Abstract
RING-finger E3 ligases are instrumental in the regulation of inflammatory cascades, apoptosis, and cancer. However, their roles are relatively unknown in TGF-β/SMAD signaling. SMAD3 and its adaptors, such as β2SP, are important mediators of TGF-β signaling and regulate gene expression to suppress stem cell-like phenotypes in diverse cancers, including hepatocellular carcinoma (HCC). Here, PJA1, an E3 ligase, promoted ubiquitination and degradation of phosphorylated SMAD3 and impaired a SMAD3/β2SP-dependent tumor-suppressing pathway in multiple HCC cell lines. In mice deficient for SMAD3 (Smad3+/−), PJA1 overexpression promoted the transformation of liver stem cells. Analysis of genes regulated by PJA1 knockdown and TGF-β1 signaling revealed 1,584 co-upregulated genes and 1,280 co-downregulated genes, including many implicated in cancer. The E3 ligase inhibitor RTA405 enhanced SMAD3-regulated gene expression and reduced growth of HCC cells in culture and xenografts of HCC tumors, suggesting that inhibition of PJA1 may be beneficial in treating HCC or preventing HCC development in at-risk patients.
Keywords: E3 ligase, PJA1, hepatocellular carcinoma, Smad3, TGF-β, β2SP
Introduction
The incidence of hepatocellular carcinoma (HCC) in the U.S. has increased three- to four-fold in recent years, and overall five-year survival rates are only 11% (1,2). Despite 4 new therapeutics and genomic analyses from The Cancer Genome Atlas (TCGA), HCC remains incurable with few effective genomic-based targeted therapeutics (3,4). Moreover, HCC is a heterogeneous cancer, and most patients present with underlying cirrhosis or decompensated liver disease; consequently, they are difficult to treat with standard doses of chemotherapeutics (5,6). Better strategies for HCC diagnosis, prevention, and therapy are urgently needed.
Liver is a regenerative organ with multiple regions in a lobule containing stem cells and niches regulating regeneration (7). The TGF-β/SMAD pathway is a key regulator of liver stem cells (LSCs) and liver regeneration (8-13). Some forms of HCC have characteristics of cancer stem cells (14) and may arise through transformation of cells in the LSC population. Mechanistic insight into the pathways that drive stem cell transformation could lead to the development or identification of targeted therapeutics for these cancers. Through an analysis of 9,125 cancers, we discovered that cancers with a cancer stem cell signature have altered TGF-β signaling (15). We observed that expression patterns of genes encoding components of this complex pathway are decreased in HCCs (~ 30-40% of HCCs), suggesting an inhibitory role of TGF-β in cancer stem cell transformation. An inhibitory role for TGF-β in this process is consistent with TGF-β–deficient mouse models of HCC (16-18).
TGF-β represents a large family of growth and differentiation factors that mobilize a complex signaling network to control cell fate by regulating differentiation, proliferation, motility, adhesion, and apoptosis (19-21). TGF-β superfamily signals are conveyed by specific intracellular mediators, the SMAD proteins. Vertebrates possess at least eight SMAD proteins, which are divided into three functional classes: (i) receptor-activated SMADs (R-SMADs): SMAD1, SMAD2, SMAD3, SMAD5, and SMAD8 (also known as SMAD9); (ii) co-mediator SMAD: SMAD4; and (iii) inhibitory SMADs (I-SMADs): SMAD6 and SMAD7. Upon phosphorylation by TGF-β receptors, R-SMADs translocate to the nucleus together with SMAD4 to regulate transcriptional targets. A negative feedback loop involving transcriptional upregulation of I-SMADs limits SMAD-mediated signaling (21). Transcriptional output of R-SMADs is influenced by positive and negative regulators of the TGF-β-SMAD pathway, interacting proteins, and the activity of other signal transduction pathways (22,23).
Here, we focused on SMAD3-dependent signaling, because SMAD3 controls stem cell function and contributes to liver regeneration (24-26). Impaired function of SMAD3 is associated with a human stem cell syndrome (27) that has an increased risk of developing cancer (28). The adaptor protein beta spectrin (β2SP), which is encoded by SPTBN1, genetically and biochemically interacts with SMAD3 to promote expression of tumor-suppressing genes in mice (18).
PJA1 of the PRAJA family of E3 ubiquitin ligases represents a negative regulator of SMAD3 and β2SP, because PJA1 promotes ubiquitination of SMAD3 and β2SP (29). The expression of PJA1 is increased in some cancers, including those of the gastrointestinal tract (29). Thus, we hypothesized that PJA1 impairs a tumor-suppressing TGF-β response mediated by SMAD3 and β2SP, thereby functioning as an oncogene or contributor to tumorigenesis.
To investigate this hypothesis, we manipulated PJA1 abundance in cultured HCC cells and determined the effect on phenotypes associated with cancer, such as proliferation, anchorage-independent growth, and tumor growth in mouse xenograft studies. We established LSCs from mice with compromised SMAD3 activity (Smad3+/−) and overexpressed PJA1 and tested their ability to form tumors when injected into mice. Because E3 ubiquitin ligases represent attractive potential therapeutic targets, we demonstrated that small molecule inhibitors of PJA1 increased SMAD3 and β2SP abundance in cultured HCC cells, increased SMAD3-dependent reporter gene expression, reduced HCC cell viability, and impaired xenografted tumor growth. Our results indicated that PJA1 functions as a tumor-promoting E3 ubiquitin ligase by inhibiting SMAD3/β2SP-regulated genes and suggested that PJA1 represents a potentially useful therapeutic target in HCC associated with impaired TGF-β signaling.
Materials and Methods
Study approval
All animal experiments were performed according to the guidelines for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of The University of Texas MDACC.
Plasmids, reagents, and antibodies
The PCR-generated DNA fragments of the human PJA1, mouse PJA1(30), human SMAD3, and human SPTBN1 genes (29) were subcloned into pCMV5 to generate constructs with a Flag tag or HA tag, or into pcDNA™6 to generate constructs with a V5 tag. Human PJA1 ring domain deletion (dR) (350aa–395aa deletion) constructs were generated in a Flag-tagged or HA-tagged vector. Mouse PJA1 1aa – 150aa or 150aa – 300aa mutants were generated in a Flag-tagged or V5-tagged vector. Mouse PJA1 cDNA was obtained from GE Dharmacon, Inc. and subcloned into the pB513B vector. Lentiviral particles containing shRNA of PJA1 (sc-91297) and control shRNA (sc-108080) were purchased from Santa Cruz Biotechnology. The 4xSBE luciferase reporter and 3TP luciferase reporter plasmids were from addgene, Inc., the Renilla plasmid was from Promega. MG132 (M7449, Sigma), and TGF-β1 (Sigma, T1654) were purchased from Sigma. G-418 was from (4727878001, Sigma).
Antibodies used were V5 (R961-25, Invitrogen), Flag (M2, Sigma, F3165), His (2366, Cell Signaling), ACTIN (A2066, Sigma), TUBULIN (T8328, Sigma), SMAD3 (9523, Cell Signaling), p-SMAD3 (9520, Cell Signaling), PJA1 (customized from BioSythesis), β2SP (customized from BioSythesis), Ki6 (2586, Cell Signaling), caspase-3 (ab2302, abcam), human IgG (2729, Cell Signaling), and CD133 (130-090-851, Miltenyi Biotec). Goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with Alexa-488 or Alexa-555 were from Molecular Probes. 4’,6-Diamidino-2-phenylindole (DAPI) or DRAQ5 (4084, Cell Signaling) was used to label nuclei. Propidium iodide (P1304MP, Thermo Fisher) was used to distinguish live and dead cells. Annexin V-FITC (ab14085) was purchased from abcam, Inc.
