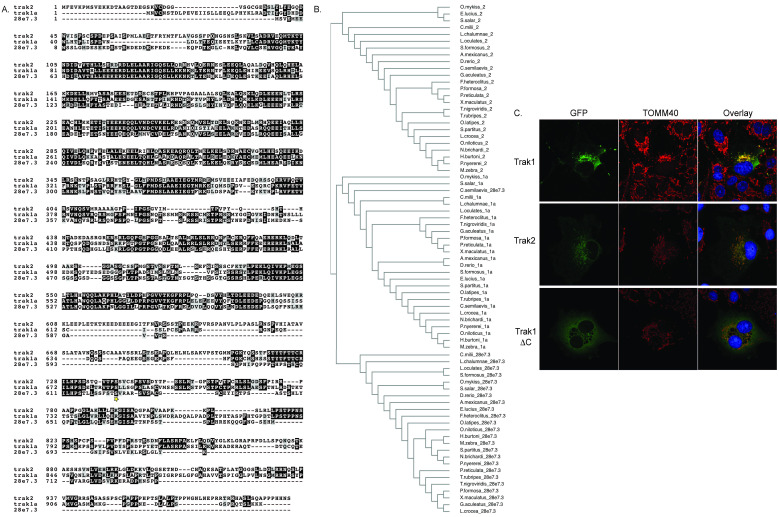
Figure 1. The zebrafish Trak proteins are paralogous to each other, and EGFP-tagged Trak1a and 2 proteins localize to the mitochondria when expressed in mammalian tissue culture cells.
A. D. rerio sequences used for alignment: trak1a (GenBank XM_001921277.3), si:dkey-28e7.3-201 (ENSDARG00000074508), and trak2-201: (ENSDARG00000102471). The star represents the position of an intron in the cloned trak1a gene to generate a truncated protein (Trak1 ΔC). B. Various fish species also have three distinct genes which predominantly group with trak1a, trak2, and si:dkey-28e7.3. C. COS7 cells were transfected with zebrafish Trak1a/pEGFP, Trak2/pEGFP, or Trak1 ΔC/pEGFP plasmids using Lipofectamine 3000 (GFP). After expression, cells were fixed with methanol and immunostained to identify mitochondria using a human TOMM40 primary antibody and TRITC secondary antibody. Nuclei were counterstained with DAPI. Overlay is presented at far right. Images were collected at 630X magnification on a Zeiss LSM 700 confocal microscope, and representative images are displayed.
Description
Inside a cell, mitochondria are organelles that exhibit dynamic locomotion and spatial rearrangement (Cai and Sheng 2009; Sheng 2017). This movement is necessary for a cell to maintain basic metabolic functions, and disruption of this motility often results in cell death. In fruit flies and mammals, one protein complex is primarily responsible for trafficking mitochondria along microtubules. This protein complex typically, but not always, consists of three proteins: Miro, Trak, and a motor protein (Stowers et al. 2002; Guo et al. 2005; Glater et al. 2006; Macaskill et al. 2009; Koutsopoulos et al. 2010; Brickley and Stephenson 2011; van Spronsen et al. 2013; Barel et al. 2017; López-Doménech et al. 2018; Henrichs et al. 2020).
