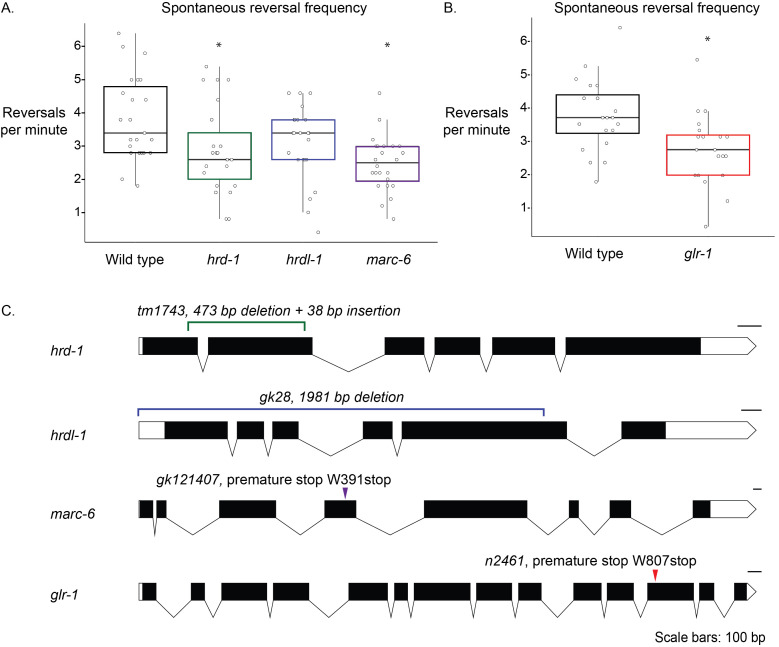
Figure 1. Mutations in E3 ubiquitin ligase genes and glr-1 affect spontaneous reversal frequencies.
A. Spontaneous reversal frequency assays were performed using C. elegans strains harboring mutations in E3 ubiquitin ligases. Box and whisker plots show the average reversals per minute, bounded by quartiles; the line in each box represents the median of the average reversals per minute for each genotype. N=23 individual animals measured for WT, hrd-1, and hrdl-1; N=24 individual animals for marc-6. * = p <0.05 (p=0.00069 for marc-6; p=0.032 for hrd-1); for hrdl-1, p= 0.14. Significance was calculated using the Tukey-Kramer test following a one-way ANOVA. The data is normally distributed (Shapiro-Wilk test, p=0.37) and groups show equal variance (residuals vs. fitted plot). B. Spontaneous reversal frequency for wild-type and glr-1 (n2461) animals. N=20 individual animals for both genotypes; significance was calculated using Student’s t-test (p=0.0063). The data is normally distributed (Shapiro-Wilk test, p=0.52) and groups show equal variance (residuals vs. fitted plot). C. Schematics of E3 ubiquitin ligase genes and glr-1 (Oswald et al., 2020). Schematics were made and annotated using http://wormweb.org/exonintron. The mutant E3 ubiquitin ligase strain hrd-1 contains an indel, hrdl-1 contains a deletion and marc-6 contains a nonsense mutation resulting in a premature stop codon.
Description
Secretory and membrane-bound proteins must be properly folded and matured at the endoplasmic reticulum (ER). Despite the molecular machinery dedicated to these processes, up to 1/3 of proteins are destroyed within minutes of their synthesis (Hirsch et al. 2009; Schubert et al. 2000).Misfolded proteins in the endoplasmic reticulum can accumulate and disrupt proteostasis, which can contribute to neurodegenerative diseases. The Endoplasmic Reticulum Associated Degradation (ERAD) pathway relies on E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases to ubiquitylate misfolded proteins, signaling for degradation of these misfolded proteins by the proteasome (Vembar and Brodsky 2008). Three putative E3 ligases that are expected to be involved in ERAD in C. elegans are HRDL-1, HRD-1, and MARC-6 (Sasagawa et al. 2007). We used strains harboring mutations in hrdl-1, hrd-1 and marc-6 genes to determine if these proteins are required for regulating spontaneous reversal behavior in C. elegans.
Spontaneous reversals are a C. elegans behavior whose frequency is regulated by well-defined circuitry and neurotransmitter receptors, including the glutamate receptor, glr-1 (Brockie et al. 2001; Burbea et al. 2002; Dahlberg and Juo 2014, Hart et al. 1995; Kowalski et al. 2011; Zheng et al. 1999). We hypothesized that the spontaneous reversal frequency behavior of C. elegans would be affected by E3 ligase mutations if they are important for normal spontaneous reversal behavior. glr-1 animals reverse significantly less than wild-type animals and were used as a positive control (Figure 1B) (Hart et al. 1995; Kowalski et al. 2011). Animals harboring mutations in marc-6 and hrd-1 also reverse significantly less than wild-type animals (Figure 1A). Animals lacking full-length hrdl-1 reversed less than wild-type animals, but this was not statistically significant (Figure 1A).
The primary motivation for our work was to ask if the glutamate receptor, GLR-1, might be regulated by these E3s, but further research must be done in order to determine molecular mechanisms that cause the differences in behavior that we report. Because ERAD E2 and E3 proteins can compensate for each other’s absence, we hypothesize that in the absence of any one E3 ligase, others are upregulated either through protein activity or gene expression (Bays et al. 2001; Weber et al. 2016). For example, if HRD-1is upregulated to compensate for the putative loss-of-function of HRDL-1 in hrdl-1 mutant animals, this could explain why hrdl-1 animals did not show a statistically significant reduction in reversals/minute compared to wild-type animals. However, our results from the hrd-1 and marc-6 mutants suggest that there is not complete redundancy between the E3 ligases. Future experiments will focus on testing this hypothesis using double and triple mutants in the E3 ligase genes.
