Abstract
Glioma is defined as a common brain tumor which causes severe disability or death. As many genes are reported to relate with glioma's occurrence and development, their prognostic and therapeutic value still remains uncertain. This study aimed at investigating the association between STAT3/p-STAT3 and glioma prognosis. Nine studies (12 trials) scored ≥5 on the Newcastle-Ottawa scale were meta-analysed from the Medline, Embase, and Web of Science databases. We found that STAT3/p-STAT3 overexpression in glioma patients was associated with worse overall survival (hazard ratio (HR) = 1.40, 95%confidence interval (CI) = 1.05 ~ 1.86, P = 0.020), progression-free survival (HR = 2.05, 95%CI = 1.63 ~ 2.58, P < 0.001), and better recurrence-free survival (HR = 0.37, 95%CI = 0.15 ~ 0.95, P < 0.039). Subgroup analysis implied that STAT3/p-STAT3 overexpression was associated with worse OS in standard treatment (HR = 1.80, 95%CI = 1.06 ~ 3.04, P = 0.030), and in China (HR = 2.18, 95%CI = 1.77 ~ 2.70, P < 0.001), and metaregression analysis indicated countries (P = 0.001) may be the source of heterogeneity in our study. In conclusion, we suggested STAT3/p-STAT3 was associated with poor prognosis in patients with glioma, which indicated that STAT3/p-STAT3 might be a valuable prognostic biomarker and a promising therapeutic target for glioma.
1. Introduction
Brain malignancy is a grievous type of brain tumor with high incidence and mortality, while the concentration on brain malignancy is still poor [1, 2]. Glioma, starting in the glial cells of the brain or the spine [3], is the most common primary intracranial tumor, which represents about 30% of all brain tumors and central nervous system tumors, and 80% of all malignant brain tumors [2, 4]. Although the etiology of glioma remain unclear, researches on monogenic Mendelian disorders [4], hereditary genetic disorders such as neurofibromatoses (type 1 and type 2), and tuberous sclerosis complex underscore that genetic factors are strongly associated with the development and progression of glioma [5]. Results from genome-wide association studies have identified common genetic variation in 7 genes (TERT [6], EGFR [7, 8], CCDC26 [9], CDKN2B [6], PHLDB1 [10], TP53 [11, 12], and RTEL1 [6]) and germ-line (inherited) polymorphisms of the DNA repair genes ERCC1, ERCC2, and XRCC1 increase the risk of glioma [13]. Recent study found that mutations in IDH1/2 may result in the development of glioma and be independent prognosis factors of glioma [14]. These indicate altered or deficient repair of DNA damage and different oncogenes contribute to the development of glioma [15, 16]. Diet, radiation, and infection with cytomegalovirus are regarded as potential pathogenic factors [17, 18]. In addition, certain occupations, such as farmers, are more susceptible to this disease [19, 20]. The potential influences of occupational exposures and cell phones have also been examined, with inconclusive results [2].
Although glioma is relatively rare compared with cerebral vascular diseases, they both cause severe mortality and morbidity [21]. Symptoms of glioma depend on the brain region where the tumor locates and show as headaches, vomiting, seizures, and cranial nerve disorders as a result of mass effect and increased intracranial pressure. Although glioma treatment options have developed, the prognosis remains poor. Multiple genes have been identified as glioma biomarkers that may predict patient's susceptibility and prognosis [21]. STAT3 is a latent cytosolic transcription factor and activates genes in human chromosome 12 (q13 to q14-1) by phosphorylating the tyrosine705 in the SH2 domain [22]. STAT3 plays an important role in multiple malignant cases, especially in glioma [23–25]. Phosphorylated STAT3 (p-STAT3) dimerizes spontaneously, migrates into cell nucleus, and activates the expression of downstream genes to regulate the tumor cell growth, proliferation, differentiation, and metastasis [26, 27]. In addition, p-STAT3 was reported to affect the occurrence, development, and even prognosis of glioma [28–31]. Emerging studies have shown STAT3 and its phosphorylation have similar functions in tumors [32–34], including in glioma [35–37], and given this consideration, we conducted a meta-analysis of STAT3/p-STAT3 to identify any direct correlations with glioma patient prognosis.
2. Materials and Methods
2.1. Study Search Strategy
We comprehensively searched for potential studies from the Medline, Embase, and Web of Science databases using relevant key words through December 1, 2018, without any language restrictions. The detailed literature search strategy in Medline was ((STAT3 Transcription Factor [Mesh]) OR (((((((APRF Transcription Factor) OR (Signal Transducer and Activator of Transcription 3)) OR IL6 Response Factor) OR LIF Response Factor) OR STAT3b Transcription Factor) OR STAT3a Transcription Factor) OR Transcription Factor, STAT3 [All fields]) OR ((((phosphorylated signal transducer and activator of transcription 3)) OR phosphorylated stat3 transcription factor) OR phospho-STAT3 [All fields])) AND ((Glioma [Mesh]) OR ((((((Glial Cell Tumors [All fields]) OR Glial Cell Tumor [All fields]) OR Mixed Glioma [All fields]) OR Tumor, Glial Cell [All fields]) OR Malignant Glioma [All fields]) OR Malignant Gliomas [All fields])).
