Abstract
Accurate DNA repair and replication are critical for genomic stability and cancer prevention. RAD51 and its gene family are key regulators of DNA fidelity through diverse roles in double-strand break repair, replication stress, and meiosis. RAD51 is an ATPase that forms a nucleoprotein filament on single-stranded DNA. RAD51 has the function of finding and invading homologous DNA sequences to enable accurate and timely DNA repair. Its paralogs, which arose from ancient gene duplications of RAD51, have evolved to regulate and promote RAD51 function. Underscoring its importance, misregulation of RAD51, and its paralogs, is associated with diseases such as cancer and Fanconi anemia. In this review, we focus on mammalian RAD51 structure and function and highlight the use of model systems to enable mechanistic understanding of RAD51 cellular roles. We also discuss how misregulation of the RAD51 gene family members contributes to disease and consider new approaches to pharmacologically inhibit RAD51.
Keywords: RAD51, RAD51 paralog, double-strand break repair, homologous recombination, replication, Shu complex
INTRODUCTION TO DOUBLE-STRAND BREAK REPAIR
Double-strand breaks (DSBs) are one of the most cytotoxic DNA lesions, and their misrepair leads to mutations and translocations. DSBs can arise from exogenous sources, such as radiation and chemotherapy, as well as from endogenous sources, such as metabolic byproducts, reactive oxygen species, replication stress, and even scheduled endonucleolytic activity [V(D)J recombination and meiosis]. Homologous recombination (HR) and nonhomologous end joining (NHEJ) are two major DSB repair pathways. NHEJ is a fast, although potentially error-prone, mechanism that religates the DNA ends. NHEJ is active during all phases of the cell cycle and is the preferred DSB repair pathway in higher eukaryotes (91). On the other hand, HR uses a homologous DNA template for repair and in mammalian cells is most active during the S and G2 phases of the cell cycle (61). HR is favored over NHEJ at DSBs with dirty ends or when only one DNA end is available, such as in replication-associated DSBs (53).Thus, HR offers a high-fidelity and versatile alternative for DSB repair.
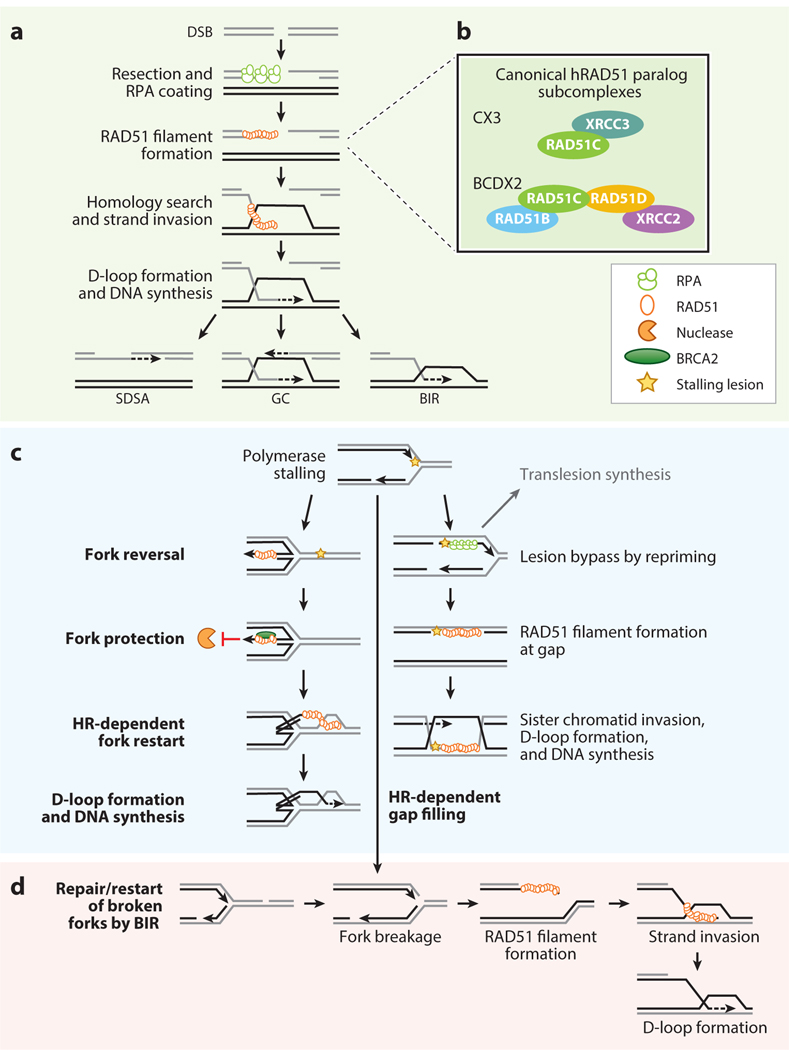
HR, and the recombinase activity of RAD51, is central to three main DSB repair pathways: gene conversion (GC), synthesis-dependent strand annealing (SDSA), and RAD51-dependent break-induced replication (BIR) (Figure 1a). The defining feature of these pathways is the strand exchange of homologous sequences that serve as a template to restore broken DNA. The initial steps of these pathways are shared. Briefly, the MRE11-RAD50-NBS1 (MRN) complex with CtIP recognizes and binds to DSBs (128). This enables short-range resection to expose 3′ single-stranded DNA (ssDNA) overhangs. Subsequent long-range resection is achieved by the 5′ to 3′ exonuclease activity of EXO1 or with the combined activities of DNA2 and BLM (72). This ssDNA is rapidly coated by replication protein A (RPA), preventing the formation of ssDNA secondary structures and degradation (27). RAD51 then displaces RPA to assemble nucleoprotein filaments with the 3 ssDNA ends. This central HR step is highly regulated to prevent unscheduled recombination. RAD51 filament assembly is stimulated by RAD51 mediators such as the RAD51 loader, BRCA2, and the RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3, and SWSAP1) (Figure 1b). The RAD51 nucleoprotein filaments invade a homologous region, forming a displacement loop (D-loop). The way that this heteroduplex DNA intermediate is resolved following DNA synthesis determines which pathway, GC, SDSA, or BIR, occurs (49) (Figure 1a). Most recombination events are likely resolved through SDSA, where after DNA synthesis the D-loop is disrupted, allowing the newly synthetized DNA end to anneal to the other end of the broken DNA molecule. During GC, the second end of the DSB is captured, forming a Holliday junction. This structure is then processed by endonucleases, helicases, and topoisomerases to allow separation of the DNA molecules. Alternatively, when the second end of the DSB is not captured, then DNA synthesis at the D-loop proceeds, resulting in BIR.
Figure 1.
Potential models for the RAD51 gene family roles in mitotic cells. (a) Schematic of the initial steps of DSB repair through HR. Upon DSB formation, the 5 strands of the DNA ends are resected by the MRN complex. Further resection is performed by the exonuclease EXO1 and/or DNA2 together with the BLM helicase. These exposed 3 ssDNA regions are immediately coated by the RPA complex (green circles), thus preventing the formation of secondary structures. RAD51 (orange circles) displaces RPA, aided by the RAD51 mediators (i.e., BRCA2 and the RAD51 paralogs) to form a RAD51 nucleoprotein filament on the ssDNA. The RAD51 filaments perform homology search and strand invasion, leading to D-loop formation. After the D-loop is extended by DNA synthesis, the repair process can be completed by SDSA, GC, or BIR, depending on whether the D-loop is disrupted, the second end is captured, or it is not captured, respectively. (b) Schematic of the canonical hRAD51 paralog subcomplexes BCDX2 (consisting of RAD51B, RAD51C, RAD51D, and XRCC2) and CX3 (consisting of RAD51C and XRCC3). The lines indicate where BCDX2 and CX3 function during HR. (c) Schematic of the roles of RAD51 during replication stress response. Replicative polymerases can be stalled by DNA lesions such as methylation adducts or abasic sites (yellow star). Fork reversal (left) occurs by the annealing of the newly synthetized strands and is dependent on RAD51 and other enzymes such as translocases or helicases (i.e., SMARCL1 and RAD54). This chicken-foot structure protects stalled forks and allows the rescue of the fork by an incoming replication fork or by bypassing the lesion. Protection of the reversed forks from nuclease digestion (orange pacman) prevents ssDNA accumulation and depends on the formation of stable RAD51 filaments at the ssDNA of the reversed arm, which requires BRCA2 (green oval). Finally, reversed forks can be restarted by direct reversal (not shown) or HR. Alternatively (right) polymerase can resume replication by repriming. This leads to the formation of ssDNA gaps behind the fork, which are RPA coated. These gaps can be filled by translesion synthesis or HR-dependent gap filling. During HR-dependent gap filling, RAD51 displaces RPA in the gap and mediates sister chromatid invasion and D-loop formation. DNA synthesis enables the gap to be filled, enabling error-free lesion bypass. (d) Broken forks generated by persistent stalling or encountering of a ssDNA break by the replisome, leading to one-ended DSBs. These breaks can be repaired by RAD51-dependent HR where RAD51 filaments form on the broken DSB end, which then invade the newly synthesized sister chromatid, leading to D-loop formation, which can be extended by BIR. Abbreviations: BIR, break-induced replication; BLM, bloom syndrome protein; D-loop, displacement loop; DSB, double-strand break; GC, gene conversion; HR, homologous recombination; MRN, MRE11-RAD50-NBS1; RPA, replication protein A; SDSA, synthesis-dependent strand annealing; ssDNA, single-stranded DNA.