Methods
Reversal assay protocol: Each C. elegans strain was grown at 21.4°C in the same bin on separate NGM agar plates seeded with OP50. Before performing the reversal assays, young adulthermaphroditic nematodes from each strain were picked onto separate seeded NGM agar plates and coded. This allows the later analysis of videotaped trials to be blind in order to reduce potential bias when scoring reversal assays. One animal at a time was picked from its coded seeded plate onto an unseeded NGM agar plate using halocarbon oil (a non-food substance) to induce food-seeking behavior. The animal was allowed to move around on the plate for two minutes before videotaping began. If the animal did not move away from its initial position on the plate after two minutes it was discarded and not counted in the data set. Each animal was recorded for five minutes and then discarded. At least one of each strain was observed during each experimental session in order to allow for any potential variations in temperature and humidity to be accounted for equally across all strains. The total N values listed represent measurements from multiple experimental sessions.
Scoring reversals: After recording all reversal assay trials for an experiment, the recorded coded trials were viewed and scored blindly by one researcher. A reversal was only counted if the animal moved backwards at least 1/6th of its body-length. The posterior pharyngeal bulb position was used as a marker for that distance.
Recording setup: Videos were recorded using an Olympus SZ61 microscope attached to a TLB 4000 Series Substage Illuminator base. The microscope was connected to The Imaging Source DFK 31AF03 color camera, which was connected to a computer running Windows OS. The software used to record the videos was Debut Professional by NCH Software.
Genotyping: The genotype of KP4 was confirmed using DNA sequencing. The genotypes of FJ861 and FJ863 were confirmed by PCR. The genotype of CLD33 was confirmed using PCR and restriction digest mapping using EarI (method noted on Million Mutation Project website, http://genome.sfu.ca/mmp/about.html).
Note on background transgene in CLD33: although the marc-6 mutation is in a GLR-1::GFP background, GLR-1::GFP animals show similar responsiveness to nose-touch assays (another GLR-1 dependent behavior) compared to wild-type animals (Rongo et al. 1998), and we do not expect that it affected the response of the animals.
Reagents
Table 1. C. elegans strains used.
| Strain Name | Genotype | Description | Reference |
| N2 | Wild-type | ||
| KP4 | glr-1(n2461) III | glr-1 putativeknockout, nonsense mutation | Hart AC et al. 1995 |
| FJ861 | hrd-1(tm1743) V*6 | E3 ubiquitin ligase gene hrd-1 putativeknockout, indel, backcrossed 6 times | C. elegans Deletion Mutant Consortium 2012 |
| FJ863 | hrdl-1(gk28) I*6 | E3 ubiquitin ligase gene hrdl-1 putativeknockout, deletion. Original strain VC35, backcrossed 6 times | C. elegans Deletion Mutant Consortium 2012 |
| CLD33 | nuIs24(IV);marc-6(gk121407) I*6 | Transgene Pglr-1::GLR-1::GFP crossed with E3 ubiquitin ligase gene marc-6 putative knockout, nonsense mutation, backcrossed 6 times. Original marc-6 strain is VC20284. Please also see the note in Methods. | WormBase ID: WBTransgene00001321 WormBase ID: WBVar00344650. |
Acknowledgments
Acknowledgments
We thank the students of Western Washington University’s Biology 487 course (Spring 2020) and Dr. Jacqueline Rose for critical readings of this manuscript.
Funding
Partial funding was provided by a Research and Creative Opportunities Grant for Undergraduate Students from the Office of Research & Sponsored Programs at Western Washington University.
References
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001 Jan 01;3(1):24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001 Aug 30;31(4):617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002 Jul 01;35(1):107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium. large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2012 Nov 01;2(11):1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg CL, Juo P. The WD40-repeat proteins WDR-20 and WDR-48 bind and activate the deubiquitinating enzyme USP-46 to promote the abundance of the glutamate receptor GLR-1 in the ventral nerve cord of Caenorhabditis elegans. J Biol Chem. 2013 Dec 19;289(6):3444–3456. doi: 10.1074/jbc.M113.507541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995 Nov 01;378(6552):82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009 Mar 26;458(7237):453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J Neurosci. 2011 Jan 26;31(4):1341–1354. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoldt A, Benthe HF, Haberland G. Digitoxin metabolism by rat liver microsomes. Biochem Pharmacol. 1975 Sep 01;24(17):1639–1641. [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998 Sep 18;94(6):751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Sasagawa Y, Yamanaka K, Ogura T. ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells. 2007 Sep 01;12(9):1063–1073. doi: 10.1111/j.1365-2443.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000 Apr 13;404(6779):770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008 Nov 12;9(12):944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Cohen I, Popp O, Dittmar G, Reiss Y, Sommer T, Ravid T, Jarosch E. Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol Cell. 2016 Aug 25;63(5):827–839. doi: 10.1016/j.molcel.2016.07.020. [DOI] [PubMed] [Google Scholar]
- WormBase web site, http://www.wormbase.org. WormBase ID: WBTransgene00001321.
- WormBase web site, http://www.wormbase.org. WormBase ID: WBVar00344650.
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999 Oct 01;24(2):347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]