In contrast to the single Drosophila protein Milton (Stowers et al. 2002), there are two mammalian genes of the Trak protein family: trak1 and trak2 (Koutsopoulos et al. 2010; Brickley and Stephenson 2011; van Spronsen et al. 2013). Both proteins have also been called huMilt1 and huMilt2, OIP106 and OIP98, or ALS2CR3/KIAA0549 and GRIF-1, respectively (Beck et al. 2002; Iyer et al. 2003; Brickley et al. 2005; Gilbert et al. 2006). Overexpression of either human Trak protein in mammalian cells generates abnormal clumping of the mitochondria, indicating that these proteins regulate mitochondrial motility and maintain the normal network of mitochondria in the cell (Koutsopoulos et al. 2010). In contrast, by reducing Trak protein levels in rat hippocampal neurons, Trak1 was identified as necessary for mitochondrial movement, yet Trak2 was not. However, Trak2 appears to be partially redundant in function with Trak1 since increasing Trak2 protein levels can rescue the loss of Trak1 protein (Brickley and Stephenson 2011). Discrepancy in the structure of the two paralogs may allow them to perform unique functions within the mitochondrial trafficking process (van Spronsen et al. 2013). For example, Trak1 associated with both the kinesin and dynein motor protein complex. In contrast, the Trak2 protein adopts a different structure that interferes with kinesin binding, only permitting interaction with dynein (van Spronsen et al. 2013; Loss and Stephenson 2015). In this way, these two similar proteins are distinct in their cellular functions, since Trak1/kinesin interactions mediate mitochondrial transport towards the axon, and the Trak2/dynein interactions cause mitochondrial transport toward the dendrites (van Spronsen et al. 2013; Loss and Stephenson 2017). Overall, mitochondrial trafficking and cell viability is highly sensitive to the concentration of Trak proteins (Stowers et al. 2002; Webber et al. 2008; Brickley and Stephenson 2011; Barel et al. 2017).
Assuming conserved function, our research sought to characterize the Trak proteins in zebrafish, D. rerio. Zebrafish are an excellent vertebrate model system. Their entire genome has been sequenced, annotated, and revised multiple times. They reproduce and develop quickly in comparison to other species, permitting observation of the roles of Trak proteins throughout developmental stages. Additionally, they are fertilized externally and are transparent, simplifying the ability to study Trak in live animals. In zebrafish, the most recent Ensembl genome assembly (April 2018) suggests that there are three paralogs of trak: trak1a, trak2, and si:dkey-28e7.3 (Figure 1A). trak1a is located on chromosome 16; trak2 is on chromosome 6; and si:dkey-28e7.3 is on chromosome 11. The fact that there are more than two genes is consistent with whole genome duplication in the teleost fish lineage (Meyer and Schartl 1999; Taylor et al. 2001; Taylor 2003; Woods et al. 2005). After genome duplication, redundant genes were pseudogenized, resulting in three instead of four genes (Meyer and Schartl 1999; Taylor et al. 2001). Indeed, three putative Trak genes are found in twenty-five other sequenced fish species, consistent with this hypothesis (Figure 1B and Table 1).
Based on findings about Trak in mammals and fruit flies, we aimed to understand this zebrafish protein family. As a first step, we co-localized each of the zebrafish Trak proteins with mitochondria in a heterologous system by cloning and overexpressing EGFP-tagged Trak proteins in easy-to-transfect and image mammalian tissue culture cells. trak1a and trak2 were amplified from cDNA from pooled embryos (1, 2, and 5 days post fertilization (dpf)). trak2 transcripts were expected to be abundant in this sample, based on high-throughput Expression Atlas data (Busch-Nentwich lab); there are no current expression data for either trak1a or si:dkey-28e7.3. For primers to trak1a, we used the original gene sequence from GenBank (XM_001921277.3), which has a longer N-terminus than the most recent version of the trak1a gene from Ensembl; a starting ATG occurs 225 nucleotides upstream of the ATG noted in trak1a-202 (atga to acag). We could not amplify an intact si:dkey-28e7.3 transcript, suggesting it might not be expressed or that the transcript may be of low abundance at these developmental time points. The trak1a and trak2 genes were cloned into pEGFP vectors and transfected into COS7 cells, and cells were immunostained for the endogenous outer mitochondrial protein TOMM40 (Figure 1C). In both cases, overexpressed Trak1a- and Trak2-EGFP proteins co-localized with TOMM40 and caused mitochondrial clumping, like their mammalian orthologs (Koutsopoulos et al. 2010). Notably, we also cloned a trak1a gene that had an intron at position 686 (starred in Figure 1A) that caused a frameshift nonsense mutation, resulting in a truncated C-terminus (Trak1 ΔC). This aberrant protein was predominantly cytosolic, rather than mitochondrial (Figure 1C), suggesting that the C-terminus is important for appropriate mitochondrial localization. These data are consistent with data regarding the human Trak1 protein where a protein containing only amino acids 1-734 (of 953) was cytosolic instead of mitochondrial (Koutsopoulos et al. 2010). Although we could not obtain a full-length si:dkey-28e7.3 transcript to test in our system, we predict that si:dkey-28e7.3 is unlikely to localize to the mitochondria, given that we demonstrate that the C-terminus of trak1a seems to anchor the protein to the mitochondria and this region is the most divergent in si:dkey-28e7.3 (Figure 1A). si:dkey-28e7.3 is also less similar to the mammalian orthologs that align well with D. rerio trak1a and trak2.