RECA/RAD51 ORIGIN AND EVOLUTION
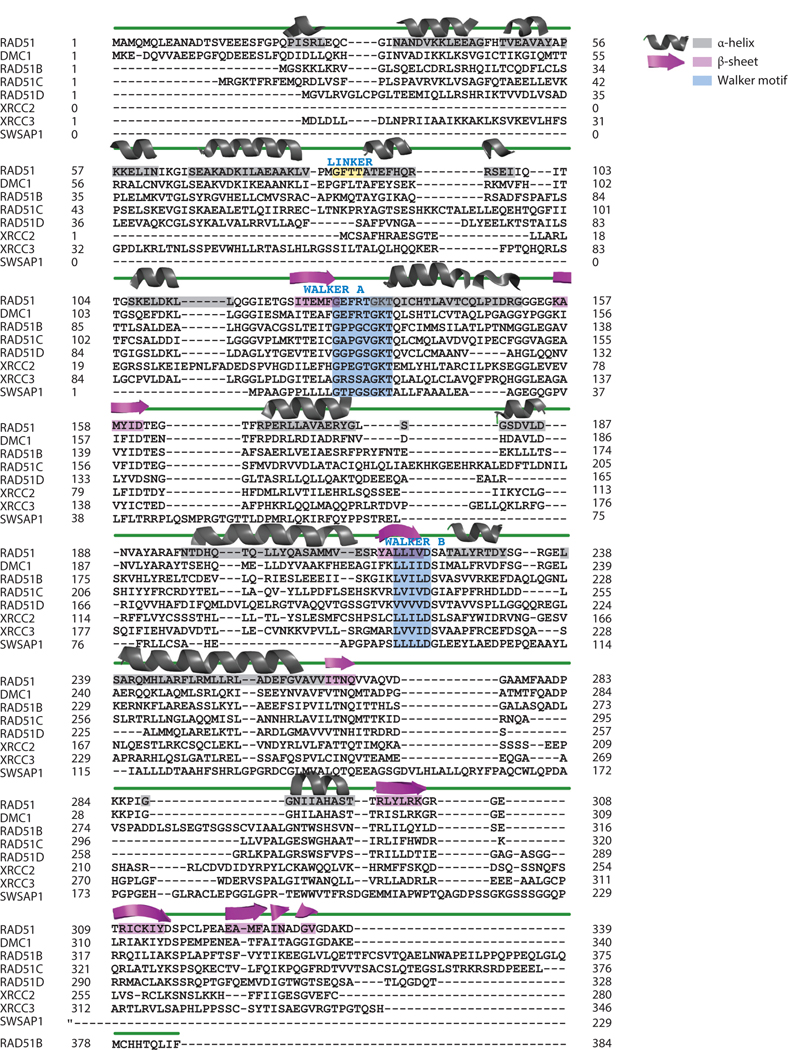
The Escherichia coli RecA and eukaryotic RAD51 superfamily of recombinases is present across all domains of life, with the only exceptions being some intracellular bacteria with extremely small genomes (87, 111). In fact, the universal distribution of this gene group has led to its use as an alternative to the 16S ribosomal RNA in phylogenetic analysis (142).The recA/RAD51 superfamily originates from an ancient common ancestor before the appearance of Archaea and Eukarya. Early seminal work by Lin et al. (70) divided this family into three groups: recA, RADα, and RADβ (28). The recA group includes all bacterial recA genes as well as eukaryotic recA genes present in plants, protists, and some fungi (70). The RADα group includes the primary recombinases in eukaryotes (RAD51 and DMC1) and Archaea (radA). Vertebrate RAD51 share ~74% amino acid sequence identity with yeast and plants, while RAD51 from humans and mice are 99% identical (3). The RADβ group includes the canonical eukaryotic RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2,and XRCC3) and the archaeal radB. Members of the RADβ group have typically evolved distinctive functions that have yet to be fully characterized (44, 123). The RADβ group exhibits a great deal of diversity, with highly divergent and rapidly evolving genes that share little sequence identity with yeast and plants, while RAD51 from humans and mice are 99% identical (3). The RADβ group includes the canonical eukaryotic RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) and the archaeal radB. Members of the RADβ group have typically evolved distinctive functions that have yet to be fully characterized (44, 123). The RADβ group exhibits a great deal of diversity, with highly divergent and rapidly evolving genes that share little sequence homology. In light of this, several proteins from different organisms have been proposed as RAD51 paralogs based on small conserved motifs, structural and/or functional conservation. These are often referred to as noncanonical RAD51 paralogs and include Schizosaccharomyces pombe rlp1 and rdl1 (63); Saccharomyces cerevisiae CSM2, PSY3, and SHU1 (76, 104, 150); human SWSAP1 (73); and Caenorhabditis elegans rip-1 and rfs-1 (131). Sequence alignment of the RADα, RADβ, and noncanonical paralog SWSAP1 highlights that while sequence is highly variant, key regions like the Walker A and B motifs are conserved (Figure 2).
Figure 2.
Sequence alignment of human RAD51 protein and its family, including DMC1, RAD51B, RAD51C, RAD51D, XRCC2, XRCC3, and SWSAP1. Secondary structure of human RAD51 [Protein Data Bank (PDB) ID 5H1B] is shown above the aligned protein sequence with α-helices shown in gray and β-sheets shown in purple. The Walker A and B sequence motifs are in blue boxes, and the RAD51 linker (amino acids 85-GTFF-88) is in a yellow box.
The current diversity observed in the recA/RAD51 superfamily in Archaea and eukaryotes is a result of ancestral gene duplications followed by diversification in function as well as horizontal gene transferring after endosymbiotic events (70). Most bacteriophages have proteins that perform DNA recombination, with several of them being recA homologs (UvsX, SAR1) (74, 142). More recent studies propose that the bacterial sms (also known as radA) are recA paralogs (30), as well as a group of archaeal radA paralogs named radC (50). This analysis suggests that there are likely additional undiscovered members of the RADβ gene group. Given the limits of phylogenetic analyses, functional and structural criteria will be critical to further define additional RAD51 gene family members.
Throughout this article, we use RAD51 when referring to general properties common across species, whereas species-specific properties use a species-designated name (i.e., Homo sapiens RAD51 as hRAD51, murine RAD51 as mRAD51, and Saccharomyces cerevisiae Rad51 as scRad51). Similarly, proteins that are species specific are not indicated (i.e.,BRCA2),whereas species-specific observations for complexes that are shared between species use a species designation (i.e., hShu complex versus scShu complex).
RAD51 AND THE RAD51 FILAMENT: STRUCTURE AND FUNCTION
The defining feature of HR is the use of a homologous DNA molecule as a repair template. To achieve this, the broken DNA molecule needs to find and invade homologous DNA. The RAD51 recombinase, assembled on ssDNA as an oligomeric nucleoprotein filament, is responsible for carrying out these activities. In this section, we describe the structure of the RAD51 filament, its properties, the molecular basis of its formation, and how it performs homology search and strand invasion.