2.2. Study Selection
Studies included in this analysis must meet some criteria. The participants must have been diagnosed with glioma via imaging, pathology, or the latest clinical diagnostic criteria. There was immunohistochemical analysis with the expression of STAT3/p-STAT3 in the glioma tissue. The association between STAT3/p-STAT3 and patients' prognosis, regardless overall survival (OS), progression-free survival (PFS), or recurrence-free survival (RFS), was investigated, and the adjusted or crude hazard ratio (HR) values could be calculated. Only studies with a sample size more than 60 were included. When the overlapping or even same data appears in different studies, the most complete or up-to-date study was included. The review and abstract were excluded [38].
2.3. Data Extraction
Two reviewers independently extracted the following data from remaining studies: the information about study characteristics (such as the first author, publication date, country, sample size, treatment, and analytic method), demographic characteristics (including age, sex, and diagnostic methods), the expression of STAT3/p-STAT3, and outcomes. Then, another two reviewers checked the received data. Inconsistent data were addressed by open discussion, and consensus was achieved. Finally, all extracted data were stored in the predesigned excel spreadsheet.
2.4. Study Quality Assessment
Two investigators conducted studies quality assessments of the included studies according to the Newcastle-Ottawa scale (NOS) developed for nonrandomized controlled trials [39]. The NOS included three domains: selection, comparability, and outcome, eight items with nine scores in total [40, 41].
2.5. Statistical Analysis
For all analyses, we used fixed and random effect models to reduce heterogeneity through Stata 14.0 (Stata Corp., College Station, TX, USA). Depending on the type of extracted data, we applied subgroup analysis by country, sample size, study subject (STAT3 or p-STAT3), treatment, and HR estimate. Moreover, sensitivity analysis was conducted to assess heterogeneity. We would assign adjectives of low, moderate, and high to I2 values of 25%, 50%, and 75% as described previously [38, 40, 42]. Lastly, the funnel plot was implemented to assess the publication bias and metaregression analysis was applied to trace the origin of heterogeneity [41].
3. Results
3.1. Study Search Results and Characteristics
The combined search yielded 1,509 potentially relevant studies after removing 753 duplicates, and 12 were retained while the title and abstract were screened. Three studies were close to meet the threshold but still excluded due to their lack of detailed data and sample size. Nine studies [43–51] (12 trials) were included in this study (Figure 1), among which 3 trials evaluated STAT3, and other three-fourths detected p-STAT3. As for countries, almost half of studies came from China [43–46, 51], and one each from Brazil [50], Greece [48], Bulgaria [47], and America [49] (Table 1). With the exception of one study (NOS score = 5 [49], all other studies had NOS scores of 7 or above, which were considered a high quality (or low-bias risk) studies [40, 41] (Table 1).
Figure 1.
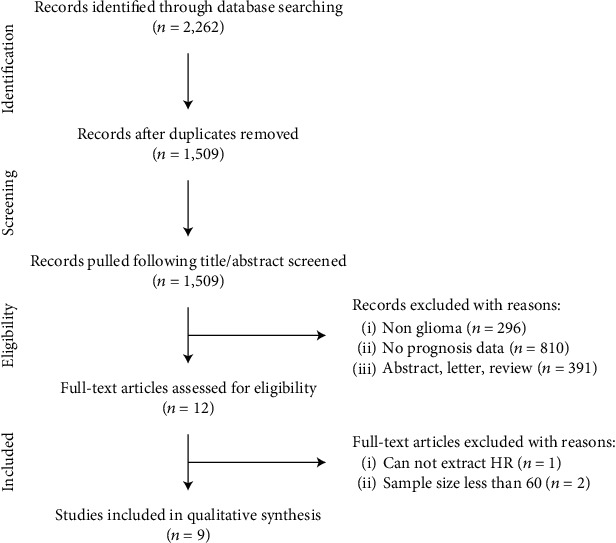
The flow chart.
Table 1.