Using the preliminary data generated from this project, we hope that the zebrafish Trak protein family can be further analyzed in vivo, allowing for better understanding of how Trak proteins contribute to mitochondrial movement in a live vertebrate animal. A previous study in zebrafish did not report a phenotype after injection of a splicing morpholino targeted to trak1 (Choksi et al. 2014). This result is not surprising given data from the knockdown of Miro where a morphant phenotype is only observed when all three paralogs are depleted, suggesting redundant functions (Hollister et al. 2016). CRISPR technology utilized in the context of zebrafish expressing mitochondrial fluorescent proteins may be a means to stably generate double and triple mutants to measure the effects at a cellular and organismal level (Fichi et al. 2019; Arribat et al. 2019). Greater knowledge of these motility mechanisms may eventually be extrapolated to neurodegenerative diseases, such as ALS and spastic paraplegia, where mitochondrial trafficking plays a significant role.
Methods
Phylogenetics
Alignments were created from Ensembl sequences from zebrafish GRCz11 (Yates et al. 2020) for si:dkey-28e7.3-201 (ENSDARG00000074508) and trak2-201 (ENSDARG00000102471) and GenBank for trak1a (XM_001921277.3; https://www.ncbi.nlm.nih.gov/nuccore/XM_001921277.3). Sequences were inputted into Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/; Madeira et al. 2019) and formatted using BoxShade (https://embnet.vital-it.ch/software/BOX_form.html) or Guide Tree Cladogram to show the relationships among the various species. Sequence data from other fish species were compiled through examining various fish genomes available on Ensembl and utilizing Ensembl’s BLAST/BLAT tool to search for Trak orthologs. A complete list of the sequence files is in Table 1.
Table 1. Sequences used for the cladogram in Figure 1B.
| species | transcipt name | gene ID | assembly |
| Astyanax mexicanus | si:dkey-28e7.3-201 | ENSAMXG00000018307 | Astyanax_mexicanus-2.0 (September 2017) |
| Astyanax mexicanus | trak1a-201 | ENSAMXG00000029426 | Astyanax_mexicanus-2.0 (September 2017) |
| Astyanax mexicanus | trak2-201 | ENSAMXG00000012437 | Astyanax_mexicanus-2.0 (September 2017) |
| Callorhinchus milii | si:dkey-28e7.3-202 | ENSCMIG00000017409 | Callorhinchus_milii-6.1.3 (December 2013) |
| Callorhinchus milii | trak2-202 | ENSCMIG00000009046 | Callorhinchus_milii-6.1.3 (December 2013) |
| Callorhinchus milii | trak1a-202 | ENSCMIG00000006222 | Callorhinchus_milii-6.1.3 (December 2013) |
| Cynoglossus semilaevis | Unnamed (listed in tree as C.semilaevis_28e7.3) | ENSCSEG00000006363 | Cse_v1.0 (January 2014) |
| Cynoglossus semilaevis | trak2-201 | ENSCSEG00000014003 | Cse_v1.0 (January 2014) |
| Cynoglossus semilaevis | trak1a-201 | ENSCSEG00000003758 | Cse_v1.0 (January 2014) |
| Danio rerio | si:dkey-28e7.3-201 | ENSDARG00000074508 | GRCz11 (May 2017) |
