Abstract
For more than four decades, the free-living nematode Caenorhabditis elegans has been extensively used in anthelmintic research. Classic genetic screens and heterologous expression in the C. elegans model enormously contributed to the identification and characterization of molecular targets of all major anthelmintic drug classes. Although these findings provided substantial insights into common anthelmintic mechanisms, a breakthrough in the treatment and control of parasitic nematodes is still not in sight. Instead, we are facing increasing evidence that the enormous diversity within the phylum Nematoda cannot be recapitulated by any single free-living or parasitic species and the development of novel broad-spectrum anthelmintics is not be a simple goal. In the present review, we summarize certain milestones and challenges of the C. elegans model with focus on drug target identification, anthelmintic drug discovery and identification of resistance mechanisms. Furthermore, we present new perspectives and strategies on how current progress in C. elegans research will support future anthelmintic research.
Keywords: Caenorhabditis elegans, Parasitic nematode, Anthelmintic drug, Anthelmintic resistance, Mode of action
Graphical abstract
1. Caenorhabditis elegans - can a single species serve as surrogate for a whole phylum?
Parasitic nematodes have a tremendous impact on global health and socio-economic development (Hotez et al., 2014). Several parasitic nematode species directly affect large portions of the human population. Soil-transmitted nematodes are the most common helminth infections of humans with more than one billion people infected worldwide (Prichard et al., 2012). Other severe nematode infections such as onchocerciasis or lymphatic filariasis have estimated global infection numbers of around 40 million and 120 million people, respectively (Prichard et al., 2012; Turner et al., 2016). Most nematode infections, although rarely lethal, lead to chronic ailments, including physical disabilities and delayed development (Hotez et al., 2014; Lustigman et al., 2012). Yet, these direct implications on human health do not cover all aspects of the global nematode burden. For instance, parasitic nematodes are of veterinary importance because they are a major threat to the health of livestock and companion animals (Charlier et al., 2014; Kaplan, 2004; Whittaker et al., 2017). Although no reliable data for global estimates are available, infections of livestock cause dramatic economic losses each year (Charlier et al., 2014). In combination with plant parasitic nematodes, these parasites are a threat to global food security and cause substantial economic losses worldwide (Torto et al., 2018; Trudgill and Blok, 2001).
Molecular phylogenetic analyses split the highly diverse phylum Nematoda into five major clades in which parasitism to animals and plants has arisen multiple times independently (Blaxter and Koutsovoulos, 2015; Blaxter, 2003; Dorris et al., 1999; Quist et al., 2015). Despite the enormous impacts of parasitic nematodes, their control relies only on an astonishingly small repertoire of chemical drug classes (Kotze et al., 2014). Unfortunately, extensive usage of these drugs has led to drug resistance, especially in veterinary medicine and agriculture (Kotze et al., 2014) Furthermore, the threat of resistance in human parasites is increasing as well (Diawara et al., 2013; Krücken et al., 2017; Schwab et al., 2005). Although these alarming trends highlight the pressing need for the introduction of new and innovative drugs, their development is hampered by multiple challenges. Compound evaluation against parasitic nematodes is limited by multiple factors, including insufficient access to relevant lifecycle stages, host-dependent and cost-intensive laboratory life cycles, often highly complex and fragile in vitro culture systems, and a limited molecular tool kit. To circumvent some of these limitations, the free-living nematode Caenorhabditis elegans is an elegant system to study nematode-related scientific questions. C. elegans is one of the best-characterized organisms in the world. Since its broad introduction as a genetic model system by Sydney Brenner (1974), discoveries using C. elegans have revolutionized various disciplines of biology, including genetics, gene regulation, development and cell fate, and neurobiology (Corsi et al., 2015). Some features of C. elegans are the simple and rapid life cycle, the well annotated genome, and the extensive number of molecular tools available. Its importance in scientific discovery is exemplified by multiple Nobel prizes awarded to S. Brenner, H. R. Horvitz, and J. Sulston (in 2002); A. Fire and C. C. Mello (in 2006); and M. Chalfie (in 2008) for their research using C. elegans.
