Various types of somatic mutations occur in cells of the human body and cause human diseases including cancer and some neurological disorders1. Recently, Lee et al.2 (hereafter “the Lee study”) reported somatic copy number gains of the APP gene, a known risk locus of Alzheimer’s disease (AD), in 69% and 25% of neurons of AD patients and controls. The authors argue that the mechanism of these copy number gains was somatic integration of APP mRNA into the genome, creating what they called genomic cDNA (gencDNA). Our reanalysis of the data from the Lee study and two additional whole exome sequencing (WES) datasets by the authors of the Lee study3 and Park et al.4 revealed evidence that APP gencDNA originates mainly from exogenous contamination by APP recombinant vectors, nested PCR products, and human and mouse mRNA, respectively, rather than from true somatic integration of endogenous APP. We further present our own single-cell whole genome sequencing (scWGS) data that show no evidence for somatic APP retrotransposition in AD neurons or in neurons from normal individuals of various ages.
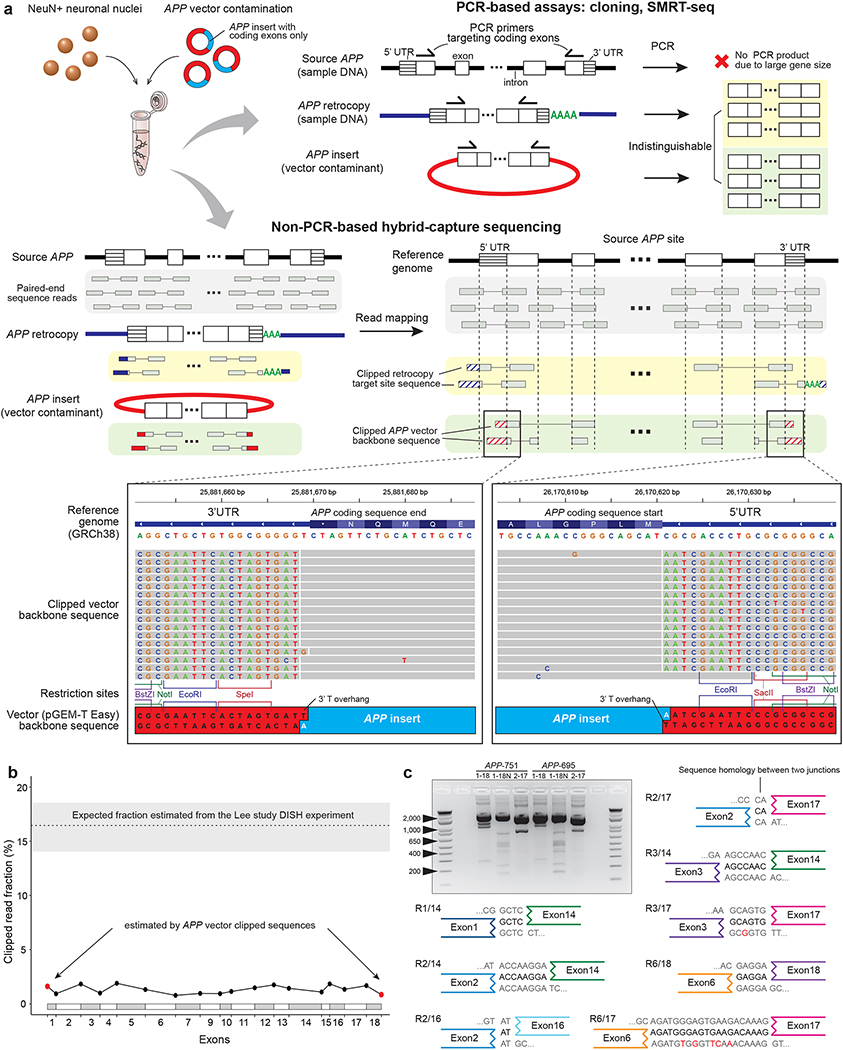
We examined the original APP-targeted sequencing data from the Lee study to investigate sequence features of APP retrotransposition. These expected features included (a) reads spanning two adjacent APP exons without intervening intron sequence, which would indicate processed APP mRNA, and (b) clipped reads, which are reads spanning the source APP and new genomic insertion sites, thus manifesting partial alignment to both the source and target site (Extended Data Fig. 1a). The first feature is the hallmark of retrogene or pseudogene insertions, and the second is the hallmark of RNA-mediated insertions of all kinds of retroelements, including retrogenes as well as LINE1 elements. We indeed observed multiple reads spanning two adjacent APP exons without the intron; however, we could not find any reads spanning the source APP and a target insertion site. Surprisingly, we found multiple clipped reads at both ends of the APP coding sequence (CDS) containing the multiple cloning site of the pGEM-T Easy Vector (Promega), which indicates external contamination of the sequencing library by a recombinant vector carrying an insert of APP coding sequence (Fig. 1a). The APP vector we found here was not used in the Lee study, but rather had been used in the same laboratory when first reporting genomic APP mosaicism5, suggesting carryover from the prior study.
