Graphical abstract
Keywords: Organ-on-a-chip, COVID-19, SARS-CoV-2, Microphysiological system, Gastrointestinal infection
Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global pandemic. Clinical evidence suggests that the intestine is another high-risk organ for SARS-CoV-2 infection besides the lungs. However, a model that can accurately reflect the response of the human intestine to the virus is still lacking. Here, we created an intestinal infection model on a chip that allows the recapitulation of human relevant intestinal pathophysiology induced by SARS-CoV-2 at organ level. This microengineered gut-on-chip reconstitutes the key features of the intestinal epithelium-vascular endothelium barrier through the three-dimensional (3D) co-culture of human intestinal epithelial, mucin-secreting, and vascular endothelial cells under physiological fluid flow. The intestinal epithelium showed permissiveness for viral infection and obvious morphological changes with injury of intestinal villi, dispersed distribution of mucus-secreting cells, and reduced expression of tight junction (E-cadherin), indicating the destruction of the intestinal barrier integrity caused by virus. Moreover, the vascular endothelium exhibited abnormal cell morphology, with disrupted adherent junctions. Transcriptional analysis revealed abnormal RNA and protein metabolism, as well as activated immune responses in both epithelial and endothelial cells after viral infection (e.g., upregulated cytokine genes), which may contribute to the injury of the intestinal barrier associated with gastrointestinal symptoms. This human organ system can partially mirror intestinal barrier injury and the human response to viral infection, which is not possible in existing in vitro culture models. It provides a unique and rapid platform to accelerate COVID-19 research and develop novel therapies.
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global epidemic. As of late August 2020, it has caused 22 million infections and more than 790,000 deaths. SARS-CoV-2 infection is characterized by a process ranging from asymptomatic and mild disease to severe systemic symptoms involving multiple organs, most notably the lung and gastrointestinal tract, and finally, multi-organ failure [1]. Respiratory symptoms dominate the clinical manifestations of COVID-19, yet obvious gastrointestinal symptoms are observed in 20% to 50% of patients. These include abdominal pain, diarrhea, hematochezia, and even intestinal perforation [2], [3], [4], [5]. These symptoms sometimes appear before the onset of respiratory symptoms [6]. In addition, biopsy samples show a large number of interstitial edema plasma cells and lymphocytes infiltrated into the lamina propria of the stomach, duodenum, and rectum [7]. It has been reported that SARS-CoV-2 can cause acute hemorrhagic colitis, thus providing evidence implicating the gastrointestinal tract in the transmission of SARS-CoV-2 infection [8]. Moreover, viral RNA has been identified in the stool samples of COVID-19 patients, and typical coronavirus virions have been observed in rectal tissue using electron microscopy, implying that SARS-CoV-2 may potentially be transmitted via the fecal-oral route [9], [10], [11]. These clinical evidences suggest that the intestine is another high-risk organ for SARS-CoV-2 infection besides the lungs, but the pathogenesis of the intestinal infection in COVID-19 is not known.
The human intestine contains complex multicellular components and host-pathogen interactions in a physiological flow microenvironment with mechanical cues. Currently, SARS-CoV-2 infection in the intestine is studied based on monolayer cultures of intestinal epithelial cells [12], [13] and human organoids [14], [15]. However, these in vitro models have limitations. Monolayer cell culture systems are oversimplified and cannot recapitulate the multiple cellular components, complex structure, and functions of the native intestine. Moreover, they lack cell–cell/matrix interactions and the tissue-specific dynamic microenvironment that exist in vivo. Recently, intestinal organoids have provided a new in vitro three dimensional (3D) model of studying SARS-CoV-2 infection, by providing multiple cell types and supporting viral replication in gut enterocytes [14]. However, these organoids are still limited by a lack of the typical characteristics of the intestinal barrier, the extracellular matrix (ECM), immune cells, and physiological flow, which are key features of the intestinal microenvironment. As such, it is highly desirable to develop alternative in vitro models to better reflect the pathophysiology of SARS-CoV-2 infection in human organs.
Organ-on-a-chip technology has evolved to provide the possibility to reproduce the complex structures and physiological functions of human organs in an engineered microfluidic culture device [16], [17], [18]. It has been used to represent organ-level physiology and pathology, and applied in various biomedical applications, including organ engineering, disease studies, and drug testing [19], [20], [21], [22]. For example, human gut-on-chip systems have been used to study drug metabolism [23], host-microbiome interactions [24], [25], [26], and coxsackie B1 virus infection [27]. In this study, we built an intestinal infection model on chip that allows the simulation of the response of the human intestine to SARS-CoV-2 at the organ level. The microengineered gut-on-chip device consists of a human intestinal epithelial layer and a vascular endothelial layer separated by an ECM-coated porous polydimethylsiloxane (PDMS) membrane, in which human colon adenocarcinoma (Caco-2) cells and human colorectal adenocarcinoma grade II (HT-29) cells are co-cultured in the upper channel, while human umbilical vein endothelial cells (HUVECs) and circulating immune cells line the lower channel under fluid flow. Using this system, we examined the infection and replication of SARS-CoV-2 in epithelial cells. We then used confocal imaging to systematically analyze the changes in the intestinal epithelium and endothelium induced by viral infection. We also characterized the pathological changes and immune responses of the intestinal barrier after viral infection via RNA-sequencing analysis. This human disease model on a chip offers a novel strategy and platform for organ-level COVID-19 research and potential therapeutic development.