However, from a helminthologist's point of view an important question remains: How well can a single species represent the overall biology of a whole phylum? Consequently, this question has been addressed in various review and opinion articles (Geary and Thompson, 2001; Gilleard, 2004; Holden-Dye and Walker, 2014; Salinas and Risi, 2018). Undeniably, C. elegans shares the main characteristics of the nematode body plan such as the cuticle and the organization of the nervous system along with other physiological aspects. The neuromuscular system for example shows a high degree of conservation within the phylum Nematoda, allowing us to draw generally valid hypotheses and conclusions on motility, egg laying, and feeding. For instance, the highly conserved neurotransmitters acetylcholine, glutamate, and gamma-aminobutyric acid (GABA) were found to play central roles in most nematode species (Harder, 2016). However, besides “classical” neurotransmitters, several nematode-specific neuropeptide families (McVeigh et al., 2012) within the phylum were found to modulate muscular function when applied in vitro (Franks et al., 1994; Geary et al., 1995; Maule et al., 1995). Finally, not only the intrinsic molecular control but also the common effects of multiple anthelmintics on the neuromuscular system reflect the similarities between C. elegans and major parasite members of the phylum. Moreover, other aspects include a significant genetic overlap with parasitic nematodes (International Helminth Genomes Consortium, 2019). For instance, Ascaris suum and Haemonchus contortus share around 67–69% of their predicted genes with C. elegans (Jex et al., 2011; Schwarz et al., 2013). Together, these similarities support the use of the C. elegans model in anthelmintic research.
However, C. elegans has a much different evolutionary history than parasitic nematodes. Consequently, genes and gene families with functions linked to a parasitic life style, such as host infection or immune evasion, are completely absent or fulfill different biological roles in C. elegans (International Helminth Genomes Consortium, 2019; Viney, 2018). Among these parasite-specific genes are different groups of proteases/peptidases and protease inhibitors (International Helminth Genomes Consortium, 2019; Viney, 2018) like astacins, a group of metallopeptidases especially found in clade IV and clade V parasitic nematodes (Hunt et al., 2016; Williamson et al., 2006) or cathepsins, linked to digestion of host tissues (Trap et al., 2006; Williamson et al., 2003). Another group that shows a strong expansion in parasites is the apyrase family. Members of this family are postulated to fulfil immunomodulatory functions by hydrolyzing host ATP, which can act as a host immune-stimulatory “danger” signal (International Helminth Genomes Consortium, 2019; Nisbet et al., 2011). If such parasite-specific proteins play key roles in the adaption of parasites to their hosts, then these proteins or processes might be the most promising targets for anthelmintics.
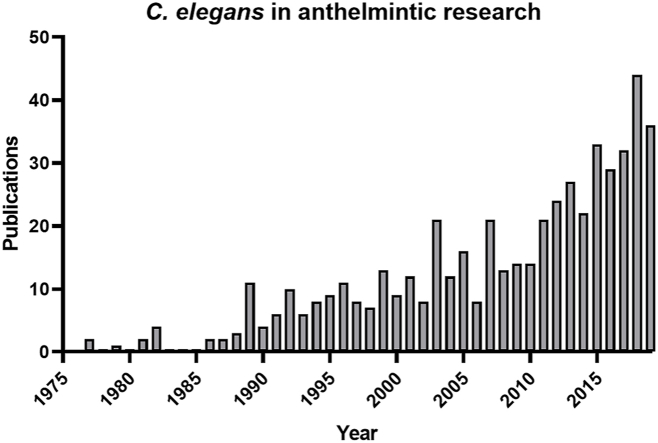
Many helminthologists hoped that the increased availability of genome sequencing data from many species would enable the transfer of findings across different nematode species. However, robust strategies to process the genetic diversity in the nematode phylum and to mine genomic data sets for shared processes between different nematode species are still in development (International Helminth Genomes Consortium, 2019). Nevertheless, it seem to be apparent that the genetic diversity identified in the phylum not only applies to the comparison of free-living versus parasitic nematodes but might be the same when it comes to direct comparisons among different parasite species (Holden-Dye and Walker, 2014). Despite the diversity within the phylum Nematoda, C. elegans consistently finds its way onto the laboratory benches of parasitologists (Fig. 1). Approximately 500 publications have been published in peer-reviewed journals in which C. elegans was used as a model to study anthelmintic drugs and drug targets (Sup Table 1). These numbers reflect the crucial role of C. elegans in anthelmintic research and motivated us to reflect on the importance of the system to helminthology but also to present future perspectives for the most famous non-parasitic nematode in parasitological research.