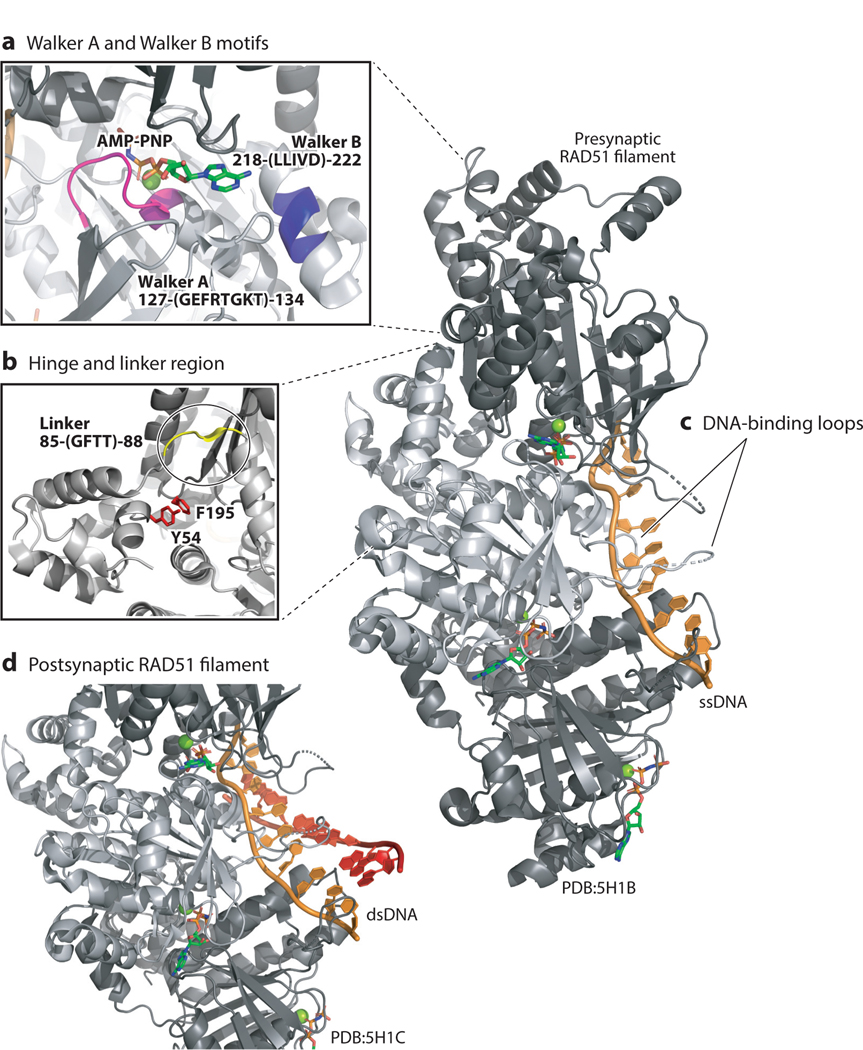
The RAD51 protein itself contains an α/β ATPase core domain similar to those present in helicases and other proteins that hydrolyze nucleotide triphosphates. This domain comprises a Walker A motif (GXXXXGKT/S) and a Walker B motif (R/K-XXX-G-XXX-LhhhD), which are important for ATP binding and hydrolysis (114, 143) (Figure 3a). hRAD51 ATP hydrolysis is dependent on DNA, with observed kcat of 0.16 to 0.21 ATP/min and 0.05 ATP/min in the presence of ssDNA and double-stranded DNA (dsDNA), respectively (135). Compared to RecA, hRAD51 is a weak ATPase, with 150–200-fold lower activity even when stimulated by ssDNA and dsDNA. However, the significance of this difference remains unknown. Monomeric hRAD51 protein binds three nucleotides of DNA. However, maximal ATPase activity is observed when there are six to eight nucleotides of ssDNA per hRAD51 monomer, suggesting multiple binding sites (135). Other conserved domains in hRAD51 include an N-terminal domain containing five α-helices and a β-strand that mediates monomer–monomer interactions, two disordered loops that bind DNA (Figure 3c), and a flexible interdomain linker (Figure 3b).
Figure 3.
Structural view of human RAD51 presynaptic and postsynaptic filament complexes (PDB IDs 5H1B and 5H1C). RAD51 protomers are shown in dark and light gray, Mg2+ ions are in green, AMP-PNP is depicted in stick form, ssDNA is shown in orange, and the dsDNA template is shown in orange and red. (a) AMP-PNP is buried between two RAD51 protomers. The Walker A (pink; amino acids 218-LLIVD-222) and Walker B (blue; amino acids 127-GEFRTGKT-134) motifs are highlighted. (b) Key residues for promoter–protomer interfaces are the linker region (circled; amino acids 85-GFTT-88 in yellow) and Y54 in one protomer with F195 in an adjacent protomer. (c) The ssDNA-bound RAD51 filament viewed from the side highlights the phosphate backbone (orange) directly engaging DNA-binding loops of RAD51 (arrows). (d) The postsynaptic filament highlights the initial bound dsDNA is in a similar conformation as the presynaptic filament shown in panel c. Abbreviations: AMP-PNP, adenylyl-imidodiphosphate; dsDNA, double-stranded DNA; PDB, Protein Data Bank; ssDNA, single-stranded DNA.
Helical Parameters and Dynamic Rearrangements of RAD51 Nucleoprotein Complexes
Since the RAD51 filaments are dynamic structures, different biophysical approaches such as single-molecule scanning force microscopy, cryo-electron microscopy (cryoEM), and X-ray crystallography have captured RAD51 filaments in several conformations. Moreover, the conformations of the RAD51 filament are influenced by experimental conditions such as DNA substrate, nucleotide cofactors, and cations (100, 138).
The DNA in the hRAD51 filament is nonuniformly extended 1.5-fold compared to the canonical B-form DNA confirmation (100, 114, 143). The experimental helical parameters obtained for RAD51 filaments across several studies vary depending on the bound nucleotide analog (114). While crystallography and cryoEM structures capture ordered conformations of the hRAD51 nucleoprotein complex, experiments using scanning force microscopy show that hRAD51 rarely displays regular ordered complexes on ssDNA in solution (100). The observation of irregular filaments supports the idea that hRAD51 on ssDNA samples many conformations and is dynamic in solution. All atomic structures have several conserved features. For example, ATP or analog molecules bind between two hRAD51 monomers, and the RAD51-ATP/analog complexes demonstrate several possible hRAD51 conformations (Figure 3a). The flexibility of the interdomain linker likely enables the observed conformational states, since the free energy difference between the two states is small (4 kBT per hRAD51 protomer), and interconversion between states is proposed to occur independently of ATP hydrolysis (17).The hRAD51 monomers interact with each other in an antiparallel manner through a β-strand (143) (Figure 3b). Recent cryoEM structures of the hRAD51-ssDNA filament bound to ATP analog adenylyl-imidodiphosphate (AMP-PNP) revealed an open conformation of the filament with 6.4 monomers per turn on ssDNA (pitch 103 Å and rise 16.1 Å) (114, 143) and 6.3 hRAD51 monomers per turn on ssDNA (pitch 110 Å and rise 15.8 Å) in the presence of the meiosis-specific cofactor mHop2-mMnd1 (143). In the presence of nonhydrolyzable ATP analogs or ssDNA, captured RAD51 conformations have similar helical parameters (114, 143, 146). These conformations with similar helical parameters are likely due to dynamic changes upon ATP hydrolysis (118).
RAD51 DNA Binding and Nucleation
hRAD51 binds both ssDNA and dsDNA to form a right-handed helical nucleoprotein filament where each hRAD51 monomer binds three nucleotides of DNA (114, 143). Filament assembly begins by nucleation of hRAD51 dimers at multiple discontinuous and irregular sites along the ssDNA and not exclusively at the DNA end (100, 121). After nucleation, the hRAD51 filament is elongated bidirectionally by the addition of hRAD51 dimers (121). In this dynamic, stepwise elongation process, assembly and disassembly occur at similar rates (121). Interestingly, scRad51 filaments are elongated by both monomers and dimers in a 5′–3′ direction, whereas bacterial RecA grows by the bidirectional addition of monomers (9,98). The hRAD51 assembly on dsDNA occurs at multiple nucleation positions via the binding of two to three hRAD51 monomers (57).
RAD51 Mediators
RAD51 displaces RPA-coated ssDNA with the aid of the RAD51 mediators. The main RAD51 mediator in vertebrates is BRCA2 (96), whose function is performed by Rad52 in yeast (124). scRad52 and BRCA2 are responsible for accelerating the rate-limiting step of recruiting and nucleating RAD51 on RPA-coated ssDNA (122). Other mediators in vertebrates include the RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) and the Shu complex, which contains the RAD51 paralog SWSAP1 (123). The function of the RAD51 paralogs is discussed separately in the section titled RAD51 Paralogs.
BRCA2 is recruited to DSBs through its interaction with PALB2, which itself interacts with BRCA1, binds DNA, and associates with chromatin (96). BRCA2 then binds to ssDNA through its oligonucleotide/oligosaccharide-binding (OB)-fold-domain-containing C-terminal DNA-binding domain (145). BRCA2 interacts with RAD51, which facilitates its recruitment to ssDNA, through eight conserved BRC domains and a C-terminal motif (141). Binding of the BRCA2 BRC motif to RAD51 inhibits RAD51 ATPase activity, which in turn enhances RAD51’s affinity for ssDNA (24). Additionally, the fifth to eighth BRC motifs preferentially bind the RAD51 filament, promoting its stability (25). Also contributing to RAD51 filament stabilization are additional interactions between RAD51 and PALB2, BRCA1, and BARD1 (22, 35, 151). RAD51 is also post-translationally modified by the phosphorylation of tyrosine 54 (121). hRAD51 phosphorylation enhances the recombinase activity of hRAD51 by increasing its ability to compete with RPA for ssDNA and stimulating its strand exchange activity. This modification likely enables a conformation of the hRAD51 filament that is optimal for recombination (Figure 3b).