| Danio rerio | trak2-201 | ENSDARG00000102471 | GRCz11 (May 2017) |
| Danio rerio | trak1a-202 | ENSDARG00000041304 | GRCz11 (May 2017) |
| Esox lucius | si:dkey-28e7.3-201 | ENSELUG00000009890 | Eluc_v4 (April 2019) |
| Esox lucius | trak2-202 | ENSELUG00000002196 | Eluc_v4 (April 2019) |
| Esox lucius | trak1a-204 | ENSELUG00000004615 | Eluc_v4 (April 2019) |
| Fundulus heteroclitus | si:dkey-28e7.3-202 | ENSFHEG00000001760 | Fundulus_heteroclitus-3.0.2 (January 2015) |
| Fundulus heteroclitus | trak2-201 | ENSFHEG00000007156 | Fundulus_heteroclitus-3.0.2 (January 2015) |
| Fundulus heteroclitus | trak1a-201 | ENSFHEG00000016654 | Fundulus_heteroclitus-3.0.2 (January 2015) |
| Gasterosteus aculeatus | si:dkey-28e7.3-201 | ENSGACG00000018327 | BROAD S1 (February 2006) |
| Gasterosteus aculeatus | trak1a-201 | ENSGACG00000006003 | BROAD S1 (February 2006) |
| Gasterosteus aculeatus | trak2-201 | ENSGACG00000014156 | BROAD S1 (February 2006) |
| Haplochromis burtoni | si:dkey-28e7.3-201 | ENSHBUG00000000586 | AstBur1.0 (December 2011) |
| Haplochromis burtoni | trak1a-203 | ENSHBUG00000011007 | AstBur1.0 (December 2011) |
| Haplochromis burtoni | trak2-202 | ENSHBUG00000016106 | AstBur1.0 (December 2011) |
| Larimichthys crocea | si:dkey-28e7.3-202 | ENSLCRG00005003733 | L_crocea_2.0 (November 2018) |
| Larimichthys crocea | trak2-203 | ENSLCRG00005003901 | L_crocea_2.0 (November 2018) |
| Larimichthys crocea | trak1a-204 | ENSLCRG00005020918 | L_crocea_2.0 (November 2018) |
| Latimeria chalumnae | si:dkey-28e7.3-201 | ENSLACG00000007300 | LatCha1 (September 2011) |
| Latimeria chalumnae | trak2-201 | ENSLACG00000006273 | LatCha1 (September 2011) |
| Latimeria chalumnae | trak1-201 | ENSLACG00000001844 | LatCha1 (September 2011) |
| Lepisosteus oculates | si:dkey-28e7.3-201 | ENSLOCG00000013653 | LepOcu1 (December 2011) |
| Lepisosteus oculates | trak1a-201 | ENSLOCG00000001329 | LepOcu1 (December 2011) |
| Lepisosteus oculates | trak2-201 | ENSLOCG00000010723 | LepOcu1 (December 2011) |
| Maylandia zebra | si:dkey-28e7.3-201 | ENSMZEG00005008443 | M_zebra_UMD2a (April 2018) |
| Maylandia zebra | trak2-201 | ENSMZEG00005006192 | M_zebra_UMD2a (April 2018) |
| Maylandia zebra | trak1a-202 | ENSMZEG00005002331 | M_zebra_UMD2a (April 2018) |
| Neolamprologus brichardi | si:dkey-28e7.3-201 | ENSNBRG00000023010 | NeoBri1.0 (December 2011) |
| Neolamprologus brichardi | Unnamed (listed in tree as N.brichardi_1a) | ENSNBRG00000010142 | NeoBri1.0 (December 2011) |
| Neolamprologus brichardi | trak2-201 | ENSNBRG00000014231 | NeoBri1.0 (December 2011) |
| Oncorhynchus mykiss | si:dkey-28e7.3-205 | ENSOMYG00000018119 | Omyk_1.0 (June 2017) |
| Oncorhynchus mykiss | Unnamed (listed in tree as O.mykiss_1a) | ENSOMYG00000039092 | Omyk_1.0 (June 2017) |
| Oncorhynchus mykiss | trak2-201 | ENSOMYG00000011517 | Omyk_1.0 (June 2017) |
