Abstract
Endocasts are increasingly relied upon to examine avian brain evolution because they can be used across extant and extinct species. The endocasts of birds appear to be relatively faithful representatives of the external morphology of their brains, but it is unclear how well the size of a surface feature visible on endocasts reflects the volume of the underlying brain region. The optic lobe and the Wulst are two endocast structures that are clearly visible on the external surface of avian endocasts. As they overlie two major visual regions of the brain, the optic tectum and hyperpallium, the surface areas of the optic lobe and Wulst, respectively, are often used to infer visual abilities. To determine whether the surface area of these features reflects the volume of the underlying brain regions, we compared the surface areas of the optic lobes and Wulsts from digital endocasts with the volumes of the optic tecta and hyperpallia from the literature or measured from histological series of brains of the same species. Regression analyses revealed strong, statistically significant correlations between the volumes of the brain regions and the surface areas of the overlying endocast structures. In other words, the size of the hyperpallium and optic tectum can be reliably inferred from the surface areas of the Wulst and optic lobe, respectively. This validation opens the possibility of estimating brain‐region volumes for extinct species in order to gain better insights in their visual ecology. It also emphasizes the importance of adopting a quantitative approach to the analysis of endocasts in the study of brain evolution.
Our results show that the size of the optic lobe, the structure highlighted on these endocasts in orange, correlates with the size of the optic tectum, the underlying brain structure also highlighted in orange on this histologically stained cross‐section of a brain. We found that a similar relationship exists between the size of the brain structure called the hyperpallium and its overlying endocast structure, the Wulst. Both the optic tectum and the hyperpallium are involved in the visual pathways of birds, so researchers may be able to make inferences about visual capabilities in extinct birds based on the sizes of their optic lobes or Wulsts.
1. INTRODUCTION
Birds are one of the most speciose groups of vertebrates, and this diversity is reflected in their behavior and brain anatomy (Walsh and Milner, 2011a; Jetz et al., 2012; Cooney et al., 2017; Billerman et al., 2020; Pigot et al., 2020). Understanding the evolution of the avian brain has typically relied upon the study of extant species (Boire and Baron, 1994; Northcutt, 2002; Iwaniuk et al., 2004; Iwaniuk and Hurd, 2005; Gutiérrez‐Ibáñez et al., 2009, 2014; Corfield et al., 2012; Wylie et al., 2015), as well as brain endocasts preserved in the fossil record (Edinger, 1929, 1951; Wiman and Edinger, 1942). In recent years, there has been increased interest in examining brain evolution in extinct species through the use of virtual endocasts derived from computed tomography (CT) images (Witmer et al., 2008; Walsh and Milner, 2011a; Balanoff et al., 2016). In a similar fashion to mammals (Jerison, 1973), the size of the avian endocranial cavity is a reasonably accurate estimate of actual brain size (Iwaniuk and Nelson, 2002) and shape (Watanabe et al., 2019) (Figure 1), and so avian endocasts can yield major insights into the evolution of the avian brain in both extant and extinct species.
FIGURE 1.
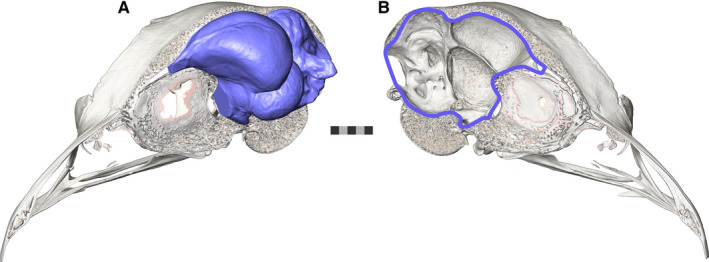
The skull and brain endocast of a ruffed grouse. (a) The brain endocast (three‐dimensionally modelled in blue) of a ruffed grouse (Bonasa umbellus, AMNH Birds SKEL 21616) in its hemisected skull as compared to its (b) endocranial cavity (outlined in blue). In birds, the endocast has been shown to be a faithful representative of the surface structure of the brain itself
The close relationship between endocranial size and brain size, as well as the increasingly widespread use of CT scanning to generate virtual endocasts, has led to a large number of paleoneurological studies of extinct birds (Domínguez Alonso et al., 2004; Ashwell and Scofield, 2008; Milner and Walsh, 2009; Scofield and Ashwell, 2009; Walsh and Milner, 2011b; Zelenitsky et al., 2011; Ksepka et al., 2012; Smith and Clarke, 2012; Balanoff et al., 2013; Walsh et al., 2013, 2016; Kawabe et al., 2014; Tambussi et al., 2015; Gold et al., 2016; Proffitt et al., 2016; Early et al., 2020). The most popular approach to studying these virtual endocasts is inspired by the Principle of Proper Mass, which states that the size of a brain region is proportional to its ability to process information (Jerison, 1973). Many researchers have studied patterns in relative sizes of brain regions and their potential functional implications in the endocasts of extant and extinct birds. For example, some species of elephant birds and moa have relatively smaller optic lobes than most extant birds, leading researchers to infer reduced visual capabilities (Torres and Clarke, 2018; Early et al., 2020). Other researchers have hypothesized that the expansion of the Wulsts of penguins throughout evolutionary time may correlate with changes in locomotion and prey choice (Ksepka et al., 2012; Proffitt et al., 2016).
Although these studies have provided some insights into avian brain evolution and the sensory ecology of extinct species, they are dependent on an untested assumption: the surface area of an endocast structure represents the volume of the underlying brain region. This is problematic because the surface area of features on the endocast could be affected by factors other than the volume of the directly underlying brain region, such as the size of neighboring endocast structures or the size of deeper brain regions. In addition, species that differ in morphology or volume of a brain region may be very similar in the surface area of the corresponding endocast structure. Determining the degree of correspondence between endocast features and brain‐region volumes is therefore essential for an effective understanding of avian, and more generally archosaurian, brain evolution throughout the fossil record and for relating endocast morphology to behavior.
Unlike the mammalian cortex in which sulci and gyri can be used to delineate individual cortical regions (Edinger, 1948; Radinsky, 1969, 1971; Falk, 1987), avian brains do not have many brain regions that can be potentially defined by surface area features on the endocast. Two structures that are readily apparent on most avian endocasts are the optic lobes and the Wulst. The optic lobes are rounded, bilateral expansions of the midbrain (Figure 2b,c). The optic lobes overlie the optic tract and optic tectum, which together receive the majority of retinal afferents (Hunt and Webster, 1975; Remy and Güntürkün, 1991; Mpodozis et al., 1995). The tectofugal pathway is the primary visual pathway in most birds (Wylie et al., 2009, 2015; Wylie and Iwaniuk, 2012) and, as such, variations in its relative size seem to reflect aspects of visual abilities. For example, kākāpō (Strigops habroptila) and kiwi (Apteryx spp.), two flightless, nocturnal taxa endemic to New Zealand, have relatively small optic tecta, as well as reductions in other parts of their visual systems (Martin et al., 2007; Corfield et al., 2011), indicating relatively poor visual abilities compared with other species. Reductions in the optic lobes of endocasts have similarly been interpreted as indications of reductions in the visual system (Smith and Clarke, 2012; Early et al., 2014, 2016, 2020; Torres and Clarke, 2018), based on the assumption that the surface area of the optic lobes reliably estimates the volume of the underlying optic tectum. However, beneath the optic tract and optic tectum are the inferior colliculus and tegmental nuclei, and changes in the sizes of these regions could affect the surface area of the optic lobes. In addition, expansion of cerebral hemispheres could obscure enough of the optic‐lobe surface area that it might not be a faithful proxy of the optic tectum (Figure 2b,c).
FIGURE 2.
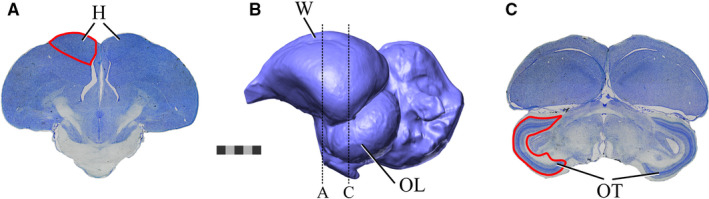
The relationship between the endocast structures and the brain regions. The brain section images have been scaled up relative to the scale bar according to the specimen's calculated shrinkage value. (a) A thionin‐stained axial section of the brain of a ruffed grouse (Bonasa umbellus, RUGR 344) sectioned by A. Iwaniuk's laboratory. The hyperpallium is outlined in red on the left hemisphere. (b) A lateral view of the endocast of a ruffed grouse (AMNH Birds SKEL 21616) showing the location of the Wulst and optic lobe. The position along the brain's rostrocaudal axis from which the slices shown in (a) and (c) would have been extracted are indicated by a dotted line with the appropriate letter. (c) A thionin‐stained axial section of the brain of a ruffed grouse sectioned by A. Iwaniuk's laboratory. The optic tectum is outlined in red on the left side. Abbreviations: H, hyperpallium; OL, optic lobe; OT, optic tectum; W, Wulst. Scale bar = 5 mm
The Wulst can also be observed readily on the dorsal surface of the telencephalic region of most avian endocasts and brains (Figure 2a,b). The name of the endocast structure refers to the bulge that is created by the hyperpallium and part of the hippocampal formation of the dorsal telencephalon (Puelles et al., 2007). The majority of the Wulst is hyperpallium, which is analogous, and potentially homologous, to the mammalian primary visual, somatosensory, and motor cortices (Funke, 1989; Deng and Wang, 1993; Wild, 1997; Medina and Reiner, 2000; Wild and Williams, 2000; Manger et al., 2002; Reiner et al., 2005). The hyperpallium is a strictly avian brain region that is not present in the forebrains of nonavian reptiles or other vertebrates (Jarvis, 2009), so the presence and expansion of the Wulst is considered to be a hallmark of avian brain evolution (Walsh and Milner, 2011a; Balanoff et al., 2013; Beyrand et al., 2019). Although the hyperpallium is multifunctional, based on both hodology (i.e., neural pathways) and neurophysiology (Funke, 1989; Deng and Wang, 1993; Wild, 1997; Medina and Reiner, 2000; Wild and Williams, 2000; Manger et al., 2002; Reiner et al., 2005), the majority of studies have focused on its functions in visual processing. Across species, birds with a large degree of binocular visual field overlap tend to have relatively larger hyperpallia than other groups of birds (Iwaniuk and Wylie, 2006; Iwaniuk et al., 2008). The size and position of the Wulst are frequently assessed in comparative studies of the endocasts of extant and extinct birds as if the size of the endocast structure represented the underlying hyperpallium (Ashwell and Scofield, 2008; Ksepka et al., 2012; Smith and Clarke, 2012; Early et al., 2014, 2016; Kawabe et al., 2014; Carril et al., 2016; Gold et al., 2016; Proffitt et al., 2016). However, the dorsoventral depth of the hyperpallium is quite variable across taxa, which could occur independently of the visible surface area. As mentioned previously, the Wulst also includes part of the hippocampal formation. Although the avian hippocampal formation typically occupies a small fraction of total telencephalic volume (Ward et al., 2012), it could contribute to Wulst surface area.
