showed that Ser46 is phosphorylated in response to DNA damage [17], we tested if crotonylation at Ser46 are inversely related to each other. As shown in Fig. 2J, etoposide induced p53 level, whereas CA, but not AA, induced p53 crotonylation and reduced Ser46 phosphorylation in HCT116 cells. A recent proteomic study suggested that several lysine residues of p53 might be crotonylated in cells [18]. However, a mutant p53 with the substitution of 8 lysines with arginines (p53–8KR) (Fig. S2B) was still crotonylated in HCT116p53−/− cells (Fig. S2C), excluding the possibility of these lysine residues as the target sites for crotonylation. Taken together, these results demonstrate that CA can reduce p53 level in human, but not mouse, cells by inducing Ser46 crotonylation (Fig. S3B).
Crotonic acid regulates glycolysis and mitochondrial activity via suppression of p53.
Next, we wanted to determine what cellular functions of p53 are affected by CA treatment. Among several pathways tested, we found that CA regulates the glycolysis pathway by reducing p53 protein level. When doing so, we treated MCF7 cell with different concentrations of CA and detected the level of proteins involved in glycolysis. As shown in Fig. 3A, CA reduced p53 level, but increased the levels of PFKP and PKM2, two enzymes involved in glycolysis as downstream targets of p53 in a dose-dependent manner (Fig. 3A). Since PKM2 was identified as a key regulator of cancer metabolism and tumor growth [19], its controversial role was observed in different cell types and mouse models [20–22]. This effect with 2mM of CA was similar to that in response to serum starvation, as 1% serum also caused the induction of PFKP and PKM2 while reduced the level of p53 (Fig. 3B). This result indicates that the effect of CA on the expression of PFKP and PKM2 is p53-dependent, as this was observed only in HCT116p53+/+ cells, but