| Oreochromis niloticus | si:dkey-28e7.3-205 | ENSONIG00000018805 | O_niloticus_UMD_NMBU (June 2018) |
| Oreochromis niloticus | trak1a-202 | ENSONIG00000007240 | O_niloticus_UMD_NMBU (June 2018) |
| Oreochromis niloticus | trak2-202 | ENSONIG00000011992 | O_niloticus_UMD_NMBU (June 2018) |
| Oryzias latipes | si:dkey-28e7.3-203 | ENSORLG00000025102 | ASM223467v1 (July 2017) |
| Oryzias latipes | trak1a-201 | ENSORLG00000005943 | ASM223467v1 (July 2017) |
| Oryzias latipes | trak2-201 | ENSORLG00000024460 | ASM223467v1 (July 2017) |
| Poecilia formosa | si:dkey-28e7.3-201 | ENSPFOG00000003564 | Poecilia_formosa-5.1.2 (October 2013) |
| Poecilia formosa | trak1a-201 | ENSPFOG00000018652 | Poecilia_formosa-5.1.2 (October 2013) |
| Poecilia formosa | trak2-201 | ENSPFOG00000001467 | Poecilia_formosa-5.1.2 (October 2013) |
| Poecilia reticulata | si:dkey-28e7.3-203 | ENSPREG00000000453 | Guppy_female_1.0_MT (April 2014) |
| Poecilia reticulata | trak1a-204 | ENSPREG00000013360 | Guppy_female_1.0_MT (April 2014) |
| Poecilia reticulata | trak2-201 | ENSPREG00000007459 | Guppy_female_1.0_MT (April 2014) |
| Pundamilia nyererei | si:dkey-28e7.3-201 | ENSPNYG00000002974 | PunNye1.0 (December 2011) |
| Pundamilia nyererei | trak2-201 | ENSPNYG00000009328 | PunNye1.0 (December 2011) |
| Pundamilia nyererei | trak1a-201 | ENSPNYG00000011722 | PunNye1.0 (December 2011) |
| Salmo salar | si:dkey-28e7.3-201 | ENSSSAG00000044165 | ICSASG_v2 (June 2015) |
| Salmo salar | trak2-201 | ENSSSAG00000031517 | ICSASG_v2 (June 2015) |
| Salmo salar | trak1a-201 | ENSSSAG00000004358 | ICSASG_v2 (June 2015) |
| Scleropages formosus | si:dkey-28e7.3-201 | ENSSFOG00015002821 | fSclFor1.1 (April 2019) |
| Scleropages formosus | trak1a-208 | ENSSFOG00015001806 | fSclFor1.1 (April 2019) |
| Scleropages formosus | trak2-201 | ENSSFOG00015004411 | fSclFor1.1 (April 2019) |
| Stegastes partitus | si:dkey-28e7.3-201 | ENSSPAG00000018438 | Stegastes_partitus-1.0.2 (May 2014) |
| Stegastes partitus | trak1a-202 | ENSSPAG00000017135 | Stegastes_partitus-1.0.2 (May 2014) |
| Stegastes partitus | trak2-201 | ENSSPAG00000001720 | Stegastes_partitus-1.0.2 (May 2014) |
| Takifugu rubripes | si:dkey-28e7.3-201 | ENSTRUG00000003718 | fTakRub1.2 (June 2019) |
| Takifugu rubripes | trak2-201 | ENSTRUG00000010804 | fTakRub1.2 (June 2019) |
| Takifugu rubripes | trak1a-202 | ENSTRUG00000002103 | fTakRub1.2 (June 2019) |
| Tetraodon nigroviridis | si:dkey-28e7.3-201 | ENSTNIG00000004864 | TETRAODON 8.0 (March 2007) |
| Tetraodon nigroviridis | trak2-201 | ENSTNIG00000010353 | TETRAODON 8.0 (March 2007) |
| Tetraodon nigroviridis | trak1a-201 | ENSTNIG00000006230 | TETRAODON 8.0 (March 2007) |
| Xiphophorus maculatus | si:dkey-28e7.3-202 | ENSXMAG00000000197 | X_maculatus-5.0-male (December 2017) |
| Xiphophorus maculatus | trak1a-202 | ENSXMAG00000008737 | X_maculatus-5.0-male (December 2017) |
| Xiphophorus maculatus | trak2-201 | ENSXMAG00000008010 | X_maculatus-5.0-male (December 2017) |
