SUMMARY
Salivary proteins are essential for maintaining health in the oral cavity and proximal digestive tract, and they serve as potential diagnostic markers for monitoring human health and disease. However, their precise organ origins remain unclear. Through transcriptomic analysis of major adult and fetal salivary glands and integration with the saliva proteome, the blood plasma proteome, and transcriptomes of 28+ organs, we link human saliva proteins to their source, identify salivary-gland-specific genes, and uncover fetal- and adult-specific gene repertoires. Our results also provide insights into the degree of gene retention during gland maturation and suggest that functional diversity among adult gland types is driven by specific dosage combinations of hundreds of transcriptional regulators rather than by a few gland-specific factors. Finally, we demonstrate the heterogeneity of the human acinar cell lineage. Our results pave the way for future investigations into glandular biology and pathology, as well as saliva’s use as a diagnostic fluid.
Graphical Abstract
In Brief
Saitou et al. present a detailed analysis of transcriptome variation among human fetal and adult salivary glands. Their analysis reveals specific developmental and regulatory processes, as well as cell-line heterogeneity, that shape the gland-specific functional variation.
INTRODUCTION
Saliva is the quintessential gatekeeper at the entry to the gastrointestinal tract (Ruhl, 2012). Itisa complex biofluid and exertsa multitude of important functions in the oral cavity and beyond that depend upon its repertoire of proteins. These functions include breakdown of dietary starch by the salivary enzyme amylase, provision of calcium phosphate to maintain mineralization of tooth enamel, and host defense against pathogenic microorganisms (Heo et al., 2013; Walz et al., 2009) while maintaining a beneficial commensal microbiome in the mouth (Cross and Ruhl, 2018; Dawes et al., 2015). Saliva also possesses physicochemical properties keeping the oral cavity moist, well lubricated, and forming a barrier against environmental and microbial insult, functions that are equally provided by saliva proteins, especially mucins (Frenkel and Ribbeck, 2015; Tabak, 1995). Thus, variation in the saliva proteome will have important biomedical consequences (Dawes and Wong, 2019; Helmerhorst and Oppenheim, 2007). At the extreme, malfunctioning of the salivary glands because of, for instance, radiation treatment of head and neck cancer or the relatively common autoimmune disease Sjögren’s syndrome, results in severe complications in oral health that debilitate patient quality of life (Mavragani and Moutsopoulos, 2020; Vissink et al., 2015). Therefore, understanding how the composition of the saliva proteome is attained and regulated remains an important avenue of inquiry.
A major complication in studying saliva and harnessing its proteome profile for diagnostic applications is the complexity of this oral biofluid, because it is a mixture of components derived from multiple sources. Saliva is predominantly synthesized and secreted by three major pairs of anatomically and histologically distinct craniofacial secretory organs: the parotid, submandibular, and sublingual salivary glands (Figure 1A). Each of these gland types produces a characteristic spectrum of salivary proteins that are thought to be predominantly based on their composition of mucous and serous acinar cells. These intrinsic proteins sustain most major functions of the saliva. In addition, saliva contains extrinsic proteins that originate in other organs and systems, including the bloodstream and cells lining the oral integuments (Ruhl, 2012; Yan et al., 2009). A multitude of studies have been conducted to catalog salivary proteins and distinguish intrinsic from extrinsic protein components (Grassl et al., 2016), including those that specifically investigated ductal secretions (Denny et al., 2008; Walz et al., 2006). These studies include proteomic analyses comparing whole saliva or saliva collected from the ducts of the parotid or submandibular/sublingual glands to body fluids such as plasma, urine, cerebrospinal fluid, and amniotic fluid (Loo et al., 2010). However, discrepancies among published datasets, likely due to variation in collection procedures, sample integrity, storage conditions, sample size, and analytical methods (Helmerhorst et al., 2018), have impaired the establishment of a robust catalog of saliva-typic proteins and their salivary gland origin—an outcome that has so far severely hampered the use of saliva as a physiological and pathophysiological research tool and as a reliable fluid for disease diagnosis (Ruhl, 2012).
Figure 1. Overview of the Transcriptome Analysis.
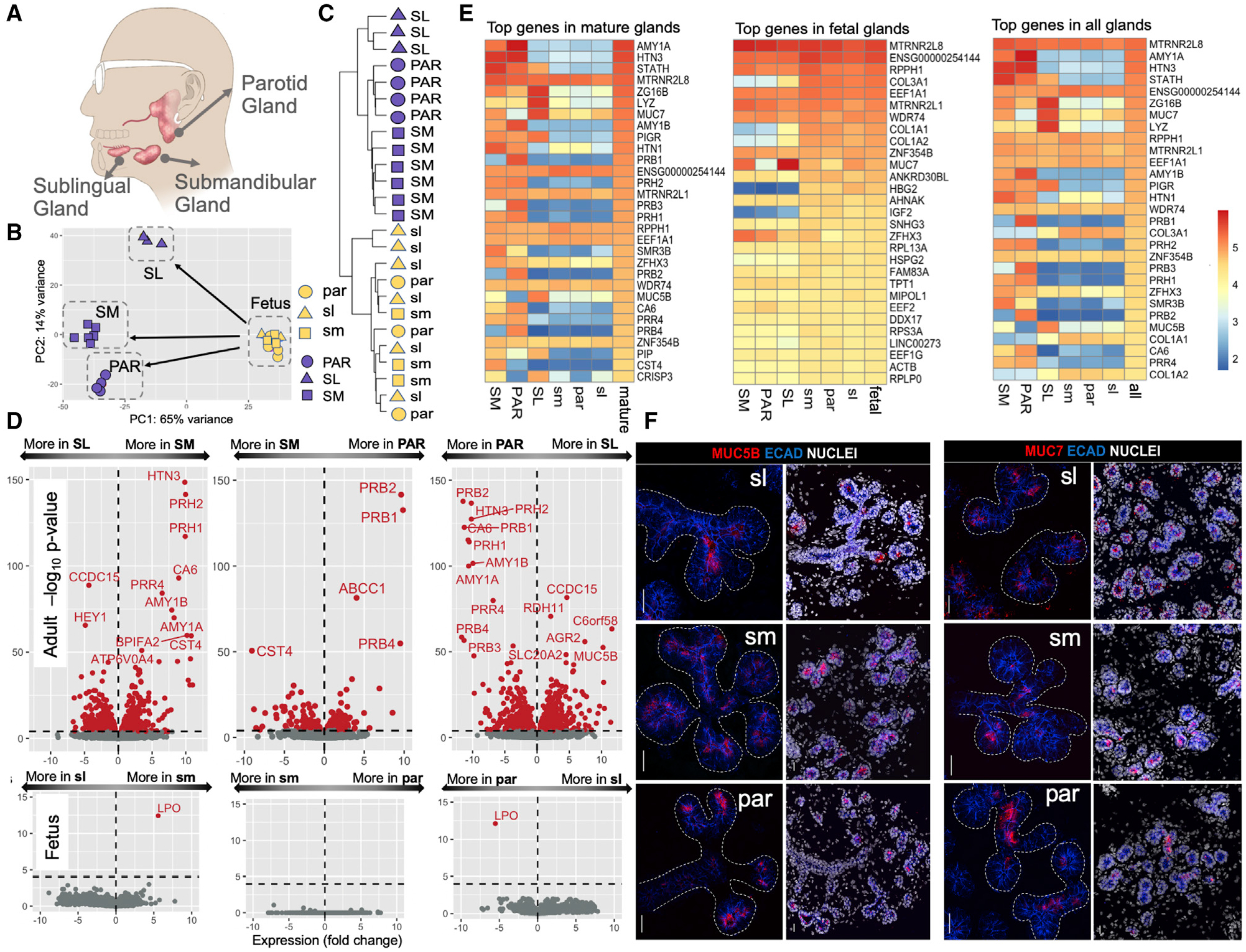
(A) Anatomical location of the three major glands in humans.
(B) Principal-component analysis of gene expression levels in adult and fetal salivary glands. Blue symbols, adult samples; yellow symbols, fetal samples. Triangle, square, and circle shapes represent the parotid (PAR), submandibular (SM), and sublingual (SL) glands, respectively (fetal gland types in lowercase letters).
(C) Hierarchical clustering analysis transcriptome data from the different adult and fetal gland types without a priori clustering information.
(D) Volcano plots showing the expression differences among gland types in a pairwise fashion for adult (top) and fetus (bottom). The x axis indicates gene expression log2 fold changes (log2). The y axis indicates a −log10 value of the adjusted p value. Genes with significantly different expressions among glands are indicated in red (adjusted p < 0.0001).
(E) Heatmaps of the log10 normalized expression values of gland-specific genes showing the 30 most highly expressed genes in the different mature and fetal gland types. The top gene, RN7SL1, was excluded because of its role as a housekeeping gene.
(F) Immunofluorescent localization of MUC5B (left panel) and MUC7 (right panel) in fetal glandular tissues. Left-side images of each panel show mucin (red) and E cadherin (blue) immunostaining without nuclei, and right-side images show a lower magnification of the same glandular region and include nuclei. ECAD, E cadherin. Scale bars, 25 μm.
To address this gap in knowledge, we sequenced the total RNA of 25 salivary gland samples collected from human adult and fetal submandibular (adult, SM; fetal, sm), sublingual (adult, SL; fetal, sl), and parotid (adult, PAR; fetal, par) glands. This dataset allowed us to analyze transcriptomes across gland type and developmental stage, compare the salivary gland transcriptome to that of other organ systems, and integrate the salivary transcriptome with available proteome data.
RESULTS
The Functional Specialization of Adult Salivary Glands Occurs during Late-Stage Development
To comprehensively identify gene expression differences among the three major salivary gland types, we conducted a transcriptome analysis of multiple healthy male and female SL, sl, SM, sm, PAR, and par glandular tissues (Figure 1A; Table S1). Human salivary gland development begins within 6–8 weeks, with the formation of a branched structure with clearly defined end buds (pre-acini) by 16 weeks and lumenized acini by 20 weeks (Kumagai and Sato, 2003). The period after 20 weeks is associated with cytodifferentiation and the presence of intercalated/striated ducts and is characterized as the last stage of salivary gland development (Ianez et al., 2010). Salivary glands are considered fully differentiated by 28 weeks, as noted by the presence of secretory vesicles and the expression of secretory protein BPIFA1/SPLUNC1 (Zhou et al., 2006). Here we used glandular tissues taken from 22 to 23 weeks of age and, based on these previous studies, define this age group as late-stage development.
We comparatively analyzed the expression levels of 167,278 transcripts consolidated into 40,882 coding and noncoding genes (Table S2). We could clearly differentiate mature glands from fetal glands without any a priori hypothesis based only on the first principal component of transcriptome data (Figure 1B). The second principal component of the transcriptome data evidently separated the mature gland types. However, the same analysis could not differentiate among fetal gland types. We verified these results using a hierarchical clustering analysis (Figure 1C), in which the transcripts of the mature glands clustered into a major branch distinct from that of the fetal glands. Moreover, we found that the transcripts of the mature glands branched up according to their glandular origin, whereas those of the fetal glands did not. We quantified these observations using Pearson correlation analysis (Figure S1). As expected for tissues composed of similar cell types, the PAR gland exhibited greater similarity in global gene expression to the SM than the SL gland.
To determine which transcripts account for the differences among the gland types, we conducted a comparative analysis of glandular transcripts (Figure 1D; Table S2) and found hundreds of transcripts that were differentially expressed among mature glands. Gene Ontology (GO) analysis of these transcripts showed that genes found predominantly expressed in the fetal tissues were significantly enriched in categories linked to growth and development, including cell cycle, cell division, and other fundamental cellular processes (Table S3). Differences among mature gland types mainly resulted from genes that, as defined by the Human Protein Atlas (https://www.proteinatlas.org/), code for secreted proteins.
We confirmed the gland-type-specific and abundant expression of a limited number of genes coding for secreted proteins that are also found in saliva in high abundance (Ruhl, 2012) (Figure 1E). Several other genes encoding abundant secreted proteins were expressed by two gland types only or by one gland type exclusively (Table 1). For example, MUC7 was highly enriched in the SL and to a lesser extent in the SM but virtually absent from the PAR gland. We also identified several genes coding for secretory proteins that had not been previously described to differ among gland types. One example is cysteine-rich secretory protein 3 (CRISP3), an early response gene that may participate in the pathophysiology of the autoimmune lesions of Sjögren’s disease (Tapinos et al., 2002) and is expressed by human labial glands (Laine et al., 2007), was highly expressed by the SL and to a lesser extent by the SM but absent from the PAR glands.
Table 1.