Cell culture, transfection, and shRNA silencing
All cells were grown in 5% CO2 in a humidified environment at 37°C. Human liver cancer cell lines HepG2 (ATCC, B8065), Hep3B (ATCC, HB8064), SNU449 (ATCC, CRL-2234), SNU475 (ATCC, CRL-2236), SNU398 (ATCC, CRL-2233) were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA), and Huh7 was gifted from Dr. Mien-Chie Hung’s lab, MD Anderson Cancer Center. All cells (gift from Dr. Mien-Chie Hung’s lab, MD Anderson Cancer Center) were cultured in DMEM/F-12 medium (Sigma-Aldrich, D5671) supplemented with 10% fetal bovine serum (Sigma-Aldrich, F2442). The cells were authenticated by short tandem repeat (STR) profiling and examined for Mycoplasma routinely by Mycoplasma Detection Kit (ThermoFisher Scientific, Catalog No. M7006). All cells were preserved in our lab between passages 2 and 20. Human normal hepatocytes THLE-3 (ATCC, CRL-11233) were purchased from ATCC, and directly lysated for Western Blot analyses. HepG2 and Hep3B cells were transfected with tagged PJA1, PJA1-dR, or SMAD3 plasmids using Lipofectamine 2000 or LTX (Invitrogen) according to the manufacturer’s instructions. For generating stable cell lines, cDNA-expressing PJA1-dR fragments were cloned into PcDNA3.1+ (Invitrogen), and the plasmids were transfected into HepG2 and HepG3 cells. The transfectants were selected with G-418 at 800 mg/ml for 2 weeks. The stable cell lines, PJA1-dR-c1 and PJA1-dR-c2, were cloned by a limiting dilution method (31). For PJA1 knockdown by shRNA silencing, HCC cells were exposed to 200 μl lentiviral particles containing shPJA1 or shCtrl (Santa Cruz Biotechnology) and incubated for 5-7 hours; medium was then replaced. After 48 hours, stable HCC cell lines expressing shPJA1 or shCtrl were generated by selection with 10 μg/ml puromycin for 5 days.
CD133+ LSCs were grown on poly-D-lysine/laminin-coated plates in “Liver Cell Medium”: DMEM/F-12 media with 10% heat-inactivated serum, rHGF (hepatocyte growth factor; 50 ng/mL), rEGF (epidermal growth factor; 20 ng/mL), insulin-transferrin selenium (1×), rFGF (fibroblast growth factor; 20 ng/mL), dexamethasone (1 × 10−7 mol/L), and nicotinamide (10 mmol/L) (32).
Cell proliferation and viability assay
PJA1-dR-c1, PJA1-dR-c2, and control cells were seeded onto 6-well plates (1 × 104 cells/well). The cultures were incubated for 6 days. Cell numbers were measured daily by Trypan blue staining (0.4%) (ThermoFisher, T10282) using the Countess™ Automated Cell Counter (Invitrogen). All assays were performed in triplicate and repeated at least three times. PJA1/wt/LSCs and PJA1/Smad3+/−/LSCs were isolated from single mice and immediately seeded and cultured onto 96-well plates (3 × 103 cells/well) in suitable supplement-containing medium. Cell proliferation was measured using CyQUANT NF Cell Proliferation Assay reagent at day 1, day 3, and day 6 as described previously (32). Data are from three independent experiments. HepG2, Hep3B, and Huh7 cells were seeded onto 96-well plates (3 × 103 cells/well). Twenty-four hours later, the cells were exposed to RTA402 or RTA405 at the indicated concentration in serum-free DMEM. Viability was measured by MTS assay after 24 hours, 48 hours, or 72 hours. Control cells were treated with DMSO. Data presents the viable cells as a percent of control cells.
Luciferase assay
HepG2 (1×104 cells/well) were added to 24-well dishes. The next day, the cells were co-transfected with either 4×SBE luciferase reporter or 3TP luciferase reporter plasmids, Renilla, HA-PJA1, and Flag-SMAD3, or both plasmids using Lipofectamine 2000. After 24 hours, half of the cells were exposed to TGF-β1 (200 pM) for 4 hours. Non-transfected cells served as the control. SNU449, Huh7, or SNU475 cells (1×104 cells/well) were added to 24-well dishes. The next day, the cells were co-transfected with 4×SBE luciferase reporter and Renilla using Lipofectamine 2000. After 24 hours, cells were exposed to RTA402 (1 mM), RTA405 (1 mM), or TGF-β1 (200 pM) for 2 hours. Control cells were exposed to equivalent volume of DMSO.
In all transfections, the expression plasmid Renilla (Promega) served as an internal control to correct for transfection efficiency and samples were collected and analyzed according to the manufacturer’s instructions for the Luciferase Assay System (Promega). The cells were extracted using 100 μL of luciferase cell culture lysis reagent. Ten microliters of cell extract were used for measuring Renilla enzyme activity; 20 μl were used for the luciferase assay. After subtracting the background (non-transfected cell control or DMSO-treated), luciferase activity was normalized to Renilla activity (arbitrary units) for each sample.
Immunoblotting and immunoprecipitation
For analysis of total cell content, cells were lysed with Nonidet P-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1% NP-40, 1 mM EDTA), protease inhibitor cocktail (Roche Applied Science), 1 mM PMSF, 1 mM NaF, and 1 mM sodium orthovanadate. For separate analysis of nuclear and cytoplasmic protein content, nuclear and cytoplasmic samples were prepared as previously described (Supplementary Materials and Methods) (33).
Immunohistochemistry and immunofluorescence analyses
For immunohistochemical analysis, tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Cryosections (5 μm thick) were stained for caspase-3, Ki67, or PJA1. Diaminobenzidine was used as a chromogen, and haematoxylin and eosin (HE) was used for counterstaining. Quantitative analysis was performed by manually counting positive staining cells in samples from multiple tumors or normal liver tissue and HCC tissue. The researcher scoring the samples was blinded to the sample identity.
For immunofluorescence analysis of CD133+ stem cells, cells derived from wild-type mice injected with PJA1 plasmids (PJA1/wt/LSCs) or from Smad3+/− mice injected with PJA1 plasmids (PJA1/Smad3+/−/LSCs) were isolated and seeded onto chamber slides at 2 × 104 cells per well for 12 h. For confocal imaging of HepG2 cells exposed to RTA402 or RTA405, 2 × 104 cells were plated onto cover slips in 6-well plates. After 12 hours, the cells were exposed to TGF-β1, RTA402, or RTA405 for 2 hours. Control cells were exposed to DMSO. For all immunofluorescence analysis, cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked in 10% normal goat serum in PBS. The cells were incubated with primary antibodies, washed 3 times in PBS, and then incubated with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with Alexa-488 or Alexa-555. DAPI or DRAQ5 was used for nuclear staining. For confocal analysis, slides were examined using Zeiss LSM 710. Quantitative analysis of the cells with nuclear SMAD3 was performed by manually counting the number of foci in positive-staining cells. The researcher scoring the samples was blinded to the sample identity.
Hydrodynamic tail vein injections and liver cancer stem cell formation assays in mice
Ten- to 12-week-old female wild-type and Smad3+/− mice were prepared for hydrodynamic tail vein injection using sleeping beauty (SB) ransposon/transposase system. The PiggyBac Transposon System (pB513B transposon and SB transposase vector) was purchased from System Biosciences, Inc., Palo Alto, CA (32). Mouse PJA1 cDNA (GE Dharmacon, Inc.) was subcloned into the pB513B vector. PJA1 plasmids or control plasmid DNA (empty pB513B) (5 μg in 1.5 ml of 0.45% saline solution per mouse) in an equivalent volume of saline solution, along with SB transposase in a ratio of 25:1, were diluted in 1.5 ml of 0.45% saline solution, filtered through 0.22 μm filter (Millipore), and injected hydrodynamically through the tail vein into wild-type or Smad3+/− mice twice per month for three months. At 3 months, the mice were humanely killed, and samples of liver tissue were fixed in 10% formalin, embedded in paraffin, and stained for PJA1. Single-cell suspensions from the mouse livers were prepared as described previously (32). CD133+ LSCs were isolated from each liver preparation using a magnet-activated cell-sorting column with an antibody recognizing CD133. Isolated CD133+ LSCs from each mouse were assessed for anchorage-independent growth, colony formation, and proliferation assays or were injected into mice to evaluate tumor formation.
Whole-transcriptome sequencing and database analyses
Whole-transcriptome RNA sequencing was performed and analyzed at MDACC DNA core facility. Gene expression profiling data were gathered from human liver cancer cell lines: HepG2-shRNA-Ctrl (cells stable expressing control shRNA), HepG2-shRNA-PJA1 (cells stable expressing shRNA targeting PJA1), HepG2-TGF-β1 (cells were treated with TGF-β1 at 200 pM for 1 hour), and HepG2-Ctrl (cells were treated with vehicle). We identified differentially-expressed mRNAs between the four experimental conditions using the standard comparison mode with multiple testing corrections (adjusted p-value < 0.05). Differentially regulated genes in each condition were determined and compared (GEO: GSE132723; Supplementary Table S1).