Figure 1. APP vector contamination in the Lee study.
a. APP vector contamination and its manifestation in genome sequences. PCR-based assays in the Lee study fail to distinguish between APP retrocopy and vector APP insert. Hybrid-capture sequences from the Lee study shows clipped reads with a vector backbone sequence (pGEM-T Easy), including restriction sites at the multiple cloning site and a 3’ T-overhang. b. Estimated fractions of cells with APP gencDNA at the exon junctions in the Lee hybrid-capture data. All exon junction fractions (black dots) are comparable to the fraction at the coding sequence ends with vector backbone sequences (red dots). The dotted line on the top represents the conservative estimate of expected fraction based on the Lee DISH experiment (see Supplementary Methods); shaded area, 95% confidence interval. c. Electrophoresis and sequencing of PCR products from the vector APP inserts (APP-751/695) showing novel APP variants as artifacts. Eight out of 12 IEJs found both in our APP vector PCR sequencing and the Lee study RT-PCR results are shown (see also Extended Data Fig. 3).
Recombinant vectors with inserts of gene coding sequences (typically without introns or untranslated regions (UTRs)) are widely used for functional gene studies. Recombinant vector contamination in next-generation sequencing is a known source of artifacts in somatic variant calling, as sequence reads from the vector insert confound those from the endogenous gene in the sample DNA6. We have identified multiple incidences of vector contamination in next-generation sequencing datasets from different groups, including our own laboratory (Extended Data Fig. 1b), demonstrating the risk of exposure to vector contamination. In an unrelated study on somatic copy number variation in the mouse brain7, from the same laboratory that authored the Lee study, we found contamination by the same human APP pGEM-T Easy Vector in mouse single-neuron WGS data (Extended Data Fig. 1c). We also observed another vector backbone sequence (pTripIEx2, SMART cDNA Library Construction Kit, Clontech) with an APP insert (Extended Data Fig. 1c, magnified panel) in the same mouse genome dataset, indicating repeated contamination by multiple types of recombinant vectors in the laboratory.
PCR-based experiments with primers targeting the APP coding sequence (e.g., Sanger sequencing and SMRT sequencing) are unable to distinguish APP retrocopies from vector inserts (Fig. 1a, upper panel). Therefore, to definitively distinguish the three potential sources of APP sequencing reads (original source APP, retrogene copy, and vector insert), it is necessary to study non-PCR-based sequencing data (e.g., SureSelect hybrid-capture sequencing) and examine reads at both ends of the APP coding sequence. Such data can help to assess whether the clipped sequences map to a new insertion site or to vector backbone sequence (Fig. 1a, lower panel). From the SureSelect hybrid-capture sequencing data in the Lee study, we directly measured the level of vector contamination by calculating the fraction of the total read depth at both ends of the APP coding sequence comprised by clipped reads containing vector backbone sequences (Fig. 1b, red dots). Similarly, we measured the clipped read fraction at each APP exon junction, which indicates the total amount of APP gencDNAs (either from APP retrocopies or vector inserts) (Fig. 1b, black dots). The average clipped read fraction at coding sequence ends that contained vector backbones (1.2%, red dots) was comparable to the average clipped read fraction at exon junctions (1.3%, black dots; P=0.64, Mann-Whitney U test), suggesting vector contamination as the primary source of the clipped reads across all the exon junctions. Even including these vector-originating reads, all the fractions at every junction are far below the conservative estimate of 16.5% gencDNA contribution based on the Lee study’s DISH experimental results, which are from the same samples (see Supplementary Information for more details on the discrepancy between sequencing and DISH results). It is incumbent on the authors to provide explanation for this significant inconsistency. Moreover, if the clipped reads were from endogenous retrocopies, the clipped and non-clipped reads would be expected to be of similar insert (DNA fragment) size distribution; however, we observed that in the Lee study, the clipped reads were of significantly smaller and far more homogeneous insert size distribution than the non-clipped reads that were from original source APP, thus demonstrating the foreign nature of the clipped reads (P < 2.2×10−16, Mann-Whitney U test; Extended Data Fig. 2a–c, see Supplementary Information). Finally, we found no direct evidence supporting the existence of true APP retrogene insertions, such as clipped and discordant reads near the APP UTR ends that mapped to a new insertion site, or clipped reads with polyA tails at the 3’ end of the UTR although the sequencing depth of UTRs was over 500x. Given that the hybrid capture experiment appears properly designed to detect APP gencDNA, the absence of any bona fide insertion signal suggests the absence of true APP gencDNA and that a majority of APP-gencDNA-supporting reads originated from APP vector contamination.