Fig. 1.
Overview on scientific publications using Caenorhabditis elegans for anthelmintic research.
The bar plot presents an overview of scientific studies that used the free-living model nematode C. elegans for anthelmintic research, sorted by the year of publication (1975–2019). Abstracts of relevant publications were collected by an automated literature search using the bibliographic databases CAB Direct (www.cabdirect.org), provided by Centre for Agriculture and Bioscience International (CABI), and EMBASE (www.emabse.com). For initial abstract extraction, terms were used that combined “C. elegans” with relevant key words, including “anthelmintic drugs”, “nematode parasite species” or “anthelmintic resistance”. The data set was manually curated to obtain a final number of 475 abstracts. For this final selection, publications were considered that included the experimental usage of C. elegans for anthelmintic drug discovery, target characterization, and mode of resistance elucidation. Moreover, selected review articles have a focus on different aspects of C. elegans in anthelmintic research. All selected abstracts are listed in Sup Table 1.
Table 1.
Examples for major anthelmintic drug targets identified by mutant screens in C. elegans.
| Drug/drug class | Major C. elegans target gene(s) | Protein family | Genetic screen |
|---|---|---|---|
| Benzimidazole | ben-1 | beta-tubulin | Driscoll et al. (1989) |
| Macrocyclic lactones | avr-14 | GluCl subunit | Dent et al. (2000) |
| Macrocyclic lactones | avr-15 | GluCl subunit | Dent et al. (2000) |
| Macrocyclic lactones | glc-1 | GluCl subunit | Dent et al. (2000) |
| Imidazothiazoles | lev-1 | nAChR subunit | Lewis et al. (1980), Fleming et al. (1997) |
| Imidazothiazoles | lev-8 | nAChR subunit | Lewis et al. (1980), Towers et al. (2005) |
| Imidazothiazoles | unc-29 | nAChR subunit | Brenner (1974), Fleming et al. (1997) |
| Imidazothiazoles | unc-38 | nAChR subunit | Brenner (1974), Fleming et al. (1997) |
| Imidazothiazoles | unc-63 | nAChR subunit | Brenner (1974), Culetto et al. (2004) |
| AADs | acr-23 | nAChR subunit | Kaminsky et al. (2008a) |
| AADs | acr-20 | nAChR subunit | Baur et al. (2015) |
| Cyclooctadepsipeptides | slo-1 | voltage-gated potassium channel | Guest et al. (2007) |
Abbreviations - GluCl: glutamate-gated chloride channel; nAChR: nicotinic acetylcholine receptor; AADs: amino-acetonitrile derivatives.
2. C. elegans in anthelmintic drug target identification and characterization
For more than four decades, the C. elegans model contributed to the elucidation of the modes of action (MoA) of all major anthelmintic drug classes. C. elegans easily grows under laboratory conditions, and its outstanding genetic tractability and molecular toolkit makes it the model of choice to study anthelmintic MoA. The use of large-scale genetic screens led to the selection of mutants with high-levels of resistance against anthelmintic drugs and subsequently to the identification of the molecular drug targets (Table 1) (Dent et al., 2000; Driscoll et al., 1989; Fleming et al., 1997; Kaminsky et al., 2008a; Lewis et al., 1980). Additionally, C. elegans elegantly and successfully served as a heterologous expression system to characterize potential drug target genes from parasite species (Table 2) (Cook et al., 2006; Courtot et al., 2015; Glendinning et al., 2011; Kwa et al., 1995; Law et al., 2015; Sloan et al., 2015; Welz et al., 2011). Retrospectively, the success of the C. elegans model in anthelmintic target characterization contributed to the hypothesis that highly conserved nematocidal drug targets exist and ultimately caused a shift from phenotypic screens towards target-based screening approaches (Woods et al., 2007) (see section 3).