For evaluating stemness indices and TGF-β pathway activity, we used TCGA pan-cancer and liver hepatocellular carcinoma (LIHC) transcriptome sequencing data (bam files downloaded on May 2019); their related clinical data were obtained from the Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/) and TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). Paired-end FASTQ files for each sample were extracted from bam files using bam2fastq (http://www.hudsonalpha.org/gsl/information/software/bam2fastq). The number of fragments per kilobase of non-overlapped exon per million fragments mapped (FPKM) was generated at MDACC. Stemness indices (mRNAsi score) were generated by a one-class logistic regression (OCLR) machine-learning algorithm program (15). TGF-β pathway activity was determined as described previously (15,17). TCGA HCC samples (n=368) were stratified by stemness index, defined by ranking the samples by their RNA-based stemness index (mRNAsi) and dividing the samples into top, intermediate, and bottom thirds.
To evaluate the relationship between PJA1 expression and gene expression in HCC, we downloaded the Affymetrix mRNA microarray data GSE9843 (n = 91, HCC) from NCBI’s Gene Expression Omnibus (GEO). This data set was divided into four quartiles according to PJA1 mRNA abundance and differentially expressed genes. We used an mRNA z-scores threshold of ± 2.0 to determine whether a gene is significantly increased or decreased compared to the normal samples. Transcriptomes of the highest PJA1 quartile were compared to those of the lowest PJA1 quartile using Nexus Expression 3.0 (BioDiscovery). Upregulated or downregulated genes in samples expressing a high level of PJA1 are provided in Supplementary Table S2.
To analyze mRNA abundance of FOS, SERPINE1, and PJA1 in HCC patients, Wurmbach Liver (GSE6764), Roessler liver (GSE14520), and Mas Liver (GSE14323) Affymetrix mRNA microarray datasets were downloaded from Oncomine. To determine the relative difference in PJA1 expression in normal liver and HCC tissue, TCGA transcriptome sequencing data were downloaded from the Cancer Genomics Hub (https://cghub.ucsc.edu), Affymetrix mRNA microarray datasets from Roessler liver 2 (GSE14520) and Wurmbach liver (GSE6764) were downloaded from Gene Expression Omnibus database, and these gene expression profiles were analyzed using Oncomine analysis tools (www.oncomine.org).
Results
PJA1 functions as a tumor promoter and enhances the function of liver cancer stem cells
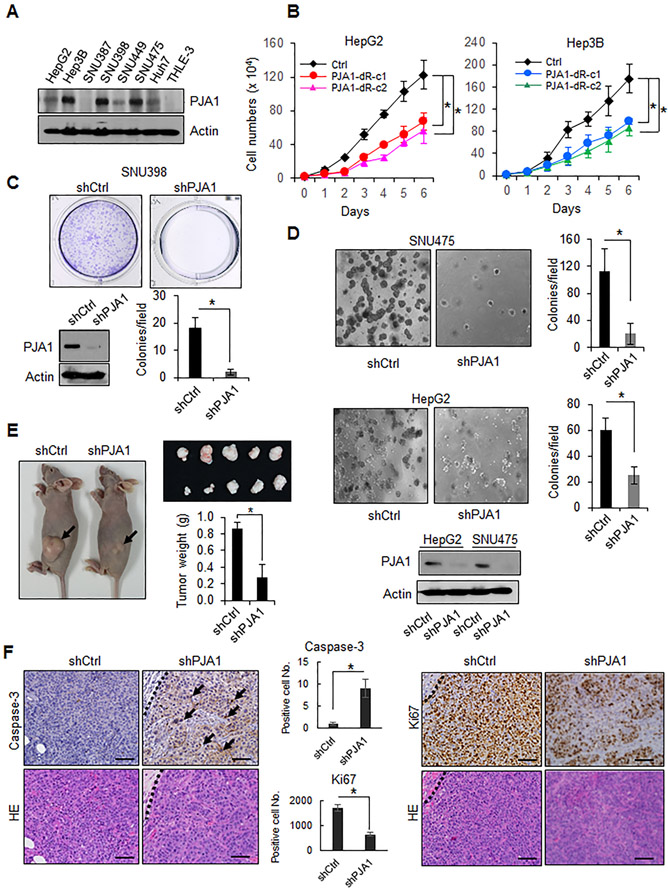
We first detected the amount of PJA1 in human liver cancer cell lines including HepG2, Hep3B, SNU387, SNU398, SNU449, SNU475 and Huh7, and compared the analyses to a normal hepatocyte cell line, THLE-3. We found that higher levels of PJA1 were present in most liver cancer cell lines, whereas PJA1 was not detected in normal hepatocytes (Fig. 1A). We inhibited PJA1 function by knocking down PJA1 with shRNAs or by expressing a dominant-negative mutant lacking the RING domain (34) in 4 human liver cancer cell lines (Hep3B, HepG2, SNU398, and SNU475). Each cell line overexpressing the RING domain-deleted PJA1 exhibited significantly reduced proliferation compared with control cells (Fig. 1B). Knockdown of PJA1 significantly reduced colony formation in all 4 liver cancer cell lines (Fig. 1C; Supplementary Fig. S1) and anchorage-independent growth of SNU475 and HepG2 cells (Fig. 1D). Knockdown of PJA1 impaired tumor growth in a xenograft model of subcutaneously injected SNU475 cells in nude mice (Fig. 1E). Knockdown of PJA1 resulted in reduced numbers of Ki67-positive cells and increased numbers of cells positive for the apoptosis effector caspase3, suggesting that PJA1 promoted HCC cell proliferation and protected against apoptosis (Fig. 1F). These data indicated that PJA1 functions as a tumor promoter and that reducing its activity in liver cancer cells impairs malignant phenotypes.
Figure 1.
PJA1 functions as a tumor promoter. (A) Abundance of PJA1 in liver cancer cell lines and normal hepatocytes. Cell lysates from a panel of liver cancer cell lines and a normal hepatocyte cell line THLE-3 were used. (B) Proliferation of stable cell lines overexpressing RING domain-deleted PJA1. Two stable lines (dR-c1 and dR-c2) were generated.*: P-value < 0.05, one-way analysis of variance. (C) Effect of PJA1 knock down on colony formation of SNU398 cells. A representative image of a well for each shRNA (upper panel) and quantitative data (lower right graph) from three independent experiments are shown. The effect of shRNA on PJA1 protein levels is shown (lower left). (D) Effect of knockdown of PJA1 on anchorage-independent colony formation of SNU475 and HepG2 cells grown in soft agar. The effects of shRNA on PJA1 protein levels in HepG2 and SNU475 cells are shown (lower panel). (E) Effect of PJA1 knockdown in SNU475 cells on tumor growth when xenografted subcutaneously into nude mice. Mice were injected with SNU475-shCtrl (n = 5 mice) or SNU475-shPJA1 cells (n = 5 mice). Representative mice bearing xenografts from each group, photographs of the tumors, and quantitative analysis of tumor weight are shown. (F) Histology and immunohistochemistry of xenografts in nude mice. Quantification of immunohistochemistry staining is shown in the bar graphs. Scale bars indicates 100 μm. For B-E, data are presented as mean ± standard deviation, and each result shown is representative of three independent experiments. For C-F, statistical analysis was performed by two-tailed Student’s t- tests (*, P-value < 0.05).
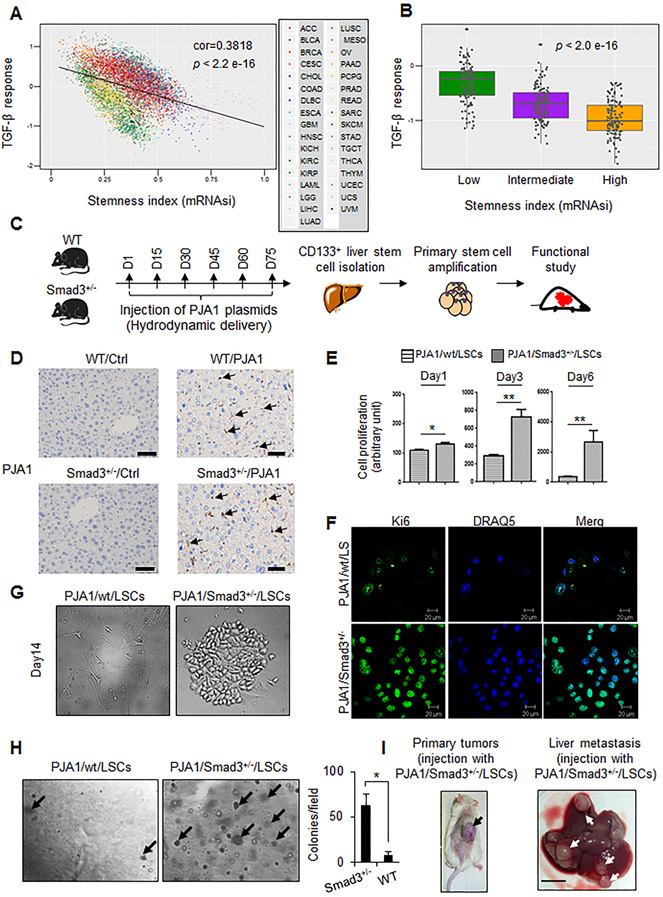
Previously, we identified a range of stem cell signatures in 9,125 samples from 33 tumor types, including 368 TCGA HCCs, in the TCGA databases (15) . Here, we assessed the relationship between TGF-β pathway activity (17) and stem cell signatures in the 33 tumor types (Fig. 2A; Supplementary Fig. S2), which revealed that low TGF-β pathway activity correlated with a higher stem cell index. Additionally, we stratified the HCC samples in this data set according to those with a high, intermediate, and low stem cell index and found a negative correlation between TGF-β pathway activity and stem cell-like character (Fig. 2B). Thus, these results indicated that impairment of TGF-β signaling, as would be expected if PJA1 activity is high, could contribute to stem cell-like properties of cancers, including HCC.