2. Experimental
2.1. Device fabrication
The human intestinal model consisted of upper and lower layers fabricated using conventional soft lithography procedures. The PDMS pre-polymer was prepared by mixing 10:1 (wt/wt) PDMS base with a curing agent (184 Silicone Elastomer, Dow Corning Co., Midland, MI, USA) and casted on molds to produce a molded device with channels by thermal curing at 80 °C for 45 min. The channels were 1.5 mm wide × 0.25 mm high, and the length of overlapping channels was 8 mm. The two channels were separated by a thin (~25 μm) through-hole PDMS membrane (pore size: 5 μm, gaps between pores: 70 μm) to construct tissue-tissue interfaces. The density of the porous PDMS membrane was approximately 12,000 pores/cm2. The membrane was sandwiched in the gut-on-chip device by oxygen plasma bonding. Finally, the devices were exposed to ultraviolet light for disinfection and sterilization. Prior to cell seeding, both sides of the porous membrane were coated with rat tail collagen type I (200 μg/mL, Corning) and incubated at 37 °C for 48 h.
2.2. Cell culture
Caco-2 were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco). HT-29 were purchased from Procell Life Science & Technology Co., Ltd. (CL-0118; Wuhan, China) and were maintained in growth medium (Procell, CM-0118) supplemented with 10% FBS. HUVECs were isolated from human umbilical cords and cultured in Endothelial Cell Medium (ScienCell Research Laboratories, Inc., Carlsbad, CA, USA). Human peripheral blood mononuclear cells were isolated from fresh human blood using Ficoll (Stem Cell Technologies, Vancouver, Canada) density centrifugation. Isolated PBMCs were resuspended in RPMI 1640 medium containing 10% FBS and 50 IU/mL IL-2 and then used for adhesion assays on the chip. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. To create the intestinal model, HUVECs (~1 × 105 cells) were initially seeded on the bottom side of the collagen-coated porous PDMS membrane and allowed to attach to the membrane surface under static conditions for 2 h. Subsequently, cells were washed with fresh medium to exclude unattached cells. Caco-2 (~1 × 105 cells) and HT-29 cells (~1 × 104 cells) were then mixed and seeded into the upper channel under static culture. After cell attachment, cells were grown to confluence for 5 days and the chips were maintained in an incubator with 5% CO2 at 37 °C.
2.3. Virus
A clinical isolate, SARS-COV-2 strain 107, was obtained from the Guangdong Provincial Center for Disease Control and Prevention, China, and propagated in Vero E6 cells. Virus titers (infectious titers of virus) were determined using a 50% tissue culture infective dose assay on Vero cells. The multiplicity of infection (MOI) was calculated according to the formula: MOI = TCID50 × 0.7/cell number. All work involving live SARS-CoV-2 was performed at the Chinese Center for Disease Control and Prevention-approved BSL-3 Laboratory of the Kunming Institute of Zoology, Chinese Academy of Sciences.
2.4. SARS-CoV-2 infection
Caco-2 cells were seeded in 24-well plates (≈2 × 105 cells per well) in high-glucose DMEM containing 10% FBS. After reaching approximately 80% confluence, cells were infected with SARS-CoV-2 at the indicated MOI (0.04, 0.4, or 2). After 1 h of infection, cells were washed three times with PBS to remove residual virus and then kept in fresh medium for 3 d. At day 3 post-infection, cells were washed with PBS and then fixed with 4% paraformaldehyde (PFA) before analysis.
For SARS-CoV-2 infection in the human intestinal model, the upper intestinal channel of the chip device was infused with 30 μL of high-glucose DMEM containing the virus at an MOI of 1. After 1 h of infection, cells were washed three times with PBS and kept in fresh medium. At day 3 post-infection, the epithelial cells and endothelial cells cultured on the chip were fixed for immunofluorescence analysis or lysed for RNA-sequencing analysis.
2.5. Immunostaining
Caco-2 cells cultured in a 24-well plate were washed with PBS and fixed with 4% PFA at 4 ℃ overnight. Cells were then permeabilized with 0.2% Triton X-100 in PBS (PBST buffer) for 5 min and blocked with PBST buffer containing 5% normal goat serum for 30 min at room temperature. Antibodies were diluted with PBST buffer. Cells were stained with corresponding primary antibodies at 4 ℃ overnight and with secondary antibodies (Table S1 online) at room temperature for 1 h. After staining with secondary antibodies, cell nuclei were counterstained with DAPI. For immunofluorescent imaging of the intestinal model, cells were washed with PBS through the upper and lower channels and fixed with 4% PFA. The fixed tissues were subjected to immunofluorescence staining using the procedure described above. All images were acquired using a confocal fluorescence microscope system (FV-1000; Olympus, Tokyo, Japan). Image processing was performed using ImageJ (National Institutes of Health, Bethesda, MD, USA).
2.6. RNA extraction, library preparation, sequencing and Real-time quantitative PCR
Intestinal epithelial cells and endothelial cells were collected from the chips separately. Total RNA was extracted from samples using TRIzol (Invitrogen, Carlsbad, CA, USA) and RNA quality was determined using a NanodropTM OneC spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). RNA integrity was confirmed by 1.5% agarose gel electrophoresis. Qualified RNAs were quantified using a Qubit3.0 instrument, with the QubitTM RNA Broad Range Assay kit (Life Technologies, Carlsbad, CA, USA). One hundred nanograms of total RNA was used for stranded RNA sequencing library preparation using the KC-DigitalTM Stranded mRNA Library Prep Kit for Illumina® (catalog No. DR08502; Wuhan SeqHealth Co., Ltd., Wuhan, China), according to the manufacturer’s instructions. The kit eliminates duplication bias introduced by the PCR and sequencing steps using a unique molecular identifier (UMI) of eight random bases to label the pre-amplified cDNA molecules. The library products corresponding to 200–500 bp were enriched and quantified, finally, sequenced on a HiSeq X Ten sequencer (Illumina, San Diego, CA, USA). qRT-PCR was performed using TB Green®Premix Ex Taq™ II (Takara, PR820A) in a PCR System (Thermo Scientific, PikoReal 96). PCR amplification was performed under the following conditions: 95 ℃ 1 min, followed by 40 cycles consisting of 95 ℃ for 5 s, 58 ℃ for 30 s, 72 ℃ for 30 s. The primers are shown in Table S1 (online).