Main characteristics of all included studies.
| Trials | Year | Country | Sample size | Treatment | HR for OS (95% CI) | HR for RFS (95% CI) | HR for PFS (95% CI) | NOS | References |
|---|---|---|---|---|---|---|---|---|---|
| Bruna (STAT3) | 2016 | Brazil | 85 | Nonstandard | 1.050 (0.540-2.040) | 2.560 (0.006-110.040) | NA | 8 | [50] |
| Bruna (pSTAT3-Tyr705) | 2016 | Brazil | 85 | Nonstandard | 0.850 (0.520-1.390) | 0.380 (0.120-1.190) | NA | 8 | [50] |
| Bruna (pSTAT3-Ser727) | 2016 | Brazil | 85 | Nonstandard | 0.990 (0.590-1.670) | 0.290 (0.050-1.470) | NA | 8 | [50] |
| Lin (pSTAT3-Tyr705) | 2014 | China | 90 | Nonstandard | 2.120 (1.070-4.203) | NA | 2.158 (1.136-4.098) | 8 | [44] |
| Lin (pSTAT3-Ser727) | 2014 | China | 88 | Nonstandard | 1.797 (1.028-3.142) | NA | 1.830 (1.090-3.074) | 9 | [45] |
| Liang (STAT3) | 2013 | China | 68 | Nonstandard | 1.480 (0.460-4.730) | NA | 1.850 (0.530-6.520) | 9 | [51] |
| Liang (pSTAT3-Tyr705) | 2013 | China | 68 | Nonstandard | 2.301 (1.726-3.021) | NA | 2.001 (1.509-2.877) | 9 | [51] |
| Wang (pSTAT3-Tyr705) | 2011 | China | 68 | Standard | 2.402 (1.268-4.550) | NA | 2.629 (1.362-5.073) | 9 | [46] |
| Piperi (pSTAT3-Tyr705) | 2011 | Greece | 97 | Nonstandard | 0.611 (0.332-1.124) | NA | NA | 9 | [48] |
| Tu (STAT3) | 2011 | China | 96 | Nonstandard | 2.360 (1.370-10.980) | NA | NA | 7 | [43] |
| Birner (pSTAT3-Tyr705) | 2010 | Bulgaria | 111 | Standard | 1.400 (0.790-2.460) | NA | NA | 7 | [47] |
| Mohamed (pSTAT3-Tyr705) | 2008 | America | 129 | Nonstandard | 1.200 (0.640-2.220) | NA | NA | 5 | [49] |
Note: HR: hazard ratio; OS: overall survival; RFS: recurrence-free survival; PFS: progression-free survival; NOS: Newcastle-Ottawa scale; NA: not applicable.
3.2. Association of STAT3/p-STAT3 with OS
The combined analysis of 12 trials showed that STAT3/p-STAT3 overexpression in glioma was associated with worse OS (HR = 1.40, 95% confidence interval (CI) = 1.05 ~ 1.86, P = 0.020) (Figure 2). Obviously, there was significant heterogeneity among trials (I2 = 64.2%, Ph = 0.001), so we conducted subgroup and metaregression analysis to investigate the possible source of the heterogeneity. As for STAT3 and its phosphorylation, there was no association with OS (Figure 3(a)). Furthermore, we conducted subgroup analysis of STAT3 phosphorylation site (Tyr705 and Ser727), and the results were consistent (Figure S1). Treatment subgroup analysis results implied that STAT3/p-STAT3 overexpression was associated with worse OS in standard treatment (HR = 1.80, 95%CI = 1.06 ~ 3.04, P = 0.030), and there was no significant association in nonstandard treatment (HR = 1.33, 95%CI = 0.95 ~ 1.84, P = 0.095) (Figure 3(b)). As for country subgroup analysis, STAT3/p-STAT3 overexpression was associated with worse OS in China (HR = 2.18, 95%CI = 1.77 ~ 2.70, P < 0.001), but no evidences in other countries (HR = 0.98, 95%CI = 0.78 ~ 1.23, P = 0.852) (Figure 3(c)). Subgroup analysis results of sample size showed that STAT3/p-STAT3 expression was not associated with OS (Figure 3(d)). Moreover, metaregression analysis indicated countries (P = 0.001) may be the source of heterogeneity in this study, while STAT3/p-STAT3 (P = 0.863), treatment (P = 0.423), and sample size (P = 0.996) were not.
Figure 2.
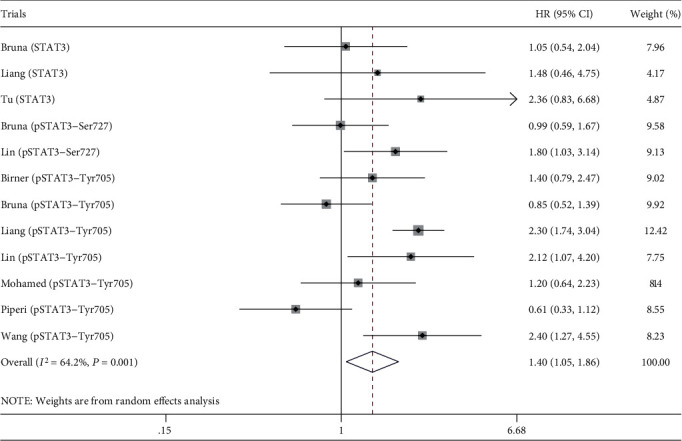
Association of STAT3/p-STAT3 with OS.