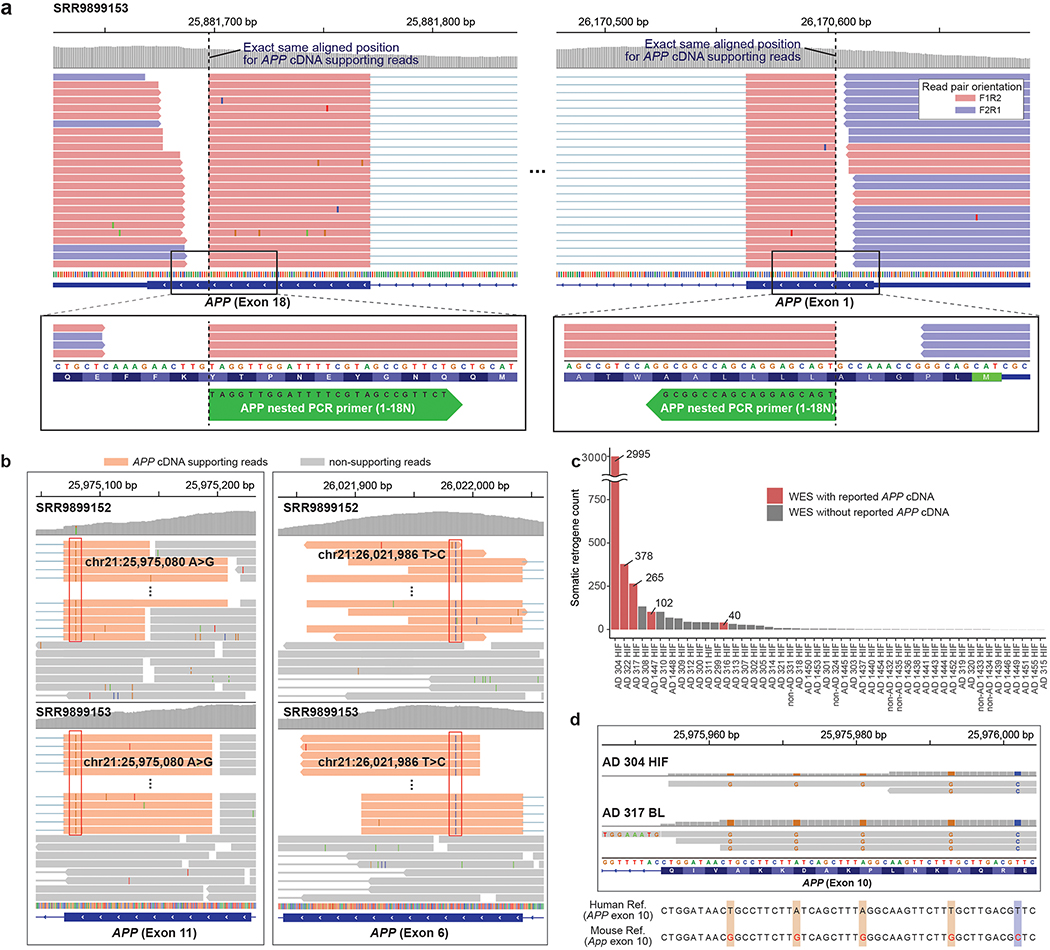
The authors of the Lee study have subsequently generated WES datasets from the brain samples of six AD patients and one non-AD control (SRA Accession: PRJNA558504), and reported multiple reads spanning APP exons without introns as evidence of somatic APP gencDNA3. We confirmed this in the data, but again, found not a single read spanning the source APP and any insertion sites. Instead, the data revealed anomalous patterns in a subset of reads supporting APP gencDNA. Those reads spanning exons 1 and 18 were aligned to the exact same start and end positions with the same read pair orientation (Fig. 2a), which is unlikely to occur in non-PCR-based exome capture sequencing. We found that the two aligned positions within exons 1 and 18 exactly match the target sites of the nested PCR primers used in the original Lee study (1–18N, Supplementary Table 1 in the Lee study). The only explanation for this observation is the contamination of the WES library by nested PCR products from the original APP study. This finding raises serious concerns that APP PCR products may also have contaminated the genomic DNA samples and were fragmented and sequenced together, generating more gencDNA-compatible reads for which we are unable to clarify the source. We also identified two unannotated (i.e., absent in the gnomAD) single-nucleotide variants in all APP-cDNA-supporting reads in the two independent WES libraries pooled from six AD patient samples, which is very unlikely to be observed in different individuals, thus supporting the possibility that the APP cDNA originated from the same external source (Fig. 2b).