In order to fully understand the evolution of the optic tectum or hyperpallium, fossils need to be included in analyses of the relative size of these regions through the use of endocasts. Here, we tested the assumption that endocast‐structure surface area is an accurate estimate of underlying brain‐region volume for the optic lobe/optic tectum and Wulst/hyperpallium across a range of extant bird species. We used phylogenetic generalized least squares (PGLS) regressions to compare the scaling relationships of the endocast structures to endocast size with the scaling relationships of the brain regions to brain size. We also evaluated the relationships between the endocast and brain structures directly by assessing both relative and absolute sizes of these structures. Based on the spatial relationship between these brain and endocast structures, we predicted that our results would also show a strong relationship between the sizes of these complementary structures. If our prediction is correct, these relationships and the surface areas of the endocast structures of extinct birds could be then be used to predict the volumes of their brain regions.
2. MATERIALS AND METHODS
2.1. Species sampling
Overall, we compared endocast‐structure surface areas and brain‐region volumes across 34 avian species, representing 21 orders (Table 1). Species were selected based on: (a) covering a diversity of clades; (b) availability of the same species for both histological data (Iwaniuk et al., 2004, 2006, 2008, 2010; Corfield et al., 2011, 2012, 2016; Gutiérrez‐Ibáñez et al., 2013, 2014) and microCT scanning. This study did not involve any live animal specimens.
TABLE 1.
The endocast‐structure surface areas (SA) and brain‐region volumes (Vol) analyzed
| Species | Optic Lobe SA (mm2) | Optic Tectum Vol (mm3) | Wulst SA (mm2) | Hyperpallium Vol (mm3) | Endocast SA (mm2) | Brain Vol (mm3) |
|---|---|---|---|---|---|---|
| Agelaius phoeniceus 7,vii | 104.4 | 50.8 | 115.5 | 106.1 | 832.0 | 1614.9 |
| Amazilia tzacatl 7,vii | 35.4 | 12.8 | 27.7 | 11.3 | 215.4 | 182.27, 175.9vii |
| Anas platyrhynchos 3,iii | 164.1 | 251.5 | 211.1 | 572.2 | 2098.4 | 5738.0 |
| Apteryx australis mantelli 8,v | 56.9 | 32.0 | 174.4 | 224.0 | 2629.4 | 8293.08, 5299.0v |
| Apus apus 2 | 56.9 | 42.8 | — | — | 439.8 | 668.0 |
| Ardea cinerea 3,iii | 296.7 | 697.8 | 233.3 | 520.4 | 2453.7 | 8446.0 |
| Bonasa umbellus 2,vii | 192.7 | 182.3 | 97.8 | 171.9 | 1204.7 | 3136.07, 2548.3vii |
| Bubo virginianus 6,vi | 238.5 | 277.0 | 1145.3 | 2617.4 | 3900.5 | 17199.1 |
| Bubulcus ibis 3,iii | 199.5 | 211.0 | 147.7 | 220.7 | 1323.1 | 4025.0 |
| Buteo swainsoni 4 | 266.8 | 450.1 | — | — | 2334.3 | 8099.0 |
| Colinus virginianus 3,iii | 104.6 | 112.3 | 50.8 | 69.2 | 652.8 | 1091.0 |
| Columba livia 3,iii | 128.1 | 198.3 | 57.0 | 187.4 | 953.3 | 2093.0 |
| Crypturellus tataupa 1,i | 111.7 | 159.3 | 35.0 | 66.2 | 669.1 | 1583.0 |
| Dacelo novaeguineae 3,iii | 271.6 | 333.9 | 85.2 | 176.3 | 1607.3 | 3515.0 |
| Dromaius novaehollandiae 8,v | 446.0 | 494.2 | 913.26 | 4388.0 | 5861.3 | 21029.38, 21830.0v |
| Fulica americana 2,vii | 130.3 | 127.7 | 94.5 | 227.5 | 1118.3 | 2719.02, 2797.0vii |
| Junco hyemalis 7,vii | 70.1 | 50.5 | 53.8 | 61.0 | 433.8 | 879.17, 834.9vii |
| Larus philadelphia 7 | 140.9 | 160.3 | — | — | 1034.2 | 2512.6 |
| Meleagris gallopavo 3,iii | 373.0 | 771.1 | 238.1 | 469.0 | 2671.2 | 7990.0 |
| Melopsittacus undulatus 3,iii | 67.1 | 59.6 | 88.1 | 84.4 | 677.4 | 1220.0 |
| Mergus serrator 4 | 154.5 | 188.9 | — | — | 1622.2 | 4754.0 |
| Nyctidromus albicollus 3,iii | 70.2 | 37.0 | 46.3 | 66.0 | 525.3 | 910.0 |
| Passer domesticus 3,iii | 63.3 | 62.7 | 65.4 | 117.1 | 557.6 | 989.0 |
| Phalacrocorax auritus 2,ii | 259.0 | 187.2 | 534.3 | 1381.2 | 3163.2 | 7323.02, 7149.4ii |
| Picoides pubescens 7 | 78.1 | 50.1 | — | — | 708.6 | 997.5 |
| Psittacus erithacus 5,iv | 251.6 | 155.1 | 424.8 | 668.6 | 3071.0 | 6405.4 |
| Ptilinopus superbus 4 | 93.8 | 66.2 | — | — | 671.1 | 1052.1 |
| Rhea americana 3,iii | 514.7 | 1286.6 | 594.6 | 2295.1 | 4758.7 | 19228.0 |
| Scolopax rusticola 7,vii | 97.2 | 104.9 | 75.6 | 130.4 | 1232.5 | 2593.67, 2678.6vii |
| Spheniscus magellanicus 3,iii | 290.4 | 672.3 | 546.9 | 2362.6 | 3877.4 | 16757.0 |
| Sphyrapicus varius 7 | 71.6 | 65.1 | — | — | 676.2 | 888.4 |
| Steatornis caripensis 3,iii | 97.6 | 104.7 | 214.8 | 749.5 | 1355.8 | 3900.0 |
| Thalassarche melanophris 7 | 261.1 | 246.4 | — | — | 4330.9 | 14129.3 |
| Tyto alba 3,iii | 92.8 | 136.5 | 616.3 | 1605.4 | 2249.3 | 6149.0 |
Sources for optic tectum volumes: 1Bee de Speroni and Pirlot, 1987; 2Iwaniuk et al., 2006; 3Iwaniuk et al., 2008; 4Iwaniuk et al., 2010; 5Corfield et al., 2011; 6Gutiérrez‐Ibáñez et al., 2013; 7Gutiérrez‐Ibáñez et al., 2014; 8Corfield et al., 2016 and Corfield pers. comm. Sources for hyperpallium volumes: iBee de Speroni and Pirlot, 1987; iiBoire 1989; iiiIwaniuk et al., 2008; ivCorfield et al., 2011; vCorfield et al., 2012; viGutiérrez‐Ibáñez et al., 2013; viipresent study. In cases where two different specimens were measured, both brain volumes are listed with the appropriate superscript to indicate source. For example, the brain volume of the Amazilia tzacatl specimen for which optic tectum volume was measured is 182.2 mm3, and that of the specimen for which hyperpallium volume was measured is 175.9 mm3.
2.2. Brain‐region volumes
The term “optic lobe” has been used to define a general brain region in some previous publications (Cobb, 1964; Dubbeldam, 1968; Iwaniuk et al., 2006; Walsh and Milner, 2011a), and the terms “Wulst” and “hyperpallium” have been used almost interchangeably to refer to either the endocast structure or brain region in the literature (Budzynski and Bingman, 2004; Iwaniuk and Hurd, 2005; Iwaniuk et al., 2008; Witmer et al., 2008; Wylie et al., 2009; Gutiérrez‐Ibáñez et al., 2013). We use the term “optic lobe” to refer strictly to the endocast structure, whereas “optic tectum” refers to the underlying brain region. Similarly, the term “Wulst” refers solely to the endocast structure, and “hyperpallium” refers to the underlying brain region.
