Abstract
Every type of nucleic acid in cells may undergo some kind of post-replicative or post-transcriptional chemical modification. Recent evidence has highlighted their importance in biology and their chemical complexity. In the following pages, we will describe new discoveries of modifications, with a focus on tRNA and mRNA. We will highlight current challenges and advances in modification detection and we will discuss how changes in nucleotide post-transcriptional modifications may affect cell homeostasis leading to malfunction. Although, RNA modifications prevail in all forms of life, the present review will focus on eukaryotic systems, where the great degree of intracellular compartmentalization provides barriers and filters for the level at which a given RNA is modified and will of course affect its fate and function. Additionally, although we will mention rRNA modification and modifications of the mRNA 5’-CAP structure, this will only be discussed in passing, as many substantive reviews have been written on these subjects. Here we will not spend much time describing all the possible modifications that have been observed; truly a daunting task. For reference, Bujnicki and co-workers have created MODOMICS, a useful repository for all types of modifications and their associated enzymes. Instead we will discuss a few examples, which illustrate our arguments on the connection of modifications, metabolism and ultimately translation. The fact remains, a full understanding of the long reach of nucleic acid modifications in cells requires both a global and targeted study of unprecedented scale, which at the moment may well be limited only by technology.
Keywords: modifications, metabolism, translation, mRNA, tRNA
Introduction
The existence of modified nucleotides dates back to before the very origins of life on earth and although the prebiotic synthesis of nucleobases and ribose sugar vastly differs from modern enzyme catalyzed reactions, it is implied that modified nucleotides are not a new thing and that they have been around forever (Levy and Miller, 1999). What has always been a rate-limiting step in the discovery of “new” nucleotide modifications is truly the general inadequacy of our detection methods and the difficulty to simultaneously and accurately determine the chemical structure of modifications in a position-specific and molecule-specific manner; especially true with complex mixtures. Nonetheless, we have made steady progress and by now more than 140 different modified nucleotides have been described and these may occur in essentially any, and every, type of nucleic acid in cells (Boccaletto et al., 2018). What has been underappreciated, and critically not well discussed, are the unusual connections between the synthesis and incorporation of modified nucleotides into nucleic acids and how this relates to the synthesis of canonical nucleotides and other aspects of metabolism.
The de novo biosynthesis of nucleotides in cells involves a set of stepwise enzymatic reactions leading to the formation of the nucleobases: Purines and pymidines (Lane and Fan, 2015). For purines, ten distinct enzymatic steps are needed to ultimately generate IMP, which later is interconverted into the other mono-phosphorylated forms of purines (XMP, GMP and AMP) (Figure 1). Pyrimidine biosynthesis involves 6 distinct reactions that again occur stepwise leading to the formation of UMP; 3 additional reactions ultimately are required to make CTP, using UTP as an intermediate (Lane and Fan, 2015). Most cells, with few exceptions (e.g. trypanosomes and a number of other protists), have the ability to synthesize nucleotides de novo, but additionally, all cells can salvage preformed nucleobases from their growth media (Zollner, 1982). Historically, it was well accepted that once the five main nucleotides were incorporated into intracellular pools they would, in their basic chemical form, partake in various transactions ranging from polynucleotide synthesis (DNA and RNA) to generation of cofactors important for numerous metabolic reactions (for example in the case of NAD/NADP, NADH/NADPH, SAM, etc). Then in the early 50s, studies by the Allen laboratory revealed the presence of an isomer of uridine in RNA (Davis and Allen, 1957; Yu and Allen, 1959). This isomer had clearly different chemical properties from its canonical cousin, including a unique C-C bond joining the base to the ribose sugar. Further studies then led to the characterization of this isomer termed pseudouridine, yet its biological importance remained a mystery. However, that single observation led many groups to look for more of these unusual nucleotides leading to the discovery of several methylated and thiolated species and eventually inosine in tRNA (Holley et al., 1965). The latter representing the first connection between modified nucleotides and translation; well captured in Francis Crick’s “wobble hypothesis” (Crick, 1966). Together these findings effectively gave birth to the now blossoming field of post-transcriptional modifications.
Figure 1. Canonical and modified nucleotide synthesis share common building blocks.
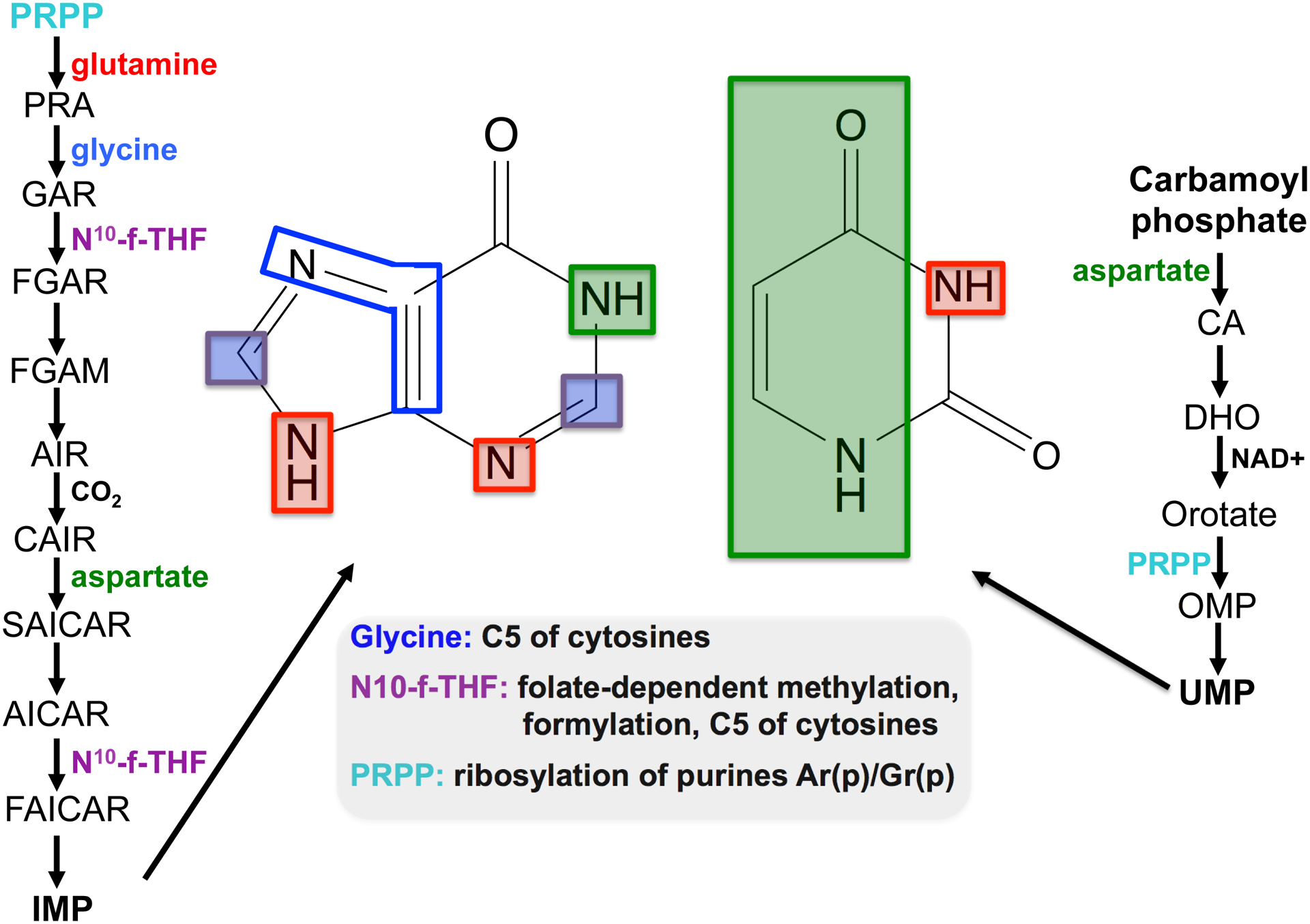
The figure shows the de novo purine (right side) and pyrimidine biosynthetic (left) pathways, leading to the formation of inosine monophosphate (IMP) and uridine monophosphate (UMP), the core nucleobases (the base portion is depicted in the middle of the figure). The different portions of the purine and pyrimidine rings are color coded to match the given substrate used for their biosynthesis. The inset shows substrates in common with modified nucleotides. “Purple squares” denotes F10-formyl-tetrahydrofolate, in “green” is aspartate, “red” is glutamine and blue denotes glycine. PRPP, 5’-phospho-D-ribosyl-1-pyrophospahte; PRA, 5-phosphoribosylamine; GAR, 5-phosphoribosylglycinamide; FGAR, 5-phosphoribosyl-N-formylglycinamide, FGAM, 5-phosphoribosyl-N-formylglycinamidine; AIR, 5-phosphoribosylaminoimidazole; CAIR, -(5-phosphoribosyl)-5-amino-4-carboxyimidazole; SAICAR, (5-phosphoribosyl)-4-(N-succinocarboxamide)-5-aminoimidazole; AICAR, 1-(5-phosphoribosyl)-5–4-imidazolecarboxamide; FAICAR, 1-(5-phosphoribosyl)-5-formaide-4-imidazolecarboxamide. CA, carbamoyl aspartic acid; DHO, dihydroorotic acid; OMP, orotidine 5’-monophosphate.