Genes Expressed in Abundance in Salivary Glands
| Top Transcribed Genesa Specifically Expressed in Salivary |
|---|
| STATH, HTN3, HTN1, AMY1, SMR3B, PRH2, ENSG00000254144, CST4, RPPH1, CST1, PRB1, PRB3, PRB4, C6orf58, MUC19, ENSG00000225840, CD24P4, RIMBP3C, LINC00273 |
| Top 20 Transcribed Genes in the SLb |
| RN7SL1, LYZ, ZG16B, MUC7, MTRNR2L8, PIGR, MUC5B, ENSG00000254144, CRISP3, STATH, C6orf58, RPPH1, MTRNR2L1, EEF1A1, PIP, FCGBP, WDR74, DMBT1, ZNF354B, IGHA1 |
| Top 20 Transcribed Genes in the PARb |
| RN7SL1, AMY1A, HTN3, AMY1B, PRB1, MTRNR2L8, PRB3, STATH, PRB2, PRH1, PRH2, PRB4, HTN1, CA6, ENSG00000254144, PIGR, MTRNR2L1, PRR4, EEF1A1, RPPH1 |
| Top 20 Transcribed Genes in the SMb |
| RN7SL1, STATH, HTN3, MTRNR2L8, HTN1, AMY1A, SMR3B, ZG16B, PIGR, PRH2, MTRNR2L1, ENSG00000254144, MUC7, CST4, RPPH1, ZFHX3, PRH1, AMY1B, CST1, EEF1A1 |
| Additional Highly Transcribed Genes of Reported Functional Relevance in Gland Development, Physiology, or Pathologyc that Show Salivary-Gland-Type-Specific Expression |
| KLK1, LRP1B, MUCL1, CNTN5, SPP1, LRP2, EDN3, AGR2, TFF1, GALNT12, GALNT13, FSTL1, COL3A1, SPARC, PLXND1, POSTN |
| Additional Highly Transcribed Genes of Reported Functional Relevancec Coding for Non-Secreted Protein Products |
| RDH11, MAOB, FEZF2, LIM1XB, SLC5A5, DNER, PGR, ABCC1, GALNT13, NKX2-3, MCFD2, TCN1, FURIN |
| Top 10 Proteins Abundantly Found in Whole-Mouth Saliva that Likely Originate from Extrinsic Sources Such as Blood Plasma or Epithelial Linings of the Oral Cavity |
| ALB, IGHG2, AZGP1, IGHG1, ACTG1, IGKV3-20, IGKV4-1, IGKV4-5, S100A9, KRT1 |
| Top 10 Transcribed TF Genesa in Salivary Glands |
| ZFHX3, ZNF354B, LTF, XBP1, TFCP2L1, EHF, FEZF2, SON, NFIB, FOXO3, JUN, ETV1, FOS |
| Additional Highly Transcribed TF Genes of Reported Functional Relevancec in Gland Development, Physiology, or Pathology |
| NKX3-1, BHLHA15, FOXA1, NKX2-3, HEY1, YAP1, TP63, SOX2, SOX2, SOX9, SOX10, FOXC1, FOXD3, CBX2, SOX11, ZBTB16, KLF9 |
Listed are the top 10 genes expressed in each major gland type. Because some of these genes overlap, the total of genes listed here is lower than 30. For a more systematic look into their expression in salivary glands, see Tables S2 and S4.
Underlined gene names designate those that are predominantly expressed in the respective gland category (log2 fold change > 2).
A more detailed description of these genes, including references, is provided in the main text.
We found that kallikrein 1 (KLK1), low-density lipoprotein receptor-related protein 1B (LRP1B), mucin-like 1 (MUCL1/SBEM; Miksicek et al., 2002), carbonic anhydrase (CA6; Parkkila et al., 1990), and C6orf46/SSSP1 (skin and saliva secreted protein 1; cell origin of protein is unknown; Gerber et al., 2013) were expressed by the PAR and SM glands and absent from the SL glands, whereas contactin 5 (CNTN5) and secreted phosphoprotein 1/osteopontin (SPP1) were restricted to the PAR and SL glands, respectively. A small fraction of secreted genes was also found to be exclusively expressed by only one adult gland type. For example, transcripts for low-density lipoprotein receptor-related protein 2 (LRP2/megalin), a multiligand uptake receptor that is involved in protein reabsorption (Christensen and Birn, 2002), were only found in PAR tissue; endothelin 3 (EDN3; Gurusankar et al., 2015) was highly enriched in the SM; and the mucous components FCGBP (Pelaseyed et al., 2014), AGR2 (Park et al., 2009), and trefoil factor 1 (TFF1; Chaiyarit et al., 2012), along with a gene of unknown function enriched in mucous tissues, C6orf58/LEG1 (Pelaseyed et al., 2014), were restricted to the SL. Some proteins that these genes encode are found in saliva (e.g., C6orf58/LEG1; Ramachandran et al., 2008), whereas others have a negligible presence (e.g., LRP2/megalin), although whether this deficiency results from protein degradation or whether they are simply not secreted into the ductal lumina is yet to be determined.
We found several genes that are not secreted but still show remarkable gland-type specificity. For example, the SL gland was enriched in transcripts for retinol dehydrogenase 11 (RDH11) compared with the PAR and SM glands, whereas transcripts encoding enzymes such as the dopamine-degrading monoamine oxidase B (MAOB), transcription factors (TFs) FEZF2 (a regulator of cell differentiation; Takaba et al., 2015; Zhang et al., 2014) and LIM1XB (LIM homeobox TF 1 beta), co-transporter SLC5A5, and growth factor or steroid receptors such as DNER (Delta and Notch-like epidermal growth factor-related receptor) and progesterone receptor (PGR) were almost exclusively expressed by the SM and PAR glands. The multi-drug resistance gene ABCC1 was highly enriched in the PAR; GALNT13, an initiator of O-linked glycosylation of mucins, was enriched in the SM; and the TF NKX2–3, which is required for murine sublingual gland development (Biben et al., 2002), was almost exclusive to the SL. A few of these protein-coding genes were previously reported to be specific for other organ systems not included in the genotype-tissue expression (GTEx) database, e.g., placenta-specific protein (PLAC4).
Adult gland types also differed significantly in the expression of immune-related secretory genes. Through GO analysis (Table S3), we found distinct complement cascades and immunoglobulin production pathways (e.g., IGHV1–58 and C6) that were shared by the PAR and SM but were different from those shared by the SM and SL. The SM and SL showed enhanced levels of transcripts for genes assigned by GO to the categories “acquired immunity” (secretory immunoglobulin A [S-IgA] and immunoglobulin G [IgG]) and “innate immunity” (lysozyme, BPI, BPI-like, and PLUNC proteins; cystatins; mucins; peroxidases; statherin [STATH]; and others). This observation raises the possibility that the SM and SL glands may provide a constant background level of acquired and innate immunity in the oral cavity that is maintained independently of salivary flow stimulation through food intake and chewing activity. In that context, it is of interest that glandular inflammatory conditions (sialadenitis) show a predilection for certain gland types. For example, Heerfordt syndrome causes parotitis (Takahashi and Horie, 2002), whereas chronic sclerosing sialadenitis predominantly affects the SM glands (Gupta et al., 2015).
The proportion of total gene transcripts encoding secreted proteins was significantly higher in mature glands than in their fetal counterparts (p < 0.05, Mann-Whitney test) (Figure S2). Yet several secretory genes present in saliva were also expressed at significant levels in fetal glands, albeit at lower levels than in adult glands (Figures 1E and 1F). Many of those genes did not match the tissue-specific expression patterns of the adult organs. For example, transcripts for MUC7 and MUC5B, which are expressed exclusively by the SM and SL glands, were expressed by all fetal gland types (Figure 1E, left panel). Such an outcome hints at the possibility of unknown functions of mucin genes during fetal development.
There Is Extensive Retention of Gene Transcripts from Fetal to Adult Stages in All Mature Gland Types but Most Pronouncedly in the SL Gland
We next analyzed our RNA sequencing (RNA-seq) datasets for genes that were retained or depleted during maturation of the salivary gland types. Overall, we found 7,166 genes were expressed at similar levels at both fetal and adult stages (Figure 2A; also see Figure 1E). These globally expressed genes are enriched for functions related to organ development and adult homeostasis and physiology (Table S3). Among the highly retained gene transcripts, we identified factors, e.g., fibroblast growth factors 1, 7, and 10 (Mattingly et al., 2015), that in mice are reduced in expression during salivary gland formation and are known to promote salivary gland development in these animals, suggesting species variation in gene retention during gland development.
Figure 2. Categorization of Genes Based on Their Expression Trends in Fetal and Mature Salivary Glands.
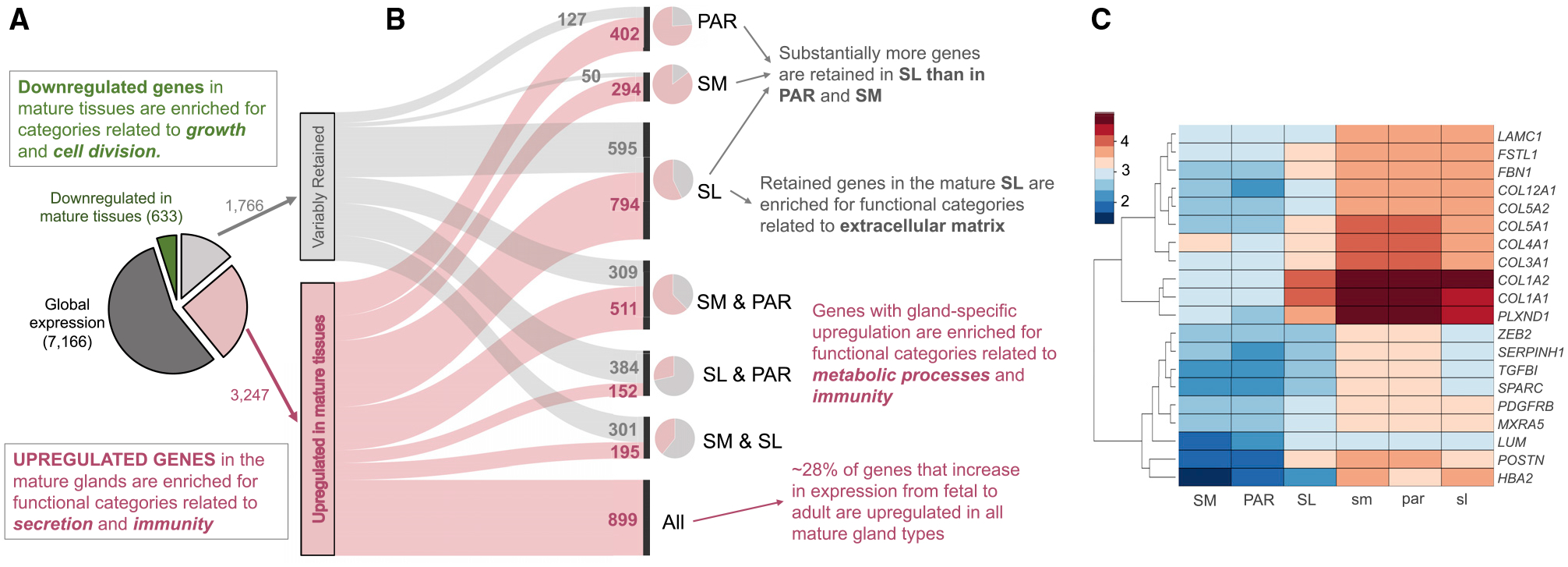
(A) Pie chart on the left indicates the proportions and numbers of genes (i.e., expressed >100 DESeq2 normalized counts [NCs]) that showed no significant differences (adjusted p > 0.0001, dark gray), were downregulated (adjusted p < 0.0001, green), or were either retained or upregulated in mature salivary glands compared with their fetal counterparts.
(B) Parallel set graph to summarize the breakdown of genes that show variable gene expression in adult glands, indicating how the differential transcriptome repertoires of mature glands are a product of gland-specific retention and upregulation of gene expression. Smaller pie charts at the right side of the parallel set graph indicate the proportion of retained and upregulated genes for each mature gland type.
(C) Heatmap showing genes highly expressed in fetal glands (>1,000 NCs) shown in relative abundance (Z score) that are retained in only one mature gland type from its fetal counterpart, but not in the other two mature gland types.
Despite extensive similarities in gene retention among all adult gland types, the SL gland stands out, because it retains a group of additional 595 genes from fetal to adult stages (Figure 2B). The most highly expressed genes in this group are primarily related to extracellular matrix formation and function (Figure 2C; Table S3). Some genes, including those coding for collagen 1 and 3 isoforms (e.g., COL3A1), as well as SPARC/osteonectin, were retained at fetal-like transcript levels (10- to 100-fold higher than in the SM and PAR). These results suggest that the SL retains a more fetal-like extracellular matrix that may guide stem cell-mediated repair, as was suggested for other organ systems.
We also identified several highly abundant genes not related to the extracellular matrix that were retained in the SL gland compared with the PAR and SM glands. These included the extracellular glycoprotein Fst-SPARC family member follistatin-like 1 (FSTL1), which is an essential regulator of tracheal formation and lung epithelial cell maturation (Geng et al., 2011), and the receptor for semaphorin class 3 ligands plexin D1 (PLXND1), which has multiple roles during development (e.g., synaptogenesis, heart formation, and vasculogenesis) and is heavily associated with Moebius syndrome, a developmental neurological disorder that is characterized by paralysis of the facial nerves and variable other congenital anomalies (Tomas-Roca et al., 2015). In regard to Moebius syndrome, patients show salivary gland dysfunction (Martins Mussi et al., 2016), although whether the tissues are affected at morphological levels is unknown. In addition, periostin (POSTN), which is highly retained in the SL, has been implicated in stem cell regulation in multiple tissues, including bone (Niklason, 2018), heart (Hudson and Porrello, 2017), pancreas (Hausmann et al., 2016), and tendon (Noack et al., 2014).