Optic tectum, hyperpallium, and brain volumes were obtained for 34 species in total (Table 1). The majority of the data were compiled from the literature (Iwaniuk et al., 2004, 2006, 2008, 2010; Corfield et al., 2011, 2012, 2016; Gutiérrez‐Ibáñez et al., 2013, 2014). Details of the source of specimens and histology are provided in those previous papers, but briefly, all brains were immersion‐fixed and sectioned in the coronal (axial) plane at a standard thickness (40 μm) and stained with thionin. Brain regions were delineated and measured according to previous studies and published stereotaxic brain atlases (Karten and Hodos, 1967; Stokes et al., 1974; Matochik et al., 1991; Boire and Baron, 1994; Puelles et al., 2007). Hyperpallial volumes were not published for all 34 species, so where possible, we measured the hyperpallium in the same specimens from which optic tectum data were taken. Sections throughout the telencephalon were digitized with an Olympus VS120 Virtual Slide Microscope (Olympus Corporation of the Americas) or photographed with a macro lens with a Nikon Coolpix digital camera. Brain regions were delineated in ImageJ (National Institutes of Health, Bethesda, MD) with the borders of the regions based on the same studies and atlases cited above, as well as De Groof et al. (2016). 3D brain‐region volumes and shrinkage factors were calculated for each specimen, and the volumes were corrected for shrinkage following Iwaniuk et al. (2010).
2.3. Generating endocasts
The specimens were CT scanned at slice thicknesses ranging from 25 µm to 96 µm, depending on specimen size, on a Bruker Skyscan 1173 µCT scanner, a TriFoil Imaging eXplore CT 120 Small Animal µCT scanner, or a GE Phoenix v|tome|x s240 µCT scanner (Table S1). The images were then segmented in Avizo (Thermo Fisher Scientific). Natural openings and breaks in the bone were patched and smoothed following best practices protocols for generating avian endocasts (Balanoff et al., 2016). Care was taken to remove endocranial vasculature such as the carotid artery and the transversotrigeminal and transversooccipital veins at their boundaries with the endocast, but the occipital sinus was not removed as it is sometimes a diffuse structure that obscures the cerebellar folia and could not be segmented consistently across all species.
Some of the openings in the braincase allow the passage of cranial nerves into their respective canals. Although nerves comprise part of the overall neuroanatomy of birds, they were considered as separate entities from the brain in this study, so their canals were cut off from the endocasts using points of inflection as indicators of the brain‐nerve boundary (e.g., Figure 3a). For all the endocasts that were contributed by colleagues, only one investigator (CE) performed the removals of the cranial nerves to ensure consistency across specimens. The fossa and canals of the trigeminal nerve (CN V) make up the ventral surface of the bony fossa tecti mesencephali in many birds, which means that CN V is continuous with the optic lobe on the endocast (Figure 3). The ventrolateral curvature of the fossa tecti mesencephali and the angle of the brainstem in axial view guided removal of CN V. The resulting boundary was smoothed in sagittal slices of CT scan data using an interpolation‐based approach until the optic lobe and CN V on the endocast closely resembled the shape of these structures in diffusible iodine‐based contrast‐enhanced CT (diceCT; Gignac et al., 2016) and histological datasets of the brains of birds.
FIGURE 3.
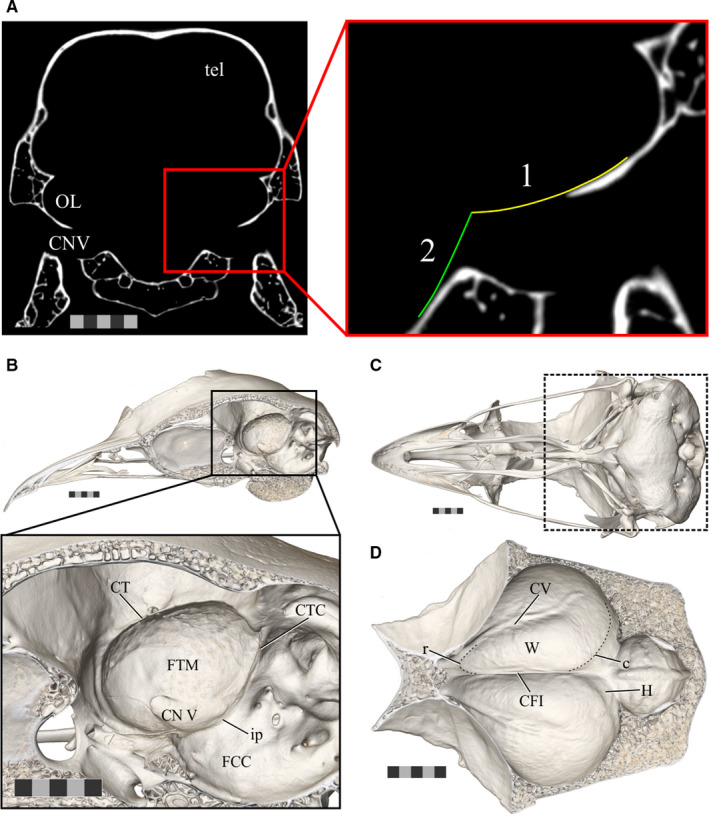
The boundaries of the endocast structures of interest. (a) An axial slice of CT scan data of Anas platyrhynchos (USNM Birds 631963) demonstrating the boundaries used to guide removal of the trigeminal ganglion from the optic lobe when generating endocasts for the present study. (1) Indicates the ventrolateral curvature of the fossa tecti mesencephali and (2) indicates the angle of the brainstem. (b) The sagittally hemisected skull of a ruffed grouse (Bonasa umbellus, AMNH Birds SKEL 21616) in a dorsolateral oblique view, with an inset zoomed in to focus on the fossa tecti mesencephali in which the optic lobe sits. (c) Ventral view of the skull of a ruffed grouse to provide orientation for (d). The area of focus of (d) is indicated by the box outlined in a dashed line. (d) A ventral view of the dorsal surface of the inside of the braincase of a ruffed grouse showing the Wulst on both hemispheres. Scale bars = 5 mm. Abbreviations: c, caudal boundary of the Wulst (subtle inflection between the Wulst and impression of hippocampus); CFI, crista frontalis interna; CTC, crista tecti caudalis; CT, crista tentorialis; CV, crista vallecularis; FCC, fossa cranii caudalis; FTM, fossa tecti mesencephali; H, impression of the hippocampus; ip, inflection point between the FCC and FTM; OL, optic lobe; r, rostral boundary of the Wulst; tel, telencephalon; TrO, impression of the tractus opticus; CN V, trigeminal nerve; W, Wulst. Scale bars = 5 mm
All CT scan image stacks and endocast surface files are available on MorphoSource (www.morphosource.org; see Table S1 for citeable DOIs).
2.4. Measuring endocast‐structure surface areas
Endocasts were simplified and remeshed to 600,000 faces in Avizo, exported from Avizo as little‐endian STL files, and imported into Maya modeling software (Autodesk). The optic lobes were isolated on each side of the endocast based on the boundaries of the fossa tecti mesencephali. The anatomical criteria used to bound the fossa tecti mesencephali were the crista tentorialis dorsally, the inflection point between the fossa cranii caudalis and the ventral floor of the fossa tecti mesencephali (where CN V had been removed) ventrally, the impression of the tractus opticus rostrally, and the crista tecti caudalis caudally (Figures 3 and 4). The Wulst was isolated from the dorsal surface of each cerebral hemisphere of the endocast using the anatomical criteria of the crista frontalis interna as the medial boundary, the crista vallecularis as the lateral boundary, the point at which the crista vallecularis and crista frontalis interna intersected as the rostral boundary, and the subtle inflection marking the caudal boundary of the Wulst (Figures 3 and 5). Identifying this caudal boundary based on surface features can be challenging, so 3D avian brain atlases based on MRI data were used to ensure the most accurate delimitation possible (De Groof et al., 2016). The remaining portions of the endocast were assigned to the “endocast remainder” material. These selections (“endocast remainder,” “optic lobes,” and “Wulst”) were separated, exported as OBJ files, and converted to STL files with MeshLab (Visual Computing Laboratory, Tuscany, Italy). The surface areas of the resulting STL files were measured in Avizo with its “Surface Area Volume” feature (Table 1). Although separating parts of mesh files could result in warping along the “cut” edge, this does not occur in Maya, which extracts faces without recomputing, smoothing, or redistributing the borders across vertices. To confirm this, we measured the surface area of a few endocasts before they had been exported from Avizo and after they had been treated in Maya and did not find a noticeable difference. The general steps of our method for measuring isolated endocast structures are similar to the few studies that have collected similar measurements (Walsh and Milner, 2011b; Torres and Clarke, 2018).
FIGURE 4.