Questionably then, what is the connection between de novo canonical nucleotide synthesis and the synthesis of modified nucleotides? At a bird’s eye view, the connection may seem tenuous, however, upon careful inspection one would appreciate that both may utilize a number of similar building blocks derived from seemingly unrelated metabolic reactions within cells (Fig. 1). For instance, the biosynthesis of nucleobases requires the amino acids glycine, aspartate and glutamine for purines; pyrimidines, likewise, require glutamine and aspartate. In addition, purine biosynthesis also requires N10-formyltetrahydrofolate and as such is tightly linked to folate metabolism. Remarkably, some of these substrates are also utilized in the synthesis of a number of modified nucleotides, linking metabolite pools to the synthesis of both canonical and modified nucleotides (Fig. 1) (Helm and Alfonzo, 2014).
Previously and partly based on the preceding observations, we raised the possibility that the connection between central metabolism and modifications is not a coincidental one and in fact may be part of a well-orchestrated coordination network between metabolism and protein synthesis (Helm and Alfonzo, 2014). In addition, we have also highlighted how intracellular RNA transport systems (particularly in eukaryotes) further provide a regulatory step that may not only affect substrate availability, but also ultimately lead to a great level of variegation in the chemical composition of nucleic acids in the amounts and types of modified nucleotides they contain (Kessler, Silveira d’Almeida, and Alfonzo, 2018).
In terms of cell physiology, it is clear that modification content (as recent data shows) in various nucleic acids, can indeed change both as a response to, and consequence of, not only environmental stress but with fluctuations in nutrient levels during normal growth. The fact that the most notable effects in mammals, upon defects on certain modifications, are generally phenotypically revealed when cells have to cope with high metabolic demand may in fact serve as the ultimate corollary for the connection between modifications, nucleotide biosynthesis and metabolism via protein synthesis outputs.
Modification diversity and the limits of detection
The recent discoveries of various group substitutions in mRNA as discussed in the following section, has created much excitement. This is partly due to the apparent novelty of it, but also increased interest arises from the implications of the discoveries. For example, established correlations with various disease states (Dorn et al., 2019; Hsu, Shi, and He, 2017) and even impacting cell development (Zhao et al., 2017). Still, however, it is in tRNA where the most bewildering diversity of modified nucleotide chemistry exists. These include deamination, isomerization, amino acid addition, transglycosylation, methylation, acetylation, thiolation, and a seemingly never-ending myriad of group-transfer reactions (Boccaletto et al., 2018). The number of new modifications reported keeps increasing and not surprisingly the ultimate limit will be set by the chemistry of what is enzymatically possible with the 5 canonical nucleotides in the context of a polynucleotide chain. To date, every available position in the purine and pyrimide nucleotides have been seen naturally modified, including the ribose sugar.
Answering the question of “are we there yet?” with modifications is really not an easy job. Perhaps more important is what keeps us from answering such question. Despite major advances in modification detection methods, the task of establishing both the position and chemical structure of a given modification is still a tall order. In general, there are two major approaches: 1) indirect detection and inference of a modification based on sequencing methods; 2) the use of direct detection methods, such as the time-honored Thin-layer chromatography (TLC)(which will not be discussed further) and the rapidly progressing mass spectrometry approaches. The latter are in fact the true golden standards of the modification detection field.
Various molecular biology-based sequencing methods have been established, these methods have proven of value given their requirement for small sample size and can efficiently reveal which nucleic acid is potentially modified at a whole transcriptome or genome scale. For example, the detection of 5-methylcytosines (m5C) exploits the ability of bisulfite to efficiently deaminate unmodified cytosines in nucleic acids causing a C to U change easily detectable by sequencing, while m5C remains protected from deamination (Singhal, 1971; Bhanot and Chambers, 1977). Downstream analysis, then dictates that every cytosine that remains unchanged (read as guanosine upon reverse transcription and sequencing), when compared to an untreated sample, must be modified (Edelheit et al., 2013). Other approaches have been implemented with modern sequencing technology to map pseudouridines, these take advantage of an early observation by Ofengand and co-workers who in the early 90s introduce the use of N-cyclohexyl-N’-beta-(4-methylmorpholinium) ethylcarbodiimide p-tosylate (CMCT) for detection of pseudouridines in RNA (Bakin and Ofengand, 1993). CMCT forms a bulky adduct with uridines and pseudouridines, with uridines the CMCT adduct is efficiently removed by alkaline treatment; pseudouridines retain it. Traditionally, CMCT treatment was followed by primer extension-type assays and relies on the fact that the “bulky” adduct creates a strong stop on sequencing lanes. Modern uses of this approach have simply coupled CMCT treatment to downstream reverse transcription and library preparation (Carlile et al., 2014). Clearly, with modern sequencing methods the roadblock appears as a pile up of sequence reads that end at pseudouridine-containing positions. In this approach, however, there is a word of caution that many researchers not familiar with the original technique fail to realize, even after careful titration during the alkaline treatment, many uridines may remain CMCT-modified generating false positives during sequencing. Similar approaches of chemical modification followed by RNAseq have also been implemented for the detection of inosines (Okada et al., 2019). The approaches above have proven very powerful in inferring the presence of some modifications in bulk RNA and assigning potentially modified positions. However, in all cases the chemicals used are rather harsh and undoubtedly lead to RNA damage leading to an overall underrepresentation of the entire modified pool.
A bigger challenge has been posed by base and ribose methylations. In the case of base methylation the detection of m6A is particularly difficult and most approaches involve the fragmentation of the RNA followed by enrichment of m6A containing RNA fragments by antibody immunoprecipitations and RNAseq (Linder et al., 2015; Dominissini et al., 2012; Meyer et al., 2012; Chen et al., 2015). However, the reliance on antibodies for enrichment is also full of complications given that depending on the specificity of the antibodies, the assignment of m6A to particular mRNAs may yield a lot of false positives. Most recently, the relative affinity of the m6A-binding YTH domain has been taken advantage of, by cleverly fusing it to the catalytic domain of the C to U RNA editing deaminase apobec-1 (Meyer, 2019). The latter if positioned at a given distance from a cytosine, will deaminate it and yield a C to U change easily detectable by sequencing. Thus, C to U changes mediated by the fusion protein become proxy for the presence of m6A. Again, an indirect method that is promising but one that undoubtedly will suffer from off-target binding of the YTH domain and the distance constrain of the targeted cytosine.
The detection of 2’-O-methylations in RNA by molecular biology approaches has also always been challenging. Chemically, it has been clear for quite sometime that the 2’-OH in RNA is quite reactive and upon activation can act as a strong nucleophile, nature has indeed taken advantage of this property as revealed by the mechanisms of ribozymes and many RNA specific nucleases. Recently, new approaches that couple the differential resistance of 2’-O-methyl to either cleavage or chemical treatment have been implemented with some success (Galvanin et al., 2019; Krogh and Nielsen, 2019).
Most recently, while attempting to improve the use of bisulfite sequencing for the detection of m5C in total RNA, several important observations were made: 1) that the inclusion of formamide and its improvement on RNA denaturation greatly increases the reliability of m5C detection by bisulfite sequencing, 2) the use of alkaline conditions led to deletion at sites containing pseudouridine and 3) the same conditions led to Dimroth rearrangement at positions containing m1A, which were then converted to m6A (Khoddami et al., 2019). The former can be easily detected via a characteristic nucleotide mis-incorporation signature during reverse transcription, yet the latter becomes silent and easily read by reverse transcriptase as an A. Thus the improved bisulfite sequencing technique permits the simultaneous detection of 3 different modifications, and because no bulky adduct is added, it detects pseudouridines even at adjacent sites. Still, however, the readers must appreciate the cautionary nature of the statements above and realize that all of these indirect approaches are full of complications and do require further validation downstream. Taken together, one must emphasize that all sequencing approaches ultimately provide inferences or models for the occurrence of a modification, but none directly reveals the true chemical nature of the modified nucleotides.
An alternative more direct approach has been recently introduced with Nanopore technology, which uses changes in electrical output generated by an oligonucleotide as it passages through a pore of diminishing size. The signal called “squiggle” can differentiate the 4 canonical nucleotides, however, it cannot still differentiate the signals from differentially modified RNAs de novo without prior historical knowledge of the site or type of modification. Beyond this, it also relies on computer-based methods to make a call on the presence of a modification (e.g. m6A) with little recourse for direct inspection if the call is right or wrong by the investigator (Smith et al., 2019). So this technology, although promising, will not be discussed further.