Few fetal transcripts were absent from all mature gland types (Table 1). Those few that were included gene transcripts involved in fetal blood (e.g., hemoglobin gamma A [HBGA]), embryonic development (e.g., insulin-like growth factor 2 [IGF2]), and cell proliferation (e.g., topoisomerase [DNA] II alpha [TOP2A]), as well as several TFs known to regulate developmental processes in other organ systems (e.g., SOX11) (Huang et al., 2016). HBGA is a fetal globin gene known to be absent from adults. Thus, its low transcript level in adult glands (~20 transcripts in adults compared with ~2,500 in fetal tissue) ensures the rigor of our study.
The Diverse TF Repertoire of Mature Salivary Glands May Shape Hotspots of Hundreds of Genes with Salivary-Gland-Specific Expression
We next tested the hypothesis that TFs display gland-specific gene expression. To address this hypothesis, we investigated the expression patterns of hundreds of TFs that were (1) highly abundant in each gland type at each developmental stage, (2) showed salivary-gland-specific expression, or (3) had been previously implicated in salivary gland development, disease, or cancer (Figure 3A; Table 1; Table S4). More than 60% of known TFs (1,025 of 1,648) were expressed (>100 DESeq2 normalized counts [NCs]) by at least one of the salivary glands, with 64% (661) of them expressed in each of the fetal and mature glands and thus suggestive of conserved function during maturation and homeostasis.
Figure 3. The Diverse Transcription Factor (TF) Repertoire of Mature Salivary Glands May Shape Hotspots of Salivary-Gland-Specific Expression across the Genome.
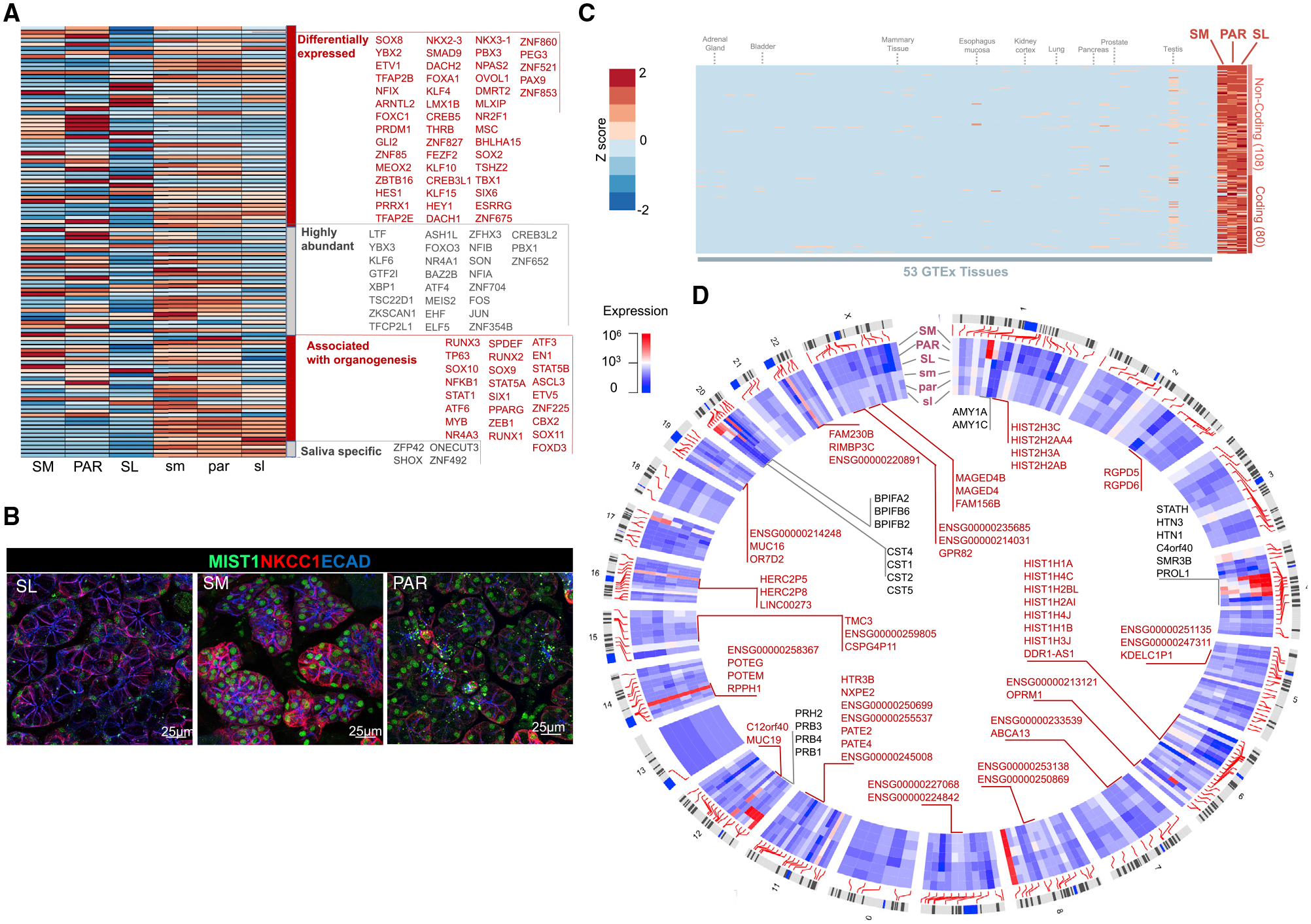
(A) Heatmap of expression levels of TF genes (as listed in TF2DNA database; Pujato et al., 2014) across fetal and mature salivary gland tissues. Four categories of TFs are shown in the heatmap: TFs that are (1) differentially expressed (p < 0.0001) among mature glands, (2) abundant (>2,000 NCs) in adult or fetal glands, (3) previously associated with organogenesis, and (4) salivary gland specific (that is, >100 NCs in the salivary glands but negligible expression in all 53 GTEx tissue [<10 transcripts per million (TPM)]). LTF, a secreted protein in saliva, is listed here because one of its isoforms, delta lactoferrin, displays TF activity (He and Furmanski, 1995; Mariller et al., 2007).
(B) Immunofluorescent analysis of TF BHLHA15/MIST1 in adult glandular tissues. The SM and PAR cells are highly enriched for MIST1 compared with the SL cells. NKCC1/SLC12A2, Na-K-Cl cotransporter 1. Scale bar, 25 μm.
(C) Heatmap of expression levels of genes in mature salivary glands (>100 NCs) that show negligible expression in other tissues and organs (expression in all 53 GTEx tissues < 10 TPM). The specific tissues in the GTEx database used for this analysis are listed in Table S5. Epithelial or secretory tissues and organs important for comparison to salivary glands are indicated on top of the heatmap. Deviation from the mean expression for each column is shown as a Z score with a scale similar to that used in (A).
(D) Circos plot showing the locations of genes with salivary-gland-specific expression. These genes show considerable expression in the salivary glands (>100 NCs) but negligible expression in all 53 GTeX tissues. Clusters of genes located within 1 Mb of one another are pointed out with gene names inside the Circos plot. Genes that previously had not been reported within the context of salivary glands are indicated in red.
Our analysis identified a host of TFs previously shown to be essential regulators of salivary gland development in mice to also be expressed in developing human glands. These include regulators of acinar cell development (e.g., SOX2, SOX9, and SOX10; Athwal et al., 2019; Chatzeli et al., 2017; Emmerson et al., 2017); targets of FGF10 signaling (e.g., ETV5); regulators of duct formation, such as TFCP2L1 (Yamaguchi et al., 2006) and YAP1 (Szymaniak et al., 2017), and of basal stem cells such as TP63 (Song et al., 2018); and a recently discovered TF that promotes salivary organoid initiation from mouse embryonic stem cells (FOXC1; Tanaka et al., 2018). A group of TFs, including FOXD3, CBX2, and SOX11, was found to be exclusively expressed in fetal glands, indicative of roles in human salivary gland development. This latter group of TFs is of high interest to those studying organ bioengineering, wound repair, and cancer, because multiple markers present in fetal tissue are also expressed in various cancers (e.g., SOX11; Yang et al., 2019) and are required for regeneration and de novo generation of tissues (Miao et al., 2019; Sock et al., 2004), yet their exact functions remained unclear due to the absence of information on fetal organs. We also found several TFs that were far less abundant at the fetal stage than at the adult stage, suggestive of adult-specific functions. Examples are BHLHA15/MIST1, the master regulator of the secretory program and secretory cell architecture (Lo et al., 2017); KLF9, a negative regulator of epithelial and tumor cell proliferation (Shen et al., 2014; Spörl et al., 2012); and ZBTB16, which affects diverse signaling pathways, including cell cycle, differentiation, programmed cell death, and stem cell maintenance (Xiao et al., 2016).
SL tissue, compared with PAR and SM tissue, demonstrated a 5- to 10-fold enrichment for transcripts of TFs known to regulate mucous cell formation (Table 1), including FOXA1 (Ye and Kaestner, 2009), NKX2–3 (Biben et al., 2002), and NKX3–1 (Schneider et al., 2000), as well as of TFs regulating cell differentiation, including HEY1, a downstream effector of NOTCH signaling (Nandagopal et al., 2018). In addition, we found TFs that are routinely used as markers to define gland maturity independent of gland type. Those include BHLHA15 (MIST1), which indeed shows mature-gland-specific expression yet exhibits differential expression at both mRNA and protein levels among mature gland types, with the SM gland showing the highest expression and the SL gland showing the lowest (Figures 3A and 3B). Altogether, our results suggest that rather than a few gland-specific TFs driving functional diversity, specific dosage combinations of dozens, if not hundreds, of TFs likely shape the transcriptome variation of individual adult salivary glands.
To identify genes that are expressed specifically in salivary glands, we compared transcript levels in salivary glands with those of 54 other tissues in the GTEx portal, including other epithelial organs that secrete fluids, such as the pancreas, mammary tissue (non-lactating), and intestine (Battle et al., 2017) (Figures S3–S5; Table S5). This analysis identified 188 transcripts (Figure 3C; Table S4) with observable gene expression (>100 NCs) in adult salivary glands but negligible (<10 TPM) expression in 53 other tissues and organs reported in the GTEx database.
Of the 188 genes identified as salivary gland specific, 80 are predicted to be protein coding based on RefSeq (O’Leary et al., 2016) (Table 1; Table S4). Besides genes encoding proteins abundantly found in saliva (e.g., HTN, MUC7, PRB, and AMY1), most of these salivary-gland-specific genes are long non-coding RNAs (108 genes) that to our knowledge have not been identified in the salivary gland context. They include LINC00273, a possible regulator of lung cancer metastasis (Jana et al., 2017), and AC092159.2, which has been suggested to play a role in metabolic processes (Hu et al., 2019). Given the multiple suggested roles of long non-coding RNAs in other organ systems, these transcripts may play a role in controlling nuclear architecture and transcription in the nucleus, as well as in modulating mRNA stability, translation, and posttranslational modifications in the cytoplasm of salivary gland cells.
We mapped dozens of gene clusters across the human genome that show salivary-gland-specific expression (Figure 3D; Table S4). Some of these, such as SCPP (Xu et al., 2016), CST (Dickinson et al., 2002), BPIFA (Zhou et al., 2006), and PRB (Stubbs et al., 1998) gene clusters, contain genes encoding proteins secreted in saliva (see also Figures 1D and 1E). This cohort of additional loci harboring salivary-gland-specific gene sets offers opportunities for investigating the regulation of gene candidates within these clusters in salivary gland development, homeostasis, and disease.
Transcriptional and Posttranslational Regulation of Abundant Salivary Secreted Proteins
To determine whether saliva protein abundance is mainly regulated at the transcriptional level, we compared transcript levels in each glandular tissue type with protein abundances in the corresponding glandular ductal secretions as they became available through the Human Salivary Protein Wiki (HSP-Wiki: https://salivaryproteome.nidcr.nih.gov/). Looking at the entirety of the data, we did not find a global correlation between transcript levels of secretory genes in any glands and corresponding protein levels in the respective glandular secretions (R2 < 0.1) or in the whole saliva (R2 < 0.1) (Figure 4A; Figure S6). We also noted differences among gland types in terms of how transcript and protein abundances were related. In SM/SL ductal saliva, a greater proportion of more highly abundant proteins were derived from genes with lower transcript levels in the SL gland (<104 NCs), a relationship that was not observed for the SM or PAR gland (as seen in the top-left quadrant of plots in Figure 4A). However, we did find that most highly abundant proteins in ductal salivary secretions are also highly expressed at the RNA level in their corresponding glandular tissues of origin (Figure 4A). Overall, our data indicate that the major salivary glands differ in posttranscriptional regulation and that transcript levels in salivary glands are not necessarily reflected by protein abundances in saliva, except for those proteins that occur in saliva at highest abundances. This finding suggests that most proteins in whole saliva are not derived from genes expressed in salivary glands or that salivary proteins are being affected by posttranscriptional regulations or modifications, likely including posttranscriptional modifications such as glycosylation, affecting the quantitative detectability of highly glycosylated proteins (e.g., mucins) by mass-spectrometric methods, and massive postsecretory enzymatic modifications known to affect protein abundances in whole-mouth saliva (Thomadaki et al., 2011; Helmerhorst and Oppenheim, 2007).
Figure 4. The Shaping of the Salivary Proteome.