Table 2.
Examples for anthelmintic drug target genes characterized in the C. elegans expression system.
| Expressed parasite target genes | Drug/drug class | Findings | Reference |
|---|---|---|---|
| tbb-iso-1 (H. contortus) | Benzimidazoles | Characterization of putative benzimidazole resistance alleles using C. elegans as a heterologous expression system. | Kwa et al. (1995) |
| Glucl_alpha3 (H. contortus) | Ivermectin | Expression of glucl_alpha3 restores motor movement in C. elegans avr-14 knock out when expressed under control of the avr-14 promoter | Cook et al. (2006) |
| avr-14b, glc-2, glc-5, glc-6 (H. contortus) | Ivermectin | Expression of H. contortus GluCl subunits rescued ivermectin sensitivity in a highly resistant triple mutant C. elegans strain (DA1316) | Glendinning et al. (2011) |
| slo-1 (A. caninum, C. oncophora) | Emodepside | Expression of A. caninum and C. oncophora slo-1 in the emodepside-resistant C. elegans slo-1 knock-out strain rescued emodepside sensitivity. | Welz et al. (2011) |
| acr-26, acr-27 (H. contortus, P. equorum) | Morantel, pyrantel | Heterologous expression of the H. contortus and P. equorum receptors drastically increased C. elegans sensitivity to morantel and pyrantel | Courtot et al. (2015) |
| unc-29, unc-38 (H. contortus), | Levamisole | H. contortus and A. suum unc-38 and unc-29 were able to (partially) rescue the levamisole-sensitivity and locomotion defects when expressed in C. elegans null mutant strains | Sloan et al. (2015) |
| unc-29 (A. suum) | |||
| Acr-8 (H. contortus) | Levamisole, pyrantel | Expression of H. contortus acr-8 in the C. elegans lev-8 null mutant rescued levamisole and pyrantel sensitivities | Blanchard et al. (2018) |
A striking example for the usefulness of C. elegans in anthelmintic research is associated with the benzimidazole (BZ) anthelmintic drug class. Development of BZ compounds began in the early 1950s when the chemical core ring system was of major interest as a lead structure for the development of novel chemotherapeutic agents (Wright, 1951). During the following years, BZ derivatives were evaluated for a broad range of applications, including antiviral (Tamm et al., 1953), cancer therapy (Mizutani et al., 1960), and also for anthelmintic treatments (Brown et al, 1961). Subsequently, thiabendazole was developed in the early 1960s as the first BZ with broad anthelmintic activity (Gordon, 1961; Brown et al, 1961). In the following years, parasitological in vivo screens identified several BZ derivatives, which facilitated treatment of nematode infections in a variety of hosts (Campbell, 1990). Nevertheless, because of their intensive use in the livestock industry, resistance against BZ developed and spread quickly after their commercialization. Since the mid-1970s, multiple studies indicated that the major molecular mechanism of BZ activity involves altering microtubule formation through binding to beta-tubulins (Barrowman et al., 1984; Dawson et al., 1984; Lacey, 1988; Lubega and Prichard, 1990). This hypothesis was further supported by mutant screens that selected for BZ resistance in yeast (Thomas et al., 1985) and C. elegans (Driscoll et al., 1989), identifying multiple resistance alleles in beta-tubulin genes. These findings were subsequently translated to parasitic nematodes. In a gastrointestinal nematode of sheep, H. contortus, it was shown that BZ-resistant and BZ-susceptible populations differed in single amino acid being encoded in the β-tubulin isotype-1 (tbb-iso-1) gene. The putative resistance allele is a substitution of a phenylalanine residue at position 200 to tyrosine (F200Y) (Kwa et al., 1994). Using C. elegans as heterologous expression system, wild-type alleles of tbb-iso-1 were able to rescue BZ susceptibility in BZ-resistant C. elegans mutants but the H. contortus tbb-iso-1 F200Y variant did not alter the resistance phenotype (Kwa et al., 1995). This combination of genetic screens and functional confirmation of a target and/or resistance gene in C. elegans is a prime example for the relevance of the model to our current understanding of anthelmintic drug MoA and their resistance development.