By far the most challenging enterprise in modification mapping deals with tRNA, which by some calculations have an average of 9–12 modified positions per molecule (Phizicky and Alfonzo, 2010). Some modifications such as m1G, m2,2G, m1A, m3C and m3U because of their placement on the Watson and Crick face of the base are powerful blockers of reverse transcription, so assessment of the full-length sequence of a native tRNA is increasingly difficult. Recently the use of a de-alkylation step (incubation with the Alk-B demethylase) prior to library separation has proven relatively useful in increasing the efficiency of full-length tRNA sequencing but has also helped map these modifications on tRNA by comparing AlkB-treated to untreated samples (Cozen et al., 2015). A similar approach combines demethylation with the nucleotide mis-incorporation signature of reverse transcriptase as it reads through he modifications (Zheng et al., 2015). The caveats with such techniques is that some modifications like m2,2G, because of the very nature of the dealkylation reaction, are recalcitrant to AlkB treatment, yielding a population of RNAs that escape sequencing in the context of a full-length molecule. Secondly, when mapping a given modification on a full-length molecule, since the reactions rely on a reverse transcription step and inevitably extension of an oligonucleotide primer in the complementary 5’−3’ direction, then one may only detect the 3’ most modification in a transcript given that during treatment all modifications, should they occur more than once, get removed. So in general, these approaches are wonderful at mapping modifications at sites where they historically occur and closer to the primer, but may not be as good at detecting the same modifications at new sites occurring further downstream from the primer annealing site.
In the end, the only true and proven method to directly map modifications is mass spectrometry. Here we will not go through an extensive discussion of the different mass spectrometry approaches, but rather we will use this space to dismiss a current growing myth in the modification field. An increasing number of researchers, especially those describing new methods of mapping modifications by molecular biology-based sequencing methods, have made the claim that currently mass spectrometry approaches cannot be used to sequence RNA and gather positional information on modifications on a sequence context. Emphatically, we point out that the use of LC MS/MS to sequence oligonucleotides dates back to the pioneering work of McLuckey (McLuckey, Van Berkel, and Glish, 1992) and independently McCloskey (Pomerantz, Kowalak, and McCloskey, 1993; Kowalak et al., 1993) who not only demonstrated the feasibility of the technique, but also highlighted its usefulness in sequencing RNA by mapping the positions of the modifications. Improvements of such approaches then have included the use of heavy isotopes and the utilization of chemically synthesized standards (Paulines and Limbach, 2017; Ross, Cao, and Limbach, 2017). Usually, with such approaches, the study starts with a total nucleoside analysis of the RNA in question, yielding a census of the modifications detectable in the sample. With the new generation of accurate mass instruments, such as the orbitrap tribrid mass spectrometers, identification and characterization of isobaric species (m4C from m5C), mass silent modifications (m3Ψ from Ψm) or possible new, unknown modifications is becoming routine. Furthermore, the ability to measure a molecule fine isotope structures allows for accurate generation of chemical formulae, for example resulting in differentiating modifications with mass differences of less that a Dalton (e.g. 5-aminomethyl-2-geranylthiouridine, 425.1984 Da from epoxyqueuosine, 425.1547 Da); when no standard is available. Nucleoside analysis is then followed by the generation of oligomers of manageable size (5-mers to 20-mers) via the use of base-specific nucleases prior to chromatographic separation and mass spectrometric analysis (Thakur et al., 2020). These approaches have been used successfully to sequence tRNAs but can easily be used with other types of RNAs. Clearly the drawback with LC MS/MS has always been the requirement for relatively large amounts of pure sample, however, as chromatographic technology continues to improve, sample consumption continues to decrease. Recently the implementation of new base-specific nucleases has permitted the analysis of modification in their natural sequence context in complex mixtures (Thakur et al., 2020). So the future for mass spectrometry based approaches is bright and in our opinion is truly the way of the future. Critically, since mass is an intrinsic quality of matter, what mass spectrometry offers above all is accuracy; unequivocally establishing the chemical structure of modifications and in this realm is without substitutes.
Multisubstrate specificity vs. sequential reactions of RNA modification enzymes: The case for modification cascades
Because reconstitution of RNA modifications in vitro inevitably requires the use of purified systems; historically, the study of modification enzyme specificity entailed one enzyme-one substrate type of analysis. This naturally led to the conclusion that RNA modification enzymes act independent of each other where each nucleotide position was modified by a single enzyme. However, many examples had disproven the general validity of this statement. Studies with the TruA enzyme revealed that it could catalyze formation of pseudourines at positions 38–40 in E. coli tRNAs (Hur and Stroud, 2007). The basis for this substrate flexibility was illustrated by the structure of TruA in complex with two different tRNAs, which coupled with functional assays, suggested that this enzyme used the flexibility of the anticodon-stem loop (ASL) of their substrates for multi-site specificity (Hur and Stroud, 2007). Such mechanisms of programmed enzyme promiscuity may provide important principles when studying the mechanisms and specificity of mRNA modifying enzymes, given the generally less structured nature of such substrates.
A clear example of this is FTO (Fat mass and Obesity-associated), which can demethylate different substrates and is also promiscuous in relation to the type of methylation it can target (Zhang et al., 2019). This enzyme was initially characterized for its ability to remove methyl groups from internal m6A sites in mRNA (Jia et al., 2011). However, subsequent studies showed that FTO only has a weak preference for m6A and is actually more efficient at demethylating m6Am (a modification specifically occurring at the mRNA CAP) under physiological conditions (Mauer et al., 2017). Although changing FTO levels does appear as a successful strategy to modulate global m6A content, genetic manipulation of FTO yields confusing results in relation to the biological role of m6Am vs. m6A on cellular function. It is currently hard to define the specific contribution of m6A versus m6Am removal. One study suggested that the main function of FTO was to destabilize mRNA by mainly removing the m6A at the CAP (Mauer et al., 2017). However, a recent study with the newly discovered m6A CAP methylase (CAPAM or PCIF1) showed that a knockout mutant for the gene encoding this enzyme, which effectively removes CAP m6A, only showed phenotypes under conditions of oxidative stress; importantly it had no impact on message stability but rather affected translation (Akichika et al., 2019). To further complicate the biology surrounding FTO, we should note that it can remove m6A from other RNA species other than messengers (i.e. snRNA, tRNA). The affinity of FTO for tRNA is particularly interesting as it can also target m1A modifications, raising the possibility that FTO could also remove methyl groups on N1 position of adenosines in mRNAs (figure 2) (Zhang et al., 2019; Wei et al., 2018; Zaccara and Jaffrey, 2020).
Figure 2. The pathway and fates of modified mRNAs.
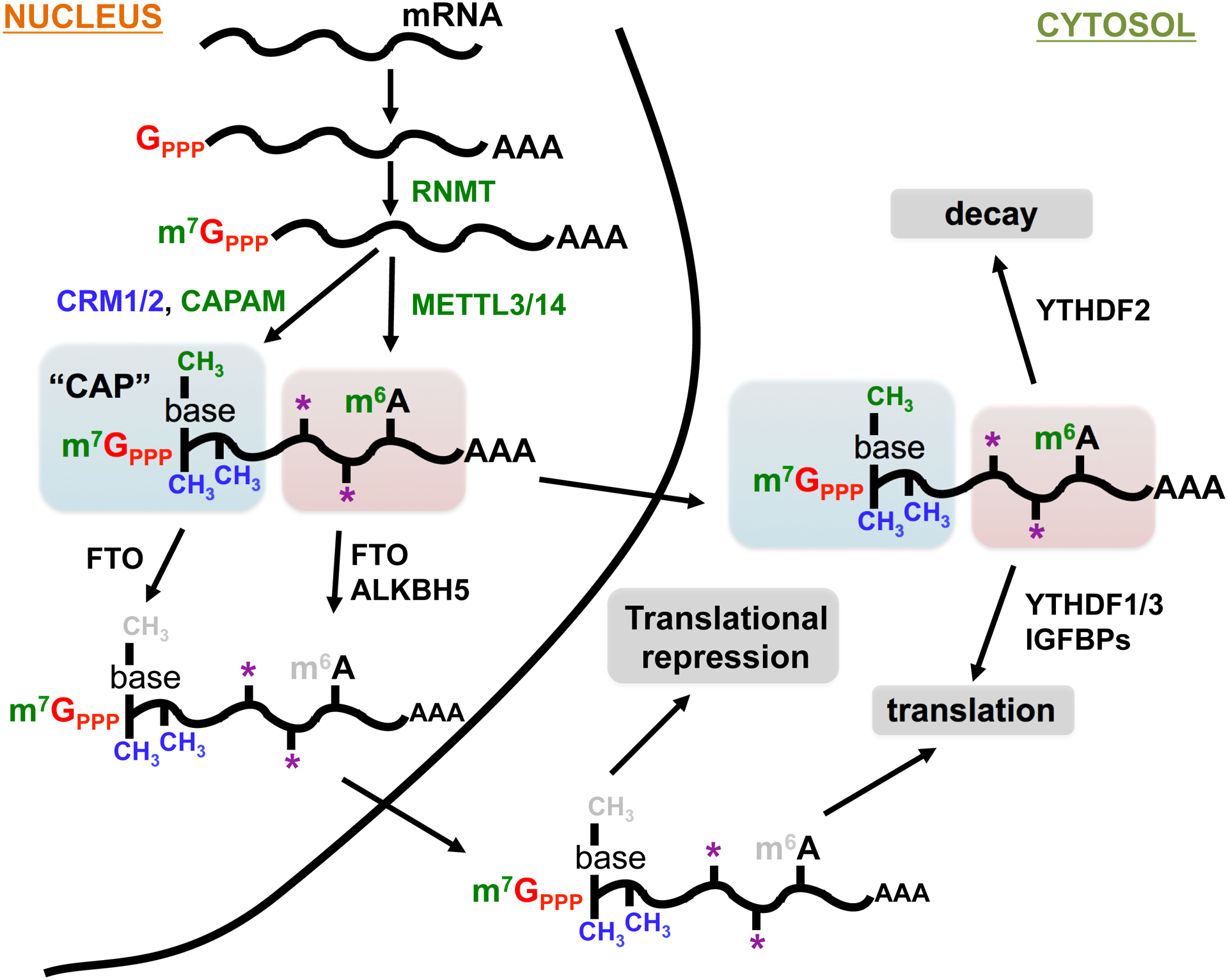
Shown are the steps an mRNA may undergo following transcription and splicing. Starting with addition of the non-templated triphosphorylated G (red), followed by sequential methylation of the cap starting with m7G (green), followed by ribose and base methylation of the first two template nucleotides. mRNAs may also undergo internal modifications such as m6A or inosine (not shown). Asterisks (*) denote less common modifications such as 5-methylcytosine (m5C), pseudouridine, N1-methyladenosine (m1A), ribose methylation, acetylation, etc. CAP formation is important for nuclear export. The mRNA can also be demethylated in the nucleus by the demetylase FTO and ALKBH5. The fate of mRNAs in the cytoplasm can be determined by their interaction with modification-specific mRNA-biding proteins such as YTHDF1/2/3 or IGFBPs.