Homology Search and Strand Exchange
After RAD51-ssDNA filament formation, the RAD51 nucleoprotein filament performs homology search and strand invasion of a homologous template. As was first shown in yeast, and subsequently in mammalian cells, the RAD51-mediated homology search is an active process where the DNA ends of multiple breaks cluster together into a repair center (4,109).In vitro single-molecule studies demonstrate that hRAD51 nucleoprotein filaments can slide along DNA and perform three-dimensional sampling to efficiently locate a homologous repair template (47). scRad51 samples DNA at distances ranging from <1 kb to >70 kb from the DSB site, and this sampling is impaired at the more distal regions by the loss of the Rad51 paralog complex Rad55-Rad57 (99). hRAD51 homology search occurs via eight nucleotide homologous tract lengths, where dsDNA strand capture occurs upon nine homologous nucleotides and commitment to perform HR with ~15 nucleotides of homology (68, 97). Strand exchange occurs in three nucleotide increments (68, 97).
While structures of hRAD51-DNA complexes are well described, few studies have captured the homology search and strand-exchange process at near atomic-level resolution. CryoEM studies by the Sung lab (143) captured hRAD51 presynaptic, synaptic, and postsynaptic states, demonstrating that it is the complementary donor dsDNA that deforms, while the DNA in the hRAD51 filaments maintains its conformation throughout this process. These findings differ from studies of scRad51, which report that scRad51 is displaced from the D-loop once the homologous sequence is found (130). In vitro assays with scRad51 show that homology search, strand invasion, and joint molecule formation are independent of ATP hydrolysis (126). However, in vivo recombination requires RAD51 ATPase activity (88, 119).
Because RAD51 bound to ADP has a lower affinity for DNA, ATP hydrolysis facilitates filament disassembly (57).scRad51 filament disassembly can be stimulated by several proteins, including the anti-recombinase Srs2. The motor protein Rad54 assists with the unloading of scRad51 filaments from D-loops and late HR structures, which promotes efficient recombination (130). Other examples include RECQL5, FBH1,and RTEL1 (123).Consistent with their suggested role in filament disassembly, the knockdown of any of these proteins results in increased chromosomal abnormalities, most likely due to aberrant or excessive recombination (7, 36, 60).
RAD51 PARALOGS
The RAD51 paralogs are conserved mediators of RAD51, supporting its function in DSB repair, meiosis, and replication (8, 44). Despite their importance, mechanistic details on how they promote HR are still unclear. In this section we describe RAD51 paralog complexes, their properties, and their general role promoting HR.
In budding yeast, the scRad51 paralogs form two distinct complexes: the Rad55-Rad57 complex and Shu complex. The canonical scRad51 paralogs Rad55-Rad57 are required for DSB repair and interact with scRad51 and scRad52 (39, 71, 125, 127). Rad55 also interacts with the other scRad51 paralog complex, the Shu complex (39, 45). This heterotetrametric complex is composed of the SWIM domain–containing protein Shu2 and the scRad51 paralogs Csm2, Psy3, and Shu1 (104,113).Importantly,bothscRad51 paralog complexes promote HR by at least two mechanisms: (a) helping scRad51 overcome the inhibitory effect of RPA during filament formation (39, 125) and (b) stabilizing scRad51 filaments by counteracting the anti-recombinase activity of Srs2 (10, 71). Notably, the Rad55-Rad57 complex also promotes scRad51-mediated strand exchange (125). In nematodes, C. elegans RAD51 (ceRAD-51) paralogs RSF-1 and RIP-1 alter the ceRAD-51 filament by increasing its flexibility, thus enhancing strand exchange and D-loop formation (131). This RAD51 paralog-mediated filament remodeling is likely conserved in other eukaryotes.
Progress in understanding hRAD51 paralog function has been particularly challenging, due to their propensity to form insoluble aggregates in vitro, their low cellular abundance, and the fact that knockout mutants show lethality in mice and noncancerous cells (123). Mutant mice lacking canonical mRAD51 paralogs die at different developmental stages, ranging from embryonic day 7.5 to 10.5 (reviewed in 123). Consistent with paralogs promoting RAD51 activities, a recent study by Garcin et al. (42) individually disrupted the canonical hRAD51 paralogs in U2OS and HEK293 cells and observed reduced RAD51 foci, growth defects, DNA damage sensitivity, and impaired HR. These observations are analogous to those made using CHO and DT40 knockout hamster and chicken cell lines (123). Unlike the rest of the hRAD51 paralogs, RAD51B disruption generally results in milder phenotypes and is actually tolerated in nontransformed MCF-10A cells (42). SWSAP1 disruption is tolerated in human cell lines and mice (1, 78). SWSAP1 disruption leads to decreased RAD 51 focus formation upon treatment with methyl methanesulfonate(MMS), increased sensitivity to mitomycin C and MMS,but no increase in sensitivity to ionizing radiation, mirroring the phenotypes of scShu-complex knockout strains (78).
The hRAD51 paralogs function as distinct subcomplexes, including the CX3 complex (RAD51C and XRCC3), the BCDX2 complex (RAD51B, RAD51C, RAD51D, and XRCC2), the RAD51C-RAD51-BRCA2-PALB2 complex, and the Shu complex (SWSAP1, SWS1 likely with SPIDR and PDS5B) (5, 73, 78, 123) (Figure 1b). Like RAD51, the RAD51 paralogs are ATPases with conserved Walker A and B motifs (Figure 2). The BCDX2 complex hydrolyzes ATP in the presence of ssDNA (observed kcat of 0.88 min−1) (80). The BCDX2 and CX3 paralog complexes bind to a diverse range of DNA substrates such as ssDNA, 3 and 5 flaps, gapped circular DNA, and nicked duplex substrates (80). Although the BCDX2 complex ssDNA binding activity is ATP-independent, ATP hydrolysis is stimulated by ssDNA. The human Shu (hShu) complex is composed of SWSAP1 and SWS1. The hShu complex also likely includes SPIDR and PDS5B, although the interaction with PDS5B may not be direct (78). Consistent with a RAD51 mediator function, SWS1-SWSAP1 promotes RAD51 recruitment into DNA repair foci, enables sister-chromatid exchange and replication restart, and counteracts FIGNL1 antirecombinase activity (76–78, 81). The CX3 complex, like hRAD51, has additional functions in mitochondrial replication and maintenance as well as the FA pathway during interstrand crosslink repair (86, 102, 103). The role of the RAD51 paralogs during replicative DNA damage is discussed below.
ROLES OF RAD51 IN REPLICATION-ASSOCIATED DNA DAMAGE
Accurate and timely DNA replication is critical to maintain genome stability. RAD51 is a central player in overcoming replication stress, which slows or stalls replication forks, threatening replication integrity (149). In this section, we summarize the roles of the RAD51 gene family during replication stress, including promoting fork reversal, protecting reversed forks, repairing and restarting broken replication forks, and postreplicative gap filling (Figure 1c,d).
RAD51 Function in Fork Reversal
Upon replication fork stalling, fork reversal can promote genome stability by (a) limiting ssDNA accumulation,(b) replacing replication-blocking lesions in the context of dsDNA to allow the subsequent repair of the lesion by other repair pathways such as base excision repair, (c) providing a template for lesion bypass through template switching, and (d) enabling HR-dependent replication restart (55, 90). Stalled replication fork reversal or regression involves the reannealing of the parental strands and annealing of the newly synthesized daughter strands, which converts a three-way DNA junction into a four-way DNA junction (Figure 1c). Migration of this Holliday-like junction extends the reversed arm, forming a so-called chicken-foot structure (90). Many proteins are able to catalyze fork reversal in vitro, such as fork remodelers (SMARCAL1, ZRANB3, HLTF, and RAD54) or helicases (BLM, FBH1, WRN, and FANCM) (13, 90). hRAD51 is similarly required for fork reversal (148). How hRAD51 promotes fork reversal is still unclear, but several models are proposed. RAD51 may assist other factors to promote fork reversal where hRAD51 binding to the ssDNA at the chicken-foot arm might drive the reaction toward the reversed products (13, 21). Alternatively, RAD51 bound to ssDNA at one of the stalled fork strands may invade the newly replicated strand, reannealing the parental DNA and thus displacing the newly synthesized strand (16, 90). Surprisingly, hRAD51’s fork-reversal role does not require its strand exchange activities and is BRCA2-independent (64, 79, 85). The length of the ssDNA at the reversed arm during fork reversal initiation is not sufficient for RPA binding, and thus RAD51 filament mediators may not be needed (13). Alternatively, the MMS22L-TONSL complex might perform mediator functions in this context (13).