Figure 2.
PJA1 promotes liver stem cell (LSC) proliferation and liver metastasis in Smad3+/− mice. (A) Negative correlation between cancer stem cell signature and TGF-β response in TCGA pan-cancer samples (n=9660). Scatter plot shows the position of each cancer sample, color indicates tumor type (abbreviations according to Supplementary Fig. S2). Significance was determined by Pearson correlation. (B) Relationship between TGF-β response score in TCGA HCC cohort and stemness status. TCGA HCC samples (n=368) were stratified based on the stemness status, defined by ranking the samples by their RNA-based stemness index (mRNAsi) and dividing the samples in top, intermediate, and bottom thirds. Statistical differences among the groups was assessed by 1-way ANOVA. (C) Diagram of the paradigm for generation and isolation of LSCs and analysis of their potential to function as cancer stem cells. Timeline shows days of tail vein injection of wild-type (WT) (n = 6) and Smad3+/− (n = 6) mice with PJA1-encoding plasmid over a 75-day period (D1, day 1; D15, day 15, and so on). (D) Presence of PJA1 in mouse livers in WT and Smad3+/− mice with hydrodynamic tail vein PJA1 injections. Representative immunohistochemical staining images of PJA1 from WT or Smad3+/− mouse injected with PJA1-encoding plasmids or control plasmid DNA (empty pB513B) for 3 months were shown. Scale bars indicates 50 μm. (E) Proliferative potential of CD133+ cells isolated from PJA1-injected wild-type mice (PJA1/wt/LSCs) and PJA1-injected Smad3+/− mice (PJA1/Smad3+/−/LSCs). CD133+ LSCs were cultured in Liver Cell Medium on lysine/laminin-coated plates. *:P < 0.05; ** :P < 0.001, one-way analysis of variance. (F) Identification of proliferative LSCs by immunostaining of Ki67. Nuclei were labeled with DRAQ5. (G) Differences in growth behavior of PJA1/Smad3+/−/LSCs and PJA1/wt/LSCs. CD133+ LSCs from wild-type or Smad3+/− mice were plated on lysine/laminin-coated plates and grown in Liver Cell Medium. Representative images are from day 14 in culture are shown. (H) Growth of LSC colonies in soft agar. Cells were grown for 4 weeks, and colonies were counted from three experiments. *, P-value < 0.05, two-tailed Student’s t- test. (I) Tumor formation by PJA1/Smad3+/−/LSCs or PJA1/wt/LSCs. LSCs were injected subcutaneously into NOD SCID gamma chain knockout (NSG) mice (n = 6, per cell line). After 30 days, mice were sacrificed and evaluated for the formation of tumors at the site of injection and in the liver. Representative images from the 2 mice injected with PJA1/Smad3+/−/LSCs that developed tumors are shown. Scale bar indicates 5 mm. For E and H, quantitative data are presented as mean ± standard deviation.
To explore the effect of PJA1 overexpression on stem cell properties and an interaction with TGF-β signaling, we investigated whether PJA1 exhibited an interaction with SMAD3 in transforming LSCs into liver cancer stem cells by comparing the effect of PJA overexpression in wild-type or Smad3+/− mice on LSCs. We introduced sleeping beauty (SB) transposase along with SB transposon expressing mouse PJA1 in the mouse liver using hydrodynamic tail vein injection as described in Materials and Methods. We confirmed the presence of PJA1 in mouse livers by immunohistochemical staining, and isolated LSCs at day 90, using the stem cell marker CD133+ (32) (Fig. 2C and D).
Compared with the CD133+ cells from the PJA1-injected wild-type mice, CD133+ cells isolated from the PJA1-injected Smad3+/− mice exhibited enhanced proliferation in culture and increased Ki67 staining (Fig. 2E and F). Furthermore, the cells from the PJA1-injected Smad3+/− mice formed colonies of cells at 14 days in culture (Fig. 2G), indicating that individual cells within the population were responsible for increased proliferation. To assess if the LSCs from the PJA1-injected Smad3+/− mice had properties of cancer stem cells, we compared the anchorage-independent growth of cells from the PJA1-injected wild-type mice and the PJA1-injected Smad3+/−. The LSCs from the PJA1-injected Smad3+/− mice exhibited an increased frequency to form colonies in soft agar (Fig. 2H) and when injected subcutaneously into immune-compromised mice, 2 of 6 mice injected with cells from the PJA1-injected Smad3+/− mice formed tumors and liver metastases (Fig. 2I). None of the mice injected with the cells from the PJA1-injected wild-type or plasmid control-injected Smad3+/− mice formed tumors. These data suggested that alone increased PJA1 did not overcome regulatory mechanisms limiting transformation of LSCs. However, in the context of compromised SMAD3 signaling, increased PJA1 activity enhanced the potential for individual LSCs to transform into highly proliferative, malignant cancer stem cells.
PJA1 promotes phosphorylated SMAD3 (p-SMAD3) ubiquitination and degradation
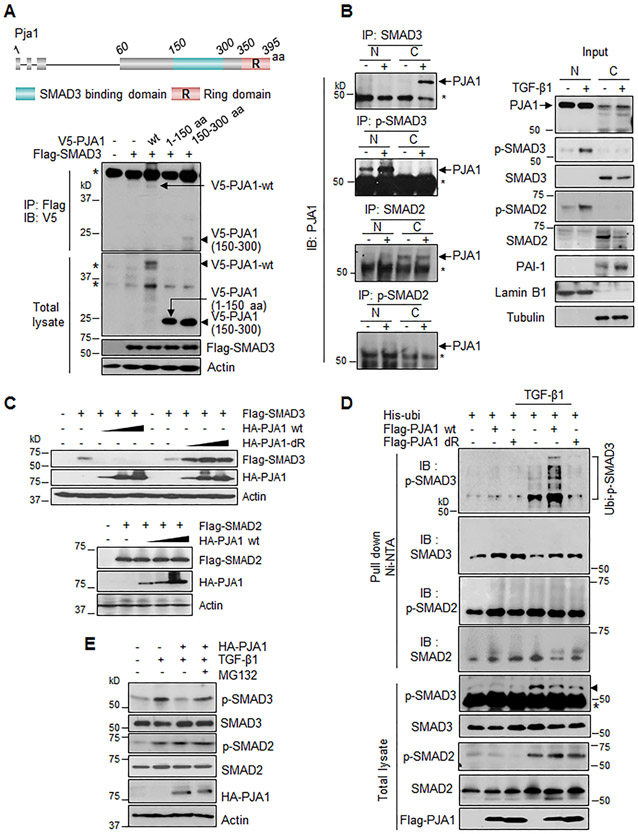
We previously reported that PJA1 overexpression in HepG2 cells promoted SMAD3 ubiquitination (29). Here, we investigated the interaction between PJA1 and SMAD3 in terms of the regions of PJA1 involved and the subcellular location of the interactions. We found that the middle region (amino acids 150-300) of PJA1 was necessary for the interaction with SMAD3 (Fig. 3A). In HepG2 cells exposed to TGF-β1, we found that TGF-β1 treatment promoted the interaction of PJA1 and SMAD3 both in the cytoplasm and the nucleus (Fig. 3B, right upper 2 panels). PJA1 also interacted with SMAD2 in cell cytoplasm in a TGF-β-independent manner, and the two exhibited a weak interaction in the nucleus (Fig. 3B, right 2 lower panels). Overexpression of PJA1 decreased co-expressed Flag-tagged SMAD3 but not SMAD2 abundance (Fig. 3C). PJA1-mediated poly-ubiquitination was only detected for phosphorylated SMAD3 (p-SMAD3) in cells exposed to TGF-β1; poly-ubiquitin-modified forms of non-phosphorylated SMAD3 were not detected in HepG2 cells in the presence or absence of TGF-β1 (Fig. 3D). Although mono-ubiquitin-modified forms were detected for non-phosphorylated SMAD2 in both presence and absence of TGF-β1, no ubiquitin-modified form was detected for phosphorylated SMAD2 (Fig. 3D). Although both SMAD3 and SMAD2 may interact with PJA1, only the phosphorylated form of SMAD3 was a substrate for ubiquitination. Inhibition of the proteasome with MG132 partially prevented the reduction in p-SMAD3, but not in p-SMAD2 abundance in cells overexpressing HA-tagged PJA1 (Fig. 3E). These data indicated that PJA1 promotes the ubiquitination and proteasomal degradation of p-SMAD3, leading us to predict that PJA1 limits SMAD3-dependent signaling downstream of TGF-β signaling.