Early studies also hinted at the fact that the one enzyme-one substrate paradigm may not be true for every RNA-enzyme pair. For instance, with CAP formation, there is a clear order to the modification events at the template nucleotides at the 5’ of mRNAs. Although m7G occurs at the 5’ CAP untemplated guanosine of mRNA in a reaction catalyzed by RNA Guanine-7 methyltransferase (RNMT), the first and second templated nucleotides are also often chemically altered (Wei, Gershowitz, and Moss, 1975; Wei, Gershowitz, and Moss, 1975; Furuichi et al., 1975). In this case, the modification is a 2’-O-methylation by CAP methyltransferase (CMTR) enzymes. Overall modification of the mRNA CAP is a coordinated and sequential process, as CMTR1, the enzyme that modifies the first templated nucleotide, can only function on m7G-modified mRNA (Belanger et al., 2010). Also, the enzymatic activity of CMTR2, which methylates the second templated nucleotide, reaches functional levels only when mRNA is already capped and methylated in the first position (Werner et al., 2011).
Studies of m1I formation in the TΨC of many archaeal tRNAs also revealed the potential sequential nature of some modifications. To form m1I57, an encoded adenosine is first methylated at the N-1 position to form m1A, which is then deaminated to m1I by an independent activity (Constantinesco, Motorin, and Grosjean, 1999). Likewise, long-range influences on backbone modifications were revealed by studies on the 2’-O-methylation at position 34 of the anticodon of tRNASer(sec). This tRNA is modified at multiple positions yet, ribose methylation at position 34 is influenced by the presence of several modifications on the same tRNA; the balance yielding two differentially modified species with predictably different functions (Kim et al., 2000).
The sequential nature of editing and modification was also shown in the first example of anticodon C to U editing of a tRNA in marsupial mitochondria (Janke and Paabo, 1993). Here, C to U editing at the second position of the anticodon (C35) in tRNAGlyGCC changes it to tRNAAspGUC (Borner et al., 1996), in addition, several other modifications were required for later forming queuosine at position 34 (Q34) (Morl, Dorner, and Paabo, 1995). More importantly deamination of C35 to U35 creates the sequence U33G34U35 and this motif is an essential recognition motif for TGT to incorporate Q into tRNA. These authors thus pointed out the sequential nature of this pathway (Morl, Dorner, and Paabo, 1995).
Despite the early studies, we were among the first to verbalize the potential role that modification cascades could play as regulatory mechanisms in what we termed “the interdependence model for editing and modification” (Rubio M.A., 2005). This idea was prompted by our discovery of the second example of C to U editing at the anticodon of tRNATrp, which is essential for decoding of the UGA codons in the mitochondria-encoded mRNAs of trypanosomes (Alfonzo et al., 1999). Our failure to reconstitute C to U editing activity with crude mitochondrial extracts led us to explore the modification content of the tRNA. This tRNA is encoded in the nucleus, but because the mitochondrial genome of trypanosomes is devoid of tRNA genes, the nucleus-encoded tRNA (as is the case for the majority of tRNA in trypanosomes) is imported into the mitochondria. We showed by using LC-MS/MS sequencing (as described in the previous session) that following import, tRNATrp undergoes a number of additional modifications in the anticodon loop and we suggested that stepwise addition of these may play a role in changing the structural landscape of the ASL, where each step may provide a slightly different structure that can be exploited for specificity by the different modification enzymes effectively creating a modification cascade. Notable among these modifications was the presence of 2-thouridine (s2U) at position 33, the previously thought “universally unmodified position” (Crain et al., 2002). We later found that s2U33 critically acts as a negative determinant for C to U editing in vivo, helping keep the balance between edited and non-edited tRNA; presumably necessary for decoding both tryptophan codons used in mitochondrial translation (Wohlgamuth-Benedum et al., 2009).
Most recently, the Phizicky laboratory showed that modifications at position 32 in several yeast tRNAs could influence modifications at position 34 and they themselves could be affected by the modification status at position 37. Creating a modification cascade that can determine the final modification content on a given molecule (Paradiso, Carney, and Freeman, 1989). Similar examples followed with Q34 formation, which stimulates formation of m5C38 in the D. discoideum system (Tuorto et al., 2018). Perhaps the most extreme case of modification interdependence comes from our studies of modification at position 32 of tRNAThr in T. brucei, where an encoded C32 is first methylated to m3C and later deaminated to m3U in a reaction whose sequence is akin to the previously mentioned m1I formation in archaeal tRNAs (Rubio et al., 2017; Rubio et al., 2006). What makes these reactions remarkable is the fact that both the methylase and deaminase are in complex and one cannot have one activity or the other without both enzymes being present. In fact, the requirement for these two seemingly independent enzymes in the reaction is so strict, that the deaminase cannot form m3U even if a previously m3C-containing transcript is provided, unless the methylase is still present. How this strict interdependence is achieved is not yet clear and will require further structural and enzymology studies. Given the increasing number of examples of interdependent modifications, the Phizicky group has proposed a very interesting model for modification circuits, which provides by extension an evolutionary look at why interdependence and circuits arise (Han and Phizicky, 2018).
Intracellular localization of RNA modifying enzymes and RNA transport dynamics affecting modifications
One of the signatures of eukaryotic cells is their intricate intracellular membrane systems, which creates distinct compartments, including the genome-containing organelles (chloroplast and mitochondria). All the RNAs used for cytoplasmic translation are encoded in the nuclear genome. While the mitochondria and/or chloroplast contain their own genomes, and, owing to their evolutionary ancestry, maintain a translational system that most resembles that of bacteria. However, a growing theme in biology is the fact that cytoplasmic tRNAs are imported into the mitochondria and chloroplast, thus these organelles may contain a mixture of nucleus-encoded and organelle-encoded tRNAs (Rubio and Hopper, 2011). What this degree of intracellular compartmentalization then implies is the existence of robust and dynamic transport systems that move many molecules around. mRNAs and tRNAs are no exception and this concept is relevant when thinking of subcellular locations of modification events. Although it has always been clear that nucleus-encoded tRNAs have to be exported to the cytoplasm to engage in protein synthesis, unlike mRNAs, many years ago, the Yoshihisa and Hopper laboratories independently reported on the remarkable discovery that tRNAs can be transported back to the nucleus by the mechanism of retrograde transport (Rubio and Hopper, 2011; Shaheen and Hopper, 2005). In turn retrograde transport could be influenced by the nutrient status of the cells and under conditions of nutrient deprivation, tRNAs are retained in the nucleus as if waiting for a better day (Hurto et al., 2007; Shaheen et al., 2007). It is now accepted that retrograde transport is truly constitutive. Along these lines, many years later, the impact nuclear transport in an out of the nucleus has on tRNA modification was first revealed in yeast. The biosynthesis of the hypermodified nucleotide wybutosine involves several sequential enzymatic reactions starting with formation of m1G37 in the anticodon loop of tRNAPhe (Figure 3). It turns out that Trm5, the enzyme catalyzing m1G formation, localizes exclusively to the nucleus of S. cerevisiae and since this enzyme cannot act on an intron-containing tRNA, tRNAPhe is first exported to the cytoplasm to get spliced, then imported to the nucleus to get m1G and finally re-exported to the cytoplasm where the next 3 enzymes in the wybutosine pathway reside (Ohira and Suzuki, 2011).
Figure 3. The connection between retrograde nuclear transport and modifications.