RAD51 Function in Fork Protection of Reversed Forks
RAD51 also protects reversed forks from uncontrolled enzymatic degradation (Figure 1c). When unprotected, the reversed arm is an entry point for CtIP-MRE11 with EXO1 or DNA2-mediated degradation (56, 64, 69, 85, 129). When BRCA2 stabilizes the hRAD51 filament on the ssDNA region of the reversed arm, fork degradation is inhibited (107, 129). Consistently, hRAD51 filament stabilization by overexpression of a catalytic dead RAD51 mutant can overcome BRCA2 loss (108). Furthermore, an hRAD51 Fanconi anemia (FA) allele, RAD51-T131P, forms unstable filaments that impair fork protection without compromising HR when heterozygous with a wild-type hRAD51 allele (139, 147). RAD51 function in fork protection is likely structural, whereas its enzymatic activity is required for HR.
RAD51 Function in Restart of Reversed Forks
When a replication fork is stalled and cannot be rescued by an incoming fork, cells rely on fork restart mechanisms to complete replication (Figure 1c).Reversed forks can be directly restarted or restored by the helicase RECQL1 or the translocase SMARCAL1 (11, 12). An alternative restart mechanism involves DNA2-WRN-mediated limited resection at the regressed arms to produce a 3′ overhang (134). RAD51 filaments formed at an ssDNA 3′ overhang regressed arm may drive an HR-directed restart by invading the homologous DNA ahead of the reversed fork (2, 94). However, a detailed mechanism for HR-directed restart remains obscure. Similarly, in fission yeast, recent work shows that forks stalled at a replication fork barrier (RTS1-RFB) can be restarted by a DSB-independent HR-mediated process (133).
RAD51 Function in Restart of Broken Forks by BIR
Replication fork breakage results in what are usually referred to as one-ended or single end DSBs (Figure 1d). As mentioned above, the absence of a second DNA end makes cells rely on HR for their repair. Several scenarios can lead to fork breakage, such as the replisome encountering an ssDNA gap or transcription-replication collisions (31).Broken replication forks can be repaired by BIR. In yeast, this process begins with DNA end resection, followed by scRad51 filament assembly on the 3′ ssDNA (2). The scRad51 filament performs homology-directed strand invasion of the sister chromatid to form a D-loop that is extended by DNA synthesis (65). Although error-prone, the range of this synthesis is limited by scMus81 cleavage of the Holliday junction at the D-loop or by merging with an incoming replication fork in the opposite direction (82). Although most of our knowledge of BIR comes from yeast,there is evidence suggesting that this process is conserved in vertebrates (32, 51). It is worth noting that an alternative scRad51-independent BIR pathway is also known, and it accounts for several processes described in human cells (65). Finally, BIR drives the alternative lengthening of telomeres; however, the contribution of the hRAD51-dependent subpathway is still debated (65).
RAD51 Function in Postreplicative Repair
In addition to its roles at replication forks, RAD51 is also central in an HR-driven postreplicative gap-filling pathway (Figure 1c). This process relies on a template switch between sister chromatids to complete replication at ssDNA gaps and serves as an error-free alternative to translesion synthesis (TLS) (95). These ssDNA gaps are generated when the replisome bypasses polymerase-stalling DNA lesions, such as those that can arise from ultraviolet or MMS treatment (95). As demonstrated in yeast, lesions on the lagging strand are bypassed due to the intrinsic discontinuous nature of lagging-strand DNA synthesis (132). Meanwhile, lesions at the leading strand can be skipped by downstream repriming by PrimPol, a specialized polymerase conserved in many eukaryotes, including mammals (14, 41). Although there is no PrimPol homolog in yeast, downstream leading strand repriming can be performed by Polα and primase (37). Details of the gap-filling pathway have been best described in yeast, where it plays a major role in tolerating replicative damage (16,95).In short, the ssDNA gaps are extended by the nucleases scExo1 and scMre11 and the helicase Pif1, followed by scRad51 filament assembly. This scRad51-coated gap invades the sister chromatid and displaces the daughter strand, which becomes paired with the free 3 end (43). After DNA synthesis occurs, the sister chromatid junctions are dissolved by the Sgs1-Top3Rmi1 complex. Importantly, this DNA damage-tolerance pathway is dependent on proliferating cell nuclear antigen (PCNA) polyubiquitination by the sequential activities of the Rad6-Rad18 and Mms2-Ubc13-Rad5 complexes. Most of these yeast factors have homologs in higher eukaryotes, suggesting that this processes is evolutionarily conserved (137). In response to replication stress, humans may rely on fork reversal and TLS, whereas yeast are thought to primarily use HR-mediated gap filling.
Role of the RAD51 Paralogs in Replication-Associated Damage
Support for a role during the replication stress response comes from findings that, in many eukaryotes, RAD51 paralog mutations lead to sensitivity to replicative DNA damage and defects in RAD51 recruitment (44, 123). In yeast, the Rad55-Rad57 complex and scShu complexes contribute to the repair and tolerance of replication-associated damage by promoting HR-dependent gap filling (44). As with scRad51, mutants lacking either of these complexes show increased mutagenesis that is TLS-dependent and delayed S-phase progression upon MMS treatment (6, 46, 54, 75,113,144).The scShu complex is not required for DSB repair from IR, and Rad55 phosphorylation promotes MMS resistance but is dispensable for DSB repair, further supporting specific roles for these Rad51 paralog complexes during replication stress (54, 113). Recently, we showed that the scShu complex preferentially binds abasic site containing substrates and promotes error-free tolerance of these lesions, primarily on the lagging strand (101).
Similarly, recent evidence suggests a role for hRAD51 paralogs in replication-associated damage. For example, like hRAD51, both the BCDX2 and CX3 subcomplexes are required to protect forks from hMRE11 degradation (117). The CX3 complex also promotes fork restart after hydroxyurea (HU) treatment (94, 117). Additionally, XRCC2 and XRCC3 are phosphorylated by the ATR DNA damage response kinase to promote fork slowdown upon nucleotide depletion and DSB repair, respectively (105, 116). We showed that, unlike the canonical RAD51 paralogs, the hShu complex contributes to fork restart following HU treatment but is dispensable for fork protection (78).
DMC1 AND THE ROLE OF THE RAD51 GENE FAMILY DURING MEIOSIS
Meiosis enables the generation of haploid cells or gametes. During meiosis, homologous chromosomes are first paired with each other. Homologous pairing requires HR-mediated homology search to bring the two chromosomes together. HR during meiosis shares many essential features with mitotic HR-mediated DSB repair, including that both processes begin with a DSB.
Meiotic DSBs are physiologically generated by the meiosis-specific and universally conserved SPO11 (reviewed in 62). These DSBs are then processed in the same fashion as during mitotic DSB repair to generate nucleoprotein filaments that are responsible for the homology search. During meiosis, there is a strong preference to use the homologous chromosome as a repair template, as opposed to a preference for the sister chromatid observed in mitotic DSB repair. This process is known as homolog bias (66). Once homologs are paired, proper homolog segregation also requires that crossovers are formed, as opposed to during mitotic DSB repair, where crossovers are less desirable (59). Another key difference between meiotic and mitotic HR is that meiosis requires the recombinase function of DMC1 at the core of the nucleoprotein filament (29).
DMC1 is a conserved meiosis-specific RAD51 paralog that arose from an early gene duplication in the eukaryotic lineage (70). Unlike the other RAD51 paralogs, DMC1 still resembles RAD51, sharing 54% sequence identity in humans and 45% in yeast. Consistent with this, the biochemical properties, nucleoprotein filament structure, and recombinase activity of DMC1 are remarkably similar to those of RAD51 (18).
scRad51 and scDmc1 have distinct functions during meiosis. scDmc1 acts as the main recombinase, while scRad51 plays an accessory role to mediate the assembly and regulate the activity of scDmc1 (29). scRad51’s recombinase activity is dispensable during meiosis and is actively inhibited, primarily by the Hed1 protein (23, 29). However, scRad51 is required for scDmc1 focus formation and for homolog bias (112). It is still not known if these distinct roles are conserved in vertebrates. scRad51 and scDmc1 are distinctively distributed within the meiotic nucleoprotein filament and tend to self-aggregate forming side-by-side homotypic filaments (18, 19, 34). Although this configuration is likely conserved, its role is still unclear. One possibility is that this would prevent Hed1-inactivated RAD51 monomers to intercalate in DMC1 filaments (34).