Figure 2. APP cDNA-supporting reads originate from exogenous PCR products and genome-wide human and mouse mRNA contamination.
a. APP nested PCR products found in the recent Lee WES data. Reads supporting APP cDNA are aligned to the target sites (dotted lines) of the nested PCR primers (green arrows at the bottom) used in the original Lee study. All these cDNA-supporting reads contain an IEJ between exon 2 and 17 (full structure not shown). b. The same unannotated variants found at two different positions (red boxes) only in cDNA supporting reads (orange) in both WES datasets by Lee et al. (SRR989152 and SRR989153). c. Total gene counts with potential somatic retrogene insertions in the Park et al. data. WES data with reported APP cDNA are marked in red. d. APP cDNA-supporting reads originating from mouse mRNA in the Park data. Mouse-specific single-nucleotide polymorphisms (colored bases) are observed in a portion of cDNA-supporting reads, including those with clipped sequences for exon-exon junctions, suggesting the reads originated from mouse mRNA rather than genomic DNA (see also Supplementary Fig. 1).
An independent study by Park et al.4 has recently presented a small fraction of reads supporting APP cDNA in deep WES datasets from AD brain samples (SRA Accession: PRJNA532465; Supplementary Fig. 12 in the study). This data was free from vector contamination, but we found evidence of genome-wide human mRNA contamination, predominantly in the WES datasets with reported APP cDNA supporting reads. We note that their analysis of somatic single-nucleotide variants (SNVs) is likely to be unaffected by this contamination due to their visual inspection and stringent filtering of known germline SNVs. For each AD brain sample, we counted the number of genes with potential somatic retrotransposition events by checking whether a gene had cDNA-supporting reads (i.e., reads connecting two adjacent exons, skipping the intervening intron) at more than two different exon junctions in the brain sample but not in the matched blood sample from the same patient (see Supplementary Methods). All WES datasets reported by the authors to have APP cDNA showed an extremely high number of other genes in addition to APP with cDNA-supporting reads (40–2,995 genes) (Fig. 2c). Considering that far less than one somatic retrogene insertion per sample would be expected for human cells, even for human cancers with a high rate of somatic LINE1 retrotransposition (e.g., lung and colorectal cancer)8, this result strongly suggests that cDNA-supporting reads could not have originated from true somatic insertions of hundreds to thousands of retrogenes but rather supports the presence of genome-wide human mRNA contamination. We also found cDNA-supporting reads, including a subset of APP cDNA-supporting reads, originating from mouse mRNA, additionally confirming mRNA contamination of the data (Fig. 2d and Supplementary Fig. 1). We observed mRNA contamination in one cell in our scWGS data (see Supplementary Information). Neither Park et al. (per personal communication) nor we had performed any mRNA experiments, suggesting the possibility of contamination from some source outside the research laboratories, such as the sequencing facility. Taken together, we found no evidence of genuine APP genomic cDNA either in the new WES data from the Lee study authors, or in the independent Park et al. data. These findings highlight pervasive exogenous contamination in next-generation sequencing experiments, even with high quality control standards, and emphasizes the need for rigorous data analysis to mitigate these significant sources of artifacts.
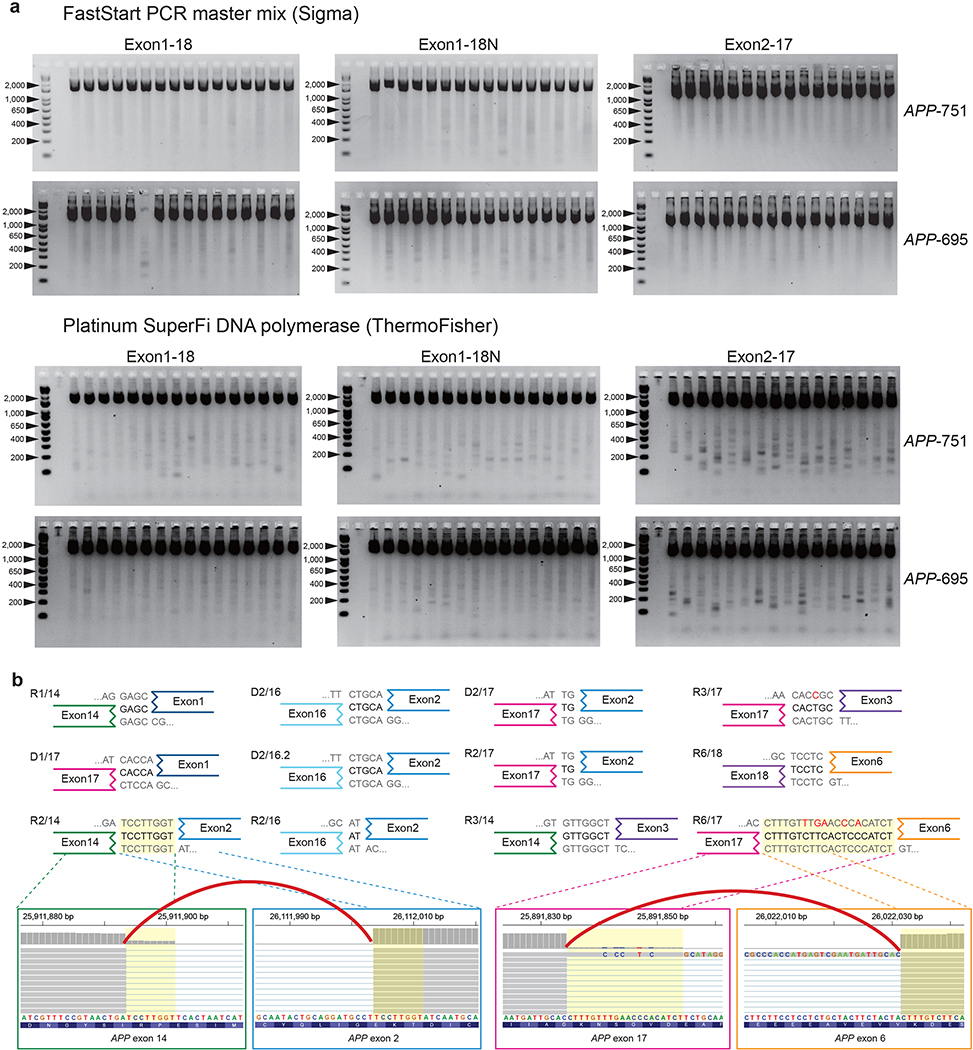