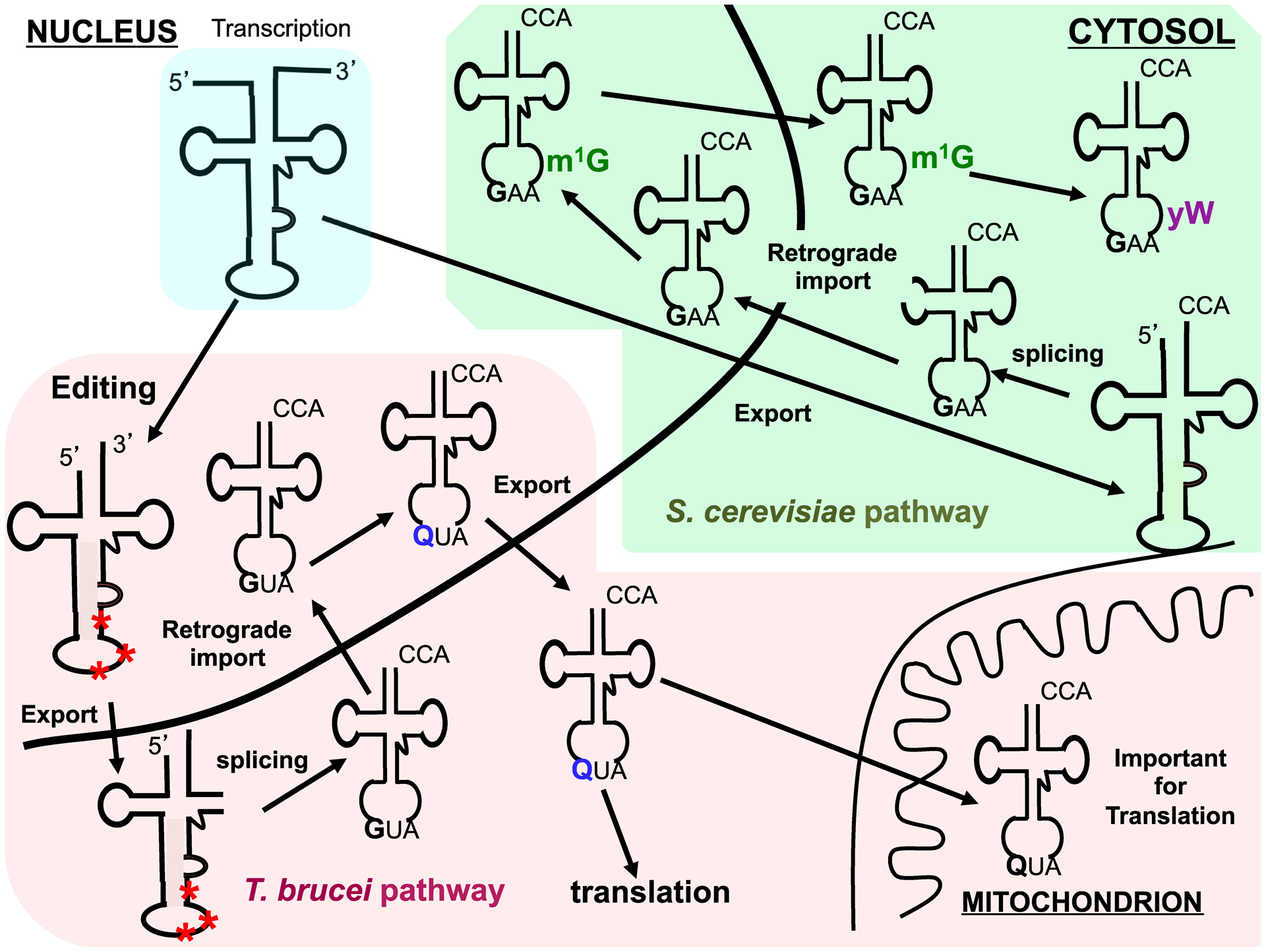
The pathways shown are exemplars of how intracellular transport may influence modification content of tRNAs. These particular examples start with an intron-containing tRNA following transcription. In both the T. brucei (highlighted in pink) and S. cerevisiae (in green) pathways. In the T. brucei tRNATyr splicing is cytoplasmic and the intron undergoes nuclear editing (red asterisk) prior to export, after cytoplasmic splicing the tRNA goes back to the nucleus to get queuosine (Q, blue). Q-containing tRNA is preferentially imported into the mitochondrion, where it is important for mitochondrial translation. In the S. cerevisiae pathway, following cytoplasmic splicing the tRNAPhe goes back to the nucleus to get N1-methylguanosine (m1G37, green) then is re-exported to the cytoplasm to undergo 4 additional sequential modifications leading to the formation of wybutosine (yW, purple) 37 as the final product.
Most recently, a similar pathway affecting queuosine formation at the first position of the anticodon of tRNATyr in T. brucei was described (Kessler et al., 2018). Like the S. cerevisiae system, tRNATyr in T. brucei has an intron and the splicing machinery is also cytoplasmic. In this case, tRNA-guanine transglycosylase (TGT), the key enzyme in queuosine incorporation into tRNA, resides in the nucleus and again is not able to modify an intron-containing tRNA. In this case the intron-containing tRNA undergoes non-canonical editing in the nucleus following transcription (Rubio et al., 2013). The edited tRNA is then exported to the cytoplasm to get spliced and then goes back to the nucleus to get Q (Figure 3). The T. brucei situation; however, is even more complicated. Since the mitochondrial genome of trypanosomes does not encode tRNA genes, the same tRNA is also imported into the mitochondria for organellar translation. Remarkably, the import pathway favors a version of tRNATyr that is fully modified with Q and almost 100% of the mitochondria-imported tRNATyr has the modification (Paris and Alfonzo, unpublished results).
Here we highlighted cases of how intracellular transport can influence the modification content of tRNAs. We note that such effects may not be unique to the two examples mentioned or for that matter to simply yeast and trypanosomes. What those two examples have in common is the fact that both modifications are easily tractable by molecular biology approaches and thus easy to detect. However, have no doubts that depending on the localization of a given modification enzyme and the maturation pathway for a particular tRNA, many more examples like this will be revealed. It is our prediction that even in systems where most tRNA processing events, other than modifications, occur in the nucleus, trafficking will still impact modification content given the fact that mechanisms such as retrograde transport appear to occur in systems where for example tRNA splicing is nuclear (Kessler et al., 2018; Schwenzer et al., 2019; Dhakal et al., 2019).
Although, based on current knowledge, nucleus-encoded mRNAs cannot reenter the nucleus after cytoplasmic export; neither can they be transported into mitochondria, still it is clear that different subcellular compartments plays unique roles in regulating their modification status. A clear example of this is m7G at the mRNA CAP. RNMT, the enzyme catalyzing this modification, is exclusively localized in the nucleus (Gonatopoulos-Pournatzis and Cowling, 2014). m7G has been considered constitutive until fairly recent work suggesting that modulation of RNMT activity plays a role in certain disease states such as cancer (Cowling, 2010; Linder and Jaffrey, 2019). It is indeed reasonable to think that regulated formation of m7G could dictate preferential processing of subsets of mRNAs in homeostatic conditions, as this modification is essential for the ability of mRNAs to be exported from the nucleus and also to engage the ribosome once in the cytoplasm. The story of possible regulatory roles of the CAP has also become far more complex with the recent discovery of cytoplasmic re-capping, where even if an mRNA reaches the cytoplasm and looses the CAP, it can be added back by cytoplasmic capping activities that are independent of the nuclear capping enzyme (Mukherjee et al., 2012). Thus, cellular sub-compartmentalization clearly increases the possibility and extent of regulated CAP formation with obvious ramifications for regulation of translation.
Taken together, be it to differentially affect the modification content of tRNA or mRNA, intracellular transport systems and their connection to environmental cues provide a nuanced way of constantly changing the diversity and quantities of modified RNAs available in the cytoplasm and organelles, in a manner that could influence codon-biased translation. For instance, it is imaginable that nuclear retention of a particular tRNA, harboring modifications that the same tRNA does not have in the cytoplasm, may directly impact translational read-out. In such a scenario, differential intracellular partitioning of the modification enzymes themselves is critical. This situation can then quickly change in response to variations in environmental cues, releasing the retained tRNAs back to the cytoplasm and providing a quick translational response based on the availability of the differentially modified species.