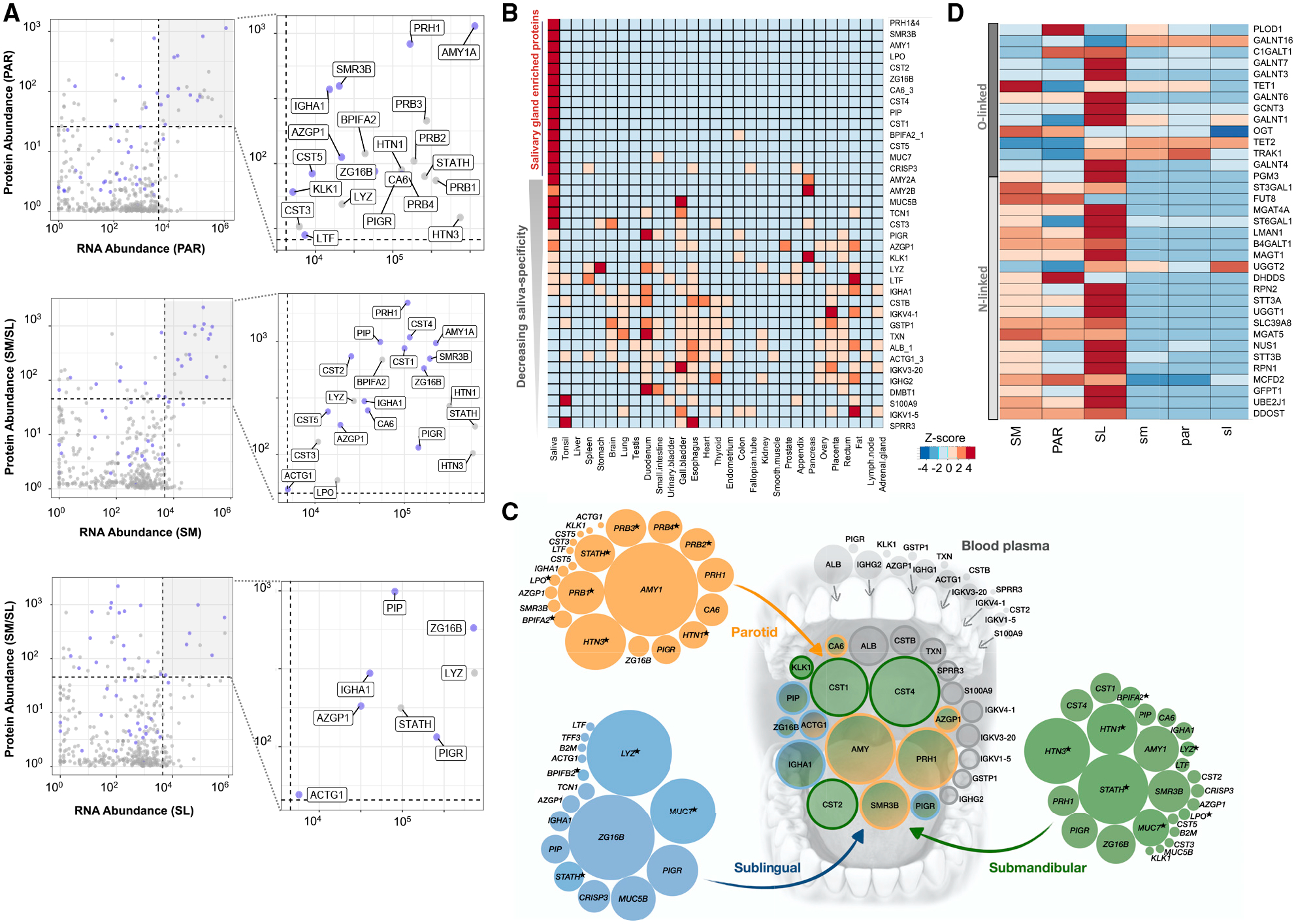
(A) Each graph represents a comparison of transcript abundances of a specific gland type, with protein abundances in that gland’s corresponding ductal saliva. x axis, log10 DESeq2 NCs; y axis, log10 normalized protein abundances. Blue dots indicate genes coding for secreted proteins. Genes showing the highest abundance (top 10%) at both the transcript and the protein level are highlighted in the top-right quadrant by a gray background and enlarged in the right panels, with their protein names indicated.
(B) Comparison of the most abundant proteins in human saliva with the protein abundances in 29 human organs from the Human Protein Atlas database (Wang et al., 2019). Genes were chosen based on their protein expression levels, according to the HSP-Wiki database, and their transcript levels, according to our salivary gland RNA-seq analysis. Heatmap colors indicate Z scores normalized for each row of data. Genes are ordered from top (highest) to bottom (lowest) based on their enrichment in salivary glands.
(C) Schematic showing the glandular origins of the most abundant saliva proteins in whole-mouth saliva. The central group of circles represents the most abundant proteins detected in whole-mouth saliva (data source: HSP-Wiki). The groups of circles on the outside represent the transcript levels in the PAR (orange), SL (blue), and SM (green) coding for the most abundant salivary proteins in the corresponding glandular secretions (data source: HSP-Wiki). The sizes (areas) of the circles symbolize relative RNA abundances normalized for each gland type. Colors in the central group of circles indicate the putative salivary gland origin of the proteins or their origin from blood plasma (gray). Blood plasma values are based on protein abundances. Data source: Human Plasma Proteome Project Data Central at PeptideAtlas http://www.peptideatlas.org/hupo/hppp/ (Schwenk et al., 2017). For blood plasma, only those proteins that were abundantly detected in whole-mouth saliva are shown. Proteins, indicated by an asterisk, are detected as secreted proteins at the glandular level but were not among the most abundant proteins detected in whole-mouth saliva.
(D) Heatmap of transcript levels for genes involved according to GO categorization in protein N-linked or O-linked glycosylation. Heatmap colors indicate Z scores normalized for each row of data. Table S2 provides the list of glycosylation-related genes and their gene expression in salivary glands.
We next compared the most abundant proteins, ranked according to protein abundance in the human salivary proteome and according to transcript abundance by our RNA-seq analysis, with publicly available mass-spectrometry-based proteomes of 29 healthy human organ tissues from the Human Protein Atlas project (Wang et al., 2019) (Table S5). Through this comparative analysis, we delineated 14 of the top 50 secreted saliva proteins to be highly enriched in salivary glands and saliva and 5 additional proteins to be highly expressed in only one or two organs other than in salivary glands (Figure 4B). These findings are supported by our comparative analysis of salivary gland transcriptomes to the 54 tissue and organ transcriptomes in the GTEx database (Figures S3–S5). However, it has to be taken into account that the transcriptomes of some secretory tissues and organs, the pancreas being the exception, are not available in the GTEx database, including the lacrimal gland and lactating mammary gland. It is known that some abundant proteins in saliva (e.g., MUC7, lactoperoxidase [LPO], and PIP) are present in other body fluids, such as tear fluid, milk, and epithelial mucus (Jung et al., 2017; Sharma et al., 1998). Examples for this are proteins, such as CST2, CST5, ZG16B, and SMR3B, which showed little to no protein or transcript expression in other tissues or organs, including the mammary gland, pituitary, prostate, pancreas, and lung, but were reported to be present in tear fluid (Jung et al., 2017).
We also found genes abundantly transcribed in salivary gland tissues (>1,000 NCs) that were not detected at the protein level in salivary secretions. This group of genes was enriched in functions related to intracellular housekeeping processes, as well as in functions typifying exocrine tissues, including vesicle-mediated transport (e.g., MCFD2), regulated exocytosis (e.g., TCN1), and cell secretion (e.g., FURIN) (Table S3). We also identified genes encoding proteins that previous proteome analyses identified in saliva but that were not detected by us at the RNA level in glandular tissues. This group of genes was enriched in functions characteristic of epithelial cells, including keratinization and cornification (e.g., KRT1 and SPRR1A). One noteworthy protein that is abundantly found in saliva (among the top 10% of proteins) but is not highly expressed (among the bottom 10%) at the RNA level in glandular tissues is albumin. This finding proves that most albumin in the whole saliva is not derived from salivary glands but rather diffuses into whole-mouth fluid via blood plasma leakage, mostly in the form of gingival crevicular fluid, as was suggested earlier (Helmerhorst et al., 2018).
Certain secreted proteins, which were abundantly detected at the mRNA level in glandular tissues and at the protein level in ductal saliva, such as STATH, LYZ, MUC7, and HTN1, were detectable by mass spectrometric analysis at lower amounts or not detectable in whole-mouth saliva (Figure 4A; Figure S6). Such reduction or loss can result from the proteins being proteolytically degraded once exposed to the mouth environment (Thomadaki et al., 2011) or through adsorption to oral surfaces after secretion from salivary glands. Indeed, multiple studies have demonstrated STATH, LYZ, and HTN1 to be selectively adsorbed from saliva onto the enamel surface in the form of the acquired pellicle (Hannig et al., 2005; Hay, 1973; Li et al., 2004). It is also possible that mass spectrometric analysis could not quantitatively detect certain proteins in saliva due to, for example, dense glycosylation that protects them from trypsin cleavage or other molecular features that impede identification of specific peptides in the mass spectrometer (Thamadilok et al., 2020; Walz et al., 2006, 2009). In that regard, a recent mass-spectrometry-based proteomic analysis of healthy PAR glands has revealed multiple proteins, including HTN1 and LYZ, to be highly expressed in the glandular tissue (Wang et al., 2019), thereby supporting our prediction of protein loss after secretion from the gland. Overall, integrating our glandular RNA-seq and mass-spectrometry-derived protein abundance data, we were able to parse out the origins of proteins present in human saliva (Figure 4C).
As stated earlier, some of the most abundant proteins in saliva, such as MUC7, MUC5B, PRB3, and S-IgA, are heavily glycosylated (Oppenheim et al., 2007) aiding in multiple functions, such as lubrication, mucus barrier formation, and microbial binding (Cross and Ruhl, 2018). Thus, we specifically investigated the expression patterns of genes that regulate O-linked or N-linked glycosylation, as per GO categorization (Table S4). We found that each salivary gland type expresses a typical repertoire of transcripts for genes that regulate glycosylation. Focusing on the most abundantly expressed glycosylation-related genes, it became clear that the SL shows dramatically increased expression of multiple GalNAc transferase genes (GALNTs). This family of enzymes is important for the initiation of O-glycosylation, a hallmark feature of mucin proteins abundantly present in salivary gland secretions. This finding makes sense biologically, given that the SL produces the major proportion of mucin proteins in human saliva. It is also worth emphasizing the magnitude in the expression of GALNTs among salivary gland types. For example, GALNT12 is expressed ~100-fold higher in SL tissue than in the other glands. We also discovered that the expression of GALNT13 was highly specific to the SM gland. GALNT genes have been reported to be non-redundant in both animals and humans and thus likely have specialized roles in catalyzing different types of glycosylation (Bennett et al., 2012; Narimatsu et al., 2019). Overall, our results will become particularly important from a biomedical perspective, because the salivary glycome forms an interface with the oral microbiome (Cross and Ruhl, 2018), and abnormalities in glycosylation are discussed as biomarkers for both Sjögren’s syndrome and oral cancers (Chaudhury et al., 2015; Nita-Lazar et al., 2009).
Cellular Heterogeneity within Gland Types Underlies Gland-Specific Protein Secretion
To consolidate our previously described findings, we conducted immunofluorescence imaging of tissue sections from the three adult gland types. We found clear concordance of gland-specific expression at the protein level with RNA transcript levels for STATH, AMY1, LPO, CRISP3, MUC7, and MUC5B (Figure 5A). The expression patterns of each of these proteins are tissue specific and are concordant with previous studies describing individual gland types or gland-specific secretions (Nielsen et al., 1996; Ruhl, 2012; Veerman et al., 2003).
Figure 5. Gland- and Cell-Specific Expression of Salivary Proteins.