DMC1 and RAD51 in yeast and humans also demonstrate differences that may explain the nearly universal need for two meiotic recombinases. Unlike RAD51, DMC1 can tolerate mismatches during heteroduplex formation (15). More recently, Steinfeld et al. (120) elegantly identified conserved residues in the L1 DNA-binding loop of DMC1 that are responsible for this mismatch tolerance. This DMC1-specific feature likely enables recombination between homologous chromosomes that contain mismatches, contributing to homolog bias.
Recent work shows that meiotic scRad51-scDmc1 filaments are more stable than mitotic scRad51 filaments. Unlike scRad51 filaments, which are readily disassembled by the antirecombinase Srs2 to promote SDSA during mitotic DSB repair, scDmc1 is a strong inhibitor of Srs2, which renders the meiotic scRad51-Dmc1 filaments resistant to Srs2 disassembly (33). An exciting possibility is that this enhanced stability of the meiotic filament contributes to the key step of crossover production.
Given the critical role of scRad51 during meiosis, it is not surprising that the scRad51 paralogs are also important for meiosis (40, 58). Deletion of RAD55 and RAD57 leads to meiotic defects that greatly resemble those observed in rad51 mutants, including impaired homolog bias and decreased spore viability (40, 110). Like the canonical scRAD51 paralogs, the scShu complex mutants also show meiotic defects, but these are not as severe as those observed upon RAD51, RAD55, or RAD57 deletion (58, 104). Interestingly, unlike during mitotic HR where deletion of any of the four scShu complex members leads to a complete loss of function, C2M2 and PSY3 are more important during meiosis than SHU1 and SHU2 (104). Consistent with their role as Rad51 mediators, deletion of RAD55-RAD57 or the scShu complex impairs recruitment of scRad51 to meiotic DSBs (104). Similarly, Rad51 paralogs in fission yeast, worms, and plants are also important for meiosis (26, 48, 83). Not much is known about the meiotic role of the RAD51 paralogs in vertebrates. However, a recent publication by Abreu et al. (1) shed light on the importance of the murine Shu complex during meiosis. Mice with either Sws1−/− or Swsap1−/− knockout mutations are sterile, cannot complete meiosis, and have decreased Rad51 and Dmc1 foci (1). These phenotypes are reminiscent of the scShu complex mutants. Despite the similarities between RAD51 and DMC1, direct physical interactions between the RAD51 paralogs and DMC1 have not yet been described. Therefore, the RAD51 paralogs are important during meiosis due to their role as RAD51 mediators.
RAD51 GENE FAMILY IN CANCER PREDISPOSITION AND FANCONI ANEMIA
Mutations in the hRAD51 gene family are associated with cancer predisposition and FA-like syndromes (123). FA is defined by bone marrow failure, congenital defects, and an increased risk for leukemia and solid cancer tumors (89). To date, there are 22 FA genes that have been identified, including RAD51 (FANCR), RAD51C (FANCO), and XRCC2 (FANCU) (89). Mutations in both alleles of RAD51, RAD51C, or XRCC2 result in FA or a FA-like syndrome, whereas monoallelic mutations in some FA genes, like RAD51C, result in cancer predisposition (84, 89). While only RAD51C and XRCC2 are associated with FA, mutations in all the hRAD51 paralogs have been identified in cancers (84, 93, 138). Most of these hRAD51 paralog mutations are associated with breast and ovarian cancer; however, there are additional mutations that correlate with other cancer types such as lung, kidney, and head and neck (84, 93, 106, 115, 138). Unfortunately, most hRAD51 and hRAD51 paralog point mutations that have been clinically identified are classified as variants of unknown significance (VUSs). Future studies to reclassify these hRAD51 gene family VUSs as pathogenic or benign are desperately needed, as many of these genes are now included on hereditary breast and ovarian cancer screening panels. Reclassification of HR-deficient VUSs would enable these patients to benefit from therapies that specifically target HR deficiency, as do poly(ADP)-ribose polymerase (PARP) inhibitors in BRCA1/2-deficient cells.
PHARMACOLOGICAL INHIBITION OF RAD51
While mutations that inactivate hRAD51 are associated with cancer, hRAD51 overexpression is also observed in cancer patients. hRAD51 is an important biomarker for cancer development, and several groups are developing strategies to pharmacologically inhibit hRAD51 functions (20, 67, 140). hRAD51 is of particular interest as a target for cancer therapeutics because it is frequently overexpressed in several tumors types, including prostate, lung, bladder, and breast cancer (20). Tumors that overexpress hRAD51 often exhibit treatment resistance and lower overall survival rates of patients (140). hRAD51 overexpression results in hyperrecombination that not only furthers cancer progression, but also enables cancer cells to resist DNA-damaging agents (38, 140). This resistance can be overcome in tissue culture by reducing hRAD51 expression, which sensitizes cancer cell lines to chemotherapeutic agents to a greater extent than noncancerous cell lines (20). hRAD51 can be targeted indirectly by downregulating hRAD51 expression or directly by impairing its protein function (67, 140). Indirect inhibitors include tyrosine kinase inhibitors, methotrexate, small interfering RNAs and microRNAs, and histone deacetylase inhibitors (140). However, clinical applications of these inhibitors are limited due to the inherent challenges that these compounds present, including incomplete suppression of hRAD51 expression, lack of specificity, and the inability to maintain high intracellular concentration (reviewed in 67). Compounds that directly target hRAD51 function either disrupt its protein interactions or modify its recombinase activity (52, 140). Direct inhibitors of hRAD51 include small molecules (halenaquinone, B02, RI-1, RI-2, IBR2, DIDS, and RS) and antibodies (such as 3E10 and Fab-F2-iPTD) (20, 52, 67, 92, 136, 140). Despite an abundance of preclinical data and the high specificity of these compounds for hRAD51, there is currently only one active clinical trial investigating the efficacy of a small molecule hRAD51 inhibitor (67). The challenge will now be to see if the results of the preclinical studies can be replicated in a clinical setting.
CONCLUDING REMARKS
The RAD51 gene family is evolutionarily conserved in all domains of life with a conserved ATPase core domain. Recent studies have identified divergent RAD51 paralogs that lack sequence homology but closely resemble the overall structure of RAD51. Future phylogenetic analysis will likely reveal additional RAD51 gene family members. RAD51 forms a nucleoprotein filament on ssDNA ends that is stimulated by its mediators. It remains unknown precisely how the RAD51 paralogs modulate RAD51 filament dynamics. It is possible that RAD51 paralogs are stably incorporated into the filament, cap the filament, or influence filament structure. Furthermore, it is debated whether the RAD51 paralogs aid in postsynaptic HR steps. While RAD51 has emerged as a key player in replication through fork reversal, protection, and restart, the role of the paralogs in regulating RAD51 in these contexts awaits elucidation. The Shu complex consists of RAD51 paralogs in all species where it has been studied. While Shu complex mutants share some of the same features of canonical RAD51 paralog knockouts, it remains to be determined why Shu complex meiotic functions are more important than roles in mitotic DSB repair. Perhaps Shu complex interaction with the cohesion protein PDS5B is more critical for meiosis, due to its role in chromosome segregation. Future studies focused on the biochemical function and structure of the hShu complex will shed light on this important distinction. Finally, misregulation of RAD51 and its gene family is associated with cancer and FA. Given the close association of RAD51 misregulation with cancer predisposition, understanding the underlying mechanisms of RAD51 gene family structure and function is critical for designing new cancer therapeutics and mechanistic insights into human disease.