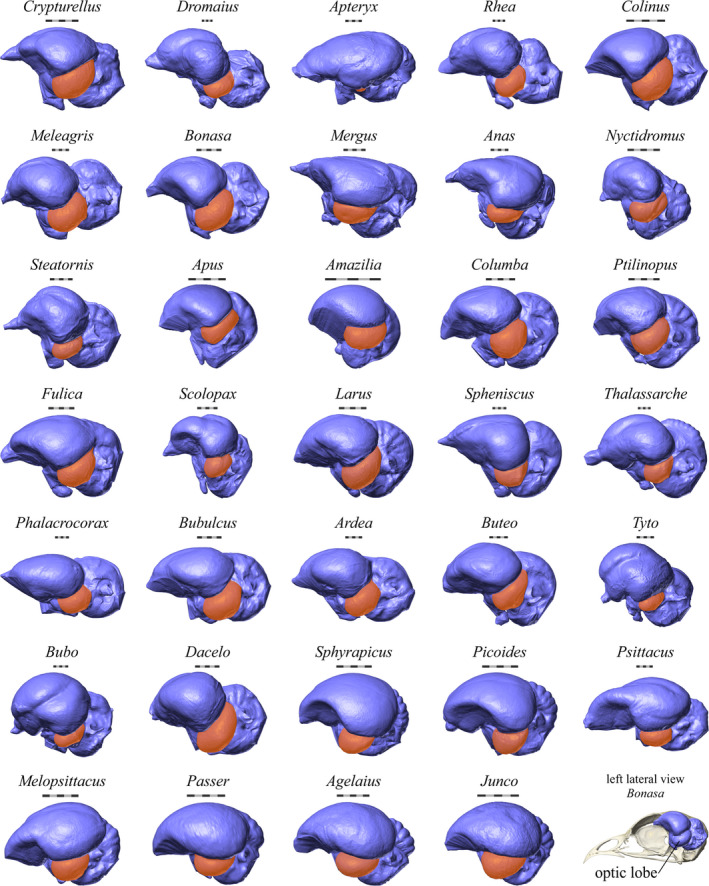
The endocasts of the species for which optic lobe surface areas were measured. The optic lobes are highlighted in orange. A skull and endocast of a ruffed grouse (Bonasa umbellus, AMNH Birds SKEL 21616) is shown in lateral view in the bottom right corner for orientation. Scale bars = 5 mm
FIGURE 5.
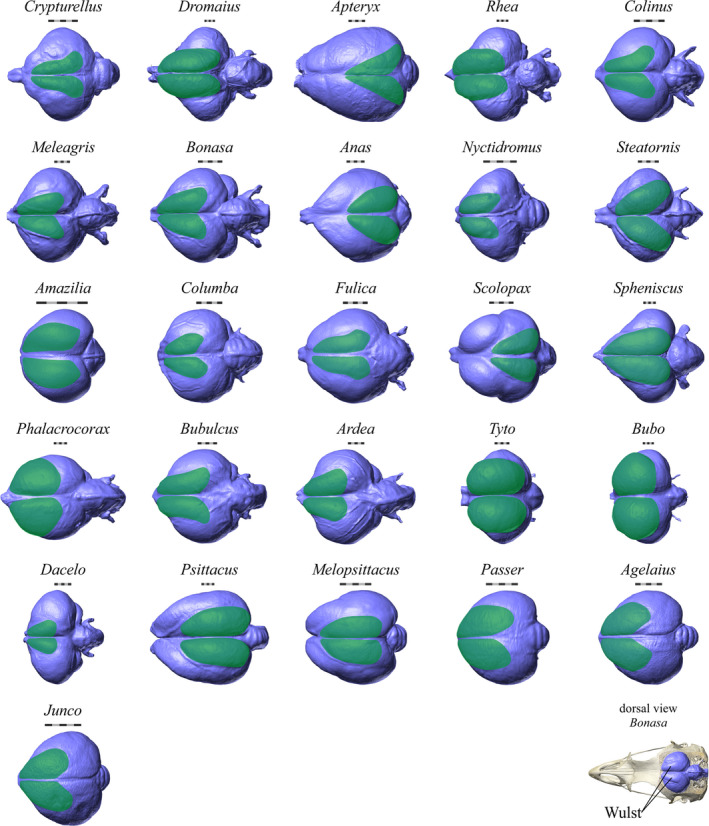
The endocasts of the species for which Wulst surface areas were measured. The Wulsts are highlighted in green. A skull and endocast of a ruffed grouse (Bonasa umbellus, AMNH Birds SKEL 21616) is shown in dorsal view in the bottom right corner for orientation. Scale bars = 5 mm
2.5. Statistical analyses
All phylogenetically informed statistical analyses were performed in R (R Foundation for Statistical Computing) running in RStudio (RStudio, Inc.). We first reconstructed a phylogenetic tree following the methods of Cooney et al. (2017). In brief, the fine‐scale relationships from Jetz et al. (2012), a complete time‐calibrated phylogeny for all extant species of birds known at the time, were mapped onto the backbone of Prum et al. (2015), a phylogeny based on next‐generation genome sequencing. The code for this procedure is available in the Supplementary Data of Cooney et al. (2017) and uses the R packages {ape} (Paradis et al., 2004) and {phytools} (Revell, 2012). As in Cooney et al. (2017), our analyses were also run with phylogenies from Ericson et al. (2006) and Hackett et al. (2008) to explore the effects of different topologies on the results. To determine the influence of phylogeny on the data, Pagel's λ was calculated for the regressions (Pagel, 1999) in {phytools} (Revell, 2012).
For all statistical analyses, we used a phylogenetic generalized least squares (PGLS) regression in {caper} (Orme et al., 2018). Tests of normality indicated that the raw data were not normally distributed, so all data were log‐transformed. For most regressions, there was at least one significant outlier, but these were not discarded as they reflect neuroanatomical diversity in the sample. 95% confidence intervals and prediction intervals for the regression lines were calculated using the {evomap} package (Smaers & Mongle, 2014).
Several PGLS analyses were performed to determine the relationship between endocast‐structure surface areas and brain‐region volumes. In cases where we were assessing relative size, we used brain or endocast remainder, that is, the total measurement of the brain or endocast minus the measurement of the region of interest, rather than the total size of the brain or endocast (Deacon, 1990), except for the regressions based on percentages. To compare the scaling of the optic lobe to that of the optic tectum, we regressed optic‐lobe surface area on endocast‐remainder surface area and optic‐tectum volume on brain‐remainder volume. To directly evaluate the relationship between endocast structures and brain regions, we regressed the percent of the brain volume occupied by the optic tectum on the percent of the brain surface area occupied by the optic lobe (Figure 6). As another evaluation of this relationship, we regressed absolute optic‐tectum volumes on absolute optic‐lobe surface areas. This set of analyses was performed on 34 species of birds. We performed the same set of analyses on Wulst surface areas and hyperpallium volumes for a subsample of 26 species of birds (Figure 7).
FIGURE 6.
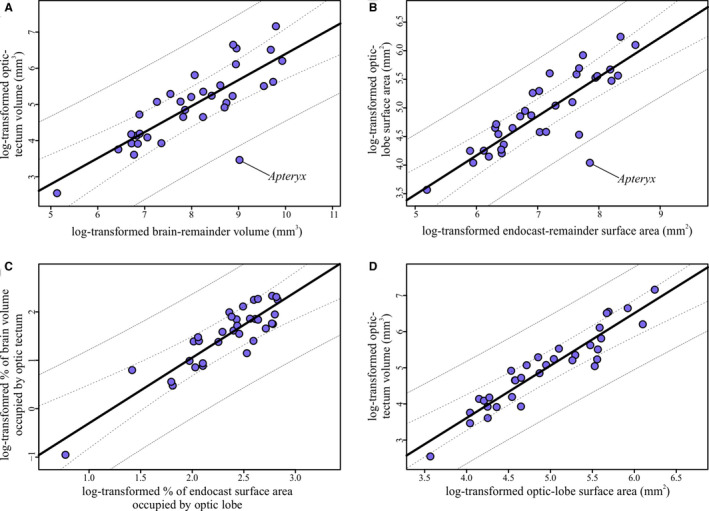
Regressions of optic tectum and optic lobe measurements. (a) Optic‐tectum volumes regressed on brain‐remainder volume (i.e., brain volume minus optic‐tectum volume). Slope = 0.72, adjusted R 2 = 0.64. (b) Optic‐lobe surface areas regressed on endocast‐remainder volume (i.e., endocast surface area minus optic‐lobe surface area). Slope = 0.69, adjusted R 2 = 0.71. (c) Relative optic‐tectum volume, as expressed as the percent of the brain occupied by the optic tectum, regressed on relative optic‐lobe surface area, as expressed as the percent of the endocast occupied by the optic lobes. Slope = 1.35, adjusted R 2 = 0.79. (d) Absolute optic‐tectum volumes regressed on absolute optic‐lobe surface areas. Slope = 1.44, adjusted R 2 = 0.88. For all regressions, the line from the PGLS regression is plotted in black, 95% confidence intervals are represented by the dashed lines, and 95% prediction intervals are represented by the dotted lines
FIGURE 7.
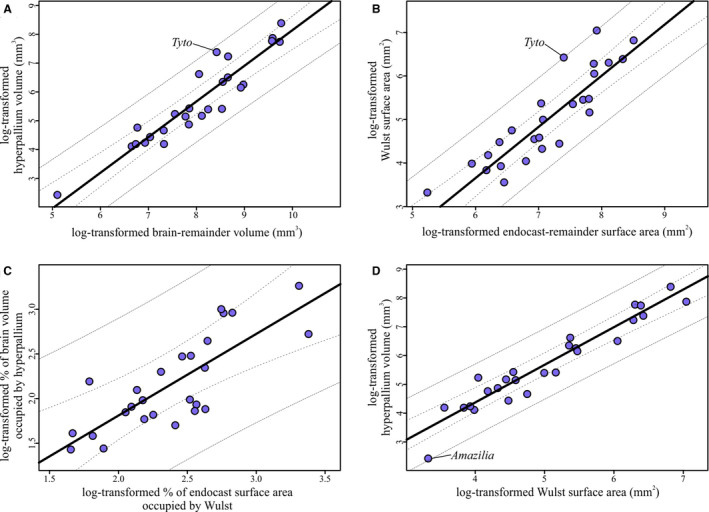
Regressions of hyperpallium and Wulst measurements. (a) Hyperpallium volumes regressed on brain‐remainder volume (i.e., brain volume minus hyperpallium volume). Slope = 1.24, adjusted R 2 = 0.87. (b) Wulst surface areas regressed on endocast‐remainder volume (i.e., endocast surface area minus Wulst surface area). Slope = 1.2, adjusted R 2 = 0.86. (c) Relative hyperpallium volume, as expressed as the percent of the brain occupied by the hyperpallium, regressed on relative Wulst surface area, as expressed as the percent of the endocast occupied by the Wulst. Slope = 0.91, adjusted R 2 = 0.61. (d) Absolute hyperpallium volumes regressed on absolute Wulst surface areas. Slope = 1.31, adjusted R 2 = 0.91. For all regressions, the line from the PGLS regression is plotted in black, 95% confidence intervals are represented by the dashed lines, and 95% prediction intervals are represented by the dotted lines
3. RESULTS
Phylogenetic signal was low and not significant in most regressions, regardless of which phylogeny was used (p = 0.05; Table S2). The slopes, intercepts, and correlation coefficients of the PGLS regressions were consistent across phylogenies (Table S2), so only the regressions corrected with the Jetz et al. (2012) + Prum et al. (2015) phylogeny are shown (Table 2; Figures 6 and 7).