Cloning and ExpressionTotal RNA was extracted from one, two, or five days post fertilization (dpf) wild-type AB zebrafish embryos and pooled. The embryos were homogenized in Trizol (Invitrogen) according to manufacturer’s directions. First strand cDNA synthesis was then carried out using 2μg of RNA and the Superscript III kit (Invitrogen). For trak1a, PCR was performed using Phusion HF DNA polymerase (NEB) and primers designed to GenBank XM_001921277.3 (zfTrak1.ECORI.F: 5’-GCCGAATTCATGAATGTGTGTAACAGCAC; zfTrak1.XHOI.R: 5’-CCGCTCGAGTCACTTTTTCTTGAGGC) to clone into pcGlobin2 (Ro et al. 2004). EcoRI and XbaI were then used to move the gene into pEGFP-C2 (Clontech). trak2 was cloned directly into pEGFP-C2 using primers (zfTRAK2.XhoI.F: 5’-CGATCTCGAGCATGTTCGAGGTGAAGCC; zfTRAK2.XmaI.R: 5’-TGGGCCCTTATGAATTATGATGTGGGG) and the restriction enzymes XhoI and XmaI. Cloned genes were sequenced through Eurofins Genomics and compared to Genbank and Ensembl sequences using primers to the vector and internal primers (Trak1.811.F: 5’- GCACTTGAAAATGAAGAG; Trak1.1798.F: 5’-GTCGTGACCAAGGGC; TRAK1.2015.R: 5′-GCTCATCTGAAGGGTG; zfTRAK2.800.F: 5’-CTCCCAGAAGAATGAGGA; and zfTRAK2.1660.R.seq: 5’-TGGTGAAGGTGTAGGTG).
COS7 cells (ATCC) were cultured in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum in a 37ºC incubator at 5% CO2. Confluent cells were split onto uncoated glass coverslips in a 6-well plate for transfection. COS7 cells were transfected with zebrafish Trak1/pEGFP-C2 or Trak2/pEGFP-C2, human Milton1/pEGFP or Milton2/pEGFP (provided by M.T. Ryan), or pEGFP-C2 using Lipofectamine 3000 (Invitrogen) following the manufacturer’s protocol. Proteins were expressed for 12 to 24 hours before processing for immunofluorescence. Transfected COS7 cells were washed in 1X PBS and fixed with 100% ice-cold methanol at -20°C for at least 10 minutes. The cells were washed three times for 5 minutes in 1X phosphate buffered saline (PBS). Fixed cells were blocked for 1 hour at room temperature on a rocker in 5% normal goat serum/0.3% Triton X/1X PBS. Rabbit polyclonal primary antibody to human TOMM40 (1:500; ULAB4; gift of C. M. Koehler) was added to blocking buffer and incubated at 4ºC overnight. The cells were washed 3 times with 1X PBS for 5 minutes and incubated with the secondary antibody goat-anti-rabbit-TRITC (1:1000; Jackson ImmunoChemicals) with 0.3% Triton X/1X PBS for 1 hour on a rocker. The cells were stained with DAPI/0.3% Triton X/1X PBS at room temperature for 5 minutes. The cells were washed twice with 1X PBS for 5 minutes and placed onto slides with Fluoromount-G (Southern Biotech). Images were collected using 630X magnification on a Zeiss LSM 700 confocal microscope. Transfection and imaging of constructs and their comparison to human Traks and untagged EGFP were performed for more than ten replicates, and representative images are shown.