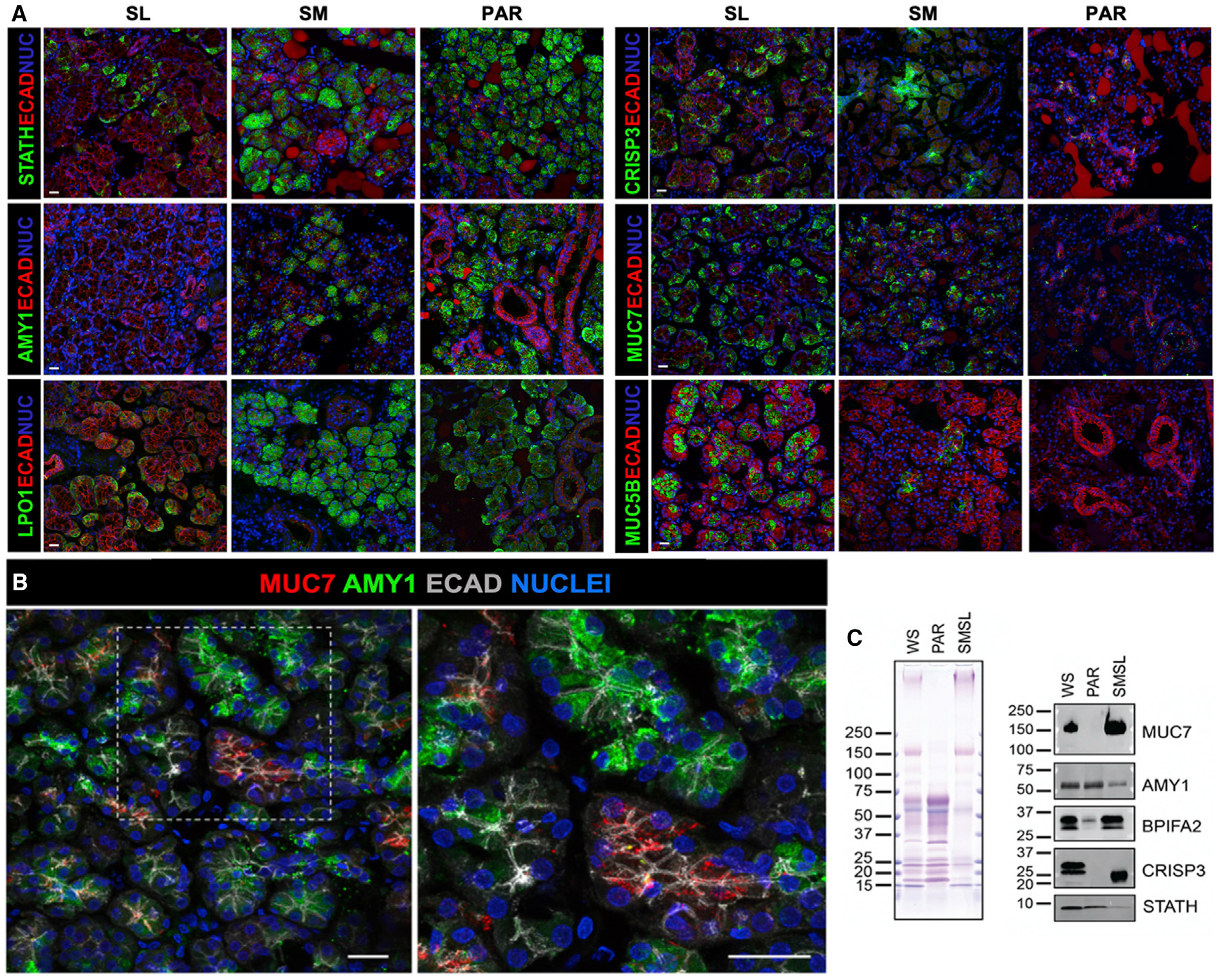
(A) Immunohistochemistry of glandular tissues. The SM and PAR acinar cells are highly enriched for amylase (AMY1), statherin (STATH), and lactoperoxidase (LPO) compared with the SL, consistent with these proteins being markers of serous cells. MUC7 and CRISP3 are expressed by a subset of acinar cells of the SM and SL, with little to no expression in the PAR. MUC5B is highly expressed by the SL mucous acinar cells, but not by the acinar cells of the PAR or SM, indicating it to be a marker of SL function. Scale bar, 25 μm.
(B) MUC7 and amylase are expressed by distinct subtypes of serous acinar cells. The mature SM tissue section was immunolabeled for MUC7 (red), AMY (green), and ECAD (gray).
(C) Gel electrophoretic separation of whole mouth saliva (WS) and glandular ductal secretions (PAR and SM/SL), followed by staining of proteins with Coomassie blue and of glycoproteins with periodic acid Schiff stain (pink bands in left panel) and probing of transfers with antibodies against MUC7, AMY, BPIFA2, CRISP3, and STATH (right panel).
One striking example for gland-specific expression is salivary amylase, an enzyme synthesized by serous acinar cells, that shows abundant expression at the protein level in PAR and SM glandular tissue while being virtually absent from the SL. A similar trend was found for STATH and LPO. The lower expression levels of these gene products in the SL likely result from the lower amount of serous acinar cells in this type of glandular tissue (Amano et al., 2012). However, the near-complete absence of amylase in serous acinar cells of the SL indicates that these cells in the SL are distinctly different from their counterparts in the SM and PAR. Our findings confirm the validity of using these proteins as key markers to discern SM- and PAR-gland-derived tissues from those of the SL.
A different example of gland- and cell-specific expression is MUC7, which shows abundant expression at the protein level in the serous cells of the SL gland and, to a lesser extent, in the serous cells of the SM gland while being absent from serous cells of the PAR gland (Figure 5A), matching MUC7 transcript levels from the respective glandular tissues (Figure 1E). Given this result illustrating the diversity of serous cells across gland types, we next asked whether there was also intraglandular variation in protein synthesis at the cellular level and pursued this question by combining immunostaining for amylase and MUC7. We found MUC7 enriched in subsets of serous acinar cells that were deficient in amylase expression, and we found AMY expression in other subsets of serous acinar cells that were deficient in MUC7 expression (Figure 5B). Our observation suggests that serous cells within the SM exist as distinct populations, each secreting its own repertoire of proteins. Recent single-cell RNA-seq of murine parotid salivary glands indicated acinar cell heterogeneity (Oyelakin et al., 2019). We propose here that human acinar cells are heterogeneous with respect to secreted protein expression.
We also discovered that for synthesis of the same salivary protein, the three major gland types use different cell lineages. For example, we found that in the SL protein expression of CRISP3 paralleled that of MUC7 in being abundantly produced by acinar cells (Figure 5A). However, in the SM, which expressed lower transcript levels of CRISP3 compared with the SL, CRISP3 protein could be located in only a few acinar cells but was found predominantly in cells of the intercalated ducts. An analogous expression pattern for CRISP3 (i.e., acinar and duct cells expressing CRISP3) has been described in the murine lacrimal gland (Reddy et al., 2008), but it was not known that these two cell populations can each produce the same protein even in different gland types.
To prove whether what we observed at the gland level by immunohistochemistry manifests at the protein level in salivary secretions, we conducted gel electrophoretic separation of glandular ductal secretions and western blot analysis for AMY1, MUC7, CRISP3, BPIFA2/SPLUNC2, and STATH (Figure 5C). As revealed by Coomassie blue and periodic acid Schiff stain, the combined secretions of the SM and SL (SM/SL) glands showed strikingly different patterns of protein and glycoprotein bands compared with PAR secretion, whereas whole mixed saliva showed a combination of both. The presence of AMY1 and MUC7 proteins in glandular secretions, as shown by western blotting, was consistent with transcriptomic and immunofluorescent analyses (Figure 5) and with previous reports (Merritt et al., 1973; Thamadilok et al., 2016; Veerman et al., 1996; Walz et al., 2009). We also found BPIFA2, a protein known to exist in whole saliva (Bingle et al., 2009), to be enriched in SM/SL secretion but weakly expressed in PAR secretion, supporting our transcriptome-based evidence that this protein is predominantly derived from the SM. We further found CRISP3, detectable in whole saliva as a doublet of bands, as previously shown (Udby et al., 2002), to be restricted solely to SM/SL secretions with no detectable protein in PAR ductal saliva, thus matching both our immunohistological and RNA-seq findings (Figure 5A). The CRISP3 band in SM/SL ductal secretion migrated farther during electrophoresis than the double bands in whole-mouth saliva. This outcome suggests that postsecretion enzymatic processing may have occurred, likely resulting in the alteration of CRISP3 sialylation by oral bacterial sialidases, which is known to lead to a loss of negatively charged sialic acid moieties, thus retarding the mobility of the protein in the electrophoretic field (Udby et al., 2002; Walz et al., 2009; Zhou et al., 2016). It is of note that we found STATH to be present in both PAR and SM/SL ductal secretions with higher abundance in PAR saliva (Figure 5C) (Gibbins et al., 2014; Proctor et al., 2005). STATH was also abundantly detected in the WS sample run on our gel. It has to be noted though that utmost precaution was taken during sample handling and preparation to minimize proteolytic degradation. When other samples, even of the same donor individual, were probed for STATH, only a faint band or no band at all could be detected in WS (data not shown) showing that enzymatic degradation affecting in particular this component can easily occur as was described earlier (Helmerhorst et al., 2010; Thomadaki et al., 2011). Overall, our combined immunohistological and immunoblotting data at the protein level in ductal secretions correlate well with transcript levels in the corresponding glands and hence intimately link the fields of human salivary gland and saliva protein research.
DISCUSSION
Our analysis of the transcriptomes of mature and fetal salivary glands identified hundreds of genes that together define mature salivary glands as specialized secretory organs. We also found that fetal glands, despite the late stage of development and the glands being anatomically separate entities, could not be distinguished based on their transcriptional profiles. This indicates that developmental differentiation of glandular function and functional specialization of the three mature gland types occur during fetal development at a time point later than when the tissue samples in our study were acquired (>22 weeks). These findings pave the way for future studies dissecting mechanisms of regulation of the transcriptome during glandular development and will have significant implications for de novo organ generation.
Our results reveal the extensive level of retention of fetal genes in adult tissues in a human epithelial organ system, strongly suggesting the involvement of these genes in both developmental and homeostatic roles. The major differences between fetal and adult tissue transcriptomes manifest in the upregulation of pathways facilitating secretory and immune function in mature tissue and in the enrichment of developmentally related genes in fetal glands. However, hundreds of fetally expressed genes are retained at significant levels in mature SL tissue while being poorly retained by SM and PAR tissue. Such differential gene retention likely contributes to gland-type differences, including the more fetal-like extracellular matrix of the SL.
Our results have identified hundreds of transcripts that are specifically expressed in salivary glands compared with other major human tissues. These genes provide a robust set for diagnostic purposes both to identify specific glandular types and test for deviations in expression under pathogenic conditions such as cancer. In an attempt to further understand the regulation of these transcripts, we verified TFs that show differential expressions in fetal and mature glands. Our analysis suggests that fine-scale dosage differences of hundreds of TFs shape the expression landscape of mature gland types. Indeed, we found dozens of clusters across the genome in which multiple genes with salivary-gland-specific expression are located in proximity. It is thus plausible that these loci harbor master regulatory sequences that are bound by a specific combination of TFs, leading to salivary-gland-specific expression. Overall, our study provides an exciting next step by having identified dozens of candidate target transcripts and TFs for investigating the mechanism of gland-specific expression regulation in these loci.
Our quantitative transcriptomic and proteomic comparison also allowed us to define relationships between the level of salivary gland gene transcripts and their corresponding proteins in salivary secretions. Similar to other studies exploring the relationship between high-abundance proteins and mRNA levels, as well as to quantitative studies in other secretory cell systems (Liu et al., 2016), we found that the abundance of most highly expressed saliva proteins is regulated mainly at the transcriptional and posttranslational level. Our data also suggest differences among gland types in terms of the glycosylation machinery that likely increases protein diversity. Furthermore, we traced the origins of abundant proteins in whole-mouth saliva to their respective glands or to extrinsic sources such as blood plasma. Collectively, our study provides a robust framework for modeling the biological makeup of a major complex secretory fluid (summarized in Figure 4C).
Finally, we provide evidence about cellular heterogeneity in human salivary glands. Specifically, we were able to identify two subsets of serous acinar cells in the submandibular gland that appeared to be specialized for expressing either AMY1 or MUC7. Acinar cells have been traditionally defined in a simple binary categorization as either serous or mucous. Our data suggest that these cell types are more heterogeneous than previously acknowledged and that subsets of them are specialized to synthesize specific salivary proteins. In addition, our data revealing CRISP3 production by serous acinar cells in the SL and by ductal and serous acinar cells in the SM demonstrate that the same saliva protein can be produced by different cell lineages. These insights will have major implications for understanding the relationship between glandular secretions and protein content of these secretory fluids, and consequently, inform saliva diagnostics. Furthermore, our transcriptomic analysis of healthy human glandular tissue will have major ramifications for de novo engineering of these organs, as well as for determining the impact of the disease on salivary gland homeostasis and function.
In future studies, it will be important to investigate how salivary gland tissue differentiation, transcriptional and posttranslational regulation, and intraglandular cellular heterogeneity play their roles in human health. We also foresee that our study will open up the possibility of asking specific questions about human evolutionary adaptations to accommodate different environmental conditions, food resources, and pathogen challenges among geographically and culturally distinct human populations with respect to their composition of saliva (Pajic et al., 2019; Perry et al., 2007; Xu et al., 2017).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources, codes, and data should be directed to and will be fulfilled by the Lead Contact, Omer Gokcumen (omergokc@buffalo.edu).
Materials availability
This study did not generate new reagents. The samples used in this study are from biopsies with specific permissions and are thus restricted in their availability.
Data and code availability
The RNA-seq data (fastq files) have been submitted to GEO https://www.ncbi.nlm.nih.gov/geo/ with the project name GSE143702. The custom codes are available through https://github.com/GokcumenLab/
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human fetal salivary glands were harvested from post-mortem fetuses obtained from elective legal abortions with the written informed consent of the patients undergoing the procedure and the approval of the Institutional Review Board at the University of California San Francisco (IRB# 10–00768). Specimens were donated anonymously at San Francisco General Hospital. Adult human salivary gland biopsies (Table S1) were collected via the UCSF Biospecimen Resources Program (BIOS) under the institutional review body approval number 17–22669. Saliva collection was performed as approved by the University at Buffalo Health Science Institutional Review Board (Study Nr. 030–505616).
METHOD DETAILS
Human tissue samples
Adult salivary gland tissues were collected with informed consent from patients aged 23 to 70 years by clinicians at the University of California, San Francisco Medical Center during routine surgeries (UCSF Biorepository, the institutional review body approval number 17–22669). The UCSF pathology lab deemed the investigated tissues to be healthy. For the RNaseq library preparation, we strictly sampled from core sections of the biopsy. Additionally, we took only every third 40 μm section (3–4 sections per sample) for analysis to ensure the specificity of our sampling.