TABLE 2.
The results of tests of phylogenetic signal, slope, intercept, adjusted R‐squared, and p‐values for all PGLS regressions run with the Prum + Jetz phylogeny
| Regression | Pagel's λ | p‐value | Slope | Intercept | Adj. R 2 | p‐value |
|---|---|---|---|---|---|---|
| OT Vol. vs. BR Vol. | 6.61e−5 | 1.00 | 0.72 | −0.80 | 0.64 | 9.65e−9 |
| OL SA vs. ER SA | 1.00 | 0.096 | 0.69 | 0.049 | 0.71 | 3.27e−10 |
| % OT Vol. vs. % OL SA | 6.61e−05 | 1.00 | 1.35 | −1.65 | 0.79 | 1.24e−12 |
| OT Vol. vs. OL SA | 7.03e−05 | 1.00 | 1.44 | −2.19 | 0.88 | <2.20e−16 |
| H Vol. vs BR Vol. | 0.75 | 0.46 | 1.24 | −4.23 | 0.87 | 2.93e−12 |
| W SA vs. ER SA | 1.00 | 2.54e−3 | 1.17 | −3.40 | 0.86 | 7.00e−12 |
| % H Vol. vs. % W SA | 6.61e−5 | 1.00 | 0.91 | −0.016 | 0.61 | 1.58e−6 |
| H Vol. vs. W SA | 0.48 | 0.68 | 1.31 | −0.85 | 0.91 | 1.50e−14 |
| OT Vol. vs. BR Vol. | 6.61e−5 | 1.00 | 0.72 | −0.80 | 0.64 | 9.65e−9 |
| OL SA vs. ER SA | 1.00 | 0.096 | 0.69 | 0.049 | 0.71 | 3.27e−10 |
| % OT Vol. vs. % OL SA | 6.61e−05 | 1.00 | 1.35 | −1.65 | 0.79 | 1.24e−12 |
| OT Vol. vs. OL SA | 7.03e−05 | 1.00 | 1.44 | −2.19 | 0.88 | <2.20e−16 |
| H Vol. vs BR Vol. | 0.75 | 0.46 | 1.24 | −4.23 | 0.87 | 2.93e−12 |
| W SA vs. ER SA | 1.00 | 2.54e−3 | 1.17 | −3.40 | 0.86 | 7.00e−12 |
| % H Vol. vs. % W SA | 6.61e−5 | 1.00 | 0.91 | −0.016 | 0.61 | 1.58e−6 |
| H Vol. vs. W SA | 0.48 | 0.68 | 1.31 | −0.85 | 0.91 | 1.50e−14 |
Phylogenetic signal is low and insignificant in most regressions. All of the relationships represented by these regressions are significant (p = 0.05). Abbreviations: BR, brain remainder; ER, endocast remainder; H, hyperpallium; OL, optic lobe; OT, optic tectum; SA, surface area; Vol., volume; W, Wulst.
Optic‐lobe surface area scaled to endocast‐remainder surface area with a similar slope to that of optic‐tectum volume relative to brain volume (Table 2, Figure 6a,b). Consistent with findings of a previous study (Martin et al., 2007), Apteryx australis mantelli is an outlier with a relatively smaller optic tectum and optic lobe. The correlation coefficients of both regressions were also similar, although this value was higher for the optic lobe‐endocast surface area regression. The regression of the relative optic‐tectum volume on relative optic‐lobe surface area expressed as percentages had a higher correlation coefficient and similarly sized confidence and prediction intervals (Table 2, Figure 6c). The slope of the line representing the relationship of absolute optic‐tectum volume to absolute optic‐lobe surface area (Figure 6d) is very similar to that of the line representing the relationship between the relative values (Figure 6c, Table 2). The correlation coefficient of the regression of absolute values was much higher than any of the regressions representing relative optic lobe or optic tectum size (Table 2). Taken together, these values suggest that optic‐lobe surface area is a good proxy for optic‐tectum volume in both relative and absolute terms.
The regressions of Wulst‐endocast surface area and hyperpallium‐brain volume had nearly identical slopes and correlation coefficients to one another (Table 2, Figure 7a,b). The distribution of species around the regression line was also similar, including the barn owl Tyto alba occupying the higher end of the 95% prediction interval. Relative Wulst surface area therefore appears to be a close approximation of relative hyperpallium volume. However, when both measurements were expressed as percentages, the correspondence between the two appears to be much weaker. As shown in Figure 7c, there is considerable scatter around the regression line, the confidence and prediction intervals are quite large, and the slope and correlation coefficients are low (Table 2). Based on this comparison, relative Wulst size and relative hyperpallium volume are not closely correlated. Finally, a comparison of the absolute measurements yielded another strong relationship (Figure 7d), with a similar slope and correlation coefficient to that of the Wulst‐endocast and hyperpallium‐brain regressions (Table 2, Figure 7a,b).
4. DISCUSSION
Our results show that optic lobe and Wulst surface areas scale similarly to endocast‐remainder surface areas as optic tectum and hyperpallium volumes scale to brain‐remainder volumes, respectively (Table 2; Figures 6 and 7). We also demonstrate that there is a strong, significant relationship between the relative volume and relative surface area of the structures of interest. Further, absolute optic lobe and Wulst surface areas are reliable proxies for optic tectum and hyperpallium volumes, respectively, with the strongest correlation coefficients of all regressions performed (Table 2). The close correspondence between these values exists in spite of the other aspects of neuroanatomy that could influence endocast‐structure surface area, such as the volume of regions deep to the underlying brain region or the degree of overlap between the endocast structure and neighboring brain regions on the endocast. Our results therefore provide support for previous studies that have inferred neuroanatomy based on relative optic lobe or Wulst size in extant and extinct birds (Ashwell and Scofield, 2008; Milner and Walsh, 2009; Scofield and Ashwell, 2009; Walsh and Milner, 2011b; Ksepka et al., 2012; Smith and Clarke, 2012; Walsh et al., 2013; Early et al., 2014, 2016, 2020; Kawabe et al., 2014; Tambussi et al., 2015; Carril et al., 2016; Gold et al., 2016; Proffitt et al., 2016; Torres and Clarke, 2018).
There are, however, a few caveats to keep in mind when interpreting the results of this study. The reported endocast measurements are based on one individual per species, as are most of the measurements based on histology. In both endocast and histological studies, this is generally due to resource constraints (e.g., access to specimens, scanner time), but it would be worthwhile to compare the endocasts of multiple adult individuals within a species to establish intraspecific variation in endocasts. A second potential source of error exists in the definition of structural boundaries, especially the boundary between the Wulst and the impression for the hyperpallium on the endocast. The difficulty inherent in distinguishing between these two structures means that any estimates of Wulst surface area should be assumed to include at least some of the hippocampal formation. In addition, other researchers may define the boundaries of the endocast structures differently, which could result in different measurements, but we have endeavored to clearly describe the boundaries we used to aid in reproducibility.
The relationships of the measured endocast‐structure surface areas with endocast‐remainder surface areas are very similar to the relationships of the volumes of their underlying brain regions with brain‐remainder volumes. The slopes of the optic lobe and optic tectum regressions are similar, as are those of the Wulst and hyperpallium regressions, although it should be noted that the intercepts are different within endocast and brain‐structure pairs (Table 2). The similarity in slopes indicates that the endocast structures scale similarly with endocast size, as do the brain regions with brain size. Thus, relative optic lobe and Wulst surface areas are reliable proxies for relative optic tectum and hyperpallium volumes, respectively.
Given the functional significance of relative brain‐region size established in histological studies (Iwaniuk et al., 2004, 2006, 2008, 2010; Corfield et al., 2011, 2012, 2016; Gutiérrez‐Ibáñez et al., 2013, 2014), if the relationship between the relative sizes of the endocast and brain regions is strong, relative endocast‐structure size could be used to make behavioral inferences in extinct birds. Regressions of the log‐transformed percentages of the brain volume occupied by the brain‐region volume on the log‐transformed percentages of the endocast surface area occupied by the endocast‐structure surface area yielded significant results (Table 2). However, the adjusted R‐squared value for the relationship between the relative hyperpallium size and relative Wulst size is low (Table 2), and there was more scatter around the regression line than in the other plots (Figure 7). Ratios are not always an appropriate way to correct for size, especially when there is an allometric rather than isometric relationship between variables (Atchley et al., 1976; Packard and Boardman, 1999), as is the case with our data (Table 2, Figures 6 and 7). That being said, the optic lobe percentage was more consistent with optic tectum percentage than was Wulst percentage with hyperpallium percentage (Figures 6 and 7). The inconsistency of using ratios for one structure versus another and the aforementioned statistical issues with ratios in general lead us to conclude that ratios should probably be avoided in quantitative analyses of endocasts, or at least interpreted with caution.