2.7. RNA-sequencing analysis
Trimmomatic (version 0.36) was used to filter the raw sequencing data (discard low-quality reads and trim the reads contaminated with adaptor sequences). Clean reads were further processed with in-house scripts to eliminate the duplication bias introduced during library preparation and sequencing. In brief, clean reads were first clustered according to their UMI sequences, in which reads with the same UMI sequence were grouped into the same cluster, resulting in 65,536 clusters. Reads in the same cluster were compared to each other by pairwise alignment, and then reads with a sequence identity greater than 95% were extracted to a new sub-cluster. After all sub-clusters were generated, multiple sequence alignment was performed to obtain one consensus sequence for each sub-cluster. After these steps, any errors and biases introduced by PCR amplification or sequencing were discarded.
RNA-sequencing analysis was performed on the de-duplicated consensus sequences, which were mapped to the Homo sapiens reference genome from the Ensembl database (ftp://ftp.ensembl.org/pub/release-87/fasta/homo_sapiens/dna/) using STAR software (version 2.5.3a) with default parameters. Reads mapped to the exonic regions of each gene were counted using featureCounts (Subread-1.5.1; Bioconductor) and RPKM values were then calculated. Genes differentially expressed between groups were identified using the edgeR package (version 3.12.1). A false discovery rate (FDR)-corrected p-value cut-off of 0.05 and a fold-change cut-off of 2 were used to define the differentially expressed genes (DEGs). Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs were both implemented using KOBAS software (version 2.1.1), with a corrected P-value cut-off of 0.05 to determine statistically significant enrichment. Alternative splicing events were detected using rMATS (version 3.2.5), with a FDR value cut-off of 0.05 and an absolute value of Δψ of 0.05.
2.8. Statistical analyses
Data were collected in Excel (Microsoft, Redmond, WA, USA). Differences between two groups were analyzed using a Student’s t-test. Multiple group comparisons were performed using a one-way analysis of variance (ANOVA) followed by post-hoc tests. The bar graphs with error bars represent the mean ± standard deviation (SD). Significance is indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3. Results
3.1. Characterization of the human gut-on-chip device
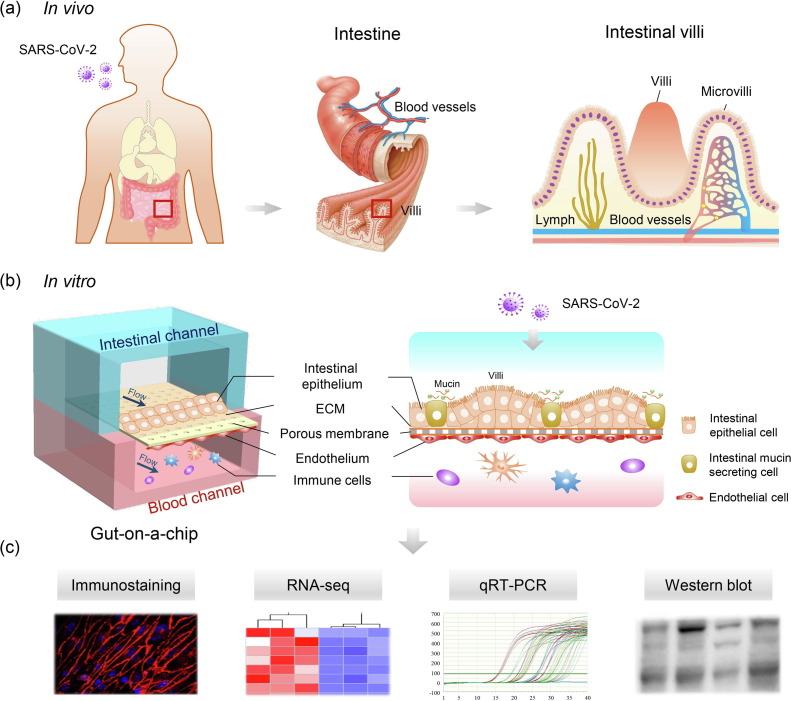
In vivo, the human intestinal epithelium is a cell monolayer that constitutes the intestinal barrier, which allows mucus secretion and nutrient absorption while preventing the invasion of pathogenic antigens or toxins in a dynamic microenvironment. To create an in vitro human model of intestinal infection by SARS-CoV-2, we initially designed and constructed a gut-on-chip using a perfusable multilayer microfluidic culture device (Fig. 1 ). Briefly, the device consists of two parallel channels separated by a thin porous PDMS membrane coated with ECM to form a bio-interface and facilitate nutrient exchange between the upper and lower cell layers.
Fig. 1.
(Color online) Schematic diagram of the construction of the human gut-on-chip infected with SARS-CoV-2. (a) Illustration of the complex structure of the human small intestine. (b) The configuration of the multilayered intestine on the chip device infected with SARS-CoV-2. The device consists of an upper intestinal epithelial channel (blue) and a lower microvascular endothelial channel (red), separated by an ECM-coated porous PDMS membrane. The intestinal barrier was established by the co-culture of intestinal epithelial Caco-2 cells and intestinal mucin-secreting HT-29 cells in the upper channel, and human umbilical vein endothelial cells (HUVECs) and immune cells in the lower channel, under fluidic flow conditions. (c) The responses of the intestinal chip to viral infection were analyzed using different methods.