Downstream effects of modifications
Considering the immediate impact that modifications may have on substrate recognition the implications for downstream RNA function and therefore translation could be significant. For instance, mRNA modifications may differentially impact potential interactions between proteins that act downstream from the modifying enzyme. These may include not only “professional” RNA binding proteins, which directly bind the modified nucleotide; these may also affect other proteins (enzymes) involved in degradation and other processing events. For example, the fate of m6A-modified mRNAs is under the control of RNA-binding proteins that can discriminate between unmodified and m6A-containing messengers (Zhao et al., 2020). The YT521-B homology (YTH) domain-containing proteins are a key family in this category. The members of this family that are not only cytoplasmic, but also ubiquitously expressed, are YTH domain family (DF) 1–3 (YTHDF1, YTHDF2 and YTHDF3), and YTHDC1 and YTHDC2. These three proteins share a highly conserved RNA binding domain, which accommodate m6A-modified RNA sequences (Xu et al., 2015). They also contain a large intrinsically disordered N-terminal domain, whose sequence is more divergent and distinctive of each YTHDF protein (Liao, Sun, and Xu, 2018). As there is no evidence for differential binding affinity of different YTHDF proteins to m6A-RNAs and their relative cellular concentration is unknown, it is currently unclear what dictates which YTHDF protein will preferentially bind a methylated mRNA site and speculations on cooperativity between different YTHDFs has been proposed. Indeed, it is predictable that one particular transcript can be bound by different YTHDF proteins on different m6A sites concomitantly occurring on an mRNA. However, it appears as each YTHDF has a quite distinct and at times opposite function, complicating our ability to interpret how a particular outcome can derive by binding of different proteins to the same mRNA (Wang et al., 2015). Indeed, YTHDF2 has clear roles in driving mRNA degradation by recruiting the CCR4-NOT deadenylase complex (Du et al., 2016), while YTHDF1 has been implicated in promoting translation by interacting with the translation initiation factor eIF3 (Liu et al., 2020). The function of YTHDF3 is still less defined and a role for this protein in strengthening either YTHDF2 or YTHDF1 function has been proposed (Shi et al., 2017). Although the overall evidence for an m6A-dependent regulation of mRNA stability and translation is strong, how this is molecularly coordinated and how changing cellular conditions affect the fate of m6A-modified mRNAs is still unclear.
Since the initial identification of the YTH domain-containing proteins as binders and regulators of m6A-modified mRNA function, the list of proteins with affinity for methylated transcripts is rapidly growing. Within these, Insulin Growth Factor 2 mRNA Binding Proteins (IGF2BP1, IGF2BP2 and IGF2BP3) are also cytoplasmic and can therefore dictate the ultimate fate of m6A-mRNAs (Huang et al., 2018). The ability of IGF2BPs to bind single stranded RNAs is mediated by two RNA recognition motifs and four K homology (KH) domains, and it has been long known. However, the discovery of IGFBPs as m6A binding proteins is more recent and mutagenesis experiments pointed at KH domain 3 and 4 as essential for this function (Huang et al., 2018). Generally, IGFBPs are proposed as mRNA stabilizers, action that could be mediated by recruitment of additional RNA binding proteins to m6A-modified mRNAs and sequestration of target mRNAs to stress granules (Huang et al., 2018). Again, how the different m6A-binding proteins compete or cooperate in vivo to overall coordinate mRNA translation is still a mystery and worthwhile of future investigation.
The question still remains, how modifications, if occurring in the coding sequence of mRNAs, affect protein synthesis, not only at the level of translational fidelity, but also at the level of translational efficiency. The same questions apply to modified tRNAs, but in this realm more is known. Relatively new techniques, like ribosome profiling, have helped define the role of some tRNA modifications at codon-level resolution. Recent studies by the Leidel laboratory have described the fact that decreases in the modifications at position U34 of tRNALysUUU, tRNAGlnUUC and tRNAGluUUG may have great impact on protein folding and then lead to activation of the unfolded protein response (Nedialkova and Leidel, 2015; Laguesse et al., 2015). Likewise, it has been shown that the same modifications, beyond any effect on translational speed, can affect reading-frame maintenance (Klassen, Bruch, and Schaffrath, 2017). Then again, even these stories can be far more complex than meets the eye given that potential overlaps between mRNA and tRNA modifications possible but yet to be uncovered. Possibly, situations where portions of an mRNA may adopt tRNA-like structures, which then would render them amenable to modifications by tRNA enzymes. Recently, it was reported that the tRNA methylase TRMT6/TRMT61A complex may also catalyzed formation of m1A of mRNAs (Safra et al., 2017). Given the aforementioned ability of modifications such as m1A to impair base pairing, when they occur in coding regions of mRNAs they may act as translational repressors. This then brings into question the importance of redundant and overlapping activities between tRNA and mRNA. However, it is worth noting, that such modifications so far occur at very low levels in mRNA, making it difficult to assess whether these are truly programmed or simply the result of off-target effects by enzyme promiscuity (Safra et al., 2017).
Connecting translation to metabolism: A case for tunable modifications
Given the fact that most modification enzymes rely on products and by-products of metabolism as a source of substrates, we previously raised the possibility of a well-coordinated connection between metabolites and translation via tRNA modifications. The same arguments could apply to modifications in just about every nucleic acid in cells. What makes tRNA and mRNA; however, such provocative targets are their direct yet transient involvement in protein synthesis. Inevitably, mRNA and tRNA are among the few nucleic acids that undergo constant flux to and from the ribosome. Implicit in this is then the fact that at each step of translational recycling, both molecules encounter, to different degrees, the cell milieu and even here their modification state may dictate their fate.
In terms of environmental effects, previously, the issue of codon-biased translation was elegantly highlighted in S. cerevisiae, where under stress, for example in the presence of alkylating agents, cells responded by increasing the levels of some modifications; these favored the translation of mRNAs encoding proteins that could potentially deal with the stressful situation (Chan et al., 2010; Chan et al., 2012; Deng et al., 2015). Thus in conditions of environmental stress, codon-biased translation may be the norm. An equally important question is what happens during “normal” growth, as defined here as growth during normal fluctuations of environmental conditions that does not necessarily constitute a situation of extreme or alarm for cells, for example, in between feedings in the daily cycle for many mammals. Recently it was reported that in S. cerevisiae there is a clear connection between the levels of sulfur-containing amino acids (cysteine and methionine) and translational efficiency (Laxman et al., 2013). Not surprisingly, as levels of these amino acids drop in the media, the levels of 2-thiouridine at the first position of the anticodon of tRNALysUUU, tRNAGluUUC and tRNAGlnUUG was also reduced. More importantly, there was an accompanying increase in the proteins responsible for the synthesis and salvage of those same amino acids. In this particular case, it can be argued that the cells are not struggling with the drops in sulfur-containing amino acids but rather sensing such drops in specific amino acid pools via tRNA modification to make compensatory adjustments to amino acid pool, indirectly affecting tRNA thiolation levels, translation and to maintain homeostasis (Laxman et al., 2013). Alas, such studies, perhaps because the enormity of the problem, only looked at translational efficiency. But as discussed below, it could also affect translational accuracy and even impact protein folding. Indeed, a later report showed that under amino acid sufficiency, thiolation-deficient mutants still showed phenotypes associated with starved cells. Strikingly, genes associated with phosphate homeostasis were down regulated, forcing a switch in sugar metabolism that allows cells to maintain homeostasis and grow optimally (Gupta et al., 2019).
Beyond effects on translational speed and reading-frame maintenance, connections exist between metabolism, modifications and translational fidelity. An interesting example is with the acetylation of tRNAs, a reaction that occurs at the first position of the anticodon of elongator tRNAMet in bacteria and position 12 in eukarya, but it is also found in 18s rRNA in eukarya. In E. coli, RNA acetylation is catalyzed by the tmcA enzyme, in eukarya by the orthologous enzymes Rra1p (S. cerevisiae) and NAT10 in mammals (Sharma et al., 2015; Taniguchi et al., 2018). In E. coli, there is interplay between ac4C34 in tRNAMet and another modification (lysidine, k2C34) in tRNAIle (Muramatsu et al., 1988). C34 acetylation in tRNAMet is important to prevent potential misreading of the AUA isoleucine codons for methionine. In turn, lysidine, as has been known for many years, occurs in a tRNAIle with anticodon CAU, which should read methionine codons. Lysidine formation converts the potential methionine tRNA to an isoleucine tRNA by not only making it a substrate for the isoleucyl tRNA synthetase, but also allowing C34 to pair like U34 (Muramatsu et al., 1988; Soma et al., 2003). Thus lysidine helps decode the AUA codons with the correct amino acid. Expectedly, based on extensive data from the Suzuki laboratory, a double mutant of both TilS (the lysidine enzyme) and tmcA (the acetylase) leads to major issues of translational fidelity. In terms of metabolism, what metabolites could affect such transactions may depend on the organism. Both E. coli and eukaryal enzymes act as true acetyltransferases, in that they use acetyl-CoA as the acetate donor (Taniguchi et al., 2018). Surprisingly, in Bacillus, the enzyme tmcAL, directly activates acetate with ATP, produces an acetyladenylate intermediate that is then added to the N4 position of C34 in a reaction akin to that performed by amino acyl tRNA synthetases; yet another example of converging evolution between synthetases and modification enzymes. Regardless, it raises the point of how changes in the levels of a small metabolite, be it acetyl CoA or acetate, may affect translational output to different degrees. As side note, it is important to remark on the fact that recently there has been much discussion on mistranslation, now accepted to occur in all cells, one must wonder how much of it is the direct result of fluctuations in tRNA modifications indirectly impacting synthetase and ribosome function (Taniguchi et al., 2018). Interestingly, ac4C has also been described in mRNAs, catalyzed by the same N-acetyltransferase (NAT10)(discussed above). Acetylation of cytidine appears to preferentially occur near the translation initiation codon in mRNA (Arango et al., 2018); however, it is unclear if the location of ac4C in mammalian cells is guided by snoRNAs (as described in yeast) (Jin et al., 2020), or if NAT10 acts in an RNA-independent manner. Whether cytidine acetylation can be reversed and/or if specific RNA binding proteins can recognize this modification is also unknown, although a role for ac4C in promoting translation efficiency has been proposed (Arango et al., 2018).