not in HCT116p53−/− cells (Fig. 3B). Consistent with these effects, CA also significantly increased glycolytic reserve (Fig. 3C) and spare respiratory capacity (Fig. 3D) as well as mitochondrial respiratory capacity in p53-positive, but not p53-deficient, HCT116p53−/− cells as measured by the Seahorse analysis (Fig. 3D). Together, in line with CA regulation of p53 in response to nutrition depletion, the glycolysis pathway is more pronounced in a p53-dependent manner.
Fig. 3. Crotonic acid increases glycolysis, and elevates cancer cell growth dependently of p53 in response to nutrition depletion.
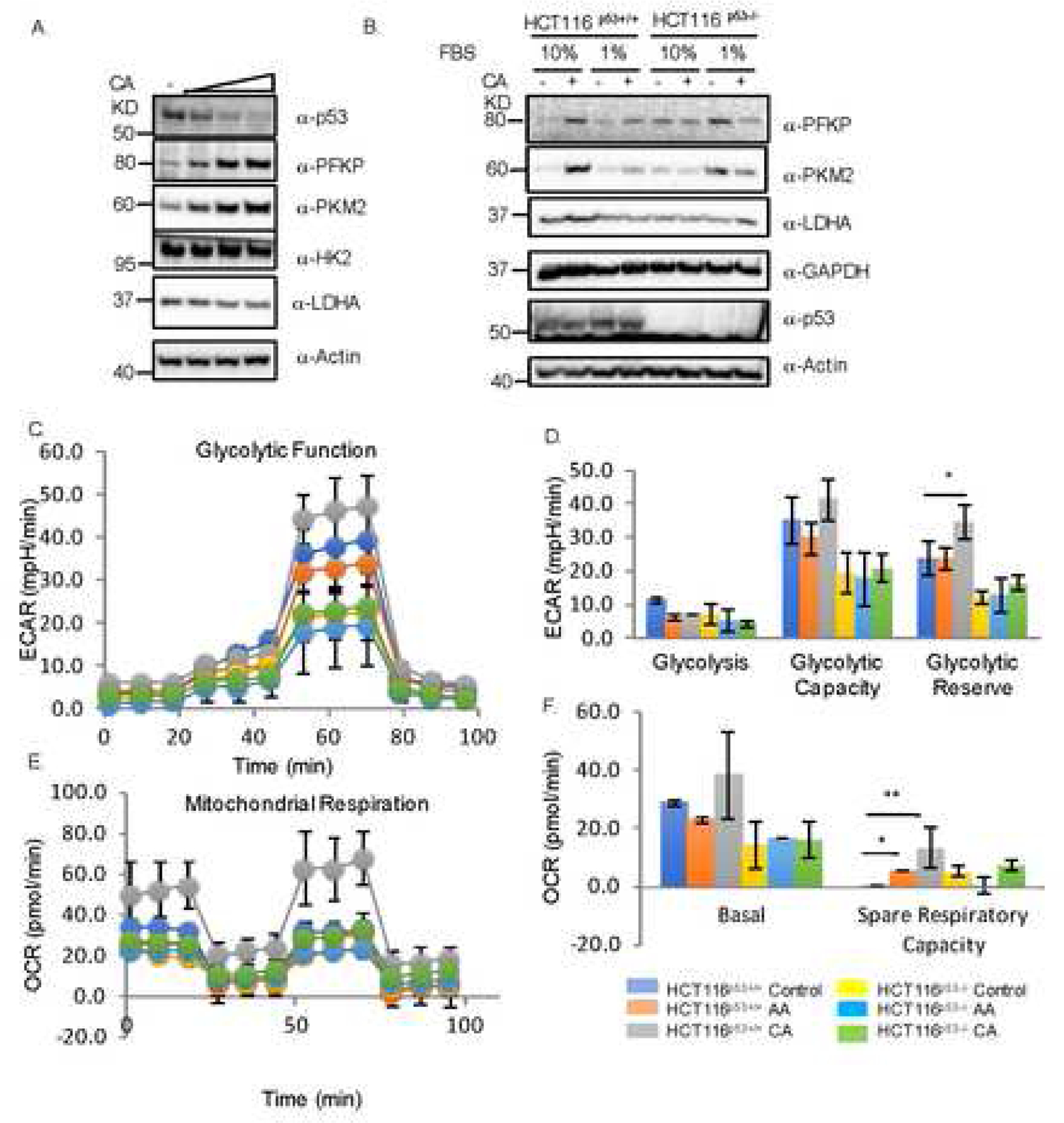
(A) CA reduces p53 level, but increases the level of PKFP and PKM2. MCF7 cells were treated with different doses of CA (2,5 and 10mM) for 4 days and harvested for IB analysis with indicated antibodies. (B) CA and serum starvation reduce p53 level, induction of PKFP and PKM2 by CA is p53-dependent. T116p53+/+ or HCT116p53−/− cells were cultured in media containing either 10% or 1% FBS in the presence or absence of 2mM of CA for 48h and harvested for IB with indicated antibodies. (C) p53-dependence of increase of extracellular acidification rate (ECAR) by CA. HCT116p53+/+ or HCT116p53−/− cells were treated with CA (Cro) or AA (Ace) for 24h and analyzed by using the Agilent Seahorse XF Analyzers to measure ECAR after sequential treatment with glucose (Glc), oligomycin (O) and 20DG. The quantification is shown in the graph below. (D) p53-dependence of increase of oxygen consumption rate (OCR) by CA. The same treatment as that in panel C was done and followed by sequential treatment with oligomycin (o), FCCP, and rotenone plus antimycin (R+A) for OCA analysis by the Seahorse Analyzers. The quantification is shown in the right graph.
Crotonic acid augments tumor cell proliferation dependently of p53 in response to glucose starvation.