Sample collection for the adult tissues took place during oral surgery procedures that were performed independent of this project. SM and PAR tissue samples were taken during oral surgeries from individuals suffering from cancers of the head and neck. The sample collection was limited to those patients who had not received radiotherapy, chemotherapy or immunotherapy. SL samples were derived from patients with salivary duct stones. Healthy tissue regions were identified and separated from inflamed or cancerous tissues by the UCSF pathology lab. Our immunofluorescent analysis further confirmed tissue health, as determined by cell and tissue morphology and the absence of lymphocytic infiltrates. It is unlikely but remains plausible that the disease status of the patients may have altered transcript levels. For example, it is possible that certain diseases, including oral cancers, can generate widespread inflammation, biasing our results for detecting higher levels of immune-system related genes. Since all adult sublingual glands were derived from female donors, we investigated the variation of gene expression among samples of the same gland type as well as sex-specific expression differences to test for any potential biases. We observed an extremely small variation in gene expression abundances of samples of the same gland type (Figures 1B and 1C) and no sex-specific trends at the global transcriptome level (data not shown). This indicates that the differences among individual samples of the same gland type are much smaller than the gland-specific transcriptome trends we are reporting. Human fetal salivary glands were harvested from post-mortem fetuses between 22 and 24 weeks of gestation with the approval of the Institutional Review Board at the University of California San Francisco (IRB# 10–00768). Tissue was identified by location and glandular appearance. Sex was confirmed through analysis of transcript levels of male-specific genes, namely, UTY and KDM5D (Staedtler et al., 2013).
Preparation of tissue samples
For RNA analysis, tissue was frozen in liquid nitrogen and stored at −80°C. RNA isolation was conducted as described previously (Emmerson et al., 2017, 2018). For immunofluorescent analysis, tissue was either flash-frozen in optimal cutting temperature compound (OCT) (Tissue-Tek) and stored at −80°C, or immediately fixed with 4% paraformaldehyde (PFA) overnight at 4°C. Fixed samples were washed with PBS, cryoprotected by immersion in a 12.5%–25% stepwise sucrose gradient, and then embedded in OCT for storage at −80°C. Tissue was sectioned (12 μm thickness) immediately before immunofluorescent analysis using a cryostat (Leica).
RNA isolation and sequencing
Human adult and fetal tissue samples were mechanically homogenized using a hand-held homogenizer (Thermo Fisher Scientific) and lysed in 500 μl RNA lysis buffer (Ambion) by sonication (1 × 2–4 s pulse, Branson SFX150). RNA was isolated from 3 × 30 μm sections of human adult and fetal tissue using the RNAqueous Micro Kit (Ambion), and total RNA samples were DNase-treated (Ambion). Sample yield and integrity was analyzed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA sequencing was performed by standard operating procedure by GENEWIZ (https://www.genewiz.com/en) using Illumina HiSeq with a 2 × 150 bp configuration. Quality control of the obtained sequences was performed using FastQC (Wingett and Andrews, 2018) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Accessed 12/10/2017). The results were further reviewed by MultiQC (Ewels et al., 2016). Adaptor sequences, low-quality bases from both sides of the read (3 bases or smaller), and reads with a length smaller than 36 bp were discarded by Trimmomatic (Bolger et al., 2014).
Sample quality control
We reasoned that if any contamination existed in our samples, we expect it to be mainly derived from muscle tissue that these glands reside adjacent to. Thus, as a further precaution, we now searched transcripts of genes that are known to be muscle tissue-specific and not expressed in salivary glands (MYH3, ACTA1, PAX7, MYH8, KLHL41, SGCA, MYBPH, MYOG, MYOZ1, XIRP1, XIRP2, LDB3, TNNT3, TTN, DMD, and MYH1). Based on this analysis, we found evidence of slight contamination in two fetal submandibular gland samples and eliminated these from the analysis reported in this study. We still provided the expression data for these two samples in Table S2 for reproducibility purposes. Also, if contamination were a major factor, we would expect to see more variation among different samples of the same gland type due to varying levels of contamination. As a further quality control step, we screened sequenced tissue for genes encoding inflammatory markers including IL1b, TNF, IL17, CXCL13, and CCL21, and did not find any of these to be upregulated in any of the samples.
Immunofluorescence imaging
The immunofluorescent analysis was performed on 12-μm-thick fixed tissue sections. Sections were imaged using a line-scanning confocal microscope (Leica Sp5). Frozen adult human salivary sections were fixed with 4 % PFA at room temperature (RT) for 20 min and subsequently washed in PBS followed by permeabilization with 0.5 % Triton-X in PBS for 10 min. Tissue sections were blocked with 10% donkey serum (Jackson Laboratories) and 1% BSA (Sigma-Aldrich) in 0.05% PBS-Tween 20 for 1 h at RT. Tissue sections were incubated with the following primary antibodies overnight at RT: rabbit anti-MUC7 (1:200; Sigma HPA006411), mouse anti-MUC7 (1:500; Abcam ab105466), mouse anti-MUC5B (1:500; Abcam ab105460), rat anti-E cadherin (1:300; Sigma U3254), rabbit anti-CRISP3 (1:200; Sigma HPA054392), rabbit anti-AMY1A (1:200; Sigma HPA045394), rabbit anti-AQP5 (1:400; Millipore AB3559), rabbit anti-statherin (1:500, Ruhl laboratory), and rabbit anti-LPO1 (1:200; Sigma HPA028688). Antibodies were detected by incubating samples with Cy2-, Cy3- or Cy5-conjugated secondary Fab fragments (1:300 in 0.05% PBS-Tween-20; Jackson Laboratories) for 2 h at RT. Nuclei were detected using Hoechst 33342 (1:1000, Sigma Aldrich). Fluorescence images were obtained using a Leica Sp5 line-scanning confocal microscope.
Saliva sample collection
Saliva from healthy humans was collected following the protocol approved by the University at Buffalo Human Subjects IRB board (study # 030–505616). Informed consent was obtained from all human participants. Stimulated whole mouth saliva (WS) was collected while chewing on parafilm. Clarified whole saliva supernatant was obtained by centrifugation at 12,000 × g for 15 min at 4°C to remove particulate matter. Ductal salivary secretions were collected following the stimulation of salivary flow by application of 2% citric acid to the dorsum of the tongue. PAR saliva was collected from the orifice of Stensons’s duct using a modified Carlsen-Crittenden device, and SM/SL saliva was collected from the floor of the mouth using disposable plastic Pasteur pipettes (VWR International, Radnor, PA) after isolation of the gland orifice with absorbent cotton rolls and blockage of the PAR duct orifice. Protein concentrations in saliva samples were determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockland, IL), using bovine serum albumin as the standard.
SDS-PAGE and immunoblotting
Saliva samples were denatured under reducing conditions. Equal amounts of total protein (15 mg per lane for Coomassie and periodic acid Schiff stain) were subjected to separation by SDS-PAGE using 8%–16% gradient Tris-glycine mini gels. Only for the detection of statherin, because of its small molecular size, Tris-tricine gels were used for better resolution (Novex, Invitrogen, Carlsbad, CA). Proteins were stained by Coomassie blue and glycans by periodic acid Schiff stain (Heo et al., 2013; Thamadilok et al., 2020). Stained gels were imaged using a flat-bed scanner in the transparent mode (ImageScanner III, GE Healthcare). Electrotransfer during immunoblotting was done using a BioRad Trans Turbo Blot apparatus. Blots were probed with the following antibodies diluted in Tris-buffered saline containing 2% milk (TBS-milk): mouse monoclonal anti-human mucin 7 (MUC7) diluted 1:500 (4D2–1D7, Abcam), rabbit polyclonal anti-human alpha-amylase 1A (AMY1) diluted 1:1,000 (HPA045394, Sigma), rabbit polyclonal anti-human parotid secretory protein (SPLUNC2B, BPIFA2) diluted 1:500 (C-20, a gift from Dr. Colin Bingle at the University at Sheffield (Bingle et al., 2009), rabbit polyclonal anti-CRISP3 diluted 1:500 (HPA054392, Sigma), and polyclonal rabbit anti-human STATH (Dundee Cell Products Ltd., Dundee, UK). As secondary antibodies, Alexa Fluor 488 tagged goat anti-rabbit or anti-mouse IgG (Life Technologies) were used diluted 1:1,000 in TBS-milk. Fluorescent bands were detected using a BioRad ChemiDoc imaging system.
QUANTIFICATION AND STATISTICAL ANALYSIS
All the downstream data analysis and comparison of our dataset with publicly available databases were conducted using custom bioinformatic pipelines written on Rstudio (v1.2.1335), R(v3.5.3), and ggplot2 (Wickham, 2009). These codes were made available through https://github.com/GokcumenLab/glabBits/tree/master/Saliva-RNASeq. We used Pearson Correlation analysis available in R base package for calculating the relationships between transcriptomes of different gland types presented in Figure S1. We used non-parametric Wilcoxon rank-sum test also available in R base package for testing differences between the expression levels of genes encoding secreted and nonsecreted proteins (Figure S2).
RNaseq analysis
Filtered reads were mapped to the human transcriptome reference (hg19) from Ensembl (Zerbino et al., 2018) and biomaRt (Durinck et al., 2005) and quantified using Kallisto (Bray et al., 2016). Differential expression analysis was performed by DESeq2 (Love et al., 2014), which calculates the fold-change of transcription of each gene using the Wald test and a correction for multiple hypotheses from the raw reads. For this analysis, we used pairwise comparison of all fetal and adult gland types for each other. Each of these categories have at least three samples. We used an adjusted (i.e., multiple-hypotheses-corrected p value of < 0.0001) to identify genes that were upregulated (fetus < adult) and downregulated (adult < fetus) during development in each type of salivary gland. Since all adult SL tissue samples were of female background, we excluded the Y chromosome from the analyses. The remaining 40,882 genes were used for subsequent analyses. The normalized RNA abundances for 40,882 genes that we interrogated as well as comparative results are provided in Table S2. The RNA-seq data (fastq files) have been submitted to GEO https://www.ncbi.nlm.nih.gov/geo/ with the project name GSE143702.
Identifying genes with salivary gland specific expression
To identify genes that are expressed in a salivary gland-specific manner, we compared our RNaseq results from the three major salivary glands to RNaseq data available through the GTEx database that were obtained from 53 tissues collected from approximately 1,000 individuals (https://gtexportal.org/home/documentationPage#AboutSamples) (GTEx Consortium, 2013; Battle et al., 2017) (Table S5). These organs include those that synthesize secreted proteins such as the pancreas, mammary gland, thyroid, and intestine. To define whether a gene was specific to salivary glands, we first identified among the non-salivary gland tissues or organs those in which the gene in question was highest expressed. We then compared this level of gene expression to the gene expression levels in each major salivary gland. To account for potential differences in the RNaseq datasets from the GTEx database due to different experimental platforms and bioinformatics data analysis processes, we compared relative expression level ranks by using a (log10 (1+normalized read counts)) transformation for each organs RNaseq dataset.
Functional categorization of genes was determined by cross-checking using Gene Ontology Resources (http://geneontology.org/) (Ashburner et al., 2000). Then, an enrichment analysis was conducted using GOrilla (Eden et al., 2009) applying default settings that enable multiple hypothesis testing (Table S3). Secreted protein gene annotations were obtained from the Human Protein Atlas (https://www.proteinatlas.org/) with the query: “protein_class:Predicted secreted proteins NOT protein_class:Predicted membrane proteins” (Uhlén et al., 2015). The glycosylated genes were defined as per Gene Ontology database. We identified genes that are as either “protein N-linked glycosylation” (GO:0006487) or “protein O-linked glycosylation” (GO:0006493), and are abundantly expressed (> 1000 normalized gene count from DESeq) in salivary gland tissues. Note that we omitted genes listed under GO subcategory “O-glycan processing” (GO:0016266), which are mostly mucin genes. The genes encode for proteins that are targets of O-glycosylation and they do not regulate glycosylation.