The regression of absolute optic‐tectum volume on absolute optic‐lobe surface area represents a strong, significant relationship between these two variables, as does the regression of absolute hyperpallium volume on absolute Wulst surface area (Table 2; Figures 6 and 7). Ultimately, these absolute endocast‐structure surface areas explain 88% (optic lobe) and 91% (Wulst) of the variation in absolute brain‐region volumes. The slope of the regression of optic‐tectum volume on optic‐lobe surface area is 1.4, which is close to what one would expect the relationship between the volume (e.g., mm3) and surface area (e.g., mm2) of a three‐dimensional object to be (1.5, or 3/2). The slope of the regression of hyperpallium volume on Wulst surface area is 1.3, which does not match the expected relationship between volume and surface area as closely. This could be due to the fact that the hyperpallium seems to be more variable in size and shape throughout its rostrocaudal extent than does the optic tectum. Regardless of the nature of the relationships between these brain and endocast structures, the relationship between absolute endocast‐structure surface areas and absolute brain‐region volumes is stronger than the relationship between these relative values, and it avoids the problems of using ratios in studies of allometry (Atchley et al., 1976; Packard and Boardman, 1999).
Studies that have identified functionally significant hyper‐ or hypotrophy in relative brain regions in extant birds have done so by comparing absolute brain‐region volume with the rest of the brain (Iwaniuk et al., 2004, 2006, 2008, 2006, 2004, 2010, 2008; Corfield et al., 2011, 2012, 2016; Gutiérrez‐Ibáñez et al., 2013, 2014). To generate a similar measure in extinct birds, absolute endocast volume could be substituted for absolute brain volume, as these values are strongly correlated in birds (Iwaniuk and Nelson, 2002). The relationship between the endocast‐structure surface areas and brain‐region volumes could be used to estimate absolute brain‐region volumes of an extinct bird from the surface area of its overlying endocast structure. The resulting brain‐region volume could then be used in conjunction with that from histological studies to estimate relative brain‐region size, infer sensory ecology and examine brain evolution more comprehensively by including the fossil record (Early et al., 2020).
Some groups of birds with enhanced tactile sensory systems have relatively smaller optic tecta (e.g., waterfowl, parrots) (Iwaniuk et al., 2010), leading some authors to hypothesize a trade‐off between the two sensory systems in these birds (Wylie et al., 2015). Relatively smaller optic tecta have also been documented in nocturnal birds like owls (Iwaniuk et al., 2010; Gutiérrez‐Ibáñez et al., 2013) and specialists like kiwi and kākāpō that have reduced their reliance on vision (Martin et al., 2007; Corfield et al., 2011) to the point of near‐blindness in the kiwi (Moore et al., 2017). Frogmouths (Podargidae) and owls, two groups of birds with significant visual field overlap and binocular vision, have relatively larger hyperpallial volumes than other groups of birds (Iwaniuk and Wylie, 2006; Iwaniuk et al., 2008). In light of our findings, similar functional capabilities could be more confidently inferred for extinct birds with reduced optic lobes or expanded Wulsts on their endocasts.
This study represents the largest sample of avian endocasts for which surface areas of optic lobes and Wulsts have been directly measured from virtual endocasts. Many previous studies comparing relative sizes of these endocast structures have done so with qualitative visual assessments, although they included quantitative measurements of other endocast traits (Milner and Walsh, 2009; Ksepka et al., 2012; Smith and Clarke, 2012; Kawabe et al., 2014; Carril et al., 2016; Gold et al., 2016; Proffitt et al., 2016). Others have estimated optic lobe or Wulst sizes from axial slices of CT scan data but have not directly measured the surface areas of the structures of interest (Ashwell and Scofield, 2008; Scofield and Ashwell, 2009; Tambussi et al., 2015). A few studies have in fact measured the surface area of these endocast structures directly from virtual models of the endocast, but these have limited samples of species (Walsh and Milner, 2011b; Early et al., 2014, 2016; Torres and Clarke, 2018). The clearly described boundaries we used to define the endocast structures of interest will make it easy for similar measurements to be taken from these endocasts. And the sample of virtual endocasts of extant birds published in this study provide the context of a comparative framework within which previously published endocasts of extinct birds could be analyzed (Early et al., 2020).
5. CONCLUSIONS
Overall, this study demonstrates a strong, significant relationship between the volume of the optic tectum and the surface area of the optic lobe and between the volume of the hyperpallium and the surface area of the Wulst. We also show that the studied endocast‐structures scale to endocast size with a similar slope as do their underlying brain regions to brain size, and that there is a significant relationship between relative brain‐region and relative endocast‐structure sizes. Now, along with endocasts being reliable estimates of overall brain size (Iwaniuk and Nelson, 2002), at least some endocast structures can also be considered faithful representatives of the size of their underlying brain regions. This finding increases the confidence with which inferences about the size of the optic tectum and hyperpallium can be made from the endocasts of extant and extinct birds. It also provides statistical relationships that can be used to calculate the volumes of the optic tectum and hyperpallium of species for which histological data are not available, potentially allowing researchers to extend studies of avian brain evolution into deep time.
AUTHOR CONTRIBUTIONS
Conceived project: CME, ANI, LMW. Collected data: CME, ANI, RCR. Analyzed and interpreted data: CME, ANI. Wrote paper: CME, ANI, RCR, LMW. Created figures: CME. All authors approved of this article and agree to be accountable for all aspects of the work.
Open Research Badges
This article has earned an Open Data Badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at https://www.morphosource.org/Detail/ProjectDetail/Show/project_id/996.
Supporting information
Table S1‐S2
R_Code
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (NSF) Graduate Research Fellowship Program under Grants No. DGE 1060934 and DGE 1645419 to CME and by NSF Grants No. IOB‐0517257, IOS‐1050154, and IOS‐1456503 to LMW. Additional funding was provided by grants from the Ohio University (OU) Graduate Student Senate, OU Office of the Vice President for Research and Creative Activity, and the Frank M. Chapman Memorial Fund of the American Museum of Natural History (AMNH) to CME and by Canada Foundation for Innovation grant CFI 30215 and support from the Canada Research Chairs Program to ANI. CT scanning performed at University of Washington's Friday Harbor Labs was supported in part by NSF DBI 1701665. Reviews by S. Kuchta, P. O’Connor, A. Stigall, S. Walsh, and anonymous reviewers improved this manuscript. We thank R. Felice and H. O’Brien for assistance with statistical analyses and phylogenetic comparative methods. S. Jordan and M. Wakui assisted with segmentation of some of the endocasts used in this study, and some CT scan data were provided by A. Balanoff, T. Gaetano, and W. R. Porter. The authors declare no conflicts of interest.
Early CM, Iwaniuk AN, Ridgely RC, Witmer LM. Endocast structures are reliable proxies for the sizes of corresponding regions of the brain in extant birds. J. Anat. 2020;237:1162–1176. 10.1111/joa.13285
DATA AVAILABILITY STATEMENT
All CT scan data of extant bird skulls and surface files of endocasts are available on MorphoSource. See Table S1 for citable DOIs for the image stacks and endocast surface files resulting from CT scans of each specimen.