Figure 3.
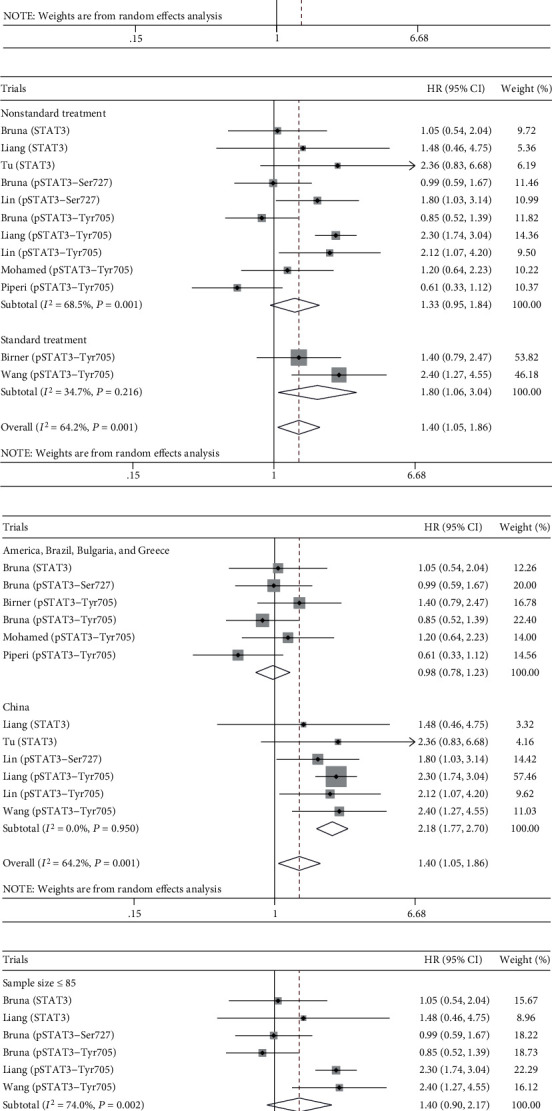
Subgroup analysis. (a) STAT3/p-STAT3. (b) Treatment. (c) Country. (d) Sample size.
3.3. Association of STAT3/p-STAT3 with PFS
The combined analysis of 5 trials suggested STAT3/p-STAT3 overexpression in glioma was associated with worse PFS (HR = 2.05, 95%CI = 1.63 ~ 2.58, P < 0.001) (Figure 4(a)). It is clear that there was no heterogeneity among trials (I2 = 0.0%, Ph = 0.938) (Figure 4(a)).
Figure 4.
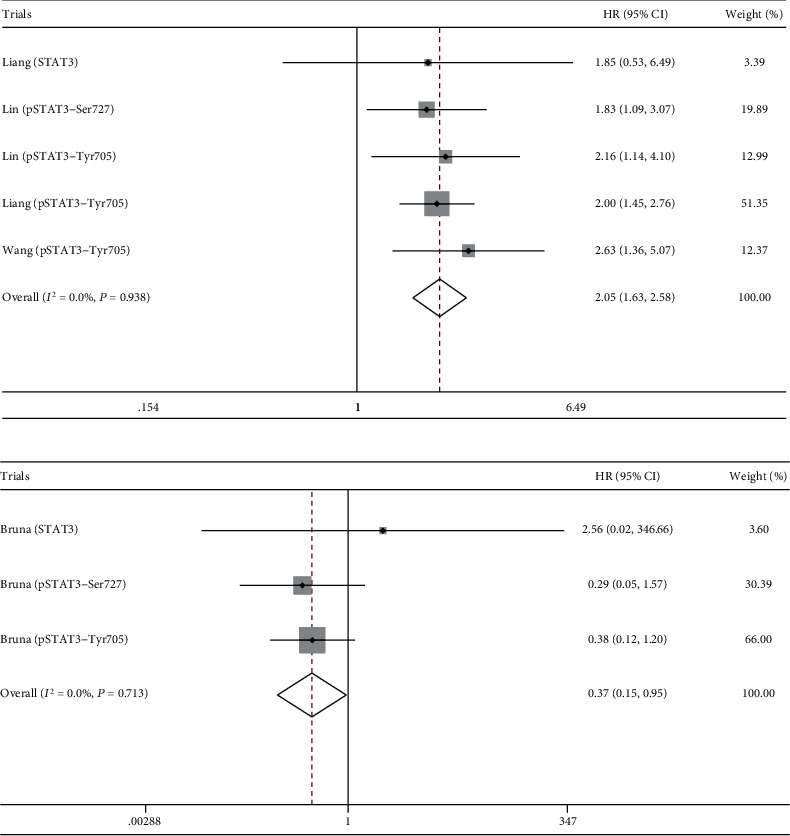
Association of STAT3/p-STAT3 with PFS and RFS. (a) PFS. (b) RFS.