The Lee study reported numerous novel forms of APP splice variants with intra-exon junctions (IEJs) with greater diversity in AD patients than controls. The authors also presented short sequence homology (2–20 bp) at IEJs suggesting a microhomology-mediated end-joining as a mechanism underlying IEJ formation. It is well known that microhomology can predispose to PCR artifacts9, and the Lee study performed a high number of PCR cycles in their experimental protocol (40 cycles). Thus, we tested the hypothesis that the IEJs in the Lee study could have arisen as PCR artifacts from the PCR amplification of a contaminant. To do so, we repeated in our laboratory both RT-PCR and PCR assays following the Lee study protocol using recombinant vectors with two different APP isoforms (APP-751, APP-695), and using the reported PCR primer sets with three different PCR enzymes as described in their study (see Supplementary Information). Indeed, with all combinations of APP inserts and PCR enzymes, we observed chimeric amplification bands with various sizes, clearly distinct from the original APP inserts (Fig. 1c, Extended Data Fig. 3a). We further sequenced these non-specific amplicons and confirmed that they contained numerous IEJs of APP inserts (Supplementary Table 1). 12 of 17 previously reported IEJs in the Lee study were also found from our sequencing of PCR artifacts (Fig. 1c and Extended Data Fig. 3b). Our observations suggest that the novel APP variants with IEJs from the Lee study might have originated from contaminants as PCR artifacts. This possibility is corroborated by the fact that IEJ-supporting reads were completely absent in the hybrid-capture sequencing data from the Lee study, and that reads supporting an IEJ in the new WES dataset by the authors originated from external nested APP PCR products (Fig. 2a).
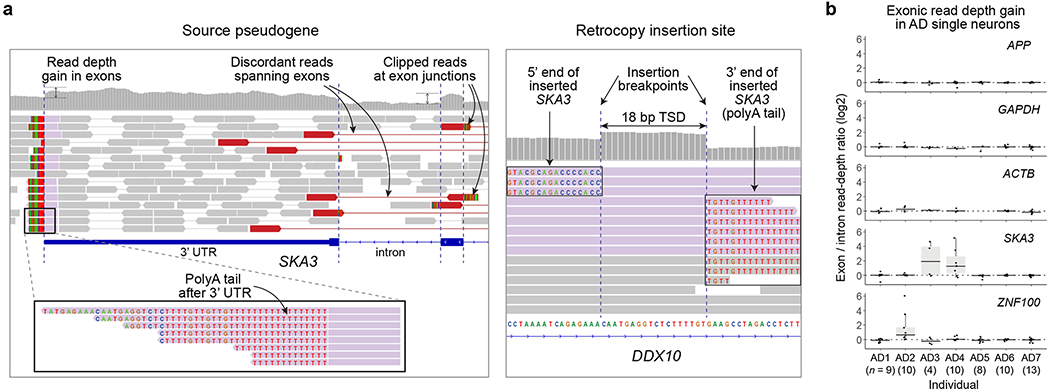
To independently investigate potential APP gencDNA, we searched for somatic APP retrogene insertions in our independent scWGS data from AD patients and normal controls. Briefly, single-neuronal nuclei were isolated using NeuN staining followed by FACS sorting, whole-genome amplified using multiple displacement amplification (MDA), and finally whole-genome sequenced at 45X mean depth10. The dataset consists of a total of 64 scWGS datasets from 7 AD patients with Braak stage V and VI disease, along with 119 scWGS datasets from 15 unaffected control individuals, some of which have been previously published11. Our previous studies and those by other groups10,12–14 have successfully detected and fully validated bona fide somatic insertions of LINE1 by capturing distinct sequence features in scWGS data, demonstrating the high resolution and accuracy of scWGS-based retrotransposition detection. Therefore, if a retrogene insertion had occurred, we should have been able to observe distinct sequence features at the source retrogene site: increased exonic read-depth, read clipping at exon junctions, poly-A tail at the end of the 3’ UTR, and discordant read pairs spanning exons (Extended Data Fig. 1a). We indeed clearly captured these features at the existing germline retrogene insertions, such as the SKA3 pseudogene insertion (Fig. 3a). If present, somatic events should be able to be detected as heterozygous germline variants in scWGS; however, our analysis revealed no evidence of somatic APP retrogene insertions in any cell. In contrast, we observed that in both patients (AD3 and AD4) carrying germline insertions of SKA3 and the patient (AD2) carrying a germline insertion of ZNF100, there was a clear increase in exonic read depth relative to introns, as would signal for polymorphic germline retrogene insertions (Fig. 3b). We observed no such read depth increase for APP in our 64 AD and 119 normal single-neuron WGS profiles, confirming that we found no evidence of APP retrogene insertions in human neurons.