REFERENCES
- Ashwell, K.W.S. and Scofield, R.P. (2008) Big birds and their brains: paleoneurology of the New Zealand moa. Brain, Behavior and Evolution, 71, 151–166. [DOI] [PubMed] [Google Scholar]
- Atchley, W.R. , Gaskins, C.T. and Anderson, D. (1976) Statistical properties of ratios. I. Empirical results. Systematic Zoology, 25, 137–148. [Google Scholar]
- Balanoff, A.M. , Bever, G.S. , Colbert, M.W. , Clarke, J.A. , Field, D.J. , Gignac, P.M et al. (2016) Best practices for digitally constructing endocranial casts: examples from birds and their dinosaurian relatives. Journal of Anatomy, 229, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanoff, A.M. , Bever, G.S. , Rowe, T.B. and Norell, M.A. (2013) Evolutionary origins of the avian brain. Nature, 501, 93–96. [DOI] [PubMed] [Google Scholar]
- Beyrand, V. , Voeten, D.F.A.E. , Bureš, S. , Bureš, S. , Fernandez, V. , Janáček, J et al. (2019) Multiphase progenetic development shaped the brain of flying archosaurs. Scientific Reports, 9, 10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billerman, S.M. , Keeney, B.K. , Rodewald, P.G. and Schulenberg, T.S. (2020) Birds of the World. Ithaca, NY, USA: Cornell Laboratory of Ornithology; https://birdsoftheworld.org/bow/home [Google Scholar]
- Boire, D. and Baron, G. (1994) Allometric comparison of brain and main brain subdivisions in birds. Journal of Brain Research, 35, 49–66. [PubMed] [Google Scholar]
- Budzynski, C.A. and Bingman, V.P. (2004) Participation of the thalamofugal visual pathway in a coarse pattern discrimination task in an open arena. Behavioural Brain Research, 153, 543–556. [DOI] [PubMed] [Google Scholar]
- Carril, J. , Tambussi, C.P. , Degrange, F.J. , Benitez Saldivar, M.J. and Picasso, M.B.J. (2016) Comparative brain morphology of Neotropical parrots (Aves, Psittaciformes) inferred from virtual 3D endocasts. Journal of Anatomy, 229, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, S. (1964) A comparison of the size of an auditory nucleus (n. mesencephalicus lateralis, pars dorsalis) with the size of the optic lobe in twenty‐seven species of birds. The Journal of Comparative Neurology, 122, 271–279. [DOI] [PubMed] [Google Scholar]
- Cooney, C.R. , Bright, J.A. , Capp, E.J. , Chira, A.M. , Hughes, E.C. , Moody, C.J.A et al. (2017) Mega‐evolutionary dynamics of the adaptive radiation of birds. Nature, 542, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield, J.R. , Gsell, A.C. , Brunton, D. , Heesy, C.P. , Hall, M.I. , Acosta, M.L et al. (2011) Anatomical specializations for nocturnality in a critically endangered parrot, the kakapo (Strigops habroptilus) E. J. Warrant, ed. PLoS ONE, 6, e22945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield, J.R. , Kolominsky, J. , Craciun, I. , Mulvany‐Robbins, B.E. and Wylie, D.R. (2016) Is cerebellar architecture shaped by sensory ecology in the New Zealand kiwi (Apteryx mantelli)? Brain, Behavior and Evolution, 87, 88–104. [DOI] [PubMed] [Google Scholar]
- Corfield, J.R. , Wild, J.M. , Parsons, S. and Kubke, M.F. (2012) Morphometric analysis of telencephalic structure in a variety of neognath and paleognath bird species reveals regional differences associated with specific behavioral traits. Brain, Behavior and Evolution, 80, 181–195. [DOI] [PubMed] [Google Scholar]
- De Groof, G. , George, I. , Touj, S. , Stacho, M. , Jonckers, E. , Cousillas, H et al. (2016) A three‐dimensional digital atlas of the starling brain. Brain Structure and Function, 221, 1899–1909. [DOI] [PubMed] [Google Scholar]
- Deacon, T.W. (1990) Fallacies of progression in theories of brain‐size evolution. International Journal of Primatology, 11, 193–236. [Google Scholar]
- Deng, C. and Wang, B. (1993) Convergence of somatic and visual afferent impulses in the Wulst of pigeon. Experimental Brain Research, 96, 287–290. [DOI] [PubMed] [Google Scholar]
- Domínguez Alonso, P. , Milner, A.C. , Ketcham, R.A. , Cookson, M.J. and Rowe, T.B. (2004) The avian nature of the brain and inner ear of Archaeopteryx . Nature, 430, 666–669. [DOI] [PubMed] [Google Scholar]
- Dubbeldam, J.L. (1968) On the Shape and the Structure of the Brainstem in Some Species of Birds: An Architectonic Study. Ph.D. Thesis. Leiden, The Netherlands: Leiden University. [Google Scholar]
- Early, C.M. , Ridgely, R.C. , Porter, W.R et al. (2016) The skull and endocranial anatomy of the extinct giant moa Dinornis robustus (Aves: Palaeognathae) and implications for the behavioral role of vision in moa. Anatomical Record Special Feature ‐ Program and Abstracts of ICVM 2016, 299, 223. [Google Scholar]
- Early, C.M. , Ridgely, R.C. and Witmer, L.M. (2014) New findings on the brain and skull structure of the recently extinct flightless giant moa (Aves: Dinornis), with implications for its behavior. Journal of Vertebrate Paleontology, 34 (supplement), 123. [Google Scholar]
- Early, C.M. , Ridgely, R.C. and Witmer, L.M. (2020) Beyond endocasts: using predicted brain‐structure volumes of extinct birds to assess neuroanatomical and behavioral inferences. Diversity, 12, 34. [Google Scholar]
- Edinger, T. (1929) Die fossilen Gehirne. Berlin/Heidelberg, Germany: Springer. [Google Scholar]
- Edinger, T. (1948) Evolution of the Horse Brain. Geological. Society of America. [Google Scholar]
- Edinger, T. (1951) The brains of the Odontognathae. Evolution, 5, 24. [Google Scholar]
- Ericson, P.G. , Anderson, C.L. , Britton, T. , Elzanowski, A. , Johansson, U.S. , Källersjö, M et al. (2006) Diversification of Neoaves: integration of molecular sequence data and fossils. Biology Letters, 2, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, D. (1987) Hominid paleoneurology. Annual Review of Anthropology, 16, 13–28. [Google Scholar]
- Funke, K. (1989) Somatosensory areas in the telencephalon of the pigeon. Experimental Brain Research, 76, 620–638. [DOI] [PubMed] [Google Scholar]
- Gignac, P.M. , Kley, N.J. , Clarke, J.A. , Colbert, M.W. , Morhardt, A.C. , Cerio, D et al. (2016) Diffusible iodine‐based contrast‐enhanced computed tomography (diceCT): an emerging tool for rapid, high‐resolution, 3‐D imaging of metazoan soft tissues. Journal of Anatomy, 228, 889–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, M.E.L. , Bourdon, E. and Norell, M.A. (2016) The first endocast of the extinct dodo (Raphus cucullatus) and an anatomical comparison amongst close relatives (Aves, Columbiformes). Zoological Journal of the Linnean Society, 177, 950–963. [Google Scholar]
- Gutiérrez‐Ibáñez, C. , Iwaniuk, A.N. , Lisney, T.J. and Wylie, D.R. (2013) Comparative study of visual pathways in owls (Aves: Strigiformes). Brain, Behavior and Evolution, 81, 27–39. [DOI] [PubMed] [Google Scholar]
- Gutiérrez‐Ibáñez, C. , Iwaniuk, A.N. , Moore, B.A. , Fernández‐Juricic, E. , Corfield, J.R. , Krilow, JM et al. (2014) Mosaic and concerted evolution in the visual system of birds. PLoS ONE, 9, e90102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez‐Ibáñez, C. , Iwaniuk, A.N. and Wylie, D.R. (2009) The independent evolution of the enlargement of the principal sensory nucleus of the trigeminal nerve in three different groups of birds. Brain, Behavior and Evolution, 74, 280–294. [DOI] [PubMed] [Google Scholar]
- Hackett, S.J. , Kimball, R.T. , Reddy, S. , Bowie, R.C.K. , Braun, E.l. , Braun, M.J et al. (2008) A phylogenomic study of birds reveals their evolutionary history. Science, 320, 1763–1768. [DOI] [PubMed] [Google Scholar]
- Hunt, S.P. and Webster, K.E. (1975) The projection of the retina upon the optic tectum of the pigeon. The Journal of Comparative Neurology, 162, 433–445. [DOI] [PubMed] [Google Scholar]
- Iwaniuk, A. , Clayton, D. and Wylie, D. (2006) Echolocation, vocal learning, auditory localization and the relative size of the avian auditory midbrain nucleus (MLd). Behavioural Brain Research, 167, 305–317. [DOI] [PubMed] [Google Scholar]
- Iwaniuk, A.N. , Dean, K.M. and Nelson, J.E. (2004) A mosaic pattern characterizes the evolution of the avian brain. Proceedings of the Royal Society of London. Series B: Biological Sciences, 271(suppl_4), S148–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaniuk, A.N. , Gutierrez‐Ibanez, C. , Pakan, J.M.P et al. (2010) Allometric scaling of the tectofugal pathway in birds. Brain, Behavior and Evolution, 75, 122–137. [DOI] [PubMed] [Google Scholar]
- Iwaniuk, A.N. , Heesy, C.P. , Hall, M.I et al. (2008) Relative Wulst volume is correlated with orbit orientation and binocular visual field in birds. Journal of Comparative Physiology A, 194, 267–282. [DOI] [PubMed] [Google Scholar]
- Iwaniuk, A.N. and Hurd, P.L. (2005) The evolution of cerebrotypes in birds. Brain, Behavior and Evolution, 65, 215–230. [DOI] [PubMed] [Google Scholar]
- Iwaniuk, A.N. and Nelson, J.E. (2002) Can endocranial volume be used as an estimate of brain size in birds? Canadian Journal of Zoology, 80, 16–23. [Google Scholar]
- Iwaniuk, A.N. and Wylie, D.R.W. (2006) The evolution of stereopsis and the Wulst in caprimulgiform birds: a comparative analysis. Journal of Comparative Physiology A, 192, 1313–1326. [DOI] [PubMed] [Google Scholar]
- Jarvis, E.D. (2009) Evolution of the pallium in birds and reptiles. Encyclopedia of Neuroscience, 1390–1400. [Google Scholar]
- Jerison, H. (1973) Evolution of the Brain and Intelligence in Vertebrates. Cambridge, MA: Academic Press. [Google Scholar]
- Jetz, W. , Thomas, G.H. , Joy, J.B. , Hartmann, K. and Mooers, A.O. (2012) The global diversity of birds in space and time. Nature, 491, 444–448. [DOI] [PubMed] [Google Scholar]
- Karten, H.J. and Hodos, W. (1967) Stereotaxic atlas of the brain of the pigeon (Columba livia). Baltimore, MD: Johns Hopkins Press. [Google Scholar]
- Kawabe, S. , Ando, T. and Endo, H. (2014) Enigmatic affinity in the brain morphology between plotopterids and penguins, with a comprehensive comparison among water birds. Zoological Journal of the Linnean Society, 170, 467–493. [Google Scholar]
- Ksepka, D.T. , Balanoff, A.M. , Walsh, S. , Revan, A. and Ho, A. (2012) Evolution of the brain and sensory organs in Sphenisciformes: new data from the stem penguin Paraptenodytes antarcticus . Zoological Journal of the Linnean Society, 166, 202–219. [Google Scholar]
- Manger, P.R. , Elston, G.N. and Pettigrew, J.D. (2002) Multiple maps and activity‐dependent representational plasticity in the anterior Wulst of the adult barn owl (Tyto alba): Somatosensory Wulst of the owl. European Journal of Neuroscience, 16, 743–750. [DOI] [PubMed] [Google Scholar]
- Martin, G.R. , Wilson, K.‐J. , Wild, J.M. , Parsons, S. , Kubke, M.F. and Corfield, J. (2007) Kiwi forego vision in the guidance of their nocturnal activities. PLoS ONE, 2, e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matochik, J.A. , Reems, C.N. and Wenzel, B.M. (1991) A brain atlas of the northern fulmar (Fulmarus glacialis) in stereotaxic coordinates (Part 1 of 2). Brain, Behavior and Evolution, 37, 215–229. [DOI] [PubMed] [Google Scholar]
- Medina, L. and Reiner, A. (2000) Do birds possess homologues of mammalian primary visual, somatosensory and motor cortices? Trends in Neurosciences, 23, 1–12. [DOI] [PubMed] [Google Scholar]
- Milner, A.C. and Walsh, S.A. (2009) Avian brain evolution: new data from Palaeogene birds (Lower Eocene) from England. Zoological Journal of the Linnean Society, 155, 198–219. [Google Scholar]
- Moore, B.A. , Paul‐Murphy, J.R. , Tennyson, A.J.D. and Murphy, C.J. (2017) Blind free‐living kiwi offer a unique window into the ecology and evolution of vertebrate vision. BMC Biology, 15, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpodozis, J. , Letelier, J.‐C. , Concha, M.L. and Maturana, H. (1995) Conduction velocity groups in the retino‐tectal and retino‐thalamic visual pathways of the pigeon (Columba livia). International Journal of Neuroscience, 81, 123–136. [DOI] [PubMed] [Google Scholar]
- Northcutt, R.G. (2002) Understanding vertebrate brain evolution. Integrative and Comparative Biology, 42, 743–756. [DOI] [PubMed] [Google Scholar]
- Orme, D. , Freckleton, R. , Thomas, G.H. , Petzoldt, T. , Fritz, S.A. and Isaac, N. (2018) Caper: comparative analyses of phylogenetics and evolution in R. R package version, 1.0.1.