Figure 3.
PJA1 enhances ubiquitination and degradation of TGF-β-induced phospho-SMAD3. (A) Interaction of PJA1 with SMAD3. Top shows a diagram of the mouse PJA1 with the RING domain and SMAD3-binding domain indicated. HepG2 cells were cotransfected with the indicated plasmids. (B) Effect of TGF-β1 on the interaction between PJA1 and SMAD3 or SMAD2. HepG2 cells were exposed to 200 pM TGF-β1 for 1 hour (+), nuclear (N) and cytoplasmic (C) fractions were isolated. (C) Effect of overexpression of PJA1 on SMAD3 and SMAD2 abundance. HepG2 cells were cotransfected with HA-tagged wild-type PJA1 (HA-PJA1 wt) or HA-tagged PJA1 lacking the RING domain (HA-PJA1-dR) and Flag-tagged SMAD3 (upper panel), or cotransfected with HA-PJA1 wt and Flag-tagged SMAD2 (lower panel). (D) Effect of PJA1 on ubiquitination of SMAD3 and SMAD2. HepG2 cells were cotransfected with His-ubiquitin plus Flag-PJA1 wild-type or Flag-PJA1 RING domain-deletion mutant. Cells were exposed to 200 pM TGF-β1 for 1 hour. Ubiquitinated proteins were isolated with Ni-NTA-agarose beads. (E) Effect of proteasome inhibition on TGF-β-induced p-SMAD3 abundance. HepaG2 cells were transfected with HA-PJA1. Cells were treated with MG132 (50 μg/mL) for 6 hours with or without 200 pM TGF-β1 for 1 hour before harvest. For A, B and D, asterisk designates nonspecific bands.
PJA1 limits SMAD3-dependent gene regulation in response to TGF-β1 stimulation
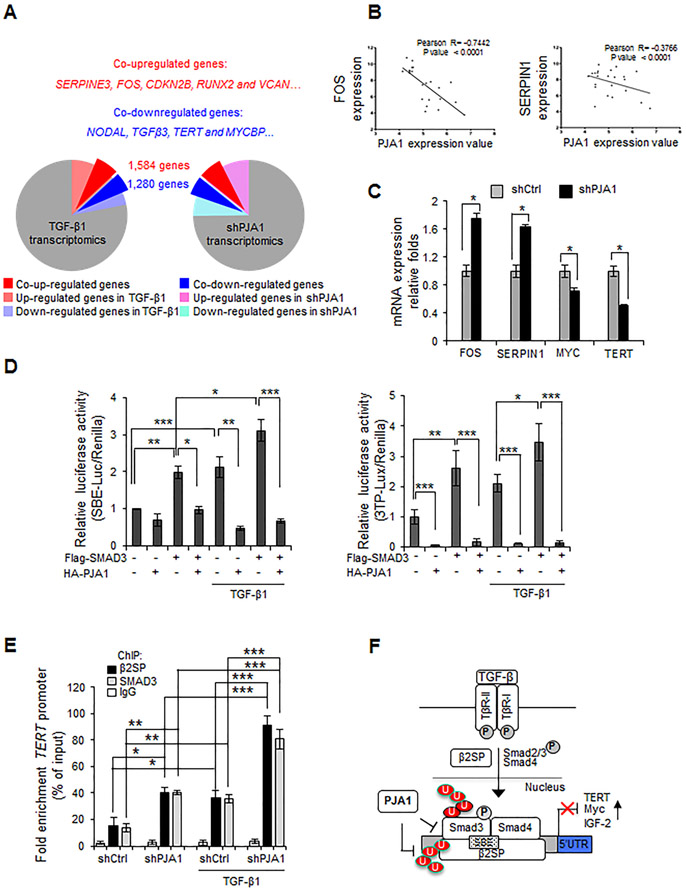
We analyzed the effect of knocking down PJA1 on the expression of SMAD3-regulated, cancer-related genes and the overlap between PJA1-regulated genes and those stimulated by TGF-β1. We performed transcriptome sequencing analysis of HepG2 cells in which PJA1 was knocked down or that were exposed to TGF-β1 and determined the differentially regulated genes (Fig. 4A; Supplementary Table S3). Among the overlapping differentially regulated genes, 48% of the genes upregulated by PJA1 knockdown overlapped with those upregulated by TGF-β1 and 52% of TGF-β1 upregulated genes overlapped with those upregulated by PJA1 knockdown. Similarly, 50% of the genes downregulated by PJA1 knockdown overlapped with those downregulated by TGF-β1 and 62% of TGF-β1 downregulated genes overlapped with those downregulated by PJA1 knockdown. The genes that showed similar regulation include: the co-upregulating genes SERPINE3 (encoding a member of the PAI1 family), RUNX2, FOS, CDKN2B, and VCAN, which are associated with cell proliferation, apoptosis, and migration, and angiogenesis; the co-downregulating genes NODAL and TGFB3 (both members of the TGF-β superfamily), TERT, and MYCBP. These data suggested that PJA1 and TGF-β1 reciprocally regulate overlapping genes and that PJA1 may play an oncogenic role by affecting TGF-β signaling.
Figure 4.
PJA1 inhibits SMAD3 transcriptional activity. (A) Genes co-regulated by PJA1 and TGF-β1. RNAseq analyses of genes regulated by PJA1 knockdown or exposure to TGF-β1 in HepG2 cells. (B) Negative association between PJA1 mRNA expression and the TGF-β target gene, FOS and SERPIN1 in HCC patients. Wurmbach Liver and Mas Liver mRNA microarray datasets from Oncomine were downloaded and analyzed. Significance was determined by Pearson correlation. (C) The expression of TGF-β-regulated genes in PJA1 knockdown cells. Transcripts were quantified by quantitative RT-PCR from HepG2-shCtrl or HepG2-shPJA1 cells (Supplementary Materials and Methods). *: P < 0.05, two-tailed Student’s t- tests. (D) Effect of PJA1 on SMAD3-dependent reporter gene activity. HepG2 cells were cotransfected with the luciferase reporter constructs controlled by 4 copies of the SBE (SBEx4) (left panel) or 3TP (right panel) along with Flag-tagged SMAD3 and HA-tagged PJA1 as indicated. Where indicated, cells were exposed to 200 pM TGF-β1 for 1 hour. (E) Effect of PJA1 knockdown on binding of SMAD3 and β2SP at the TERT promoter. HepG2-shCtrl or HapG2-shPJA1cells were treated with 200 pM TGF-β1 for 1 hour. Chromatin immunoprecipitation (ChIP) analysis was performed with antibodies against the indicated proteins and the interaction with the TERT promoter was assessed. IgG served as a negative control (Supplementary Materials and Methods). For D and E*: P < 0.05, **: P < 0.01, ***: P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. For C, D and E, data are shown as mean ± standard deviation of three independent experiments. (F) A model for the regulation of SMAD3 and β2SP by PJA1.
We verified the inverse relationship between PJA1- and TGF-β1-regulated gene expression in several independent experiments. We selected two genes (SERPIN1 and FOS) induced by the TGF-β1/SMAD3/βSPN pathway (35,36) and analyzed mRNA microarray data from Oncomine (Fig. 4B). For both SERPIN1 and FOS, there was a statistically significant inverse relationship between PJA1 expression and the mRNA abundances of these co-regulated transcripts. Consistent with a reciprocal functional relationship with the TGF-β pathway, PJA1 knockdown increased transcripts of SERPIN1 and FOS and reduced those of MYC and TERT (Fig. 4C).HepG2 cells transfected with either of 2 reporter genes controlled by TGF-β signaling— one with 4 copies of the SMAD3 binding element (4SBE) or one with 3 copies of a TGF-β-regulated sequence from tissue plasminogen promoter reporter (3TP)— exhibited significantly reduced expression in cells coexpressing Flag-SMAD3 and HA-PJA1 in comparison to the expression in cells expressing Flag-SMAD3 alone (Fig. 4D). The SMAD3/β2SP complex binds to the TERT promoter (18). PJA1 knockdown significantly increased basal and TGF-β1-induced SMAD3 and β2SP occupancy at the TERT promoter in HepG2 cells (Fig. 4E).