In this study, the intestinal epithelial cells, Caco-2, and the intestinal mucin-secreting cells, HT-29, were co-cultured to build an intestinal epithelium in the upper channel under fluid flow (200 μL/h) to simulate the intraluminal fluid flow in the intestine. HUVECs were cultured in the lower channel to form the vascular endothelium under fluid flow (50 μL/h), and circulating immune cells were added to the vascular channel to simulate the vascular microenvironment. After 5 d of co-culture, the intestinal epithelial and endothelial cells formed confluent monolayers under fluid flow, thus forming an intestinal epithelium-endothelium tissue barrier. The integrity of the epithelial barrier was determined by assessing the expression of tight junction (ZO-1) and adherent junction (E-cadherin) proteins. Immunostaining analysis showed that both ZO-1 and E-cadherin were expressed in the epithelium (Fig. 2 a). Moreover, confocal micrographs of the full-sized scan showed that the epithelial cells formed adherent junctions (E-cadherin) and endothelial cells formed conjunctions (VE-cadherin, Fig. S2 online). In particular, the epithelial cells formed villus-like structures lined with a highly polarized columnar epithelium identified by differential interference contrast (DIC) imaging. These structures were similar to intestinal villi in vivo (Fig. 2c). Moreover, the endothelial integrity was examined by ZO-1 and VE-cadherin immunostaining, which showed the regular and intact distribution of junction proteins (Fig. 2b). As the intestinal mucus layer is the crucial interface between the host and microbes, it plays a vital role in preventing microbial invasion [28]. We examined mucus production in the intestinal epithelium by immunostaining using a mucin-specific marker (MUC2). As shown in Fig. 2d, a scattered distribution of MUC2-positive cells was seen in the intestinal epithelium, suggesting the secretory function of mucus. Thus, the established gut-on-chip system exhibited intestinal villus-like structures, maintained the integrity of the tissue barrier, and had an ability to secrete mucus under fluidic flow, representing the key features of the human intestinal barrier in a physiologically relevant manner.
Fig. 2.
(Color online) Characterization of the intestinal epithelium and endothelium in the human gut-on-chip. (a) Confocal micrographs of the intestinal epithelial barrier on the chip visualized by the expression of an adhesion junction (E-cadherin) and tight junction markers (ZO-1). The intestinal villus-like structures with high levels of ZO-1 expression are indicated by white dashed lines. (b) Confocal micrographs of the vascular endothelium identified by the expression of an adhesion junction protein (VE-cadherin) and ZO-1. (c) DIC image of an intestinal villus-like structure with clumps of cells (indicated by white dashed lines). (d) Immunostaining of a mucin marker (MUC2) in intestinal epithelial cells. Scale bars: 50 μm. (e, f) The 3D reconstructed confocal image and side view of the human intestinal epithelium (E-cadherin) and endothelium (VE-cadherin). (g, h) The 3D reconstructed confocal image and side view of the intestinal epithelium, endothelium, and intestinal villus-like structures (indicated by white arrows). Each image represents three independent experiments.
3.2. SARS-CoV-2 infection in the gut-on-chip system
Prior to SARS-CoV-2 infection of the gut-on-chip, we first detected the expression levels of ACE2 and TMPRSS2 in three cell lines, Caco-2 cells, HT-29 cells, and HUVECs, by western blotting. SARS-CoV-2 is known to use ACE2 as a receptor for cellular entry and the protease TMPRSS2 for viral Spike protein priming [29], [30]. Intestinal enterocytes express a higher level of ACE2 than alveolar epithelial type II cells [31], [32], [33]. Among the three cell lines tested in this study, ACE2 and TMPRSS2 were expressed at the highest levels in Caco-2 cells (Fig. S1a online), which implied that Caco-2 cells may be more permissive to SARS-CoV-2 infection. Caco-2 cells were then inoculated with the virus at different MOI values (0.04, 0.4, or 2) to identify the ability of the virus to infect the cells. Three days after infection, immunostaining data showed that more than 80% of the Caco-2 cells were positive for the Spike protein at an MOI of 2 (Fig. S1b online).
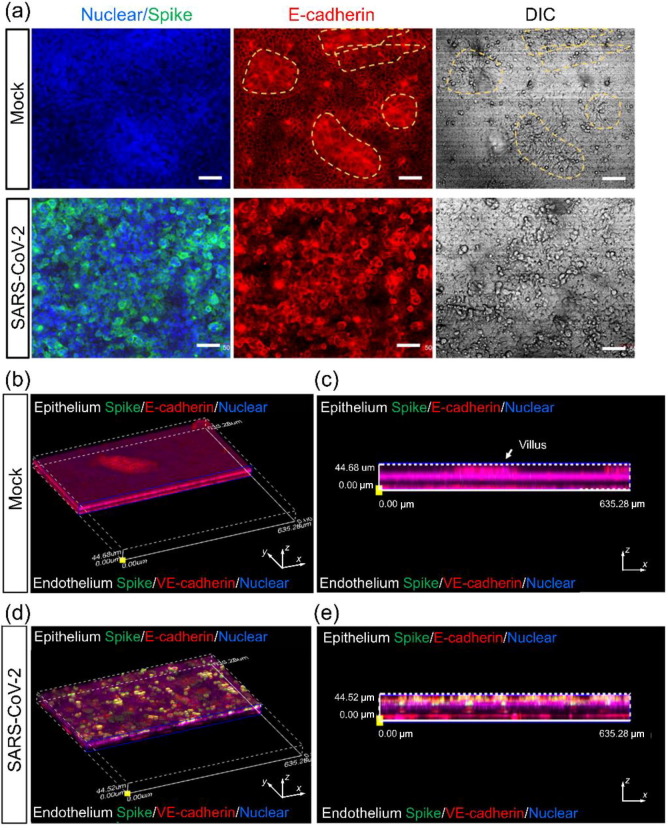
To examine SARS-CoV-2 infection of the gut-on-chip, the virus was inoculated into the intestinal channel at an MOI of 1, to allow the infection of a majority of cells, after co-culture of the intestinal epithelium and endothelium for 5 d. Viral infection in the intestinal epithelial and endothelial layers was then examined by immunostaining. Spike-positive cells were predominantly detected in the intestinal layer, indicating that the intestinal epithelial cells were more permissive to SARS-CoV-2 infection than endothelial cells (Fig. 3 ).