- Packard, G.C. and Boardman, T.J. (1999) The use of percentages and size‐specific indices to normalize physiological data for variation in body size: wasted time, wasted effort? Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 122, 37–44. [Google Scholar]
- Pagel, M. (1999) Inferring the historical patterns of biological evolution. Nature, 401, 877–884. [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. and Strimmer, K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Pigot, A.L. , Sheard, C. , Miller, E.T et al. (2020) Macroevolutionary convergence connects morphological form to ecological function in birds. Nature Ecology & Evolution, 4, 230–239. [DOI] [PubMed] [Google Scholar]
- Proffitt, J.V. , Clarke, J.A. and Scofield, R.P. (2016) Novel insights into early neuroanatomical evolution in penguins from the oldest described penguin brain endocast. Journal of Anatomy, 229, 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum, R.O. , Berv, J.S. , Dornburg, A et al. (2015) A comprehensive phylogeny of birds (Aves) using targeted next‐generation DNA sequencing. Nature, 526, 569–573. [DOI] [PubMed] [Google Scholar]
- Puelles, L. , Martinez‐De‐La‐Torre, M. , Paxinos, G. , Watson, C. and Martinez, S. (2007) The chick brain in stereotaxic coordinates: an atlas correlating avian and mammalian neuroanatomy. San Diego: Academic Press. [Google Scholar]
- Radinsky, L. (1971) An example of parallelism in carnivore brain evolution. Evolution, 25, 518–522. [DOI] [PubMed] [Google Scholar]
- Radinsky, L.B. (1969) Outlines of canid and felid brain evolution. Annals of the New York Academy of Sciences, 167, 277–288. [Google Scholar]
- Reiner, A. , Yamamoto, K. and Karten, H.J. (2005) Organization and evolution of the avian forebrain. The Anatomical Record Part A: Discoveries in Molecular, Cellular, and Evolutionary Biology, 287A, 1080–1102. [DOI] [PubMed] [Google Scholar]
- Remy, M. and Güntürkün, O. (1991) Retinal afferents to the tectum opticum and the nucleus opticus principalis thalami in the pigeon. Journal of Comparative Neurology, 305, 57–70. [DOI] [PubMed] [Google Scholar]
- Revell, L.J. (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. [Google Scholar]
- Scofield, R.P. and Ashwell, K.W.S. (2009) Rapid somatic expansion causes the brain to lag behind: the case of the brain and behavior of New Zealand's Haast’s eagle (Harpagornis moorei). Journal of Vertebrate Paleontology, 29, 637–649. [Google Scholar]
- Smaers, J.B. and Mongle, C.S. (2014) evomap: R package for the evolutionary mapping of continuous traits. GitHub, https://github.com/JeroenSmaers/evomap.
- Smith, N.A. and Clarke, J.A. (2012) Endocranial anatomy of the Charadriiformes: sensory system variation and the evolution of wing‐propelled diving A. Iwaniuk, ed. PLoS ONE, 7, e49584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.M. , Leonard, C.M. and Nottebohm, F. (1974) The telencephalon, diencephalon, and mesencephalon of the canary, Serinus canaria, in stereotaxic coordinates. The Journal of Comparative Neurology, 156, 337–374. [DOI] [PubMed] [Google Scholar]
- Tambussi, C.P. , Degrange, F.J. and Ksepka, D.T. (2015) Endocranial anatomy of Antarctic Eocene stem penguins: implications for sensory system evolution in Sphenisciformes (Aves). Journal of Vertebrate Paleontology, 35, e981635. [Google Scholar]
- Torres, C.R. and Clarke, J.A. (2018) Nocturnal giants: evolution of the sensory ecology in elephant birds and other palaeognaths inferred from digital brain reconstructions. Proceedings of the Royal Society B: Biological Sciences, 285, 20181540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, S. and Milner, A. (2011a) Evolution of the avian brain and senses In: Dyke G. and Kaiser G. (Eds.) Living Dinosaurs. John Wiley & Sons Ltd: Chichester, UK, pp. 282–305. [Google Scholar]
- Walsh, S. and Milner, A. (2011b) Halcyornis toliapicus (Aves: Lower Eocene, England) indicates advanced neuromorphology in Mesozoic Neornithes. Journal of Systematic Palaeontology, 9, 173–181. [Google Scholar]
- Walsh, S.A. , Iwaniuk, A.N. , Knoll, M.A. , Bourdon, E. , Barrett, P.M. , Milner, A.C et al. (2013) Avian cerebellar floccular fossa size is not a proxy for flying ability in birds. PLoS ONE, 8, e67176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, S.A. , Milner, A.C. and Bourdon, E. (2016) A reappraisal of Cerebavis cenomanica (Aves, Ornithurae), from Melovatka, Russia. Journal of Anatomy, 229, 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B.J. , Day, L.B. , Wilkening, S.R. , Wylie, D.R. , Saucier, D.M. and Iwaniuk, A.N. (2012) Hummingbirds have a greatly enlarged hippocampal formation. Biology Letters, 8, 657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, A. , Gignac, P.M. , Balanoff, A.M. , Green, T.L. , Kley, N.J. and Norell, M.A. (2019) Are endocasts good proxies for brain size and shape in archosaurs throughout ontogeny? Journal of Anatomy, 234, 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild, J.M. (1997) The avian somatosensory system: the pathway from wing to Wulst in a passerine Chloris chloris . Brain Research, 759, 122–134. [DOI] [PubMed] [Google Scholar]
- Wild, J.M. and Williams, M.N. (2000) Rostral Wulst in passerine birds. I. Origin, course, and terminations of an avian pyramidal tract. The Journal of Comparative Neurology, 416, 429–450. [PubMed] [Google Scholar]
- Wiman, C. and Edinger, T. (1942) Sur les cranes et les encéphales d’Aepyornis et de Mullerornis . Bulletin de L’Academie Malgache, 24, 1–47. [Google Scholar]
- Witmer, L.M. , Ridgely, R.C. , Dufeau, D.L. and Semones, M.C. (2008) Using CT to peer into the past: 3D visualization of the brain and ear regions of birds, crocodiles, and nonavian dinosaurs In: Endo H. and Frey R. (Eds.) Anatomical Imaging. Tokyo, Japan: Springer, pp. 67–87. [Google Scholar]
- Wylie, D.R. , Gutiérrez‐Ibáñez, C. and Iwaniuk, A.N. (2015) Integrating brain, behavior, and phylogeny to understand the evolution of sensory systems in birds. Frontiers in Neuroscience, 9 , 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie, D.R.W. , Gutierrez‐Ibanez, C. , Pakan, J.M.P. and Iwaniuk, A.N. (2009) The optic tectum of birds: mapping our way to understanding visual processing. Canadian Journal of Experimental Psychology/Revue canadienne de psychologie expérimentale, 63, 328–338. [DOI] [PubMed] [Google Scholar]
- Wylie, D.R.W. and Iwaniuk, A.N. (2012) Neural mechanisms underlying visual motion detection in birds In: Lazareva O.F., Shimizu T. and Wasserman E.A. (Eds.) How Animals See the World: Comparative Behavior, Biology, and Evolution of Vision. Oxford, United Kingdom: Oxford University Press, pp. 289–318. [Google Scholar]
- Zelenitsky, D.K. , Therrien, F. , Ridgely, R.C. , McGee, A.R. and Witmer, L.M. (2011) Evolution of olfaction in non‐avian theropod dinosaurs and birds. Proceedings of the Royal Society B: Biological Sciences, 278, 3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S2
R_Code
Data Availability Statement
All CT scan data of extant bird skulls and surface files of endocasts are available on MorphoSource. See Table S1 for citable DOIs for the image stacks and endocast surface files resulting from CT scans of each specimen.