In summary, these results indicated that PJA1 promotes the ubiquitination of phosphorylated SMAD3, resulting in reduced activity of a TGF-β/SMAD3/β2SP-dependent tumor-suppressing pathway in HCC cells (Fig. 4F). Thus, we predicted that inhibition of PJA1 may enhance TGF-β signaling, increasing the activity of this tumor-suppressor pathway and thus suppressing tumor growth.
Small molecules that interfere with PJA1 enhance SMAD3 signaling and impair tumor growth
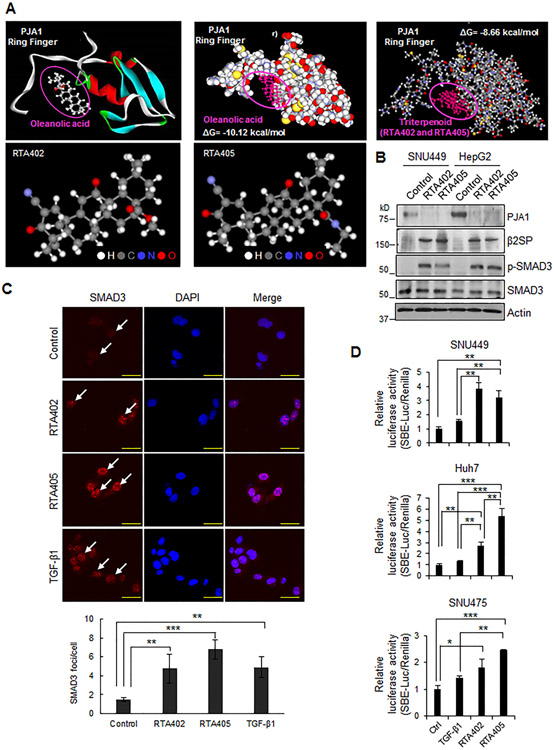
E3 ubiquitin ligases are potential targets for drug development (37). We collaborated with REATA Pharmaceuticals on a screen of small molecules for inhibitors of E3 ubiquitin ligases and identified 2 triterpenoid compounds with PJA1 inhibitory activity: RTA402 and RTA405. Synthetic triterpenoids are a subclass of antioxidant inflammation modulators, derived from oleanolic acid found in medicinal plants (38,39). Here, we performed computational molecular docking simulations with oleanolic acid, a natural triterpenoid, or the synthetic triterpenoid (RTA402/405) to the RING domain of PJA1 (Supplementary Materials and Methods) (40). Both oleanolic acid and the synthetic triterpenoid bound a pocket in the RING domain (Fig. 5A). The docking energy of oleanolic acid for the RING domain of PJA1 was predicted to be –10.12 kcal/mol and that of the synthetic triterpenoid is predicted to be –8.66 kcal/mol, suggesting that both naturally occurring and synthetic triterpenoids bind tightly to PJA1. To determine the effect of RTA402 and RTA405 on PJA1, we exposed HepG2 and SNU449 cells to each compound (0.25 μM for 1 day) and found these compounds decreased the abundance of PJA1 (Fig. 5B). Exposure of HepG2 or SNU449 cells to RTA402 increased the abundance of β2SP and p-SMAD3 (Fig. 5B).Exposure of HepG2 cells to either compound increased the proportion of cells with nuclear SMAD3 even in the absence of added TGF-β1 (Fig. 5C). Analysis of the effect of RTA402 or RTA405 on SMAD3 transcriptional activity using the SBE reporter assay revealed that these compounds enhanced SMAD3 transcriptional activity (Fig. 5D). Collectively, these data indicated that RTA402 and RTA405 enhanced SMAD3-mediated gene regulation, likely by affecting PJA1 activity.
Figure 5.
Triterpenoids RTA402 and RTA405 enhance TGF-β-SMAD3 signaling. (A) Computational molecular docking simulation analyses of oleanolic acid or triterpenoids to the PJA1 RING domain. Upper left shows the structures of oleanolic acid and the PJA1 ring finger domain. Upper middle shows the predicted interaction between oleanolic acid and PJA1 ring finger domain with the Gibbs free energy (ΔG). Upper right shows the predicted interaction between the synthetic triterpenoids and PJA1 RING domain with the ΔG of the interaction. Lower show the structures of RTA402 and RTA405. H: hydrogen, C: carbon, N: nitrogen, O: oxygen. (B) Effect of RTA402 and RTA405 on abundances of PJA1, β2SP and p-SMAD3 in HepG2 and SNU449 cells. Cells were exposed to RTA402 (0.25 μM) and RTA405 (0.25 μM) for 1 day. (C) Effect of RTA402 or RTA405 on SMAD3 nuclear translocation. HepG2 cells were treated with RTA402 (1 μM), RTA405 (1 μM), or TGF-β1 (200 pM) for 2 hours. Scale bar indicates 100 μm. Quantification of p-SMAD3 foci in nucleus is shown in the bar graph. (D) Effect of RTA402 and RTA405 on SMAD3-dependent reporter gene activity. The indicated cells were transfected with the SBEx4 luciferase reporter plasmids. After 24 hours of transfection, cells were treated with RTA402 (1 μM), RTA405 (1 μM), or TGF-β1 (200 pM) for 2 hours. For C and D, *: P < 0.05, **: P < 0.01, ***: P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test.
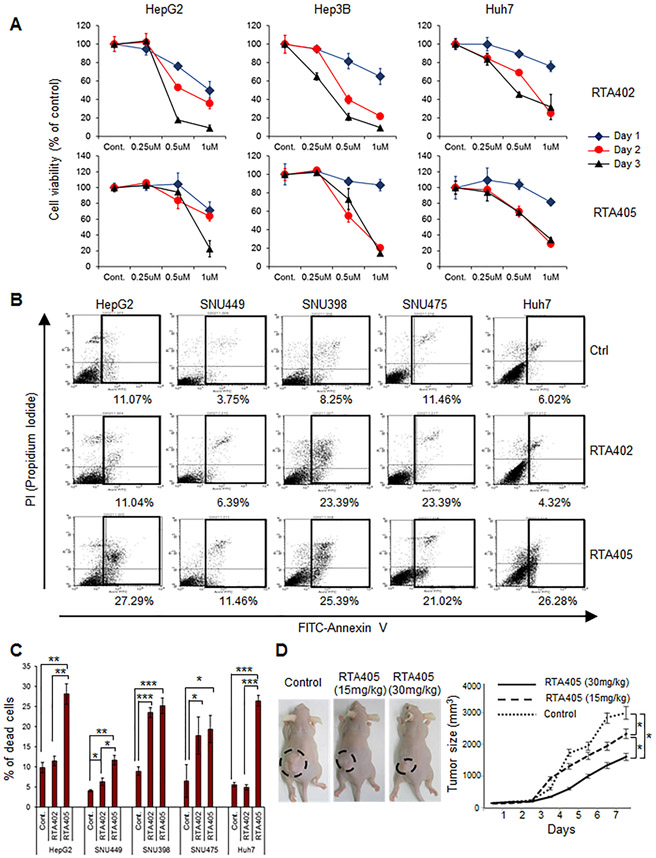
We tested the effect of RTA402 and RTA405 on liver cancer cell line growth in culture. Both compounds inhibited the growth of 3 different liver cancer cell lines in culture in a dose- and time-dependent manner (Fig. 6A). To evaluate apoptotic cell death, we exposed HCC cell lines to RTA402 or RTA405 (1 μM) for 2 hours (Fig. 6B and C). RTA405 increased the proportion of apoptotic cells in all 5 HCC cell lines, and RTA402 increased the proportion of apoptotic cells SNU449, SNU398, and SNU475 cells, but not in HepG2 or Huh7 cells. When administered to the mice at the time the tumor cells were injected, RTA405 produced a concentration-dependent reduction in growth of HepG2 tumors in immune-deficient nude mice (Fig. 6D).
Figure 6.