3.4. Association of STAT3/p-STAT3 with RFS
The combined analysis of 3 trials showed that STAT3/p-STAT3 overexpression in glioma was associated with better RFS (HR = 0.37, 95%CI = 0.15 ~ 0.95, P < 0.039) (Figure 4(b)). It is clear that there was no heterogeneity among trials (I2 = 0.0%, Ph = 0.713) (Figure 4(b)).
3.5. Sensitivity Analysis
We removed each trial and reanalysed the data, and the main findings were unchanged (Figure 5).
Figure 5.
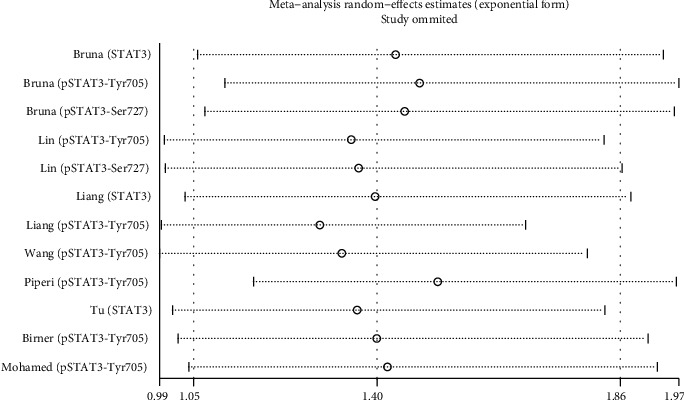
Sensitivity analysis.
3.6. Publication Bias
Funnel plot analysis showed that there was no statistical evidence of publication bias in this study (PBegg's test = 0.631 and PEgger's test = 0.290, respectively) (Figure 6).
Figure 6.
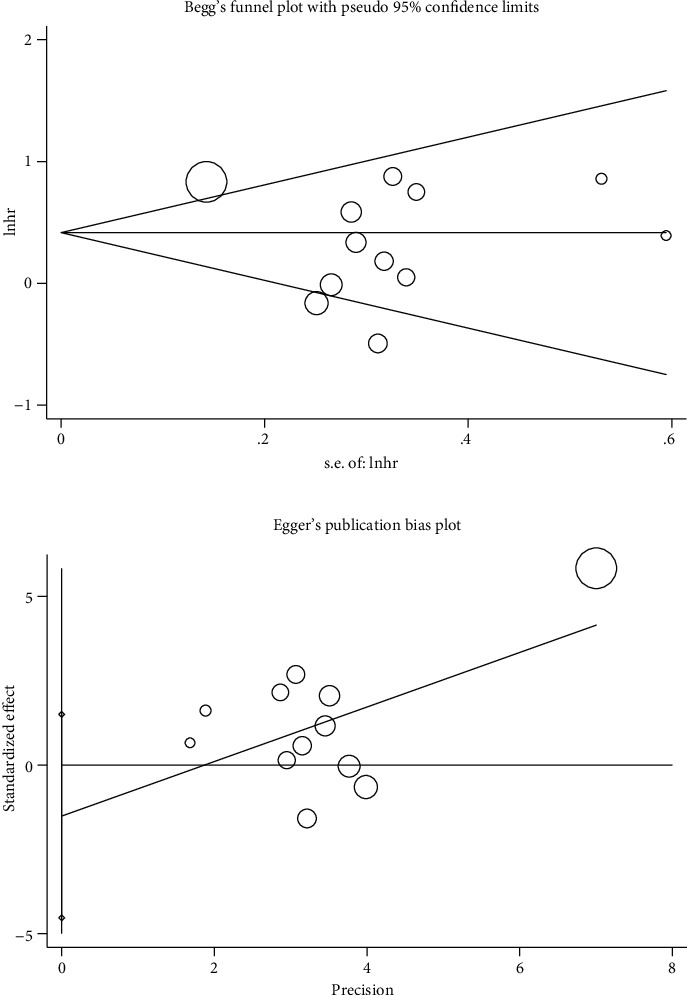
Publication bias.
4. Discussion
Few known risk factors are associated with the brain and central nervous system cancer, including glioma [52], and the only consistent correlations resulted from epidemiological studies [53, 54]. In our knowledge, this study is the most comprehensive assessment of the trials regarding STAT3/p-STAT3 expression and glioma prognosis to date. We systematically evaluated survival data for 1,070 glioma patients included in 12 different trials and came to the conclusion that the expression of STAT3/p-STAT3 may be a marker of poor prognosis in glioma. Our study shows that STAT3/p-STAT3 expression is related to poor glioma prognosis. The results of subgroup analysis further emphasize the importance and effectiveness of standard treatment and it also highlights the potential of STAT3/p-STAT3 for the development of valuable prognostic biomarkers and therapeutic agents of glioma.