Another major anthelmintic drug class is the macrocyclic lactones (ML). In the mid-1970s, the first ML, avermectin, was isolated from the soil bacterium Streptomyces avermitilis. Originally screened for antibacterial activity, a mixture of eight closely related derivatives showed activity against parasitic nematodes and finally led to the development of the semi-synthetic derivative, ivermectin, as the first commercially available ML, marketed in the 1980s for several indications and host species (Campbell, 2012; Campbell et al., 1983). Following ivermectin, multiple derivatives joined the market including abamectin, doramectin, and selamectin (avermectin sub-class), as well as moxidectin and milbemycin oxime (milbemycin sub-class). The unique characteristics of ML are their high potency against a broad range of parasites, including nematodes and arthropods, combined with their efficacy against BZ-resistant parasites (Campbell, 2012). These advantages laid the foundation for the outstanding success of ML in human and animal health in the past 40 years, which was recently honored by the 2015 Nobel prize for S. Omura and W. C. Campbell (Tambo et al., 2015). Like for the BZ, the MoA of ML remained unclear for a long time after their commercialization. From in vitro assays, it was shown that ML interfere with normal neuronal function by inducing whole-body paralysis (Campbell et al., 1983). These observations suggested a contribution of neuronal receptors like ligand-gated ion channels. Indeed, first findings pointed in the direction of GABAA receptors expressed in the nematode body-wall muscles (Campbell et al., 1983; Holden-Dye and Walker, 1990). Cully and colleagues provided first evidence for the involvement of glutamate-gated chloride channels (GluCls) in the MoA of ML by characterizing a C. elegans GluCl subunit in Xenopus laevis oocytes (Cully et al., 1994). Later, a genetic screen that selected for ML-resistant C. elegans mutants confirmed that GluCls are the major target proteins of this drug class (Dent et al., 2000). In total, three different GluCl genes, namely avr-14, avr-15, and glc-1, were identified to play a role in ivermectin resistance. Remarkably, mutagenesis screens in different genetic backgrounds revealed that only simultaneous mutations in all three genes conferred high levels of ivermectin resistance, while strains with mutations in only two of the three receptor-subunit genes showed only modest or no differences in ML susceptibility (Dent et al., 2000). Clearly, the use of C. elegans genetics was fundamental for our understanding of the complex MoA of ML and highlighted again the value of the model.
The largest and chemically most diverse group of anthelmintic drugs are summarized under the collective term modulators of nicotinic acetylcholine receptors (nAChRs). Expressed throughout the neuromuscular system of nematodes, nAChRs are composed of five subunits that form a transmembrane ion channel. Over the previous decades, various agonists, antagonists, and allosteric modulators of nematode nAChRs were discovered (Aceves et al., 1970; Aubry et al., 1970; Kaminsky et al., 2008a; Shoop et al., 1990; Thomas, 1979; Xiao et al., 2005; Zinser et al., 2002). Starting in the 1970s, levamisole (LEV) and pyrantel were shown to induce paralysis in the parasitic nematode Ascaris suum by opening nAChRs expressed in body wall muscles (Aceves et al., 1970; Aubry et al., 1970; Martin et al., 2005). Multiple independent genetic studies in C. elegans revealed a large set of genes that were associated with reduced sensitivity against LEV (Boulin et al., 2012; Brenner, 1974; Fleming et al., 1997; Gottschalk et al., 2005; Lewis et al., 1980). C. elegans not only helped to identify multiple nAChR subunits as major drug targets but also provided novel insights into neurobiological processes associated with the neuromuscular junction in nematodes (Brown et al., 2006; Holden-Dye and Walker, 2014).