RTA402 and RTA405 induces apoptosis and inhibits growth of liver cancer cell lines in vitro and in vivo. (A) Effect of RTA402 or RTA405 on HCC cell viability in culture. The indicated cells were treated with the indicated concentrations of RTA402 or RTA405 for various times. (B) Induction of apoptosis in HCC cells exposed to RTA402 or RTA405. The indicated cells were treated with RTA402 (1 μM) or RTA405 (1 μM) for 2 hours. Representative results of FACS analysis for three independent experiments are shown. Percent apoptotic cells from the boxed areas are indicated beneath each FACS plot. (C) Quantitative data of FACS analysis in (B). Data are shown as mean ± standard deviation of three independent experiments. *: P < 0.05, **: P < 0.01, ***: P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (D) Effect of RTA405 on HCC tumor growth. Mice were subcutaneously injected with 5 × 106 HepG2 cells and intraperitoneally injected with PBS (control), RTA405 (15 mg/kg), or RTA405 (30 mg/kg). Six mice were used for each treatment group. Error bars are shown as mean ± standard deviations. * P < 0.05, one-way analysis of variance.
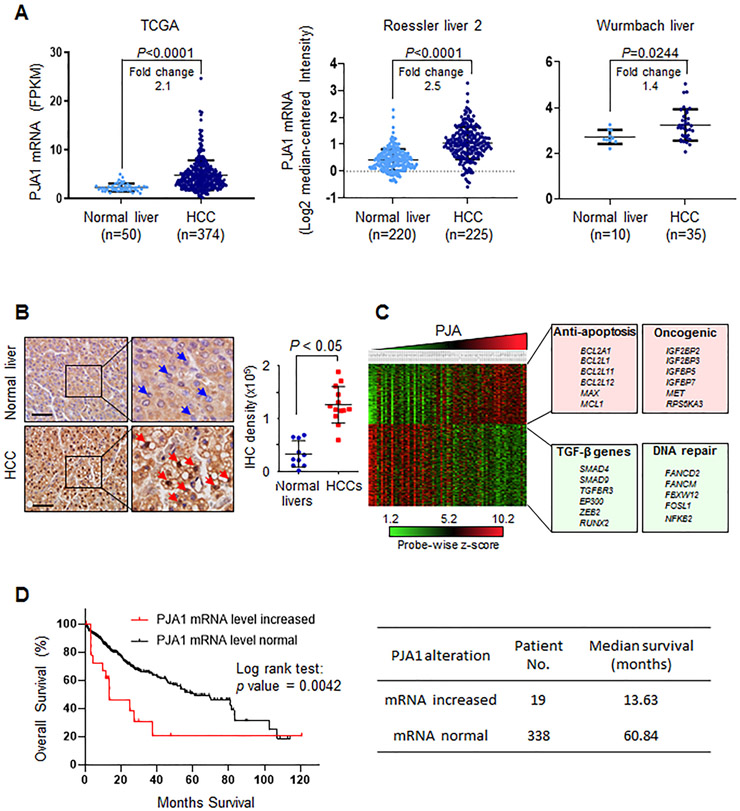
PJA1 is a potential therapeutic target for HCC
We analyzed transcriptome sequencing data of 374 HCC patient samples in TCGA. The amount of PJA1 mRNA was significantly increased in HCC compared to the amount in normal liver (Fig. 7A, left panel). An increase in PJA1 transcripts in HCC patients relative to the amount in normal liver was also detected in the Roessler liver 2 data from Oncomine and the Wurmbach liver data from Gene Expression Omnibus (41) (Fig. 7A, middle and right panels). Analysis of PJA1 protein revealed an increase in HCCs compared to its abundance in normal livers (Fig. 7B). We analyzed the transcriptomes of 91 HCC patient samples, which revealed an increase in the expression of oncogenes and genes associated with anti-apoptosis and a decrease in the expression of genes associated with TGF-β signaling and DNA damage repair in samples with high PJA1 expression (Fig. 7C). Thus, PJA1 is up-regulated in some HCC and may promote tumorigenesis or progression. We further investigated PJA1 genetic alterations in TCGA HCC data. We found that 24 HCC patients (7%) with PJA1 genetic alterations, including 19 patients with increased PJA1 mRNA expression (Supplementary Table 4). We found that increased mRNA expression of PJA1 was markedly associated with reduced overall survival of HCC patients (Fig. 7D, left panel). Median survival of HCC with high PJA1 mRNA expression is dramatically shorter than the HCC patients with normal PJA1 mRNA expression (13.63 months vs. 60.84 months) (Fig. 7D, right panel). These data suggest that high activity of PJA1 is associated with poor HCC prognosis.
Figure 7.
PJA1 is a potential target for therapeutic intervention in HCC. (A) The abundance of PJA1 mRNA in HCC compared to that in normal liver tissue from three independent cancer datasets. Fold-change compares the difference in the mean. (B) Immunohistochemical staining for PJA1 abundance in normal liver (n = 10) and HCC (n = 13). Blue arrowheads point to negative PJA1 staining in cell nucleus; red arrowheads indicate positive PJA1 staining in cell nucleus. Magnification × 10; inset magnification × 40. Scale bar is 100 μm. For A and B, mean ± standard deviation is indicated. Statistical analysis was performed by two-tailed Student’s t- tests. (C) Transcriptomic analyses of HCC patient datasets from Gene Expression Omnibus (GSE9843, n = 91). Transcriptomic data was clustered into 4 quartiles according to PJA1 expression using Nexus Expression 3.0. Representative pathways and genes associated with high PJA1 expression are listed. Green-boxed genes are those with a negative correlation with PJA1 expression and red-boxed genes are those a positive correlation with PJA1 expression. (D) Overall survival analysis of TCGA HCC patient dataset. Overall survival according to increased or normal mRNA levels of PJA1 in HCC patients shows statistically significant differences (Log rank Test, P=0.0042) (left panel). Median survival of these two groups were listed on the right panel.
Discussion
There are two major themes for roles of TGF-β in human disease: One is increased TGF-β activity as occurs in patients with fibrosis and progressive cancers, and the other is decreased TGF-β activity, as occurs in early tumorigenesis, hereditary hemorrhagic telangiectasia, developmental defects, and arteriosclerosis (42). Here, we explored a mechanism for impairment of the tumor-suppressing activity of TGF-β signaling through the ubiquitin ligase PJA1.
Following up on our previous work showing that PJA1 interacted with β2SP and mediated β2SP and SMAD3 ubiquitination (29), and that a TGF-β pathway mediated by SMAD3/ β2SP promotes tumor-suppressing gene expression (18), we focused on HCC and investigated if PJA1 functioned as a tumor promoter. We focused on HCC, because SMAD3 is a tumor suppressor that is frequently inactivated in gastrointestinal cancers including HCC (36) and because heterozygous deficiency in SPTBN results in the spontaneous development of HCC in mice between 9 and 12 months of age (18). Homozygous SPTBN deficiency is embryonic lethal (43,44).
Because SMAD3 and β2SP are implicated in regulation of stem cell-like properties and altered activity of this pathway may contribute to the formation of cancer stem cells, we examined the effect of PJA1 overexpression on the characteristics of LSCs from Smad3 wild-type and Smad3+/− mice. Excess PJA1 promoted transformation of LSCs from Smad3+/− mice such that the cells had increased proliferative potential, reduced apoptosis, and increased anchorage-independent growth. Although not a sufficiently large sample for statistical power, we observed that two of the mice injected with PJA-overexpressing Smad3+/− LSCs developed metastatic liver tumors, consistent with enhanced transformation of these cells compared with those from the PJA-overexpressing Smad3 wild-type mice. Thus, in the context of limited TGF-β/SMAD3 signaling, excess PJA1 can tip the balance toward a cancer stem cell-like phenotype. Cancers associated with impaired signaling through this pathway, for example through reduced abundance of the components or decreased release of latent TGF-β, may benefit from inhibition of PJA1 as long as the components of the TGF-β pathway are functional.
Of potentially translational importance, we showed through molecular docking simulations that synthetic triterpenoids bound PJA1 and that exposing HCC cells in culture to such compounds promoted SMAD3-mediated transcription, SMAD3 translocation into the nucleus, stabilized β2SP, and reduced cell viability by stimulating apoptosis. RTA405 was the most effective in vitro and exhibited tumor-inhibiting activity when administered to mice. Synthetic triterpenoids, including RTA405, are under investigation in chronic kidney disease and chronic liver disease (45,46), and studies are evaluating natural triterpenes for cardiovascular disease and inflammation (47). Although our results indicated that the synthetic triterpenes could bind PJA1, these compounds may have multiple cellular targets (38). Future studies will need to address the specificity of such compounds for different protein targets and optimize those for targeted therapeutic applications.