Our study also inevitably has some limitations. Summarized population-level data, rather than individual patient-level data, was only extracted from the included 9 studies. The expression and positive rate of STAT3/p-STAT3 were diverse in different studies while immunohistochemical analysis was used to detect STAT3/p-STAT3. In addition, the scope of our research results may be limited as most of included studies were from China, while our results indicated that STAT3/p-STAT3 overexpression was associated with worse OS in China. Unfortunately, this limitation cannot be improved, given the chosen inclusion criteria. Last of all, HR and 95% CI were not directly available in some included studies and we had to apply Engauge Digitizer to obtain them from the survival curve, and heterogeneity under such circumstances was so substantial that random effect models and subgroup analysis could not diminish it; thus, additional analysis is necessary to clarify these confusing problems. Glioma is classified by cell type, by grade, and by location, but we failed to conduct subgroup analysis of classification due to the limited data included in this study, as well as meta-analysis of RFS or PFS. Moreover, during the analysis, we combined STAT3 and p-STAT3 together to analyse. It is precisely for this reason that this study cannot be more in-depth and thorough.
In conclusion, our study suggested that STAT3/p-STAT3 is associated with poor prognosis in patients with glioma, which indicated that STAT3/p-STAT3 might be a valuable prognostic biomarker and a promising therapeutic target for glioma. Further studies with larger sample sizes and multicenter/countries are needed to shed more light on the more precise correlation between STAT3/p-STAT3 and glioma.
Acknowledgments
This work was funded by the Jiangsu Leading Talent Project of Traditional Chinese Medicine (Jiangsu TCM 2018 No.4), Jiangsu Universities Nursing Advantage Discipline Project (2019YSHL095), and Applied Basic Research Programs of Science and Technology Department of Sichuan Province (2020YJ0437). We are grateful to all individuals who participated in glioma.
Data Availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
BL and NG conceived, designed, or planned the study. BL, SYL, HZG, and LXW acquired the data. BL, JL, YXZ, and NG analysed the data. BL, SYL, HZG, LXW, JL, YXZ, and NG helped interpret the results. BL drafted the manuscript. All authors revised and reviewed this work, and all authors gave their final approval of the submitted manuscript.
Supplementary Materials
Figure S1: subgroup analysis of STAT3 phosphorylation site.
References
- 1.Mrak G., Korent V., Mišir Krpan A., et al. Malignant brain neoplasms incidence and mortality trends in Croatia from 2001 to 2014. Croatian Medical Journal. 2019;60(1):33–41. doi: 10.3325/cmj.2019.60.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom Q. T., Bauchet L., Davis F. G., et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro-Oncology. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mamelak A. N., Jacoby D. B. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601) Expert Opinion on Drug Delivery. 2007;4(2):175–186. doi: 10.1517/17425247.4.2.175. [DOI] [PubMed] [Google Scholar]
- 4.Goodenberger M. K. L., Jenkins R. B. Genetics of adult glioma. Cancer Genetics. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Kun S., Duan Q., Liu G., Lu J.-M. Prognostic value of DNA repair genes based on stratification of glioblastomas. Oncotarget. 2017;8(35):58222–58230. doi: 10.18632/oncotarget.17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajaraman P., Melin B. S., Wang Z., et al. Genome-wide association study of glioma and meta-analysis. Human Genetics. 2012;131(12):1877–1888. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanson M., Hosking F. J., Shete S., et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Human Molecular Genetics. 2011;20(14):2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh K. M., Anderson E., Hansen H. M., et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genetic Epidemiology. 2013;37(2):222–228. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enciso-Mora V., Hosking F. J., Kinnersley B., et al. Deciphering the 8q24.21 association for glioma. Human Molecular Genetics. 2013;22(11):2293–2302. doi: 10.1093/hmg/ddt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice T., Zheng S., Decker P. A., et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH-mutated but not IDH-normal gliomas regardless of grade or histology. Neuro-Oncology. 2013;15(5):535–541. doi: 10.1093/neuonc/nos324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enciso-Mora V., Hosking F. J., di Stefano A. L., et al. Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. British Journal of Cancer. 2013;108(10):2178–2185. doi: 10.1038/bjc.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Deimling A., Eibl R. H., Ohgaki H., et al. p53 mutations are associated with 17p allelic loss in grade II and grade III astrocytoma. Cancer Research. 1992;52(10):2987–2990. [PubMed] [Google Scholar]