Perhaps one of the most relevant connections between metabolites, tRNA and translation is provided by the modified nucleoside queuosine (Q). This modification occurs at position 34 of tRNAHisGUG, tRNAAsnGUU, tRNAAspGUC and tRNATyrGUA in Bacteria and most Eukarya (Harada and Nishimura, 1972). Only bacteria have a biosynthetic pathway to generate Q, thus all Q found in the tRNAs of eukaryotes comes via salvage either from nutrients or from their associated microbiomes (Kuchino et al., 1976). Various studies have shown that Q plays critical roles in cell physiology and it even affect development in Drosophila (Zaborske et al., 2014). Given its placement at the first position of the anticodon of the aforementioned tRNAs, the function of Q is to affect translation, but so far its role in translation seems to differ between organisms and the given growth condition; in some cases it may affect translational speed, in other organisms it may affect codon choice. A study in Drosophila showed that the Q-containing tRNAs had a preference for the C-ending codons of their cognate amino acids and argues for an important function in translational accuracy (Zaborske et al., 2014). On the other hand and as mentioned before in mammals and in S. pombe, not only Q affects m5C38 formation, it is also critical for translational speed (Muller et al., 2019). Along these lines, analogous to the case of U34 thiolation, decreased levels of Q may also cause protein-folding problems and activate the unfolded protein response (Tuorto et al., 2018). Once again arguing for modification mediated codon optimality during protein synthesis. One thing is clear, given its role as a micronutrient, Q may well be the ultimate mediator of metabolism and translation via tRNA modification. It even connects the microbiome to eukaryotic cell physiology further enhancing the global metabolic impact of modifications.
Taken together the examples above raise an important overarching question: Are some modifications tunable? It is easy to imagine that given the potential for reversibility with some modifications, for example in the case of methylations and predictably acetylation, could it be possible than in addition to simply degrading the RNA for recycling, some modifications may be actively removed and then added back as needed. The potential ability of some modification enzymes like the example of the multi-substrate specific pseudouridine synthase (mentioned before), and even the demethylase FTO, may raise the connection between overlapping substrates and tunability to levels that are not currently well understood. Establishing such connections may further enhance our understanding of a system’s level of coordination between modification pathways and indeed reveal the complexity of such circuitry. This being the case, it can then provide fertile grounds for the regulation of RNA function especially in terms of protein synthesis. The most appealing aspect of tunability may then rest with providing a rapid response to abrupt changes in environmental conditions.
Concluding remarks
We have reached a point in the modification field where the more relevant but complex question is how the immediate effects of different modifications get integrated into a much broader metabolic context at a cellular and organismal level. In this realm, we began by raising the possibility that because common building blocks derived from various aspects of metabolism are used to synthesize both canonical and modified nucleotides, changes in canonical nucleotide pools can be inevitably linked to other aspects of nucleic acid synthesis and catabolism; such changes may directly or indirectly affect modifications and help modulate translation. For instance, one could envision a scenario whereby as the availability of the amino acids glycine and aspartate drop, it may lead to a reduction in the levels of purine nucleotides leading to effects on both DNA and RNA synthesis; in turn a slow down in nucleic acid synthesis may be sensed via those same amino acids impacting certain modifications on tRNA; further sensed at the translational level. As a compensatory response one may expect increases via mechanisms that ensure nucleotide pool stability, for example increased synthesis of salvage enzymes. Alternatively nucleotide pool and amino acid instability may lead to diminished levels of protein synthesis more globally.
Similarly and beyond what has been done with global studies of the effects of modification changes on translation, other more specific but equally impactful aspects may be missed. For example another building block required for canonical nucleotide biosynthesis is folate. It is known that s-adenosyl methionine is not the only methyl donor in cells and indeed there is now a growing family of folate-dependent methyltransferases. A recent report showed that the mitochondrial folate-dependent serine hydroxymethyltrasnferase 2 (SHMT2) pays critical roles in organellar tRNA methylation and in turn is important for mitochondrial translation (Morscher et al., 2018). Again, in this particular case availability of folate may not only impact canonical nucleotide synthesis and cytoplasmic modifications, but also affect mitochondrial function.
Another important open question is that of what happens at the molecular level once a differentially modified tRNA encounters differential modified codons. As the record shows, the result could be rather unpredictable. Even in the case with conversion of stop codons to sense codon via guided-pseudouridine formation (Karijolich and Yu, 2011), it is not clear as to how the tRNAs that are predictably involved managed to read the modified codons and even in the context of the ribosome the observed base pairing escapes comprehension based on the well-accepted pairing rules.
In the end, one would argue that even minor fluctuations in a particular metabolite, will impact cell biology via changes in modifications and general effects in nucleotide pools. Such effects are far more multifaceted and difficult to assess than meets eye and one must avoid pigeonholing the problem at some level. Ultimately, to understand the full impact of such multivariate system, one will have to undertake not only the current ribosome profiling, nucleoside mass spectrometry approaches, but combine them with detailed quantitative metabolomics. All this for starters should be done in controlled conditions where perturbations can be introduced both genetically and biochemically to then establish the plasticity of the system.
Footnotes
Declaration of interest statement
No potential conflict of interest is reported by the authors. This work was supported by the NIH under Grants [number GM132254 and GM084065–11] to JDA and NIH Grant [number HL136951] to FA.
References
- 1.Levy M, Miller SL. (1999). The prebiotic synthesis of modified purines and their potential role in the RNA world. J Mol Evol, 48, 631–7 [DOI] [PubMed] [Google Scholar]
- 2.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res, 46, D303–D07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane AN, Fan TW. (2015). Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res, 43, 2466–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zollner N (1982). Purine and pyrimidine metabolism. Proc Nutr Soc, 41, 329–42 [DOI] [PubMed] [Google Scholar]
- 5.Davis FF, Allen FW. (1957). Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem, 227, 907–15 [PubMed] [Google Scholar]
- 6.Yu CT, Allen FW. (1959). Studies on an isomer of uridine isolated from ribonucleic acids. Biochim Biophys Acta, 32, 393–406 [DOI] [PubMed] [Google Scholar]
- 7.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. (1965). Structure of a Ribonucleic Acid. Science, 147, 1462–5 [DOI] [PubMed] [Google Scholar]
- 8.Crick FH. (1966). Codon--anticodon pairing: the wobble hypothesis. J Mol Biol, 19, 548–55 [DOI] [PubMed] [Google Scholar]
- 9.Helm M, Alfonzo JD. (2014). Posttranscriptional RNA Modifications: playing metabolic games in a cell’s chemical Legoland. Chem Biol, 21, 174–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessler AC, Silveira d’Almeida G, Alfonzo JD. (2018). The role of intracellular compartmentalization on tRNA processing and modification. RNA Biol, 15, 554–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn LE, Tual-Chalot S, Stellos K, Accornero F. (2019). RNA epigenetics and cardiovascular diseases. J Mol Cell Cardiol, 129, 272–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PJ, Shi H, He C. (2017). Epitranscriptomic influences on development and disease. Genome Biol, 18, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature, 542, 475–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singhal RP. (1971). Modification of Escherichia coli glutamate transfer ribonucleic acid with bisulfite. J Biol Chem, 246, 5848–51 [PubMed] [Google Scholar]
- 15.Bhanot OS, Chambers RW. (1977). Bisulfite-induced C changed to U transitions in yeast alanine tRNA. J Biol Chem, 252, 2551–9 [PubMed] [Google Scholar]
- 16.Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet, 9, e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakin A, Ofengand J. (1993). Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry, 32, 9754–62 [DOI] [PubMed] [Google Scholar]
- 18.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature, 515, 143–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada S, Ueda H, Noda Y, Suzuki T. (2019). Transcriptome-wide identification of A-to-I RNA editing sites using ICE-seq. Methods, 156, 66–78 [DOI] [PubMed] [Google Scholar]
- 20.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods, 12, 767–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485, 201–6 [DOI] [PubMed] [Google Scholar]
- 22.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell, 149, 1635–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, Han D, Dominissini D, Dai Q, Pan T, He C. (2015). High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew Chem Int Ed Engl, 54, 1587–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer KD. (2019). DART-seq: an antibody-free method for global m(6)A detection. Nat Methods, 16, 1275–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galvanin A, Ayadi L, Helm M, Motorin Y, Marchand V. (2019). Mapping and Quantification of tRNA 2’-O-Methylation by RiboMethSeq. Methods Mol Biol, 1870, 273–95 [DOI] [PubMed] [Google Scholar]
- 26.Krogh N, Nielsen H. (2019). Sequencing-based methods for detection and quantitation of ribose methylations in RNA. Methods, 156, 5–15 [DOI] [PubMed] [Google Scholar]
- 27.Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, Cairns BR. (2019). Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci U S A, 116, 6784–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M. (2019). Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One, 14, e0216709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phizicky EM, Alfonzo JD. (2010). Do all modifications benefit all tRNAs? FEBS Lett, 584, 265–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. (2015). ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods, 12, 879–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat Methods, 12, 835–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLuckey SA, Van Berkel GJ, Glish GL. (1992). Tandem mass spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom, 3, 60–70 [DOI] [PubMed] [Google Scholar]
- 33.Pomerantz SC, Kowalak JA, McCloskey JA. (1993). Determination of oligonucleotide composition from mass spectrometrically measured molecular weight. J Am Soc Mass Spectrom, 4, 204–9 [DOI] [PubMed] [Google Scholar]