Also targeting nematode nAChRs, the amino-acetonitril derivative (AADs) monepantel (MON) was developed and commercialized in 2009 for the veterinary market (Lecova et al., 2014). In contrast to the previous anthelmintic drug classes, functional assays in C. elegans played a central role in its development right from the beginning (Kaminsky et al., 2008a). Although H. contortus and Trichostrongylus colubriformis larval stages were treated in vitro with AADs to evaluate activity and derive structure-activity relationship (SAR), C. elegans was used for genetic screens during the discovery and early development phase, which led to the identification of the C. elegans nAChR subunits ACR-23 and ACR-20 as the most relevant molecular targets (Baur et al., 2015; Kaminsky et al., 2008a). Both, ACR-20 and ACR-23 were identified as novel anthelmintic drug targets that belong to the nematode-specific DEG-3 sub-group of nAChR subunits (Rufener et al., 2010). The specific binding of MON and other AADs to this sub-group represents a novel MoA, explaining their activity against nematodes resistant to other nAChR modulators like LEV (Kaminsky et al., 2008b). Today, we know that several genes in nematodes encode nAChR subunits. Different gene duplications and alternative splicing events give rise to multiple homo- and heteromeric receptor combinations with different expression patterns, pharmacological properties, and sensitivities to specific drugs (Holden-Dye et al., 2013; Kotze et al., 2014). Despite their enormous target diversity, all nAChR modulators that made it to the market as anthelmintic drugs, show a concentration-dependent phenotypic effect on C. elegans. Thus, the model has turned out to be highly successful in the MoA identification and characterization of this group of anthelmintics.
Finally, cyclooctadepsipeptides are a relatively recently developed class of anthelmintic compounds exhibiting broad-spectrum efficacy (Krücken et al., 2012). They were shown to be effective against isolates of multi-resistant (BZ, ML, LEV) parasites suggesting a novel MoA (Harder and von Samson-Himmelstjerna, 2002). The parental molecule of this chemical class is PF1022A, a fermentation product of the fungus Rosellinia spp. PF1022, which is associated with Camellia japonica (Harder and von Samson-Himmelstjerna, 2002). Its semi-synthetic derivative emodepside has been commercialized for anthelmintic treatments of cats and dogs (Altreuther et al., 2005). Emodepside acts on the neuromuscular system of nematodes by inhibiting pharyngeal pumping, motility, and egg laying (Krücken et al., 2012). Because C. elegans is highly susceptible to emodepside (Bull et al., 2007), it turned out to be a promising model to elucidate the MoA. In a genetic screen for mutants resistant to emodepside, the voltage-gated potassium channel SLO-1 was identified as essential for emodepside sensitivity (Guest et al., 2007). C. elegans strains harboring a SLO-1 loss-of-function mutation turned out to be highly resistant against emodepside and provided an excellent starting point for expression studies to rescue drug susceptibility. Both ectopic expression of C. elegans SLO-1 (Crisford et al., 2011; Guest et al., 2007) and the heterologous expression of parasitic nematode orthologs (Welz et al., 2011) were able to restore emodepside sensitivity in the otherwise resistant SLO-1 null background. Expression of C. elegans SLO-1 in Xenopus laevis oocytes unequivocally confirmed the MoA of emodepside (Kulke et al., 2014). Again, the contribution of C. elegans to anthelmintic research was impressively demonstrated.