We used the Human Salivary Proteome Wiki (https://salivaryproteome.nidcr.nih.gov/) to obtain proteome data for the whole saliva, ductal saliva from the PAR, SM, and SL, as well as from blood plasma. The database provides mass-spectrometry-based abundance data for approximately 3,000 proteins compiled from multiple studies (Murr et al., 2017), including those that focused on ductal secretions (Denny et al., 2008). Similar to the comparison approach that we used for integrating GTEx data, we used log transformation (log10 (1+normalized abundance)) for each dataset. In addition, we used mass-spectrometry-based protein abundances available through Human Protein Atlas from 29 tissues curated recently by Wang et al. (2019) (Table S5). This dataset contains abundance information on 13,000 proteins.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat polyclonal anti-E cadherin | Sigma | Cat# U3254; RRID: AB_477600 |
| Rabbit polyclonal Anti-MUC7 | Sigma | Cat# HPA006411; RRID: AB_1854204 |
| Mouse monoclonal anti-MUC7 | Abcam | Cat# 4D2-1D7; RRID: AB_10866568 |
| Mouse monoclonal anti-MUC5B | Abcam | Cat# ab105460; RRID: AB_10862195 |
| Rabbit polyclonal anti-CRISP3 | Sigma | Cat# HPA054392; RRID: AB_2682472 |
| Rabbit polyclonal anti-AMY1A | Sigma | Cat# HPA045394; RRID: AB_2679311 |
| Rabbit polyclonal anti-LPO1 | Sigma | Cat# HPA028688; RRID: AB_10601909 |
| Rabbit polyclonal anti-STATH | Dundee Cell Products Ltd., Dundee, UK | Custom |
| Rabbit polyclonal anti-BPIFA2 | Eurogentec | Bingle et al. (2009) |
| Biological Samples | ||
| Human Salivary Gland Tissues | Laboratory of Sarah Knox | UCSF Biospecimen Resources Program (BIOS) 17-22669 |
| Human Saliva - Whole Saliva | Laboratory of Stefan Ruhl | Custom |
| Human Saliva - Parotid Saliva | Laboratory of Stefan Ruhl | Custom |
| Human Saliva - Submandibular/Sublingual Saliva | Laboratory of Stefan Ruhl | Custom |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Optimal cutting temperature compound (OCT) | Tissue-Tek | Cat# 4583 |
| RNA lysis buffer, filter cartilages and collection tubes | Invitrogen | Cat# AM1912 |
| Pierce bicinchoninic acid (BCA) protein assay kit | Thermo Scientific | Cat# 23227 |
| Deposited Data | ||
| RNaseq data - Salivary glands | This study | https://www.ncbi.nlm.nih.gov/geo/[GSE143702] |
| Salivary and serum proteomics data | Human Salivary Proteome Wiki | https://salivaryproteome.nidcr.nih.gov/public/index.php/Main_Page |
| RNaseq data from multiple tissues | GTEx | https://www.gtexportal.org/home/ |
| Proteome data from multiple tissues | Human Protein Atlas | https://www.proteinatlas.org/ |
| Software and Algorithms | ||
| Custom codes for downstream analysis | Gokcumen Laboratory | https://github.com/GokcumenLab/glabBits |
| FastQC | Babraham Bioinformatics | v0.11.9 |
| Trimmomatic | Bolger et al. (2014) | v0.39 |
| Kallisto | Bray et al. (2016) | v0.46.1 |
| DESeq2 | Bioconductor | v1.28.1 |
| GOrilla | Eden et al., (2009) | vMar.8.2013 |
| ggplot2 | tidyverse | V3.3.2 |
Highlights.
Genes encoding highly abundant secreted proteins define adult gland types
Gland-specific activity of transcriptional regulators contributes to proteome diversity
Differential retention of fetal genes drives functional diversity in adult glands
Cellular heterogeneity underlies gland-specific protein secretions
ACKNOWLEDGMENTS
The authors thank William Lau for help with retrieving the protein abundance data from the NIDCR Human Salivary Proteome Wiki. We are grateful to Tasha Lea (certified pathologist assistant) and Erica Oropeza, along with the Biospecimen Resources Program (BIOS) at UCSF, for facilitating the efficient acquisition, for quality control, and for management of biospecimens. We thank Fatih Aksel for his advise on computational matters. This work was supported by the National Institute of Dental and Craniofacial Research (NIDCR) under award numbers 1R35DE028255 (to S.M.K.) and 2R01DE019807 (to S.R.), by Tobacco-Related Disease Research (TRDRP) under award 28IR-0071 (to S.M.K.), and by National Science Foundation (NSF) under award number 1714867 (to O.G).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108402.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amano O, Mizobe K, Bando Y, and Sakiyama K (2012). Anatomy and histology of rodent and human major salivary glands: overview of the Japan salivary gland society-sponsored workshop-. Acta Histochem. Cytochem 45, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal HK, Murphy G 3rd, Tibbs E, Cornett A, Hill E, Yeoh K, Berenstein E, Hoffman MP, and Lombaert IMA (2019). Sox10 Regulates Plasticity of Epithelial Progenitors toward Secretory Units of Exocrine Glands. Stem Cell Reports 12, 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Brown CD, Engelhardt BE, and Montgomery SB GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213.29022597 [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, and Tabak LA (2012). Control of mucin-type O-glycosylation: a classification of the poly-peptide GalNAc-transferase gene family. Glycobiology 22, 736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Wang C-C, and Harvey RP (2002). NK-2 class homeobox genes and pharyngeal/oral patterning: Nkx2–3 is required for salivary gland and tooth morphogenesis. Int. J. Dev. Biol 46, 415–422. [PubMed] [Google Scholar]
- Bingle L, Barnes FA, Lunn H, Musa M, Webster S, Douglas CWI, Cross SS, High AS, and Bingle CD (2009). Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein. Histochem. Cell Biol 132, 339–349. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Chaiyarit P, Utrawichian A, Leelayuwat C, Vatanasapt P, Chanchareonsook N, Samson MH, and Giraud AS (2012). Investigation of trefoil factor expression in saliva and oral mucosal tissues of patients with oral squamous cell carcinoma. Clin. Oral Investig 16, 1549–1556. [DOI] [PubMed] [Google Scholar]
- Chatzeli L, Gaete M, and Tucker AS (2017). Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 144, 2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury NMA, Shirlaw P, Pramanik R, Carpenter GH, and Proctor GB (2015). Changes in Saliva Rheological Properties and Mucin Glycosylation in Dry Mouth. J. Dent. Res 94, 1660–1667. [DOI] [PubMed] [Google Scholar]
- Christensen EI, and Birn H (2002). Megalin and cubilin: multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol 3, 256–266. [DOI] [PubMed] [Google Scholar]
- Cross BW, and Ruhl S (2018). Glycan recognition at the saliva—oral microbiome interface. Cell. Immunol 333, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, and Wong DTW (2019). Role of Saliva and Salivary Diagnostics in the Advancement of Oral Health. J. Dent. Res 98, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Pedersen AML, Villa A, Ekström J, Proctor GB, Vissink A, Aframian D, McGowan R, Aliko A, Narayana N, et al. (2015). The functions of human saliva: A review sponsored by the World Workshop on Oral Medicine VI. Arch. Oral Biol 60, 863–874. [DOI] [PubMed] [Google Scholar]
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, et al. (2008). The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res 7, 1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DP, Thiesse M, and Hicks MJ (2002). Expression of type 2 cystatin genes CST1-CST5 in adult human tissues and the developing submandibular gland. DNA Cell Biol. 21, 47–65. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, Maliskova L, Zovein AC, Shen Y, Muench MO, and Knox SM (2017). SOX2 regulates acinar cell development in the salivary gland. eLife 6, e26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Berthoin L, Cruz-Pacheco N, Nathan S, Mattingly AJ, Chang JL, Ryan WR, Tward AD, and Knox SM (2018). Salivary glands regenerate after radiation injury through SOX2-mediated secretory cell replacement. EMBO Mol. Med 10, e8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Käller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel ES, and Ribbeck K (2015). Salivary mucins in host defense and disease prevention. J. Oral Microbiol 7, 29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Dong Y, Yu M, Zhang L, Yan X, Sun J, Qiao L, Geng H, Nakajima M, Furuichi T, et al. (2011). Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc. Natl. Acad. Sci. USA 108, 7058–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber PA, Hevezi P, Buhren BA, Martinez C, Schrumpf H, Gasis M, Grether-Beck S, Krutmann J, Homey B, and Zlotnik A (2013). Systematic identification and characterization of novel human skin-associated genes encoding membrane and secreted proteins. PLoS ONE 8, e63949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbins HL, Proctor GB, Yakubov GE, Wilson S, and Carpenter GH (2014). Concentration of salivary protective proteins within the bound oral mucosal pellicle. Oral Dis. 20, 707–713. [DOI] [PubMed] [Google Scholar]
- Grassl N, Kulak NA, Pichler G, Geyer PE, Jung J, Schubert S, Sinitcyn P, Cox J, and Mann M (2016). Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Balasubramanian D, and Clark JR (2015). Salivary gland lesions: recent advances and evolving concepts. Oral Surg. Oral Med. Oral Pathol. Oral Radiol 119, 661–674. [DOI] [PubMed] [Google Scholar]
- Gurusankar R, Kumarathasan P, Saravanamuthu A, Thomson EM, and Vincent R (2015). Correlation between saliva and plasma levels of endothelin isoforms ET-1, ET-2, and ET-3. Int. J. Pept 2015, 828759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig C, Hannig M, and Attin T (2005). Enzymes in the acquired enamel pellicle. Eur. J. Oral Sci 113, 2–13. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Regel I, Steiger K, Wagner N, Thorwirth M, Schlitter AM, Esposito I, Michalski CW, Friess H, Kleeff J, and Erkan M (2016). Loss of Periostin Results in Impaired Regeneration and Pancreatic Atrophy after Cerulein-Induced Pancreatitis. Am. J. Pathol 186, 24–31. [DOI] [PubMed] [Google Scholar]
- Hay DI (1973). The isolation from human parotid saliva of a tyrosine-rich acidic peptide which exhibits high affinity for hydroxyapatite surfaces. Arch. Oral Biol 18, 1531–1541. [DOI] [PubMed] [Google Scholar]
- He J, and Furmanski P (1995). Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA. Nature 373, 721–724. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, and Oppenheim FG (2007). Saliva: a dynamic proteome. J. Dent. Res 86, 680–693. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Traboulsi G, Salih E, and Oppenheim FG (2010). Mass spectrometric identification of key proteolytic cleavage sites in statherin affecting mineral homeostasis and bacterial binding domains. J. Proteome Res 9, 5413–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst EJ, Dawes C, and Oppenheim FG (2018). The complexity of oral physiology and its impact on salivary diagnostics. Oral Dis. 24, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S-M, Choi K-S, Kazim LA, Reddy MS, Haase EM, Scannapieco FA, and Ruhl S (2013). Host defense proteins derived from human saliva bind to Staphylococcus aureus. Infect. Immun 81, 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Miao W, Chen T, Xie K, Shi A, Zhang L, Li R, and Wen J (2019). Genetic Variants in AC092159.2 and Risk of Gestational Diabetes Mellitus in a Chinese Population. DNA Cell Biol. 38, 1069–1077. [DOI] [PubMed] [Google Scholar]
- Huang H, Yang X, Bao M, Cao H, Miao X, Zhang X, Gan L, Qiu M, and Zhang Z (2016). Ablation of the Sox11 Gene Results in Clefting of the Secondary Palate Resembling the Pierre Robin Sequence. J. Biol. Chem 291, 7107–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JE, and Porrello ER (2017). Periostin paves the way for neonatal heart regeneration. Cardiovasc. Res 113, 556–558. [DOI] [PubMed] [Google Scholar]
- Ianez RF, Buim ME, Coutinho-Camillo CM, Schultz R, Soares FA, and Lourenço SV (2010). Human salivary gland morphogenesis: myoepithelial cell maturation assessed by immunohistochemical markers. Histopathology 57, 410–417. [DOI] [PubMed] [Google Scholar]
- Jana S, Jana J, Patra K, Mondal S, Bhat J, Sarkar A, Sengupta P, Biswas A, Mukherjee M, Tripathi SP, et al. (2017). LINCRNA00273 promotes cancer metastasis and its G-Quadruplex promoter can serve as a novel target to inhibit cancer invasiveness. Oncotarget 8, 110234–110256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Boontheung P, Pang E, Yan W, Wong DT, and Loo JA (2008). Comparison of N-linked Glycoproteins in Human Whole Saliva, Parotid, Submandibular, and Sublingual Glandular Secretions Identified using Hydrazide Chemistry and Mass Spectrometry. Clin. Proteomics 3–4, 80–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Ji YW, Hwang HS, Oh JW, Kim HC, Lee HK, and Kim KP (2017). Proteomic analysis of human lacrimal and tear fluid in dry eye disease. Sci. Rep 7, 13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai M, and Sato I (2003). Morphological changes in collagen fiber during development of human fetal parotid and submandibular glands. Ann. Anat 185, 45–51. [DOI] [PubMed] [Google Scholar]
- Laine M, Porola P, Udby L, Kjeldsen L, Cowland JB, Borregaard N, Hietanen J, Ståhle M, Pihakari A, and Konttinen YT (2007). Low salivary dehydroepiandrosterone and androgen-regulated cysteine-rich secretory protein 3 levels in Sjögren’s syndrome. Arthritis Rheum. 56, 2575–2584. [DOI] [PubMed] [Google Scholar]
- Li J, Helmerhorst EJ, Yao Y, Nunn ME, Troxler RF, and Oppenheim FG (2004). Statherin is an in vivo pellicle constituent: identification and immuno-quantification. Arch. Oral Biol 49, 379–385. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, and Aebersold R (2016). On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 165, 535–550. [DOI] [PubMed] [Google Scholar]
- Lo HG, Jin RU, Sibbel G, Liu D, Karki A, Joens MS, Madison BB, Zhang B, Blanc V, Fitzpatrick JAJ, et al. (2017). A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev. 31, 154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo JA, Yan W, Ramachandran P, and Wong DT (2010). Comparative human salivary and plasma proteomes. J. Dent. Res 89, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariller C, Benaïssa M, Hardivillé S, Breton M, Pradelle G, Mazurier J, and Pierce A (2007). Human delta-lactoferrin is a transcription factor that enhances Skp1 (S-phase kinase-associated protein) gene expression. FEBS J. 274, 2038–2053. [DOI] [PubMed] [Google Scholar]