Next, we tested if CA could affect cancer cell proliferation in a p53-dependent fashion when cells are under the condition of glucose starvation. To do so, we conducted a colony formation assay by starving HCT116p53+/+ and HCT116p53−/− cells in presence or absence of AA or CA. As shown in Figs. 4A and 4B, glucose starvation promoted more dramatic colony formation of HCT116p53−/− cells than that of HCT116p53+/+ cells. By sharp contrast, CA caused more dramatic increase in the colony formation of HCT116p53+/+ cells than that of HCT116p53−/− cells, while AA had no significant effect on this cellular phenotype (Figs. 4A and 4B). This result suggests that CA may promote cancer cell proliferation under the condition of glucose starvation by suppressing p53 activity, which is well in line with the effect of CA on p53 suppression of glycolysis as shown above.
Fig. 4. Crotonic acid increases tumor cell proliferation partially via suppression of p53.
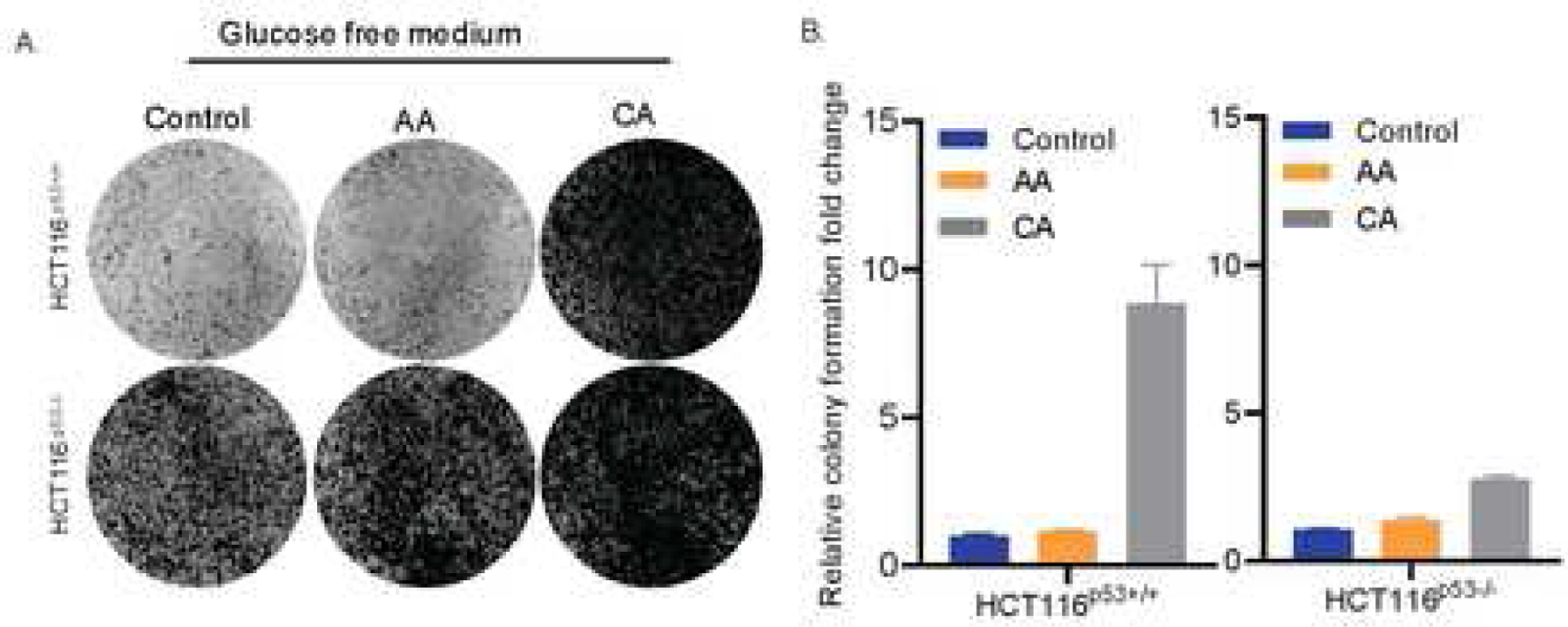
(A) CA renders p53-dependent resistance of colorectal cancer cells to glucose starvation. HCT116p53+/+ or HCT116p53−/− cells were cultured glucose free media in the presence or absence of 5mM of AA (acetic acid) or CA (crotonic acid) for colony formation assays. (B) The quantification of colonies formed from the experiments in panel A is shown here.