ACKNOWLEDGMENTS
The authors acknowledge that not all primary sources were able to be cited due to a limited number of references permitted. K.A.B. is supported by grants from the National Institutes of Health (ES030335) and an American Cancer Society Research Scholar Award (129182-RSG-16-043-01-DMC), and by a Stand Up to Cancer Innovation Research Grant (SU2C-AACR-IRG-02-16). S.R.H. is supported by an American Cancer Society Post-Doctorate Research Fellowship (133947-PF-19-132-01-DMC).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abreu CM, Prakash R, Romanienko PJ, Roig I, Keeney S, Jasin M. 2018. Shu complex SWS1-SWSAP1 promotes early steps in mouse meiotic recombination. Nat. Commun 9:3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait Saada A, Lambert SAE, Carr AM. 2018. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 71:135–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andriuskevicius T, Kotenko O, Makovets S. 2018. Putting together and taking apart: assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability. Cell Stress 2:96–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, et al. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303:92–95 [DOI] [PubMed] [Google Scholar]
- 5.Baldock RA, Pressimone CA, Baird JM, Khodakov A, Luong TT, et al. 2019. RAD51D splice variants and cancer-associated mutations reveal XRCC2 interaction to be critical for homologous recombination. DNA Repair 76:99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. 2009. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol 73:89–102 [DOI] [PubMed] [Google Scholar]
- 7.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, et al. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135:261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer FE, Deichsel S, Mahl P, Nagel AC. 2020. Drosophila Xrcc2 regulates DNA double-strand repair in somatic cells. DNA Repair 88:102807. [DOI] [PubMed] [Google Scholar]
- 9.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. 2012. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491:274–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. 2011. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell 22:1599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, et al. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol 20:347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betous R, Couch FB, Mason AC, Eichman BF, Manosas M, Cortez D. 2013. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 3:1958–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhat KP, Cortez D. 2018. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol 25:446–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, et al. 2013PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52:566–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgogno MV, Monti MR, Zhao W, Sung P, Argarana CE, Pezza RJ. 2016. Tolerance of DNA mismatches in Dmc1 recombinase-mediated DNA strand exchange. J. Biol. Chem 291:4928–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branzei D, Szakal B. 2017. Building up and breaking down: mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol 52:381–94 [DOI] [PubMed] [Google Scholar]
- 17.Brouwer I, Moschetti T, Candelli A, Garcin EB, Modesti M, et al. 2018. Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA. EMBO J. 37:e98162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MS, Bishop DK. 2014. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol 7:a016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown MS, Grubb J, Zhang A, Rust MJ, Bishop DK. 2015. Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLOS Genet. 11:e1005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budke B, Lv W, Kozikowski AP, Connell PP.2016Recent developments using small molecules to target RAD51: how to best modulate RAD51 for anticancer therapy? ChemMedChem 11:2468–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bugreev DV, Rossi MJ, Mazin AV. 2011. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 39:2153–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, et al. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol 17:1247–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22:786–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, et al. 2009The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136:1032–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carreira A, Kowalczykowski SC. 2011. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. PNAS 108:10448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlot F, Chelysheva L, Kamisugi Y, Vrielynck N, Guyon A, et al. 2014. RAD51B plays an essential role during somatic and meiotic recombination in Physcomitrella. Nucleic Acids Res. 42:11965–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Lisby M, Symington LS. 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50:589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chintapalli SV,Bhardwaj G,Babu J,Hadjiyianni L,Hong Y,et al. 2013Reevaluation of the evolutionary events within recA/RAD51 phylogeny. BMC Genom. 14:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337:1222–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper DL, Lovett ST. 2016. Recombinational branch migration by the RadA/Sms paralog of RecA in Escherichia coli. eLife 5:e10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez D. 2019. Replication-coupled DNA repair. Mol. Cell 74:866–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, et al. 2014. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343:88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crickard JB, Kaniecki K, Kwon Y, Sung P, Greene EC. 2018. Meiosis-specific recombinase Dmc1 is a potent inhibitor of the Srs2 antirecombinase. PNAS 115:E10041–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crickard JB, Kaniecki K, Kwon Y, Sung P, Greene EC. 2018. Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J. Biol. Chem 293:4191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, et al. 2010. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol 17:1255–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, et al. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol 186:655–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fumasoni M, Zwicky K, Vanoli F, Lopes M, Branzei D. 2015. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polα/Primase/Ctf4 complex. Mol. Cell 57:812–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gachechiladze M, Skarda J, Soltermann A, Joerger M. 2017. RAD51 as a potential surrogate marker for DNA repair capacity in solid malignancies. Int. J. Cancer 141:1286–94 [DOI] [PubMed] [Google Scholar]
- 39.Gaines WA, Godin SK, Kabbinavar FF, Rao T, VanDemark AP, et al. 2015. Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat. Commun 6:7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Game JC,Mortimer RK.1974A genetic study of X-ray sensitive mutants in yeast.Mutat.Res 24:281–92 [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, et al. 2013. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52:541–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcin EB, Gon S, Sullivan MR, Brunette GJ, Cian A, et al. 2019. Differential requirements for the RAD51 paralogs in genome repair and maintenance in human cells. PLOS Genet. 15:e1008355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. 2014. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol 21:884–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godin SK, Sullivan MR, Bernstein KA. 2016. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem. Cell Biol 94:407–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godin SK, Wier A, Kabbinavar F, Bratton-Palmer DS, Ghodke H, et al. 2013. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 41:4525–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Godin SK, Zhang Z, Westmoreland JW, Lee AG, Mihalevic MJ, et al. 2016The Shu complex promotes error-free tolerance of alkylation-induced base-excision repair products. Nucleic Acids Res. 30:8199–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graneli A, Yeykal CC, Robertson RB, Greene EC.2006Long-distance lateral diffusion of human Rad51 on double-stranded DNA. PNAS 103:1221–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grishchuk AL, Kohli J.2003Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165:1031–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haber JE. 2018. DNA repair: the search for homology. Bioessays 40:e1700229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haldenby S, White MF, Allers T. 2009. RecA family proteins in archaea: RadA and its cousins. Biochem. Soc. Trans 37:102–7 [DOI] [PubMed] [Google Scholar]
- 51.Hashimoto Y, Puddu F, Costanzo V. 2011. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol 19:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hengel SR, Spies MA, Spies M. 2017Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem. Biol 24:1101–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Her J, Bunting SF. 2018. How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem 293:10502–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, et al. 2006. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell. Biol 26:8396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Higgins NP, Kato K, Strauss B. 1976. A model for replication repair in mammalian cells. J. Mol. Biol 101:417–25 [DOI] [PubMed] [Google Scholar]
- 56.Higgs MR,Reynolds JJ,Winczura A,Blackford AN,Borel V,et al. 2015BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell 59:462–77 [DOI] [PubMed] [Google Scholar]
- 57.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. 2009. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. PNAS 106:361–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong S, Kim KP. 2013. Shu1 promotes homolog bias of meiotic recombination in Saccharomyces cerevisiae. Mol. Cells 36:446–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol 7:a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Islam MN, Fox D 3rd, Guo R, Enomoto T, Wang W. 2010. RecQL5 promotes genome stabilization through two parallel mechanisms—interacting with RNA polymerase II and acting as a helicase. Mol. Cell. Biol 30:2460–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karanam K, Kafri R, Loewer A, Lahav G. 2012. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47:320–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keeney S. 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab 2:81–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khasanov FK, Salakhova AF, Chepurnaja OV, Korolev VG, Bashkirov VI.2004Identification and characterization of the rlp1+, the novel Rad51 paralog in the fission yeast Schizosaccharomyces pombe. DNA Repair 3:1363–74 [DOI] [PubMed] [Google Scholar]
- 64.Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, et al. 2017. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell 67:867–81.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kramara J,Osia B,Malkova A. 2018Break-induced replication: the where, the why, and the how. Trends Genet. 34:518–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lao JP, Hunter N. 2010. Trying to avoid your sister. PLOS Biol. 8:e1000519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurini E, Marson D, Fermeglia A, Aulic S, Fermeglia M, Pricl S. 2020. Role of Rad51 and DNA repairin cancer: a molecular perspective. Pharmacol. Ther 208:107492. [DOI] [PubMed] [Google Scholar]