Fig. 3.
(Color online) Examination of SARS-CoV-2 infection in the human gut-on-chip system. (a) Confocal micrographs of SARS-CoV-2 infection (Spike protein) on the intestinal epithelium (E-cadherin) and intestinal villus-like structures (indicated by yellow dashed lines) at day 3 post-infection. Scale bars: 50 μm. (b, c) The 3D reconstructed confocal image and side view of a mock-infected gut-on-chip. (d, e) The 3D reconstructed confocal image and side view of the virus-infected intestinal model. SARS-CoV-2 infection was identified in the epithelial layer by the expression of the viral Spike protein. Each image represents three independent experiments.
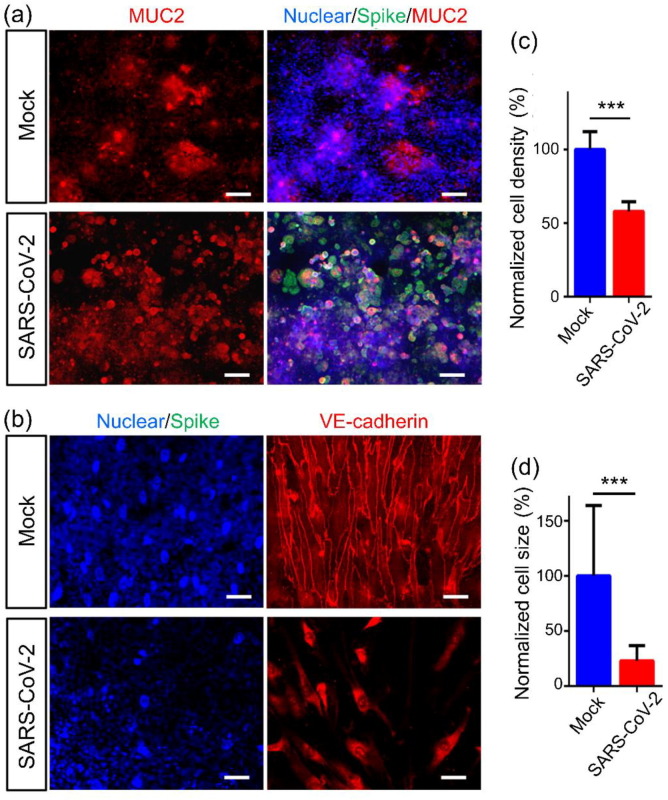
In vivo, the development and maintenance of the intestinal barrier depends heavily on the proper function of adherent junctions. Therefore, we further examined the adherent junctions of the intestinal epithelium and endothelium. Confocal micrographs of the intestinal epithelium showed that on day 3 after viral infection, the adherent junction, identified by E-cadherin expression, was severely destroyed, and this was accompanied by damage to the intestinal villus-like structures (Fig. 3a). Furthermore, the mucus-secreting cells in the intestinal epithelium were examined by immunostaining of MUC2. The clonal distribution of MUC2+ cells became dispersed after viral infection (Fig. 4 a). This suggested that the disturbed intestinal mucus layer was caused by viral infection, which may lead to virus encroachment and infection. It was noted that on the vascular side, the adherent junctions between endothelial cells were severely disrupted, as indicated by VE-cadherin immunostaining (Fig. 4b). Quantitative analysis showed that the density and size of endothelial cells were significantly decreased (Fig. 4c and d). This indicated that vascular endothelial cell injury occurred following viral infection, but there were no obvious Spike-positive cells on the vascular side. The presence of endothelial injury may partially explain the pathogenesis of COVID-19-associated coagulopathy or vascular thrombosis [34].
Fig. 4.
(Color online) Morphological changes in the intestinal barrier on the chip after viral infection. (a) Confocal micrographs of SARS-CoV-2 infection (Spike protein) and MUC2 expression in the intestinal epithelium at day 3 post-infection. Scale bars: 50 μm. (b) Confocal micrographs of viral infection (Spike protein) in the vascular endothelium (VE-cadherin). Scale bars: 50 μm. (c, d) Quantification of endothelial cell density and size for mock- and SARS-CoV-2-infected chips. Four chips were counted for cell density quantification in each group, and 100 cells were counted for cell size quantification in each group. Data are presented as the mean ± SD and were analyzed by Student’s t-test (***, P < 0.001).