DISCUSSION
As presented above, our study unveils crotonylation as a new type of PTM for p53 regulation in response to CA treatment. This finding is novel as it is the first time to learn that crotonylation can occur at a serine residue, instead of lysine, of a protein (Figs. 2 and S2). Also, by crotonylating serine 46, CA can negatively regulate p53 level and activity and consequently render the wt p53-dependent resistance of cancer cells to chemotherapeutic drugs or nutrient stress (Figs. 1, 3, and 4). This modification is specific to human p53, as this residue exists neither in mouse p53 nor in human p73 or p63 (Figs. S2D and S2F).
These findings are highly significant because even without p53 mutations, p53 activity can be inhibited by some SCFAs, such as CA. Since CA may be a byproduct or intermediate product of dietary fiber fermentation by some microflora in the human gut [23], and crotonyl-CoA is present in human cells and involved in different metabolic pathways [24], our findings implicate that p53 activity could be inactivated during the early stage of colorectal tumorigenesis even though its gene is often mutated at the late stage [25]. Thus, our findings open a window for translational research to interrogate if inactivation of p53 by microflora-produced CA is associated with the early onset of colorectal cancers that harbor wt p53 and their drug resistance late on. This question is also applicable to other types of cancers, as CA can be readily absorbed into the systemic circulation via large intestine epithelial cells. Supporting this assumption is the fact that CA inactivates p53 in lymphoma, breast and lung cancer cells as well (Figs. 1–4 and S1). Our findings also raise additional questions. For instance, it remains unclear how crotonylation at serine 46 reduces p53 protein level, as we found that this is MDM2- and ubiquitination-independent (Figs. S3C and S3D). Moreover, it is puzzling about what enzyme(s) might catalyze this crotonylation of p53, as p300 was unable to do so as shown in our study (Figs. S2E). Addressing these enticing questions would help uncover more molecular insights into the regulation of p53 by crotonylation and its biological significance in cancer development and drug resistance.
Supplementary Material
HIGHLIGHTS.
p53 in human cells is crotonylated in response to crotonic acid.
This crotonylation is detected independently by a Pan anti-crotonylated peptide antibody and a TCEP probe.
This crotonylation surprisingly occurs at the serine 46 residue of p53.
Substitution of serine 46, but not any of predicted lysines, by alanine abolishes this p53 crotonylation.
Crotonic acid increases glycolytic activity and cancer cell proliferation by impairing p53 activity in human cancer cells.
HIGHLIGHTS.
Serine 46 of p53 is crotonylated in response to crotonic acid as detected by a Pan anti-crotonylated peptide antibody and a TCEP probe.
Mutation serine 46 abolishes this p53 crotonylation.
Crotonic acid increases glycolytic activity and human cancer cell proliferation by impairing p53 activity.