Figure 3. Absence of somatic APP retrogene insertions in our single-cell WGS data.
a. A germline pseudogene insertion (SKA3) in our scWGS data showing all distinctive characteristics of true retrogene insertion. b. No read-depth gain in APP exons in our AD single neurons. Each dot represents the median of exon/intron read-depth ratios across all exons of the gene in each scWGS dataset from AD patients. AD patients who have polymorphic germline retrogene insertions of SKA3 (AD3 and AD4) and a germline insertion of ZNF100 (AD2) show clear read-depth gain; no such gain for two housekeeping genes (GAPDH, ACTB). Single cells that had poor genomic coverage for a given gene due to locus dropout are excluded. n, number of single cells in each individual; center line, median; box limits, first and third quartiles.
In summary, our analysis of the original sequencing data from the Lee study, the new WES data from the same authors, and the WES data from the independent Park study, as well as of our own scWGS data suggests that somatic APP retrotransposition does not frequently occur either in AD or control neurons. Rather, the reported evidence of APP retrocopies appears to be attributed to various types of exogenous contamination, specifically, APP recombinant vectors, PCR products, and genome-wide mRNA contamination. Our replication experiment also showed the possibility of PCR amplification artifacts creating spurious products that mimic APP gene recombination with various internal exon junctions. Thus, to support the claimed phenomenon of APP gencDNA, it would be necessary for the authors to present unequivocal evidence that cannot be attributed to contamination, such as reads supporting novel APP insertion breakpoints; however, the authors have not presented such direct evidence. In conclusion, we found no evidence of APP retrotransposition in the genomic data presented in the Lee study and further show that our own single-neuron WGS analysis, which directly queried the APP locus at single-nucleotide resolution, reveals no evidence of APP retrotransposition or insertion.
Extended Data
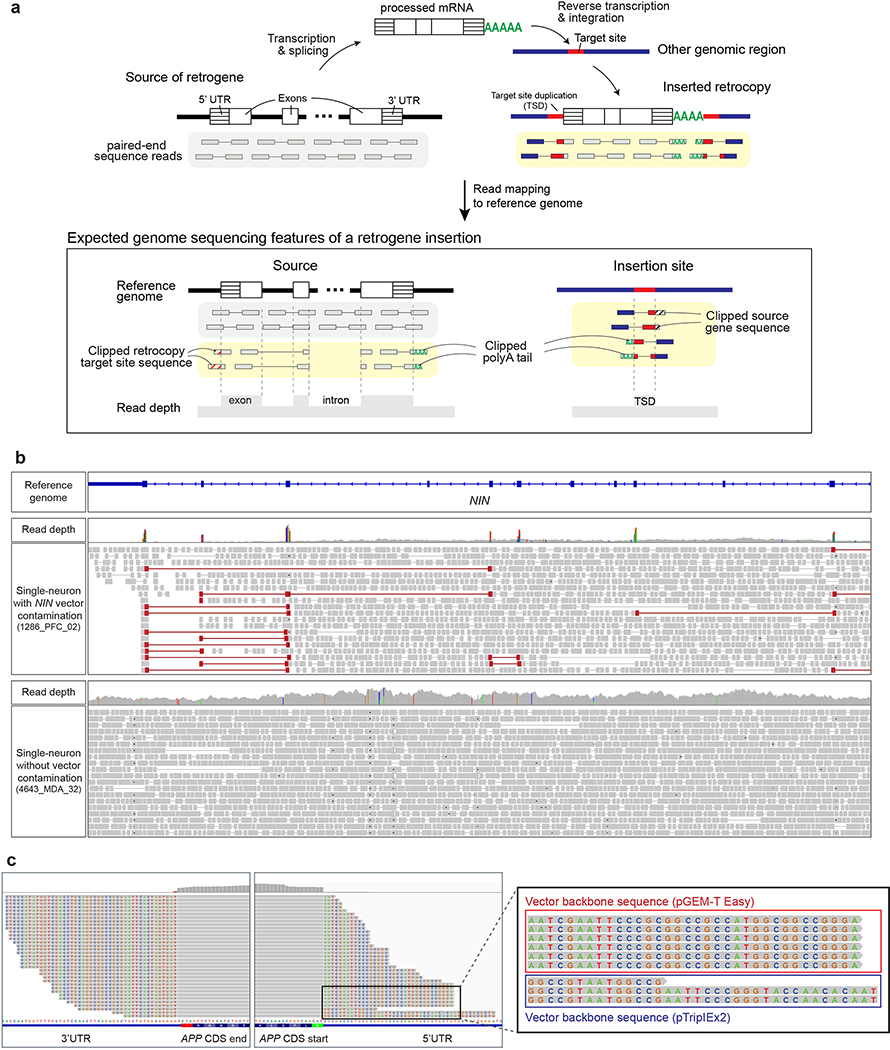
Extended Data Fig. 1. Pervasive recombinant vector contamination in next-generation sequencing.