- 34.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. (1993). A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res, 21, 4577–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulines MJ, Limbach PA. (2017). Comparative Analysis of Ribonucleic Acid Digests (CARD) by Mass Spectrometry. Methods Mol Biol, 1562, 19–32 [DOI] [PubMed] [Google Scholar]
- 36.Ross RL, Cao X, Limbach PA. (2017). Mapping Post-Transcriptional Modifications onto Transfer Ribonucleic Acid Sequences by Liquid Chromatography Tandem Mass Spectrometry. Biomolecules, 7 [Google Scholar]
- 37.Thakur P, Estevez M, Lobue PA, Limbach PA, Addepalli B. (2020). Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases. Analyst, 145, 816–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hur S, Stroud RM. (2007). How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell, 26, 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Wei LH, Wang Y, Xiao Y, Liu J, Zhang W, Yan N, Amu G, Tang X, Zhang L, Jia G. (2019). Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci U S A, 116, 2919–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol, 7, 885–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature, 541, 371–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, Suzuki T. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science, 363 [DOI] [PubMed] [Google Scholar]
- 43.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell, 71, 973–85 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaccara S, Jaffrey SR. (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell, 181, 1582–95 e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei C, Gershowitz A, Moss B. (1975). N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5’ terminal of animal cell and virus mRNAs. Nature, 257, 251–3 [DOI] [PubMed] [Google Scholar]
- 46.Wei CM, Gershowitz A, Moss B. (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell, 4, 379–86 [DOI] [PubMed] [Google Scholar]
- 47.Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. (1975). Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A, 72, 1904–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. (2010). Characterization of hMTr1, a human Cap1 2’-O-ribose methyltransferase. J Biol Chem, 285, 33037–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM. (2011). 2’-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res, 39, 4756–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Constantinesco F, Motorin Y, Grosjean H. (1999). Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res, 27, 1308–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ. (2000). Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA, 6, 1306–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janke A, Paabo S. (1993). Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res, 21, 1523–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borner GV, Morl M, Janke A, Paabo S. (1996). RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J, 15, 5949–57 [PMC free article] [PubMed] [Google Scholar]
- 54.Morl M, Dorner M, Paabo S. (1995). C to U editing and modifications during the maturation of the mitochondrial tRNA(Asp) in marsupials. Nucleic Acids Res, 23, 3380–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubio MA A JD. (2005). Editing and modification in trypanosomatids: the reshaping of non-coding RNAs, Springer, Berlin, Heidelberg [Google Scholar]
- 56.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. (1999). C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J, 18, 7056–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crain PF, Alfonzo JD, Rozenski J, Kapushoc ST, McCloskey JA, Simpson L. (2002). Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA, 8, 752–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wohlgamuth-Benedum JM, Rubio MA, Paris Z, Long S, Poliak P, Lukes J, Alfonzo JD. (2009). Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J Biol Chem, 284, 23947–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paradiso MA, Carney T, Freeman RD. (1989). Cortical processing of hyperacuity tasks. Vision Res, 29, 247–54 [DOI] [PubMed] [Google Scholar]
- 60.Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Muller M, Ehrenhofer-Murray AE, Dittmar G, Grone HJ, Lyko F. (2018). Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J, 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubio MA, Gaston KW, McKenney KM, Fleming IM, Paris Z, Limbach PA, Alfonzo JD. (2017). Editing and methylation at a single site by functionally interdependent activities. Nature, 542, 494–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubio MA, Ragone FL, Gaston KW, Ibba M, Alfonzo JD. (2006). C to U editing stimulates A to I editing in the anticodon loop of a cytoplasmic threonyl tRNA in Trypanosoma brucei. J Biol Chem, 281, 115–20 [DOI] [PubMed] [Google Scholar]
- 63.Han L, Phizicky EM. (2018). A rationale for tRNA modification circuits in the anticodon loop. RNA, 24, 1277–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubio MA, Hopper AK. (2011). Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip Rev RNA, 2, 802–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaheen HH, Hopper AK. (2005). Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A, 102, 11290–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hurto RL, Tong AH, Boone C, Hopper AK. (2007). Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics, 176, 841–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. (2007). Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci U S A, 104, 8845–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohira T, Suzuki T. (2011). Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc Natl Acad Sci U S A, 108, 10502–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kessler AC, Kulkarni SS, Paulines MJ, Rubio MAT, Limbach PA, Paris Z, Alfonzo JD. (2018). Retrograde nuclear transport from the cytoplasm is required for tRNA(Tyr) maturation in T. brucei. RNA Biol, 15, 528–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rubio MA, Paris Z, Gaston KW, Fleming IM, Sample P, Trotta CR, Alfonzo JD. (2013). Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell, 52, 184–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwenzer H, Juhling F, Chu A, Pallett LJ, Baumert TF, Maini M, Fassati A. (2019). Oxidative Stress Triggers Selective tRNA Retrograde Transport in Human Cells during the Integrated Stress Response. Cell Rep, 26, 3416–28 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dhakal R, Tong C, Anderson S, Kashina AS, Cooperman B, Bau HH. (2019). Dynamics of intracellular stress-induced tRNA trafficking. Nucleic Acids Res, 47, 2002–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonatopoulos-Pournatzis T, Cowling VH. (2014). RAM function is dependent on Kapbeta2-mediated nuclear entry. Biochem J, 457, 473–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cowling VH. (2010). Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene, 29, 930–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Linder B, Jaffrey SR. (2019). Discovering and Mapping the Modified Nucleotides That Comprise the Epitranscriptome of mRNA. Cold Spring Harb Perspect Biol, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR. (2012). Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep, 2, 674–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Y, Shi Y, Shen H, Xie W. (2020). m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol, 13, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. (2015). Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J Biol Chem, 290, 24902–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao S, Sun H, Xu C. (2018). YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics, 16, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell, 161, 1388–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun, 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, Xiao H, Li L, Rao S, Wang F, Yu J, Yu J, Zou D, Yi P. (2020). The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res, 48, 3816–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res, 27, 315–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Huttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol, 20, 285–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nedialkova DD, Leidel SA. (2015). Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell, 161, 1606–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laguesse S, Creppe C, Nedialkova DD, Prevot PP, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, Thelen N, Thiry M, Close P, Chariot A, Malgrange B, Leidel SA, Godin JD, Nguyen L. (2015). A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev Cell, 35, 553–67 [DOI] [PubMed] [Google Scholar]
- 87.Klassen R, Bruch A, Schaffrath R. (2017). Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol, 14, 1252–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature, 551, 251–55 [DOI] [PubMed] [Google Scholar]
- 89.Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. (2010). A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet, 6, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. (2012). Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun, 3, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. (2015). Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet, 11, e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. (2013). Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell, 154, 416–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta R, Walvekar AS, Liang S, Rashida Z, Shah P, Laxman S. (2019). A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL. (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res, 43, 2242–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taniguchi T, Miyauchi K, Sakaguchi Y, Yamashita S, Soma A, Tomita K, Suzuki T. (2018). Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat Chem Biol, 14, 1010–20 [DOI] [PubMed] [Google Scholar]
- 96.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. (1988). Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature, 336, 179–81 [DOI] [PubMed] [Google Scholar]
- 97.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. (2003). An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell, 12, 689–98 [DOI] [PubMed] [Google Scholar]
- 98.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, Fox SD, Zengeya TT, Andresson T, Meier JL, Coller J, Oberdoerffer S. (2018). Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell, 175, 1872–86 e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin G, Xu M, Zou M, Duan S. (2020). The Processing, Gene Regulation, Biological Functions, and Clinical Relevance of N4-Acetylcytidine on RNA: A Systematic Review. Mol Ther Nucleic Acids, 20, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harada F, Nishimura S. (1972). Possible anticodon sequences of tRNA His , tRNA Asm , and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry, 11, 301–8 [DOI] [PubMed] [Google Scholar]
- 101.Kuchino Y, Kasai H, Nihei K, Nishimura S. (1976). Biosynthesis of the modified nucleoside Q in transfer RNA. Nucleic Acids Res, 3, 393–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. (2014). A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol, 12, e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muller M, Legrand C, Tuorto F, Kelly VP, Atlasi Y, Lyko F, Ehrenhofer-Murray AE. (2019). Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res, 47, 3711–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, Rabinowitz JD. (2018). Mitochondrial translation requires folate-dependent tRNA methylation. Nature, 554, 128–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karijolich J, Yu YT. (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature, 474, 395–8 [DOI] [PMC free article] [PubMed] [Google Scholar]