- 13.Adel Fahmideh M., Schwartzbaum J., Frumento P., Feychting M. Association between DNA repair gene polymorphisms and risk of glioma: a systematic review and meta-analysis. Neuro-Oncology. 2014;16(6):807–814. doi: 10.1093/neuonc/nou003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan H., Parsons D. W., Jin G., et al. IDH1andIDH2Mutations in Gliomas. The New England Journal of Medicine. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radner H., el-Shabrawi Y., Eibl R. H., et al. Tumor induction by ras and myc oncogenes in fetal and neonatal brain: modulating effects of developmental stage and retroviral dose. Acta Neuropathologica. 1993;86(5):456–465. doi: 10.1007/BF00228580. [DOI] [PubMed] [Google Scholar]
- 16.Pei S.-J., Zhao F., Liu J.-L., Fu Q., Shang P. Association between regulator of telomere elongation helicase 1 polymorphism and susceptibility to glioma. International Journal of Clinical and Experimental Medicine. 2015;8(1):690–697. [PMC free article] [PubMed] [Google Scholar]
- 17.Dziurzynski K., Chang S. M., Heimberger A. B., et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-Oncology. 2012;14(3):246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Martinez A., Alenda C., Irles E., et al. Lack of cytomegalovirus detection in human glioma. Virology Journal. 2017;14(1):p. 216. doi: 10.1186/s12985-017-0885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efird J. T., Davies S. W., O’Neal W. T., Anderson E. Animal viruses, bacteria, and cancer: a brief commentary. Frontiers in Public Health. 2014;2(14) doi: 10.3389/fpubh.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruder A. M., Carreon T., Butler M. A., et al. Exposure to farm crops, livestock, and farm tasks and risk of glioma: the Upper Midwest Health Study. American Journal of Epidemiology. 2009;169(12):1479–1491. doi: 10.1093/aje/kwp075. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J.-G., Liang B., Jin S.-H., et al. Development and validation of an RNA-seq-based prognostic signature in neuroblastoma. Frontiers in Oncology. 2019;9:p. 1361. doi: 10.3389/fonc.2019.01361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., Levy D. E. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324(5935):1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian M.-Y., Wang S.-B., Guo X.-F., et al. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene. 2020;39(2):428–442. doi: 10.1038/s41388-019-0996-y. [DOI] [PubMed] [Google Scholar]
- 24.Zhong C.-H., Tao B., Chen Y.-T., et al. B7-H3 regulates glioma growth and cell invasion through a JAK2/STAT3/slug-dependent signaling pathway. Onco Targets and Therapy. 2020;Volume 13:2215–2224. doi: 10.2147/OTT.S237841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H.-L., Tang F., Bian E., et al. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-β. Molecular Biology Reports. 2020;47(1):433–441. doi: 10.1007/s11033-019-05146-2. [DOI] [PubMed] [Google Scholar]
- 26.Yu H., Kortylewski M., Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nature reviews Immunology. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 27.Chen X.-K., Gu C. L., Fan J.-Q., Zhang X.-M. P-STAT3 and IL-17 in tumor tissues enhances the prognostic value of CEA and CA125 in patients with lung adenocarcinoma. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2020;125, article 109871 doi: 10.1016/j.biopha.2020.109871. [DOI] [PubMed] [Google Scholar]
- 28.Xue H., Yuan G., Guo X., et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12(7):1129–1152. doi: 10.1080/15548627.2016.1178446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Susman S., Pîrlog R., Leucuța D., et al. The role of p-Stat 3 Y705 immunohistochemistry in glioblastoma prognosis. Diagnostic Pathology. 2019;14(1):p. 124. doi: 10.1186/s13000-019-0903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H.-W., Lee P. M., Bamodu O. A., et al. Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A ratios mediate the anticancer effect of garcinol in STAT3/5A-Addicted glioblastoma. Cancers. 2019;11(12):p. 1888. doi: 10.3390/cancers11121888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H., Feng H., Zhang Y. Resveratrol inhibits hypoxia-induced glioma cell migration and invasion by the p-STAT3/mi R-34a axis. Neoplasma. 2016;63(4):532–539. doi: 10.4149/neo_2016_406. [DOI] [PubMed] [Google Scholar]
- 32.Song Y.-Y., Sun L. D., Liu M. L., et al. STAT3, p-STAT3 and HIF-1α are associated with vasculogenic mimicry and impact on survival in gastric adenocarcinoma. Oncology Letters. 2014;8(1):431–437. doi: 10.3892/ol.2014.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Wang Y., Shi Z., et al. Clinicopathological and prognostic role of STAT3/p-STAT3 in breast cancer patients in China: a meta-analysis. Scientific Reports. 2019;9(1, article 11243) doi: 10.1038/s41598-019-47556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong B.-Y., Liu Q.-Q., Liu Y.-X., Xiong X., Liu Y. Expressions of STAT3, p-STAT3 and E-cadherin in colorectal cancer and clinical implications. Chinese Journal of Gastrointestinal Surgery. 2014;17(6):594–597. [PubMed] [Google Scholar]