a. Schematic of a retrogene insertion and the characteristics expected to be captured in sequencing data: increased exonic read-depth, discordant reads spanning exons, clipped reads at exon junctions, 3’ poly-A tail, target site duplication (TSD) at the new genomic insertion site, and clipped reads spanning the retrocopy and insertion sites. b. Recombinant vector contamination found in the Walsh laboratory data. Four single human neurons (1286_PFC_02, 1762_PFC_04, 5379_PFC_01, 5416_PFC_06) in our previous publication showed contamination by a mouse Nin recombinant vector15. The homologous human gene region (NIN) is visualized by the IGV browser for a vector contaminated cell (upper panel) and an unaffected control cell (lower panel). Contamination characteristics were identified, including increased exonic read-depth and exon-spanning discordant reads (reads colored in red) with numerous mismatches to the human genome reference (colored vertical bars in the read depth track). c. Mouse single-neuron WGS data from the Chun laboratory7 contaminated by the same APP recombinant vector detected in the Lee study2 and an additional APP plasmid vector (magnified panel).
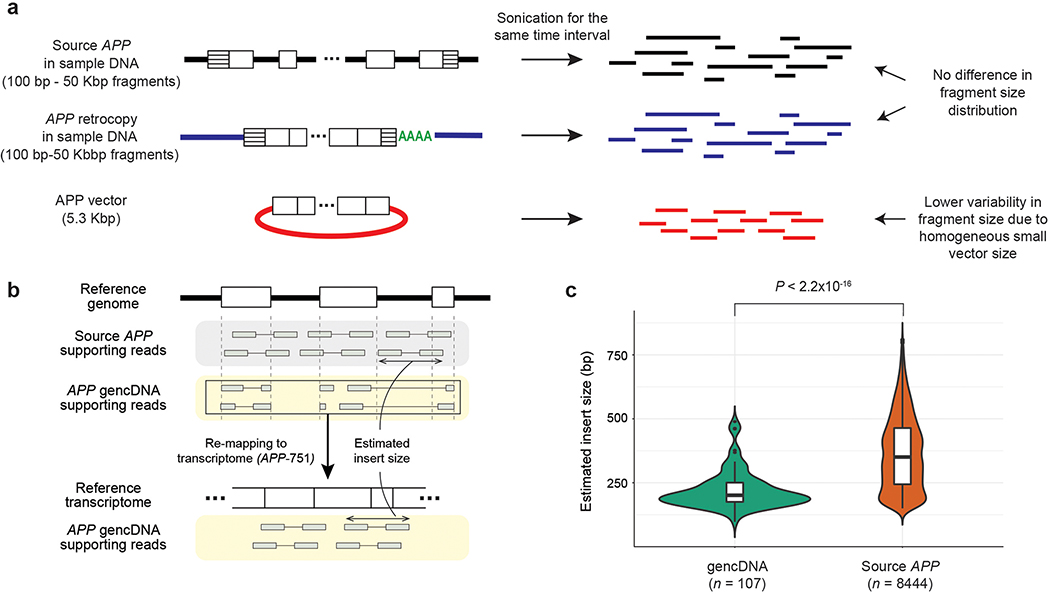
Extended Data Fig. 2. Evidence that recombinant vector contamination is the major source of APP gencDNA.
a. Schematic of the DNA fragment size distribution for each APP source (source APP, APP retrocopy, APP vector). Fragments from APP vectors are expected to be more homogeneous and smaller in size than those from other sources due to the fixed and relatively small vector size. b. DNA fragment (or insert) size estimation. Sequence reads mapped to APP exon junctions were divided into two groups: source APP (reads containing intron sequences) and APP gencDNA (reads clipped at the exon junction) supporting reads. gencDNA supporting reads were remapped to the APP reference transcript sequence (APP-751) to estimate insert sizes. c. Comparison of insert size distribution between source and gencDNA supporting reads. n, number of read pairs in each group.