In conclusion, we found that PJA1 is up-regulated in some hepatocellular tumor tissues and cell lines, that PJA1 inhibits TGF-β/SMAD3 signaling, and that the synthetic triterpene RTA 405 promoted SMAD3 activity and inhibited HCC tumor growth in nude mice. Furthermore, we identified a mechanism by which increased PJA1 could contribute to the development of HCC by inhibiting tumor-suppressing and stem cell-suppressing activities of TGF-β signaling. Targeting PJA1 in the context of TGF-β signaling may prove efficacious in battling this lethal cancer.
Supplementary Material
Acknowledgments:
We thank Mien-Chie Hung (MD Anderson Cancer Center) for the Huh7 cells. This research was supported by National Institute of Health grants R01CA106614 (L. Mishra), R01CA042857 (L. Mishra), R01AA023146 (L. Mishra), P01 CA130821 (L. Mishra) and U01CA230690 (L. Mishra). J.C. is supported by the American Association for the Study of Liver Diseases (AASLD) Foundation Pinnacle Research Award in Liver Disease, the Texas Medical Center Digestive Diseases Center Pilot/Feasibility project, and the Internal Research Grant (IRG) program in the University of Texas MD Anderson Cancer Center.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.International Agency for Research on Cancer: World Health Organization. GLOBOCAN 2014: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2014. . Lyon, France: International Agency for Research on Cancer: World Health Organization; [updated 2014; cited 2014 March 27] 2014:Available from: http://globocan.iarc.fr/Pages/fact_sheets_population.aspx. [Google Scholar]
- 2.Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477–91 e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723–50 [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [DOI] [PubMed] [Google Scholar]
- 6.Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Res 2016;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rountree CB, Mishra L, Willenbring H. Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology 2012;55:298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adebayo Michael AO, Ko S, Tao J, Moghe A, Yang H, Xu M, et al. Inhibiting Glutamine-Dependent mTORC1 Activation Ameliorates Liver Cancers Driven by beta-Catenin Mutations. Cell Metab 2019;29:1135–50 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 2015;524:180–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013;494:247–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu L, Finkelstein D, Gao C, Shi L, Wang Y, Lopez-Terrada D, et al. Multi-organ Mapping of Cancer Risk. Cell 2016;166:1132–46 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009;136:1012–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thenappan A, Li Y, Kitisin K, Rashid A, Shetty K, Johnson L, et al. Role of transforming growth factor beta signaling and expansion of progenitor cells in regenerating liver. Hepatology 2010;51:1373–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest 2013;123:1911–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018;173:338–54 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majumdar A, Curley SA, Wu X, Brown P, Hwang JP, Shetty K, et al. Hepatic stem cells and transforming growth factor beta in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2012;9:530–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Zaidi S, Rao S, Chen JS, Phan L, Farci P, et al. Analysis of Genomes and Transcriptomes of Hepatocellular Carcinomas Identifies Mutations and Gene Expression Changes in the Transforming Growth Factor-beta Pathway. Gastroenterology 2018;154:195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Yao ZX, Chen JS, Gi YJ, Munoz NM, Kundra S, et al. TGF-beta/beta2-spectrin/CTCF-regulated tumor suppression in human stem cell disorder Beckwith-Wiedemann syndrome. J Clin Invest 2016;126:527–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budi EH, Duan D, Derynck R. Transforming Growth Factor-beta Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol 2017;27:658–72 [DOI] [PubMed] [Google Scholar]
- 20.David CJ, Massague J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol 2018;19:419–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massague J TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012;13:616–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev 2005;19:2783–810 [DOI] [PubMed] [Google Scholar]
- 23.Zhang YE. Non-Smad Signaling Pathways of the TGF-beta Family. Cold Spring Harb Perspect Biol 2017;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thenappan A, Shukla V, Abdul Khalek FJ, Li Y, Shetty K, Liu P, et al. Loss of transforming growth factor beta adaptor protein beta-2 spectrin leads to delayed liver regeneration in mice. Hepatology 2011;53:1641–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Liu F, Tu W, Chang Y, Yao J, Wu W, et al. Embryonic liver fodrin involved in hepatic stellate cell activation and formation of regenerative nodule in liver cirrhosis. J Cell Mol Med 2012;16:118–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Song Y, Tu W, He X, Lin J, Liu F. beta-2 spectrin is involved in hepatocyte proliferation through the interaction of TGFbeta/Smad and PI3K/AKT signalling. Liver Int 2012;32:1103–11 [DOI] [PubMed] [Google Scholar]
- 27.Yao ZX, Jogunoori W, Choufani S, Rashid A, Blake T, Yao W, et al. Epigenetic silencing of beta-spectrin, a TGF-beta signaling/scaffolding protein in a human cancer stem cell disorder: Beckwith-Wiedemann syndrome. J Biol Chem 2010;285:36112–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBaun MR, Tucker MA. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr 1998;132:398–400 [DOI] [PubMed] [Google Scholar]
- 29.Saha T, Vardhini D, Tang Y, Katuri V, Jogunoori W, Volpe EA, et al. RING finger-dependent ubiquitination by PRAJA is dependent on TGF-beta and potentially defines the functional status of the tumor suppressor ELF. Oncogene 2006;25:693–705 [DOI] [PubMed] [Google Scholar]
- 30.Mishra L, Tully RE, Monga SP, Yu P, Cai T, Makalowski W, et al. Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene 1997;15:2361–8 [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Zhao S, Nakada K, Kuge Y, Tamaki N, Okada F, et al. Dominant-negative hypoxia-inducible factor-1 alpha reduces tumorigenicity of pancreatic cancer cells through the suppression of glucose metabolism. Am J Pathol 2003;162:1283–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra A, Yan J, Xia X, Zhou S, Chen J, Mishra L, et al. IL6-mediated inflammatory loop reprograms normal to epithelial-mesenchymal transition(+) metastatic cancer stem cells in preneoplastic liver of transforming growth factor beta-deficient beta2-spectrin(+/−) mice. Hepatology 2017;65:1222–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R, et al. Hypoxia-mediated up-regulation of Pim-1 contributes to solid tumor formation. Am J Pathol 2009;175:400–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J Biol Chem 2002;277:22541–6 [DOI] [PubMed] [Google Scholar]
- 35.Massague J, Xi Q. TGF-beta control of stem cell differentiation genes. FEBS Lett 2012;586:1953–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 1998;394:909–13 [DOI] [PubMed] [Google Scholar]
- 37.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013;154:556–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huerta C, Jiang X, Trevino I, Bender CF, Ferguson DA, Probst B, et al. Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding. Biochim Biophys Acta 2016;1860:2537–52 [DOI] [PubMed] [Google Scholar]
- 39.Cleasby A, Yon J, Day PJ, Richardson C, Tickle IJ, Williams PA, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One 2014;9:e98896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao ZX, Brown RC, Teper G, Greeson J, Papadopoulos V. 22R-Hydroxycholesterol protects neuronal cells from beta-amyloid-induced cytotoxicity by binding to beta-amyloid peptide. J Neurochem 2002;83:1110–9 [DOI] [PubMed] [Google Scholar]
- 41.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov 2012;11:790–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science 2003;299:574–7 [DOI] [PubMed] [Google Scholar]
- 44.Tang Y, Katuri V, Iqbal S, Narayan T, Wang Z, Lu RS, et al. ELF a beta-spectrin is a neuronal precursor cell marker in developing mammalian brain; structure and organization of the elf/beta-G spectrin gene. Oncogene 2002;21:5255–67 [DOI] [PubMed] [Google Scholar]
- 45.Getachew Y, Cusimano FA, Gopal P, Reisman SA, Shay JW. The Synthetic Triterpenoid RTA 405 (CDDO-EA) Halts Progression of Liver Fibrosis and Reduces Hepatocellular Carcinoma Size Resulting in Increased Survival in an Experimental Model of Chronic Liver Injury. Toxicol Sci 2016;149:111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Y, Stidham RD, Bumeister R, Trevino I, Winters A, Sprouse M, et al. The synthetic triterpenoid, RTA 405, increases the glomerular filtration rate and reduces angiotensin II-induced contraction of glomerular mesangial cells. Kidney Int 2013;83:845–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamede M, Mabuza L, Ngubane P, Khathi A. Plant-Derived Oleanolic Acid (OA) Ameliorates Risk Factors of Cardiovascular Diseases in a Diet-Induced Pre-Diabetic Rat Model: Effects on Selected Cardiovascular Risk Factors. Molecules 2019;24(2):340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.