- 68.Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, et al. 2015. Base triplet stepping by the Rad51/RecA family of recombinases. Science 349:977–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemacon D, Jackson J, Quinet A, Brickner JR, Li S, et al. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Z, Kong H, Nei M, Ma H. 2006. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. PNAS 103:10328–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. 2011. Rad51 paralogues Rad55–Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479:245–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu T, Huang J. 2016. DNA end resection: facts and mechanisms. Genom. Proteom. Bioinform 14:126–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu T, Wan L, Wu Y, Chen J, Huang J. 2011. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J. Biol. Chem 286:41758–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopes A, Amarir-Bouhram J, Faure G, Petit M-A, Guerois R. 2010. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 38:3952–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mankouri HW, Ngo HP, Hickson ID. 2007. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18:4062–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, et al. 2006. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 25:2564–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martino J, Bernstein KA. 2016The Shu complex is a conserved regulator of homologous recombination. FEMS Yeast Res. 16:fow073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martino J, Brunette GJ, Barroso-Gonzalez J, Moiseeva TN, Smith CM, et al. 2019. The human Shu complex functions with PDS5B and SPIDR to promote homologous recombination. Nucleic Acids Res. 47:1051–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mason JM, Chan YL, Weichselbaum RW, Bishop DK. 2019. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun 10:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, et al. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15:3296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsuzaki K, Kondo S, Ishikawa T, Shinohara A. 2019. Human RAD51 paralogue SWSAP1 fosters RAD51 filament by regulating the anti-recombinase FIGNL1 AAA+ ATPase. Nat. Commun 10:1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, et al. 2015. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 349:742–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McClendon TB, Sullivan MR, Bernstein KA, Yanowitz JL. 2016. Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics 203:133–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, et al. 2010. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene.Nat.Genet. 42:410–14 [DOI] [PubMed] [Google Scholar]
- 85.Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, et al. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mishra A, Saxena S, Kaushal A, Nagaraju G. 2018. RAD51C/XRCC3 facilitates mitochondrial DNA replication and maintains integrity of the mitochondrial genome. Mol. Cell. Biol 38:e00489–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moran NA,Mira A.2001The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2:research0054.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morgan EA, Shah N, Symington LS. 2002. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol. Cell. Biol 22:6336–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nalepa G, Clapp DW. 2018. Fanconi anaemia and cancer: an intricate relationship. Nat. Rev. Cancer 18:168–85 [DOI] [PubMed] [Google Scholar]
- 90.Neelsen KJ, Lopes M.2015Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol 16:207–20 [DOI] [PubMed] [Google Scholar]
- 91.Pannunzio NR, Watanabe G, Lieber MR. 2018. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem 293:10512–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pastushok L, Fu Y, Lin L, Luo Y, DeCoteau JF, et al. 2019. A novel cell-penetrating antibody fragment inhibits the DNA repair protein RAD51. Sci. Rep 9:11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pennington KP,Walsh T,Harrell MI,Lee MK,Pennil CC,et al. 2014Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res 20:764–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37:492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prado F. 2018. Homologous recombination: to fork and beyond. Genes 9:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qi W,Wang R,Chen H,Wang X,Xiao T,et al. 2015BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci 128:317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. 2013. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun 4:2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Renkawitz J, Lademann CA, Kalocsay M, Jentsch S. 2013. Monitoring homology search during DNA double-strand break repair in vivo. Mol. Cell 50:261–72 [DOI] [PubMed] [Google Scholar]
- 100.Ristic D,Modesti M,van der Heijden T,van Noort J,Dekker C,et al. 2005Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 33:3292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rosenbaum JC, Bonilla B, Hengel SR, Mertz TM, Herken BW, et al. 2019The Rad51 paralogs facilitate a novel DNA strand specific damage tolerance pathway. Nat. Commun 10:3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sage JM, Gildemeister OS, Knight KL. 2010. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem 285:18984–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S. 2004. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenet. Genome Res 104:28–34 [DOI] [PubMed] [Google Scholar]
- 104.Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, et al. 2013. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun 4:1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saxena S, Somyajit K, Nagaraju G. 2018. XRCC2 regulates replication fork progression during dNTP alterations. Cell Rep. 25:3273–82.e6 [DOI] [PubMed] [Google Scholar]
- 106.Scheckenbach K, Baldus SE, Balz V, Freund M, Pakropa P, et al. 2014. RAD51C—a new human cancer susceptibility gene for sporadic squamous cell carcinoma of the head and neck (HNSCC). Oral. Oncol 50:196–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, et al. 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwacha A, Kleckner N. 1997. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90:1123–35 [DOI] [PubMed] [Google Scholar]
- 111.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 112.Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK.1997Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination.Genes Cells 2:615–29 [DOI] [PubMed] [Google Scholar]
- 113.Shor E, Weinstein J, Rothstein R. 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169:1275–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Short JM, Liu Y, Chen S, Soni N, Madhusudhan MS, et al. 2016. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res. 44:9017–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Silva MC, Morrical MD, Bryan KE, Averill AM, Dragon J, et al. 2016. RAD51 variant proteins from human lung and kidney tumors exhibit DNA strand exchange defects. DNA Repair 42:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Somyajit K, Basavaraju S, Scully R, Nagaraju G. 2013. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol. Cell. Biol 33:1830–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Somyajit K,Saxena S,Babu S,Mishra A,Nagaraju G.2015Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43:9835–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spirek M, Mlcouskova J, Belan O, Gyimesi M, Harami GM, et al. 2018. Human RAD51 rapidly forms intrinsically dynamic nucleoprotein filaments modulated by nucleotide binding state. Nucleic Acids Res. 46:3967–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M.2002ATPhydrolysisbymammalianRAD51 has a key role during homology-directed DNA repair. J. Biol. Chem 277:20185–94 [DOI] [PubMed] [Google Scholar]
- 120.Steinfeld JB, Belan O, Kwon Y, Terakawa T, Al-Zain A, et al. 2019. Defining the influence of Rad51 andDmc1 lineage-specific amino acids on genetic recombination. Genes Dev. 33:1191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Subramanyam S, Ismail M, Bhattacharya I, Spies M. 2016. Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. PNAS 113:E6045–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sugiyama T, Kowalczykowski SC. 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem 277:31663–72 [DOI] [PubMed] [Google Scholar]
- 123.Sullivan MR, Bernstein KA. 2018. RAD-ical new insights into RAD51 regulation. Genes 9:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sung P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem 272:28194–97 [DOI] [PubMed] [Google Scholar]
- 125.Sung P.1997Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111–21 [DOI] [PubMed] [Google Scholar]
- 126.Sung P, Stratton SA. 1996Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem 271:27983–86 [DOI] [PubMed] [Google Scholar]
- 127.Symington LS. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev 66:630–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–71 [DOI] [PubMed] [Google Scholar]
- 129.Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, et al. 2017. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell 68:414–30.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tavares EM, Wright WD, Heyer WD, Le Cam E, Dupaigne P. 2019. In vitro role of Rad54 in Rad51-ssDNA filament-dependent homology search and synaptic complexes formation.Nat.Commun 10:4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor MRG, Spirek M, Chaurasiya KR, Ward JD, Carzaniga R, et al. 2015. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell 162:271–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Taylor MRG, Yeeles JTP. 2018. The initial response of a eukaryotic replisome to DNA damage. Mol. Cell 70:1067–80.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Teixeira-Silva A, Ait Saada A, Hardy J, Iraqui I, Nocente MC, et al. 2017. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun 8:1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, et al. 2015. DNA2 drives processingand restart of reversed replication forks in human cells. J. Cell Biol 208:545–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tombline G,Fishel R.2002Biochemical characterization of the human RAD51 protein.I.ATP hydrolysis. J. Biol. Chem 277:14417–25 [DOI] [PubMed] [Google Scholar]
- 136.Turchick A, Liu Y, Zhao W, Cohen I, Glazer PM. 2019. Synthetic lethality of a cell-penetrating anti-RAD51 antibody in PTEN-deficient melanoma and glioma cells. Oncotarget 10:1272–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Unk I, Hajdu I, Blastyak A, Haracska L. 2010. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 9:257–67 [DOI] [PubMed] [Google Scholar]
- 138.van der Zon NL, Kanaar R, Wyman C. 2018. Variation in RAD51 details a hub of functions: opportunities to advance cancer diagnosis and therapy. F1000Research 7:1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, et al. 2015. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell 59:478–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ward A, Khanna KK, Wiegmans AP. 2015. Targeting homologous recombination, new pre-clinical andclinical therapeutic combinations inhibiting RAD51. Cancer Treat. Rev 41:35–45 [DOI] [PubMed] [Google Scholar]
- 141.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL.1997RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem 272:31941–44 [DOI] [PubMed] [Google Scholar]
- 142.Wu D,Wu M,Halpern A,Rusch DB,Yooseph S,et al. 2011Stalking the fourth domain in metagenomics data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLOS ONE 6:e18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu J, Zhao L, Xu Y, Zhao W, Sung P, Wang HW. 2017. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol 24:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu X, Ball L, Chen W, Tian X, Lambrecht A, et al. 2013. The yeast Shu complex utilizes homologous recombination machinery for error-free lesion bypass via physical interaction with a Rad51 paralogue. PLOS ONE 8:e81371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, et al. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837–48 [DOI] [PubMed] [Google Scholar]
- 146.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. 2001. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. PNAS 98:8419–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zadorozhny K, Sannino V, Belan O, Mlcouskova J, Spirek M, et al. 2017. Fanconi-anemia-associated mutations destabilize RAD51 filaments and impair replication fork protection. Cell Rep. 21:333–40 [DOI] [PubMed] [Google Scholar]
- 148.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, et al. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol 208:563–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat. Cell Biol 16:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang S, Wang L, Tao Y, Bai T, Lu R, et al. 2017. Structural basis for the functional role of the Shu complex in homologous recombination. Nucleic Acids Res. 45:13068–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao W, Steinfeld JB, Liang F, Chen X, Maranon DG, et al. 2017. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 550:360–65 [DOI] [PMC free article] [PubMed] [Google Scholar]