Extended Data Fig. 3. Novel APP variants with intra-exon junctions as PCR artifacts.
a. Electrophoresis of PCR products from the vector APP inserts (APP-751, APP-695) showing novel APP variants as artifacts. All combinations of two PCR enzymes (FastStart PCR master mix and Platinum SuperFi DNA polymerase; OneStep Ahead RT-PCR in Fig. 1c) with three primer sets generated novel bands smaller than the expected PCR product. b. PCR-induced IEJs with homologous sequences at each junction identified by Illumina sequencing. Twelve IEJs from our vector PCR sequencing showed the exact same sequence homologies and genomic coordinates as IEJs reported in the Lee study. For two IEJs, IGV browser images show pre- (left) and post-junction sites (right) connected by split reads spanning the IEJ (red arc). Because IGV displays forward strand sequences of the human reference genome, all IEJ sequences were also reverse complemented for consistent visualization.
Supplementary Material
Acknowledgements
This work was supported by NIA grant K01AG051791 and Suh Kyungbae Foundation (E.A.L.), NINDS grant R01NS032457-20S1 (C.A.W.), NIH grants T32HL007627 and K08AG065502 (M.B.M.), and NIH grant AG054748 (M.A.L). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests
Declared none.
Code availability
Implemented custom code for the estimation of clipped read fractions and the detection of intra-exon junctions (IEJs) is available at https://sourceforge.net/projects/somatic-app-analysis/.
Data availability
APP vector PCR sequences have been deposited in the NCBI Sequence Read Archive (PRJNA577966). Single-cell whole genome sequencing data of control individuals have been deposited in the NCBI Sequence Read Archive (PRJNA245456) and dbGAP (phs001485.v1.p1). Single-cell whole genome sequencing data of AD patients will be available upon request for genomic regions of APP and source pseudogene SKA3 and ZNF100.
References
- 1.McConnell MJ et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science 356, doi: 10.1126/science.aal1641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MH et al. Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature 563, 639–645, doi: 10.1038/s41586-018-0718-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee M-H et al. Reply: Evidence that APP gene copy number changes reflect recombinant vector contamination. bioRxiv, 730291, doi: 10.1101/730291 (2019). [DOI] [Google Scholar]
- 4.Park JS et al. Brain somatic mutations observed in Alzheimer’s disease associated with aging and dysregulation of tau phosphorylation. Nat Commun 10, 3090, doi: 10.1038/s41467-019-11000-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman DM et al. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. Elife 4, doi: 10.7554/eLife.05116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J et al. Vecuum: identification and filtration of false somatic variants caused by recombinant vector contamination. Bioinformatics 32, 3072–3080, doi: 10.1093/bioinformatics/btw383 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rohrback S et al. Submegabase copy number variations arise during cerebral cortical neurogenesis as revealed by single-cell whole-genome sequencing. Proc Natl Acad Sci U S A 115, 10804–10809, doi: 10.1073/pnas.1812702115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke SL et al. Processed pseudogenes acquired somatically during cancer development. Nat Commun 5, 3644, doi: 10.1038/ncomms4644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odelberg SJ, Weiss RB, Hata A & White R Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res 23, 2049–2057, doi: 10.1093/nar/23.11.2049 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evrony GD et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron 85, 49–59, doi: 10.1016/j.neuron.2014.12.028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodato MA et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559, doi: 10.1126/science.aao4426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erwin JA et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat Neurosci 19, 1583–1591, doi: 10.1038/nn.4388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evrony GD, Lee E, Park PJ & Walsh CA Resolving rates of mutation in the brain using single-neuron genomics. Elife 5, doi: 10.7554/eLife.12966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B et al. Somatic LINE-1 retrotransposition in cortical neurons and non-brain tissues of Rett patients and healthy individuals. PLoS Genet 15, e1008043, doi: 10.1371/journal.pgen.1008043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 166, 1147–1162 e1115, doi: 10.1016/j.cell.2016.07.025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
APP vector PCR sequences have been deposited in the NCBI Sequence Read Archive (PRJNA577966). Single-cell whole genome sequencing data of control individuals have been deposited in the NCBI Sequence Read Archive (PRJNA245456) and dbGAP (phs001485.v1.p1). Single-cell whole genome sequencing data of AD patients will be available upon request for genomic regions of APP and source pseudogene SKA3 and ZNF100.