- 35.Zhang M., Sun H., Deng Y., et al. COPI-mediated nuclear translocation of EGFRvIII promotes STAT3 phosphorylation and PKM2 nuclear localization. International Journal of Biological Sciences. 2019;15(1):114–126. doi: 10.7150/ijbs.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y., Xue W., Schachner M., Zhao W. Honokiol eliminates glioma/glioblastoma stem cell-like cells via JAK-STAT3 signaling and inhibits tumor progression by targeting epidermal growth factor receptor. Cancers. 2019;11(1):p. 22. doi: 10.3390/cancers11010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiao J.-T., Zhang R., Li Z., et al. Nuclear Smad 6 promotes gliomagenesis by negatively regulating PIAS3-mediated STAT3 inhibition. Nature Communications. 2018;9(1):p. 2504. doi: 10.1038/s41467-018-04936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang B., Zhao L.-Z., Liao H.-L., Gu N. Rivaroxaban for cancer-associated venous thromboembolism: a systematic review and meta-analysis protocol. Medicine (Baltimore) 2019;98(48, article e18087) doi: 10.1097/MD.0000000000018087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 40.Wang X.-F., Liang B., Zeng D.-X., et al. The roles of MASPIN expression and subcellular localization in non-small cell lung cancer. Bioscience Reports. 2020;40(5, article BSR20200743) doi: 10.1042/BSR20200743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X.-F., Liang B., Chen C., et al. Long intergenic non-protein coding RNA 511 in cancers. Frontiers in Genetics. 2020;11:p. 667. doi: 10.3389/fgene.2020.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins J. P., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu Y., Zhong Y., Fu J., et al. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Medical Oncology. 2011;28(1):15–23. doi: 10.1007/s12032-010-9435-1. [DOI] [PubMed] [Google Scholar]
- 44.Lin G.-S., Yang L.-J., Wang X.-F., et al. STAT3 Tyr 705 phosphorylation affects clinical outcome in patients with newly diagnosed supratentorial glioblastoma. Medical Oncology. 2014;31(4):p. 924. doi: 10.1007/s12032-014-0924-5. [DOI] [PubMed] [Google Scholar]
- 45.Lin G.-S., Chen Y.-P., Lin Z.-X., Wang X. F., Zheng Z. Q., Chen L. STAT3 serine 727 phosphorylation influences clinical outcome in glioblastoma. International Journal of Clinical and Experimental Pathology. 2014;7(6):3141–3149. [PMC free article] [PubMed] [Google Scholar]
- 46.Kang C., et al. Inhibition of STAT3 reverses alkylator resistance through modulation of the AKT and β-catenin signaling pathways. Oncology Reports. 2011;26(5):1173–1180. doi: 10.3892/or.2011.1396. [DOI] [PubMed] [Google Scholar]
- 47.Birner P., Toumangelova-Uzeir K., Natchev S., Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. Journal of Neuro-Oncology. 2010;100(3):339–343. doi: 10.1007/s11060-010-0195-8. [DOI] [PubMed] [Google Scholar]
- 48.Piperi C., Samaras V., Levidou G., et al. Prognostic significance of IL-8-STAT-3 pathway in astrocytomas: correlation with IL-6, VEGF and microvessel morphometry. Cytokine. 2011;55(3):387–395. doi: 10.1016/j.cyto.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Abou-Ghazal M., Yang D. S., Qiao W., et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clinical Cancer Research. 2008;14(24):8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues B. R., Queiroz-Hazarbassanov N., Lopes M. H., et al. Nuclear unphosphorylated STAT3 correlates with a worse prognosis in human glioblastoma. Pathology, Research and Practice. 2016;212(6):517–523. doi: 10.1016/j.prp.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Liang Q.-C., Ma C.-K., Zhao Y., Gao G., Ma J. Inhibition of STAT3 reduces astrocytoma cell invasion and constitutive activation of STAT3 predicts poor prognosis in human astrocytoma. PloS One. 2013;8(12, article e84723) doi: 10.1371/journal.pone.0084723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel A. P., Fisher J. L., Nichols E., et al. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Neurology. 2019;18(4):376–393. doi: 10.1016/S1474-4422(18)30468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNeill K. A. Epidemiology of Brain Tumors. Neurologic Clinics. 2016;34(4):981–998. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Butowski N. A. Epidemiology and diagnosis of brain tumors. CONTINUUM: Lifelong Learning in Neurology. 2015;21(2 Neuro-oncology):301–313. doi: 10.1212/01.CON.0000464171.50638.fa. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: subgroup analysis of STAT3 phosphorylation site.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.