Acknowledgments
Acknowledgments
The human TOMM40 antibody was contributed by C. M. Koehler (University of California, Los Angeles), and the human Trak/Milton plasmids were provided by M.T. Ryan (Monash University). We would like to thank the Busch-Nentwich lab for providing RNA-seq data (https://www.ebi.ac.uk/gxa/experiments/E-ERAD-475/Results).
Funding
This project was funded through internal grant programs at Rollins College.
References
- Arribat Y, Grepper D, Lagarrigue S, Richard J, Gachet M, Gut P, Amati F. Mitochondria in Embryogenesis: An Organellogenesis Perspective. Front Cell Dev Biol. 2019 Nov 22;7:282–282. doi: 10.3389/fcell.2019.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel O, Malicdan MCV, Ben-Zeev B, Kandel J, Pri-Chen H, Stephen J, Castro IG, Metz J, Atawa O, Moshkovitz S, Ganelin E, Barshack I, Polak-Charcon S, Nass D, Marek-Yagel D, Amariglio N, Shalva N, Vilboux T, Ferreira C, Pode-Shakked B, Heimer G, Hoffmann C, Yardeni T, Nissenkorn A, Avivi C, Eyal E, Kol N, Glick Saar E, Wallace DC, Gahl WA, Rechavi G, Schrader M, Eckmann DM, Anikster Y. Deleterious variants in TRAK1 disrupt mitochondrial movement and cause fatal encephalopathy. Brain. 2017 Mar 01;140(3):568–581. doi: 10.1093/brain/awx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Brickley K, Wilkinson HL, Sharma S, Smith M, Chazot PL, Pollard S, Stephenson FA. Identification, molecular cloning, and characterization of a novel GABAA receptor-associated protein, GRIF-1. J Biol Chem. 2002 May 28;277(33):30079–30090. doi: 10.1074/jbc.M200438200. [DOI] [PubMed] [Google Scholar]
- Brickley K, Smith MJ, Beck M, Stephenson FA. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J Biol Chem. 2005 Jan 11;280(15):14723–14732. doi: 10.1074/jbc.M409095200. [DOI] [PubMed] [Google Scholar]
- Brickley K, Stephenson FA. Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J Biol Chem. 2011 Mar 30;286(20):18079–18092. doi: 10.1074/jbc.M111.236018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Sheng ZH. Mitochondrial transport and docking in axons. Exp Neurol. 2009 Mar 31;218(2):257–267. doi: 10.1016/j.expneurol.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Babu D, Lau D, Yu X, Roy S. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development. 2014 Sep 01;141(17):3410–3419. doi: 10.1242/dev.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichi G, Naef V, Barca A, Longo G, Fronte B, Verri T, Santorelli FM, Marchese M, Petruzzella V. Fishing in the Cell Powerhouse: Zebrafish as A Tool for Exploration of Mitochondrial Defects Affecting the Nervous System. Int J Mol Sci. 2019 May 15;20(10) doi: 10.3390/ijms20102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SL, Zhang L, Forster ML, Anderson JR, Iwase T, Soliven B, Donahue LR, Sweet HO, Bronson RT, Davisson MT, Wollmann RL, Lahn BT. Trak1 mutation disrupts GABA(A) receptor homeostasis in hypertonic mice. Nat Genet. 2005 Dec 25;38(2):245–250. doi: 10.1038/ng1715. [DOI] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006 May 22;173(4):545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005 Aug 01;47(3):379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Henrichs V, Grycova L, Barinka C, Nahacka Z, Neuzil J, Diez S, Rohlena J, Braun M, Lansky Z. Mitochondria-adaptor TRAK1 promotes kinesin-1 driven transport in crowded environments. Nat Commun. 2020 Jun 19;11(1):3123–3123. doi: 10.1038/s41467-020-16972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister BM, Oonk KA, Weiser DC, Walsh S. Characterization of the three zebrafish orthologs of the mitochondrial GTPase Miro/Rhot. Comp Biochem Physiol B Biochem Mol Biol. 2015 Oct 19;191:126–134. doi: 10.1016/j.cbpb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2002 Nov 14;278(7):5399–5409. doi: 10.1074/jbc.M209384200. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim Biophys Acta. 2010 Mar 15;1803(5):564–574. doi: 10.1016/j.bbamcr.2010.03.006. [DOI] [PubMed] [Google Scholar]