3.3. Transcriptional analysis of host cells after SARS-CoV-2 infection
To gain a comprehensive overview of the transcriptional responses to SARS-CoV-2 infection, we performed RNA-sequencing analysis of intestinal epithelial and endothelial cells in the human intestinal model. Briefly, 3 d after infection, intestinal epithelial and endothelial cells were collected separately and analyzed by RNA-sequencing. Volcano plots showed that SARS-CoV-2 infection induced dramatic transcriptome modulations in both intestinal epithelial and endothelial cells (Fig. 5 a). To identify DEGs, the critical value and p-value of the fold change in abundance were set to 2.0 and 0.05, respectively. Among the DEGs identified, 9684 genes (4022 downregulated genes and 5662 upregulated genes) were significantly modulated in intestinal epithelial cells, whereas 6713 genes (2569 downregulated genes and 4144 upregulated genes) were significantly modulated in endothelial cells following viral infection. This result suggested that SARS-CoV-2 infection had a greater effect on intestinal epithelial cells than endothelial cells, possibly due to a higher viral load in the intestinal epithelium. By combining the two data sets, we found that intestinal epithelial and endothelial cells shared 1791 overlapping upregulated DEGs (37.3% of the total number of upregulated DEGs) and 3317 overlapping downregulated DEGs (51.5% of the total number of downregulated DEGs, Fig. 5b).
Fig. 5.
(Color online) Transcriptional analysis of intestinal epithelial and endothelial cells after SARS-CoV-2 infection of the chip. (a) Volcano plots of the dysregulated genes after viral infection. Genes differentially expressed with a fold change greater than 2.0 and P < 0.05 are marked in color. P-values were calculated using a two-sided, unpaired Student’s t-test, assuming equal variances (n = 3 independent biological samples). (b) Venn diagrams depicting the shared or unique differentially expressed genes between each comparison. (c) KEGG pathway enrichment analysis of differentially expressed genes in intestinal epithelial cells following SARS-CoV-2 infection. (d) KEGG pathway enrichment analysis of differentially expressed genes in endothelial cells following viral infection. The color of the dots represents the rich factor, and the size represents the input number of each KEGG term. The horizontal axis indicates the significance of enrichment. The vertical axis indicates the enriched KEGG pathway (20 most enriched terms).
To investigate the host biological pathways modified by SARS-CoV-2 infection, KEGG pathway enrichment analysis was performed, comparing mock-infected versus SARS-CoV-2-infected gut-on-chip systems (Fig. 5c and d). In both cell types, viral infection resulted in abnormal pathway networks, including RNA metabolism pathways (e.g., spliceosome, RNA transport, RNA degradation, and mRNA surveillance) and protein metabolism pathways (e.g., ubiquitin-mediated proteolysis and protein export). Given the vital roles of RNA and protein metabolism in maintaining the normal physiological functions of cells, we reasoned that SARS-CoV-2 may seriously affect the host cells by disrupting these critical pathways. In addition, we found that some immune response-related pathways, such as tumor necrosis factor (TNF) signaling pathway and NF-kappa B signaling pathway, were particularly enriched in genes that were significantly upregulated in endothelial cells, which indicated that intestinal endothelial cells play critical roles in mediating intestinal immune responses.
3.4. Immune response of the intestinal model to SARS-CoV-2 infection
KEGG pathway enrichment analysis indicated that some immune response-related signaling pathways were activated following SARS-CoV-2 infection. We then aimed to identify the upregulated cytokine genes that mediated the subsequent inflammatory responses of the intestine to SARS-CoV-2 infection. The heat map showed that SARS-CoV-2 infection elicited extensive cytokine induction in both the intestinal epithelium and endothelium. Induced cytokines included TNF, interleukins, chemokines, and colony-stimulating factors, in both intestinal epithelial and endothelial cells (Fig. 6 a and b). Notably, in this infected intestinal model, some cytokines, including TNF, IL-6, CXCL10, CCL5, and CSF3, were significantly upregulated, which is similar to the clinical manifestation of patients with severe COVID-19 [35], [36]. These results may partially explain the presence of infiltrated immune cells and the inflammatory response in the intestine of patients with severe COVID-19.
Fig. 6.
(Color online) Assessment of immune responses in the human gut-on-chip following SARS-CoV-2 infection. (a) Heatmaps depicting significantly upregulated cytokine genes in intestinal epithelial cells. (b) Heatmaps depicting significantly upregulated cytokine genes in endothelial cells. The colored bar represents the Z-score of log2-transformed values. (c) The relative mRNA levels of the indicated genes were determined by qRT-PCR. Data are presented as the mean ± SD and were analyzed by Student’s t-test (*, P < 0.05; **, P < 0.01, ***, P < 0.001; n = 3).
We then further analyzed the expression of pro-inflammatory cytokine- and chemokine-related genes in intestinal epithelial cells by qRT-PCR. The expression levels of CCL5, CXCL1, CXCL10, and CXCL11 were significantly upregulated in SARS-CoV-2-infected epithelial cells (Fig. 6c). These data indicated that chemokine genes can be induced by SARS-CoV-2, and this may play a crucial role in the recruitment of immune cells and the modulation of the immune response to viral infection. It has previously been reported that the chemokine, CCL5, recruits T cells, dendritic cells, eosinophils, natural killer cells, mast cells, and basophils [37], CXCL1 recruits neutrophils [38]; CXCL10 recruits activated Th1 lymphocytes [39]; and CXCL11 recruits interleukin-activated T-cells [40] to sites of inflammation. This may provide clues to further identify the specific immune cells that are involved in the intestinal inflammatory responses during the progression of COVID-19.