ACKNOWLEDGEMENTS
We thank Wei Gu, Mu-shui Dai and Giannino Del Sal for providing plasmids, Zhiqiang Qin and Heather Machado for offering some cancer cell lines, as well as all of the Lu lab members for active discussion. Hua Lu and Shelya X Zeng were supported in part by National Institutes of Health (NIH)-National Cancer Institute (NCI) grants (R01CA095441, R01CA172468, R01CA127724). Binghe Wang was partially supported by the NIH-NCI grant (R01-CA180519).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES AND NOTES
- [1].Levine AJ, Oren M, The first 30 years of p53: growing ever more complex, Nat Rev Cancer, 9 (2009) 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vousden KH, Lane DP, p53 in health and disease, Nat Rev Mol Cell Biol, 8 (2007) 275–283. [DOI] [PubMed] [Google Scholar]
- [3].Kruse JP, Gu W, Modes of p53 regulation, Cell, 137 (2009) 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liao P, Zeng SX, Zhou X, Chen T, Zhou F, Cao B, Jung JH, Del Sal G, Luo S, Lu H, Mutant p53 Gains Its Function via c-Myc Activation upon CDK4 Phosphorylation at Serine 249 and Consequent PIN1 Binding, Mol Cell, 68 (2017) 1134–1146 e1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Olivier M, Hollstein M, Hainaut P, TP53 mutations in human cancers: origins, consequences, and clinical use, Cold Spring Harb Perspect Biol, 2 (2010) a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Meek DW, Anderson CW, Posttranslational modification of p53: cooperative integrators of function, Cold Spring Harb Perspect Biol, 1 (2009) a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y, Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification, Cell, 146 (2011) 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kobet E, Zeng X, Zhu Y, Keller D, Lu H, MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins, Proc Natl Acad Sci U S A, 97 (2000) 12547–12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, Qi S, Locasale JW, Roeder RG, Zhao Y, Li X, p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis, Mol Cell, 70 (2018) 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bos J, Muir TW, A Chemical Probe for Protein Crotonylation, Journal of the American Chemical Society, 140 (2018) 4757–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou X, Hao Q, Liao P, Luo S, Zhang M, Hu G, Liu H, Zhang Y, Cao B, Baddoo M, Flemington EK, Zeng SX, Lu H, Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Archer SY, Meng S, Shei A, Hodin RA, p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells, Proc Natl Acad Sci U S A, 95 (1998) 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, Allis CD, Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation, Mol Cell, 58 (2015) 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xu W, Wan J, Zhan J, Li X, He H, Shi Z, Zhang H, Global profiling of crotonylation on non-histone proteins, Cell Res, 27 (2017) 946–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lecomte M, Laneuville O, Ji C, Dewitt DL, Smith WL, Acetylation of Human Prostaglandin Endoperoxide Synthase-2 (Cyclooxygenase-2) by Aspirin, Journal of Biological Chemistry, 269 (1994) 13207–13215. [PubMed] [Google Scholar]
- [16].Roth GJ, Stanford N, Majerus PW, Acetylation of Prostaglandin Synthase by Aspirin, P Natl Acad Sci USA, 72 (1975) 3073–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y, p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53, Cell, 102 (2000) 849–862. [DOI] [PubMed] [Google Scholar]
- [18].Wei W, Mao A, Tang B, Zeng Q, Gao S, Liu X, Lu L, Li W, Du JX, Li J, Wong J, Liao L, Large-Scale Identification of Protein Crotonylation Reveals Its Role in Multiple Cellular Functions, J Proteome Res, 16 (2017) 1743–1752. [DOI] [PubMed] [Google Scholar]
- [19].Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC, The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth, Nature, 452 (2008) 230–U274. [DOI] [PubMed] [Google Scholar]
- [20].Allen AE, Locasale JW, Glucose Metabolism in Cancer: The Saga of Pyruvate Kinase Continues, Cancer Cell, 33 (2018) 337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PNM, Hollern DP, Bellinger G, Dayton TL, Christen S, Elia I, Dinh AT, Stephanopoulos G, Manalis SR, Yaffe MB, Andrechek ER, Fendt SM, Vander Heiden MG, Pyruvate Kinase Isoform Expression Alters Nucleotide Synthesis to Impact Cell Proliferation, Molecular Cell, 57 (2015) 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC, Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses, Science, 334 (2011) 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fischer CR, Tseng HC, Tai M, Prather KLJ, Stephanopoulos G, Assessment of heterologous butyrate and butanol pathway activity by measurement of intracellular pathway intermediates in recombinant Escherichia coli, Appl Microbiol Biot, 88 (2010) 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rozen R, Vockley J, Zhou LB, Milos R, Willard J, Fu K, Vicanek C, Lownang L, Torban E, Fournier B, Isolation and Expression of a Cdna-Encoding the Precursor for a Novel Member (Acadsb) of the Acyl-Coa Dehydrogenase Gene Family, Genomics, 24 (1994) 280–287. [DOI] [PubMed] [Google Scholar]
- [25].Nakayama M, Oshima M, Mutant p53 in colon cancer, J Mol Cell Biol, (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