3. Current challenges in anthelmintic drug discovery and resistance research
As stated above, despite the tremendous burden of disease caused by nematodes, the number of anthelmintic drug classes is severely limited. Moreover, most of the available drugs have been used for many decades and only few new anthelmintics entered small and economically relevant markets within recent years. In parallel, extensive use led to wide-spread resistance that further limits efficacious treatment options. This alarming trend highlights the current challenges in anthelmintic research, which are the necessity to fill the drug discovery pipeline with novel and innovative drug classes (see section 3.1) as well as the need to identify reliable resistance markers to improve existing treatment strategies and thereby the longevity of current and future drugs (see section 3.2). After particularly highlighting pivotal contributions of C. elegans in anthelmintic research in the previous section, the following section focuses on the changes and hurdles in anthelmintic drug discovery strategies as well as resistance research over the last decades.
3.1. Discovery of innovative broad-spectrum anthelmintics
Because of highly profitable markets, most anthelmintic drug classes have their origins in the animal health industry. Thus, most large-scale anthelmintic drug discovery programs, using whole-organism or target-based high-throughput screening with several million proprietary compounds, are mainly the scope of the animal health industry.
With the discovery of MLs in the 1980s, a new standard for a broad-spectrum antiparasitic was defined. Because of the outstanding pharmacology of ML, their broad-spectrum activity and safe use in a variety of hosts, the discovery of novel chemical classes that could provide a significant commercial benefit over this gold-standard became difficult to envision. Large drug discovery programs became more difficult to justify so that the discovery pipeline eventually drained. Interests in new anthelmintic drugs were rekindled only when increasing levels of ML resistance in major animal health markets became obvious (Geary et al., 1999a; Woods et al., 2007). Nevertheless, ML are still among the most important and widely used anthelmintics on the market and as such they continue to shape the conception of novel discovery programs. Modern anthelmintic drug discovery is driven by the ambitious goal to identify compounds with a novel MoA, to ensure efficacy against parasites that are resistant to existing anthelmintics. Furthermore, those molecules need to be safe (if possible, in multiple hosts) and have broad-spectrum activity against as many parasite species as possible. In order to allow return on investment in a highly competitive market, these goals need to be achieved in relatively low, profitable doses and in a dosage form that meets costumers’ convenience and compliance.
Traditionally, novel chemical entities were not tested for anthelmintic effects in the first place. Instead, after evaluating compounds for their effects against bacteria, fungi, or insects, they were also tested in small animal in vivo models for their activity against nematodes. However, with the introduction and the general acceptance of “The Three Rs” to reduce, refine, and replace animal testing, the use of in vitro assays became the “new” starting point (Eckert, 1997; Flecknell, 2002). Those assays were generally performed with whole nematodes and in a higher throughput than previous in vivo assays, enabling the testing of several derivatives of a chemical lead structure and thereby deriving meaningful SAR. It was quite common that the MoA for those compounds was fully unknown when marketed and was elucidated only in subsequent genetic studies (see section 2).
The majority of available anthelmintic drug classes was discovered in campaigns with a strong and early focus on in vivo effects. For example, the anthelmintic effects of LEV and PF1022A were identified in in vivo screens against gastrointestinal nematodes in chicken (Janssen, 1976; Sasaki et al., 1992), and ML were discovered using a mouse model infected with Heligmosomoides polygyrus (Campbell et al., 1983). By contrast, thiabendazole and AADs were discovered in in vitro assays using larval stages of parasitic nematodes (Brown et al, 1961; Kaminsky et al., 2008a). Despite the initial low success rate, in vitro screens were considered economically attractive because more compounds were tested, lower amounts of compounds were required, and testing was perceived as less labor intense. C. elegans obviously provides multiple advantages making it highly useful for whole-organism screens, including an easy to maintain life cycle under laboratory conditions. Consequently, C. elegans was considered early as an anthelmintic screening model (Simpkin and Coles, 1981) and was used among other nematodes in industry-based screening campaigns (Geary et al., 1999b).