4. Discussion
In this study, we created a biomimetic human gut-on-chip system that can mirror the pathophysiological features of the human intestine infected with SARS-CoV-2. This microfluidic human gut chip displayed an intact intestinal barrier, villus-like structures along the apical surfaces, and a mucus-secretion function by synergistically integrating multicellular components, fluidic flow, and intestinal epithelium-endothelium interactions, which are necessary to resemble the physiological microenvironment of the human intestine. SARS-CoV-2 inoculation of this gut-on-chip system caused an obvious disruption of the villi, intestinal barrier integrity, and mucus secretion and activated immune responses, demonstrating the ability of this device to mimic gut-related infection and pathogenesis in vitro. Despite the low susceptibility of endothelial cells to viral infection, the vascular endothelium exhibited significant morphological damage after SARS-CoV-2 challenge. This indicates that the injury of the intestinal barrier may be mediated by complex cross-talk between the intestinal epithelial and endothelial cells during disease progression.
The human intestine comprises the largest component of the human immune system, with large populations of scattered innate and adaptive effector cells, because it is constantly exposed to foreign antigens and other environmental agents. The protective mucus gel secreted by goblet cells is distributed on the gastrointestinal epithelium, where it provides chemical and physical defense mechanisms for the intestinal barrier and plays a crucial role in intestinal homeostasis. In this gut chip, Caco-2 cells and mucin-secreting HT-29 cells were co-cultured under continuous perfusion to form an intestinal epithelium that recapitulated the intestinal barrier in a physiologically relevant manner. In particular, we found that the secretion of intestinal mucin underwent previously unrecognized changes, from an agglomerated distribution to a scattered distribution after viral infection. This disturbance of the mucus layer may be associated with further invasion and infection by the virus. These results may also explain the increased intestinal permeability and the symptoms, such as diarrhea and hemorrhagic colitis [41], in COVID-19 patients.
Transcriptomic analysis demonstrated significant alterations in vital biological processes in both the intestinal epithelium and endothelium following SARS-CoV-2 infection. These included RNA and protein metabolism pathways, cell cycle regulation, and oxidative phosphorylation. Specifically, we identified many upregulated cytokine- and chemokine-related genes in intestinal epithelial cells, similar to the clinical manifestations of COVID-19. Because chemokines can act as chemoattractants to recruit immune cells to infected sites, these altered chemokines may contribute to the immune cell-mediated inflammatory responses in the intestine. In addition, we found that the intestinal epithelium was more permissive to SARS-CoV-2 infection than the endothelium on the chip, which is consistent with previous studies [12]. As such, we propose that the observed vascular endothelial injury may be mediated by inflammatory factors or paracrine signals released by virus-infected intestinal epithelial cells.
Certainly, this study has some limitations. One potential limitation is the use of immortalized intestinal epithelial cell lines (e.g., Caco-2) originally isolated from human colon tumors. However, Caco-2 cells have been widely used to recapitulate many physiological and pathological functions of the human intestine in vivo [42], [43]. Intestinal epithelium differentiated from primary intestinal stem cells may be selected for future studies. In addition, due to the very limited experimental conditions during the SARS-CoV-2 epidemic crisis, we were not able to conduct a comprehensive study on the complex responses of the immune cells involved in the host-virus interactions in this gut-on-a-chip system. Moreover, potential extra-respiratory transmission routes in COVID-19 patients remain uncertain. In future experiments, it is also feasible to inoculate the virus in the endothelium channel of this gut-on-chip system to further investigate the viral transmission routes associated with gastrointestinal tract symptoms.
5. Conclusions
Conclusively, this work provides the first proof-of-concept for building a human intestinal SARS-CoV-2 infection model using a biomimetic gut-on-a-chip that closely mirrors the pathophysiology and intestine-specific responses at the organ level in vitro. The novelty of this work lies in its capability to reproduce the organ-specific intestinal microenvironment and human-relevant responses to SARS-CoV-2 on a chip system. This was achieved by integrating complex multicellular components, an intestinal tissue barrier, and physiological fluid flow, which cannot be easily achieved with other in vitro culture models. This intestinal microphysiological system not only offers a unique, rapid, and low-cost platform for the study of viral infection, transmission, host-virus interactions, and disease pathogenesis in a more realistic manner, but also provides a complement to animal models. This system is expected to accelerate COVID-19 research and the development of novel therapeutics, which are critical during the SARS-CoV-2 pandemic crisis.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29050301, XDB32030200, and XDA16020900), the National Key R&D Program of China (2017YFB0405404), the National Science and Technology Major Project (2018ZX09201017-001-001), Yunnan Key Research and Development Program (202003AD150009), the National Natural Science Foundation of China (31671038, 31971373, 81703470, and 81803492), China Postdoctoral Science Foundation (2019 M660065), Innovation Program of Science and Research from the Dalian Institute of Chemical Physics, Chinese Academy of Sciences (DICP I201934). We thank Prof. Yonggang Yao (Kunming Institute of Zoology, CAS) for his strong support and suggestion on this work.
Author contributions
Jianhua Qin and Yongtang Zheng conceived of the idea for the study. Ronghua Luo, Tianzhang Song, Pengwei Deng and Min Zhang performed experiments. Yaqiong Guo and Tingting Tao provided the microfluidic chips. Peng Wang and Kangli Cui analyzed the RNA-Seq data. Yaqiong Guo and Yaqing Wang wrote the manuscript. Xu Zhang and Zhongyu Li revised the manuscript. Wenwen Chen produced the figures. All authors read and approved the manuscript.
Biographies

Yaqiong Guo is a Ph.D. candidate at Dalian Institute of Chemical Physics, Chinese Academy of Sciences. She received her B.S. degree from Central China Normal University in 2014. Her research focuses on gut-on-a-chip and its application in drug metabolism and intestinal inflammation research.