- López-Doménech G, Covill-Cooke C, Ivankovic D, Halff EF, Sheehan DF, Norkett R, Birsa N, Kittler JT. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 2018 Jan 01;37(3):321–336. doi: 10.15252/embj.201696380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss O, Stephenson FA. Localization of the kinesin adaptor proteins trafficking kinesin proteins 1 and 2 in primary cultures of hippocampal pyramidal and cortical neurons. J Neurosci Res. 2015 Feb 01;93(7):1056–1066. doi: 10.1002/jnr.23549. [DOI] [PubMed] [Google Scholar]
- Loss O, Stephenson FA. Developmental changes in trak-mediated mitochondrial transport in neurons. Mol Cell Neurosci. 2017 Mar 11;80:134–147. doi: 10.1016/j.mcn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009 Feb 26;61(4):541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 Jul 01;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999 Dec 01;11(6):699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Ro H, Soun K, Kim EJ, Rhee M. Novel vector systems optimized for injecting in vitro-synthesized mRNA into zebrafish embryos. Mol Cells. 2004 Apr 30;17(2):373–376. [PubMed] [Google Scholar]
- Sheng ZH. The Interplay of Axonal Energy Homeostasis and Mitochondrial Trafficking and Anchoring. Trends Cell Biol. 2017 Feb 20;27(6):403–416. doi: 10.1016/j.tcb.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, Mikhaylova M, Lipka J, Schlager MA, van den Heuvel DJ, Kuijpers M, Wulf PS, Keijzer N, Demmers J, Kapitein LC, Jaarsma D, Gerritsen HC, Akhmanova A, Hoogenraad CC. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 2013 Feb 01;77(3):485–502. doi: 10.1016/j.neuron.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002 Dec 19;36(6):1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001 Oct 29;356(1414):1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003 Mar 01;13(3):382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber E, Li L, Chin LS. Hypertonia-associated protein Trak1 is a novel regulator of endosome-to-lysosome trafficking. J Mol Biol. 2008 Jul 25;382(3):638–651. doi: 10.1016/j.jmb.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005 Aug 18;15(9):1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Marugán JC, Cummins C, Davidson C, Dodiya K, Fatima R, Gall A, Giron CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Lavidas I, Le T, Lemos D, Martinez JG, Maurel T, McDowall M, McMahon A, Mohanan S, Moore B, Nuhn M, Oheh DN, Parker A, Parton A, Patricio M, Sakthivel MP, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sycheva M, Szuba M, Taylor K, Thormann A, Threadgold G, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Flint B, Frankish A, Hunt SE, IIsley G, Kostadima M, Langridge N, Loveland JE, Martin FJ, Morales J, Mudge JM, Muffato M, Perry E, Ruffier M, Trevanion SJ, Cunningham F, Howe KL, Zerbino DR, Flicek P. Ensembl 2020. Nucleic Acids Res. 2020 Jan 01;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]