- Martins Mussi MC, Moffa E, Castro T, Lira Ortega A, Freitas G, Braga M, Siqueira WL, and Cury Gallottini MH (2016). Salivary parameters and oral health in the Moebius syndrome. Spec. Care Dentist 36, 265–270. [DOI] [PubMed] [Google Scholar]
- Mattingly A, Finley JK, and Knox SM (2015). Salivary gland development and disease. Wiley Interdiscip. Rev. Dev. Biol 4, 573–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavragani CP, and Moutsopoulos HM (2020). Sjögren’s syndrome: Old and new therapeutic targets. J. Autoimmun 110, 102364. [DOI] [PubMed] [Google Scholar]
- Merritt AD, Rivas ML, Bixler D, and Newell R (1973). Salivary and pancreatic amylase: electrophoretic characterizations and genetic studies. Am. J. Hum. Genet 25, 510–522. [PMC free article] [PubMed] [Google Scholar]
- Miao Q, Hill MC, Chen F, Mo Q, Ku AT, Ramos C, Sock E, Lefebvre V, and Nguyen H (2019). SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat. Commun 10, 4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksicek RJ, Myal Y, Watson PH, Walker C, Murphy LC, and Leygue E (2002). Identification of a novel breast- and salivary gland-specific, mucin-like gene strongly expressed in normal and tumor human mammary epithelium. Cancer Res. 62, 2736–2740. [PubMed] [Google Scholar]
- Murr A, Pink C, Hammer E, Michalik S, Dhople VM, Holtfreter B, Völker U, Kocher T, and Gesell Salazar M (2017). Cross-Sectional Association of Salivary Proteins with Age, Sex, Body Mass Index, Smoking, and Education. J. Proteome Res 16, 2273–2281. [DOI] [PubMed] [Google Scholar]
- Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, and Elowitz MB (2018). Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell 172, 869–880.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen Y-H, Schjoldager KT, Steentoft C, et al. (2019). An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol. Cell 75, 394–407.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PA, Mandel U, Therkildsen MH, and Clausen H (1996). Differential expression of human high-molecular-weight salivary mucin (MG1) and low-molecular-weight salivary mucin (MG2). J. Dent. Res 75, 1820–1826. [DOI] [PubMed] [Google Scholar]
- Niklason LE (2018). Understanding the Extracellular Matrix to Enhance Stem Cell-Based Tissue Regeneration. Cell Stem Cell 22, 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Noonan V, Rebustini I, Walker J, Menko AS, and Kukuruzinska MA (2009). Overexpression of DPAGT1 leads to aberrant N-glycosylation of E-cadherin and cellular discohesion in oral cancer. Cancer Res. 69, 5673–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack S, Seiffart V, Willbold E, Laggies S, Winkel A, Shahab-Osterloh S, Flörkemeier T, Hertwig F, Steinhoff C, Nuber UA, et al. (2014). Periostin secreted by mesenchymal stem cells supports tendon formation in an ectopic mouse model. Stem Cells Dev. 23, 1844–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, and Helmerhorst EJ (2007). Salivary proteome and its genetic polymorphisms. Ann. N Y Acad. Sci 1098, 22–50. [DOI] [PubMed] [Google Scholar]
- Oyelakin A, Song EAC, Min S, Bard JE, Kann JV, Horeth E, Smalley K, Kramer JM, Sinha S, and Romano RA (2019). Transcriptomic and Single-Cell Analysis of the Murine Parotid Gland. J. Dent. Res 98, 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, Pavlidis P, Dean K, Neznanova L, Romano R-A, Garneau D, Daugherity E, Globig A, Ruhl S, and Gokcumen O (2019). Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 8, e44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-W, Zhen G, Verhaeghe C, Nakagami Y, Nguyenvu LT, Barczak AJ, Killeen N, and Erle DJ (2009). The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 106, 6950–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkkila S, Kaunisto K, Rajaniemi L, Kumpulainen T, Jokinen K, and Rajaniemi H (1990). Immunohistochemical localization of carbonic anhydrase isoenzymes VI, II, and I in human parotid and submandibular glands. J. Histochem. Cytochem 38, 941–947. [DOI] [PubMed] [Google Scholar]
- Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GMH, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EEL, et al. (2014). The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev 260, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. (2007). Diet and the evolution of human amylase gene copy number variation. Nat. Genet 39, 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor GB, Hamdan S, Carpenter GH, and Wilde P (2005). A statherin and calcium enriched layer at the air interface of human parotid saliva. Biochem. J 389, 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujato M, Kieken F, Skiles AA, Tapinos N, and Fiser A (2014). Prediction of DNA binding motifs from 3D models of transcription factors; identifying TLX3 regulated genes. Nucleic Acids Res. 42, 13500–13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T, Gibbs GM, Merriner DJ, Kerr JB, and O’Bryan MK (2008). Cysteine-rich secretory proteins are not exclusively expressed in the male reproductive tract. Dev. Dyn 237, 3313–3323. [DOI] [PubMed] [Google Scholar]
- Ruhl S (2012). The scientific exploration of saliva in the post-proteomic era: from database back to basic function. Expert Rev. Proteomics 9, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Brand T, Zweigerdt R, and Arnold H (2000). Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech. Dev 95, 163–174. [DOI] [PubMed] [Google Scholar]
- Schwenk JM, Omenn GS, Sun Z, Campbell DS, Baker MS, Overall CM, Aebersold R, Moritz RL, and Deutsch EW (2017). The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays. J. Proteome Res 16, 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Dudus L, Nielsen PA, Clausen H, Yankaskas JR, Hollingsworth MA, and Engelhardt JF (1998). MUC5B and MUC7 are differentially expressed in mucous and serous cells of submucosal glands in human bronchial airways. Am. J. Respir. Cell Mol. Biol 19, 30–37. [DOI] [PubMed] [Google Scholar]
- Shen P, Sun J, Xu G, Zhang L, Yang Z, Xia S, Wang Y, Liu Y, and Shi G (2014). KLF9, a transcription factor induced in flutamide-caused cell apoptosis, inhibits AKT activation and suppresses tumor growth of prostate cancer cells. Prostate 74, 946–958. [DOI] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, and Wegner M (2004). Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol 24, 6635–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu J, and Romano R-A (2018). Genetic and scRNA-seq Analysis Reveals Distinct Cell Populations that Contribute to Salivary Gland Development and Maintenance. Sci. Rep 8, 14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spörl F, Korge S, Jürchott K, Wunderskirchner M, Schellenberg K, Heins S, Specht A, Stoll C, Klemz R, Maier B, et al. (2012). Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc. Natl. Acad. Sci. USA 109, 10903–10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedtler F, Hartmann N, Letzkus M, Bongiovanni S, Scherer A, Marc P, Johnson KJ, and Schumacher MM (2013). Robust and tissue-independent gender-specific transcript biomarkers. Biomarkers 18, 436–445. [DOI] [PubMed] [Google Scholar]
- Stubbs M, Chan J, Kwan A, So J, Barchynsky U, Rassouli-Rahsti M, Robinson R, and Bennick A (1998). Encoding of human basic and glycosylated proline-rich proteins by the PRB gene complex and proteolytic processing of their precursor proteins. Arch. Oral Biol 43, 753–770. [DOI] [PubMed] [Google Scholar]
- Szymaniak AD, Mi R, McCarthy SE, Gower AC, Reynolds TL, Mingueneau M, Kukuruzinska M, and Varelas X (2017). The Hippo pathway effector YAP is an essential regulator of ductal progenitor patterning in the mouse submandibular gland. eLife 6, e23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak LA (1995). In defense of the oral cavity: structure, biosynthesis, and function of salivary mucins. Annu. Rev. Physiol 57, 547–564. [DOI] [PubMed] [Google Scholar]
- Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, and Takayanagi H (2015). Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell 163, 975–987. [DOI] [PubMed] [Google Scholar]
- Takahashi N, and Horie T (2002). [Heerfordt syndrome]. Nihon Rinsho 60, 1822–1826. [PubMed] [Google Scholar]
- Tanaka J, Ogawa M, Hojo H, Kawashima Y, Mabuchi Y, Hata K, Nakamura S, Yasuhara R, Takamatsu K, Irié T, et al. (2018). Generation of orthotopically functional salivary gland from embryonic stem cells. Nat. Commun 9, 4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapinos NI, Polihronis M, Thyphronitis G, and Moutsopoulos HM (2002). Characterization of the cysteine-rich secretory protein 3 gene as an early-transcribed gene with a putative role in the pathophysiology of Sjögren’s syndrome. Arthritis Rheum. 46, 215–222. [DOI] [PubMed] [Google Scholar]
- Thamadilok S, Roche-Håkansson H, Håkansson AP, and Ruhl S (2016). Absence of capsule reveals glycan-mediated binding and recognition of salivary mucin MUC7 by Streptococcus pneumoniae. Mol. Oral Microbiol 31, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamadilok S, Choi K-S, Ruhl L, Schulte F, Kazim AL, Hardt M, Gokcumen O, and Ruhl S (2020). Human and Non-Human Primate Lineage-Specific Footprints in the Salivary Proteome. Mol. Biol. Evol 37, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomadaki K, Helmerhorst EJ, Tian N, Sun X, Siqueira WL, Walt DR, and Oppenheim FG (2011). Whole-saliva proteolysis and its impact on salivary diagnostics. J. Dent. Res 90, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Roca L, Tsaalbi-Shtylik A, Jansen JG, Singh MK, Epstein JA, Altunoglu U, Verzijl H, Soria L, van Beusekom E, Roscioli T, et al. (2015). De novo mutations in PLXND1 and REV3L cause Möbius syndrome. Nat. Commun 6, 7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udby L, Cowland JB, Johnsen AH, Sørensen OE, Borregaard N, and Kjeldsen L (2002). An ELISA for SGP28/CRISP-3, a cysteine-rich secretory protein in human neutrophils, plasma, and exocrine secretions. J. Immunol. Methods 263, 43–55. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Veerman EC, van den Keybus PA, Vissink A, and Nieuw Amerongen AV (1996). Human glandular salivas: their separate collection and analysis. Eur. J. Oral Sci 104, 346–352. [DOI] [PubMed] [Google Scholar]
- Veerman ECI, van den Keijbus PAM, Nazmi K, Vos W, van der Wal JE, Bloemena E, Bolscher JGM, and Amerongen AVN (2003). Distinct localization of MUC5B glycoforms in the human salivary glands. Glycobiology 13, 363–366. [DOI] [PubMed] [Google Scholar]
- Vissink A, van Luijk P, Langendijk JA, and Coppes RP (2015). Current ideas to reduce or salvage radiation damage to salivary glands. Oral Dis. 21, e1–e10. [DOI] [PubMed] [Google Scholar]
- Walz A, Stühler K, Wattenberg A, Hawranke E, Meyer HE, Schmalz G, Blüggel M, and Ruhl S (2006). Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis. Proteomics 6, 1631–1639. [DOI] [PubMed] [Google Scholar]
- Walz A, Odenbreit S, Stühler K, Wattenberg A, Meyer HE, Mahdavi J, Borén T, and Ruhl S (2009). Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics 9, 1582–1592. [DOI] [PubMed] [Google Scholar]
- Wang D, Eraslan B, Wieland T, Hallström B, Hopf T, Zolg DP, Zecha J, Asplund A, Li L-H, Meng C, et al. (2019). A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol 15, e8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2009). Ggplot2: Elegant Graphics for Data Analysis (Springer; ). [Google Scholar]
- Wingett SW, and Andrews S (2018). FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res. 7, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G-Q, Li F, Unger PD, Katerji H, Yang Q, McMahon L, and Burstein DE (2016). ZBTB16: a novel sensitive and specific biomarker for yolk sac tumor. Mod. Pathol 29, 591–598. [DOI] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Thamadilok S, Redwood E, Fox S, Blekhman R, Ruhl S, and Gokcumen O (2016). Recent evolution of the salivary mucin MUC7. Sci. Rep 6, 31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Taskent RO, Alachiotis N, Flanagan C, DeGiorgio M, Blekhman R, Ruhl S, and Gokcumen O (2017). Archaic Hominin Introgression in Africa Contributes to Functional Salivary MUC7 Genetic Variation. Mol. Biol. Evol 34, 2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Yonemura S, and Takada S (2006). Grainyhead-related transcription factor is required for duct maturation in the salivary gland and the kidney of the mouse. Development 133, 4737–4748. [DOI] [PubMed] [Google Scholar]
- Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, Cole S, Fang X, Gonzalez-Begne M, Griffin TJ, et al. (2009). Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin. Appl 3, 116–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jiang S, Lu C, Ji T, Yang W, Li T, Lv J, Hu W, Yang Y, and Jin Z (2019). SOX11: friend or foe in tumor prevention and carcinogenesis? Ther. Adv. Med. Oncol 11, 1758835919853449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye DZ, and Kaestner KH (2009). Foxa1 and Foxa2 control the differentiation of goblet and enteroendocrine L- and D-cells in mice. Gastroenterology 137, 2052–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. (2018). Ensembl 2018. Nucleic Acids Res. 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li J, Lea R, Vleminckx K, and Amaya E (2014). Fezf2 promotes neuronal differentiation through localised activation of Wnt/β-catenin signalling during forebrain development. Development 141, 4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H-D, Fan S-Q, Zhao J, Huang D-H, Zhou M, Liu H-Y, Zeng Z-Y, Yang Y-X, Huang H, Li X-L, et al. (2006). Tissue distribution of the secretory protein, SPLUNC1, in the human fetus. Histochem. Cell Biol 125, 315–324. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang J, Zhang L, Zhou X, Cisar JO, and Palmer RJ Jr. (2016). Differential Utilization of Basic Proline-Rich Glycoproteins during Growth of Oral Bacteria in Saliva. Appl. Environ. Microbiol 82, 5249–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data (fastq files) have been submitted to GEO https://www.ncbi.nlm.nih.gov/geo/ with the project name GSE143702. The custom codes are available through https://github.com/GokcumenLab/