Ronghua Luo is an experimentalist of Virology at Kunming Institute of Zoology, Chinese Academy of Sciences. His research mainly focuses on preclinical evaluation and mechanisms of antiviral compounds.

Yaqing Wang is a Ph.D. candidate at Dalian Institute of Chemical Physics, Chinese Academy of Sciences. She received her B.E. degree from Harbin Institute of Technology in 2012, followed by a M.Sc. degree majoring in Cell Biology from Peking University in 2015. Her current research focuses on organoids and organ-on-a-chip for engineering in vitro 3D tissue models.

Pengwei Deng is a Ph.D. candidate at Dalian Institute of Chemical Physics, Chinese Academy of Sciences. He received his B.S degree from Huazhong University of Science and Technology in 2016. His research mainly focuses on engineering multi-organ-on-a-chip system.

Tianzhang Song is a research associate of Immunology at Kunming Institute of Zoology, Chinese Academy of Sciences. His research mainly focuses on immunopathological mechanism study of COVID-19 and AIDS by using primate animal models.

Yongtang Zheng is a professor of Immunology and Virology at Kunming Institute of Zoology, Chinese Academy of Sciences. His research mainly focuses on virus infection and host immunity, including immunopathogenesis, primate animal models, antiviral drugs and molecular epidemiology.

Jianhua Qin is a professor at Dalian Institute of Chemical Physics, Chinese Academy of Sciences. Her research focuses on the development of engineered living model systems for biomedical applications, by combining microfluidics, organs-on-chips, biomaterials, and stem cell biology.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2020.11.015.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wiersinga W.J., Rhodes A., Cheng A.C. Pathophysiology, transmission, diagnosis, and treatment of Coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2.Lin L.u., Jiang X., Zhang Z. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 3.D’Amico F., Baumgart D.C., Danese S. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clin Gastroenterol Hepatol. 2020;18:1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng S.C., Tilg H. COVID-19 and the gastrointestinal tract: more than meets the eye. Gut. 2020;69:973–974. doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan Y., Li J., Shen L. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet Gastroenterol Hepatol. 2020;5:534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y., Liu P., Shi X.L. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 7.Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Q., Wang B., Zhang T. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am J Physiol Gastrointest Liver Physiol. 2020;319:G245–G252. doi: 10.1152/ajpgi.00148.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung K.S., Hung I.F., Chan P.P. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020;159:81–105. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson J. COVID-19: faecal–oral transmission? Nat Rev Gastroenterol Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Q., Fan L., Liu W. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanifer M.L., Kee C., Cortese M. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang R., Gomez Castro M.F., McCune B.T. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamers M.M., Beumer J., van der Vaart J. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J., Li C., Liu X. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 16.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 17.Schepers A., Li C., Chhabra A. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip. 2016;16:2644–2653. doi: 10.1039/c6lc00598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B., Korolj A., Lai B.F.L. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018;3:257–278. [Google Scholar]
- 19.Huh D., Matthews B.D., Mammoto A. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L.i., Tao T., Su W. A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice. Lab Chip. 2017;17:1749–1760. doi: 10.1039/c7lc00134g. [DOI] [PubMed] [Google Scholar]
- 21.Tao T., Wang Y., Chen W. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip. 2019;19:948–958. doi: 10.1039/c8lc01298a. [DOI] [PubMed] [Google Scholar]
- 22.Novak R., Ingram M., Marquez S. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng. 2020;4:407–420. doi: 10.1038/s41551-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y., Li Z., Su W. A biomimetic human gut-on-a-chip for modeling drug metabolism in intestine: biomimetic human gut-on-a-chip. Artif Organs. 2018;42:1196–1205. doi: 10.1111/aor.13163. [DOI] [PubMed] [Google Scholar]
- 24.Shah P., Fritz J.V., Glaab E. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun. 2016;7 doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jalili-Firoozinezhad S., Gazzaniga F.S., Calamari E.L. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer M., Gresnigt M.S., Last A. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials. 2019;220:119396. doi: 10.1016/j.biomaterials.2019.119396. [DOI] [PubMed] [Google Scholar]
- 27.Villenave R., Wales S.Q., Hamkins-Indik T. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown E.M., Sadarangani M., Finlay B.B. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P., Yang X.-L., Wang X.-G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chadchan S.B., Maurya V.K., Popli P. The SARS-CoV-2 receptor, Angiotensin converting enzyme 2 (ACE2) is required for human endometrial stromal cell decidualization. bioRxiv. 2020 doi: 10.1101/2020.06.23.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan R., Zhang Y., Li Y. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou X., Chen K.e., Zou J. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goshua G., Pine A.B., Meizlish M.L. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7:e575–e582. doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coperchini F., Chiovato L., Croce L. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques R.E., Guabiraba R., Russo R.C. Targeting CCL5 in inflammation. Expert Opin. Therapeutic Targets. 2013;17:1439–1460. doi: 10.1517/14728222.2013.837886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawant K.V., Poluri K.M., Dutta A.K. Chemokine CXCL1 mediated neutrophil recruitment: Role of glycosaminoglycan interactions. Sci Rep. 2016;6 doi: 10.1038/srep33123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasagni L., Francalanci M., Annunziato F. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokunaga R., Zhang W.u., Naseem M. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation – a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho A., Alqusairi R., Adams A. SARS-CoV-2 gastrointestinal infection causing hemorrhagic colitis: implications for detection and transmission of COVID-19 disease. Am J Gastroenterol. 2020;115:942–946. doi: 10.14309/ajg.0000000000000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H.J., Li H.u., Collins J.J. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Gunasekara D.B., Reed M.I. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.