However, despite significant efforts, no compound initially discovered in a C. elegans screen has reached the market as an anthelmintic, yet. This lack of success led to some controversy about whether C. elegans is a good model for anthelmintic drug discovery (Elfawal et al., 2019; Geary and Thompson, 2001). Indeed, some studies provide evidence that C. elegans might be less sensitive to some anthelmintic drugs compared to parasitic nematode species (Hu et al., 2013; Ruiz-Lancheros et al., 2011), which could lead to false negative conclusions (Hu et al., 2013). Hu et al. (2013) showed that C. elegans fourth-stage larvae (L4) were significantly less susceptible to albendazole or pyrantel than stages of Ancylostoma ceylanicum, Trichuris muris, and Ascaris suum in vitro. This discrepancy might be partially explained by the protective cuticle of C. elegans that is acting as a barrier for small molecules (Ruiz-Lancheros et al., 2011). Of note, C. elegans mutants with increased cuticle permeability and higher sensitivity to chemicals have been identified (Law et al., 2015; Xiong et al., 2017).
A more recent study compared the clade V nematodes C. elegans and A. ceylanicum in a drug screening approach (Elfawal et al., 2019). A 1280-compound library of approved drugs were screened against larval stages of both nematode species. Positives were further evaluated for their in vitro activity against evolutionarily distant T. muris adults (clade II) and finally in an A. ceylanicum hamster model. The authors results suggest that A. ceylanicum larval stages were superior to C. elegans based on both a reduced false negative rate and overall quality of hits, which was defined by lower overall toxicity (based on PubChem cytotoxicity assays) and a broader anthelmintic activity (Elfawal et al., 2019). However, the relevance of these findings for large-scale phenotype screens remains unclear. It is important to point out that C. elegans was not the only nematode species used for pharmaceutical in vitro whole-organism screens. With respect to the generally low success rate of this approach with thiabendazole and AADs as two of the few successful outcomes, the translation from in vitro to in vivo activity might be of higher impact than the selection of any single nematode species for in vitro screening. Encouragingly, in recent years different studies reemphasized the power of C. elegans as valid anthelmintic screening model, especially with respect to throughput and its genetic tractability (see section 4). Therefore, it seems to be beneficial to integrate C. elegans into a screening platform with multiple, parasitic and phylogenetically diverse nematode species. A prime example for this strategy was provided by Burns et al. (2015). They proved the value C. elegans in a well-designed high-throughput approach that included re-screening of promising drug candidates against two related parasite species, Cooperia oncophora and H. contortus, to confirm inter-species activity (Burns et al., 2015). To further prove broad-spectrum activity for identified hits, an evaluation in phylogenetically more distant species like T. muris, as done by Elfawal et al. (2019), would be of help to define promising “species combinations” for future whole-organism screening approaches.
More recently, following a general trend for the pharmaceutical industry, animal health drug discovery also moved from whole-organism-based screenings towards more target-centralized approaches. This trend is motivated by the desire to choose pharmacologically relevant and validated drug targets as starting points of a drug discovery program but is also inspired by technical innovations such as miniaturizing (cellular-) assay systems. More than traditional whole-organism screens, which are often limited by the availability of specimens or larger amounts of compounds, cellular target-based approaches allow fast and efficient screening of large compound libraries for hit identification and optimization. Furthermore, this strategy provides the hope that the spectrum of activity can be characterized in a much earlier phase of the discovery project. However, the success of target-based approaches strongly depends on the selection of the right molecular target, which should be conserved and possess a key function in all species of interest (Woods et al., 2007). A suitable molecular target can be novel, which means it has not yet been pharmacologically evaluated, or it can be “precedented”, a term used by Woods and colleagues (Woods et al., 2007). “Precedented” targets are described to be major targets of known anthelmintic drugs. Compared to novel targets, they are considered less risky because their relevance as a valid drug target has been already proven. However, choosing “precedented” targets has a higher risk that novel drug candidates are affected by the same target-based resistance mechanisms as the existing drug. MoA elucidation with the C. elegans laboratory model and the increasing amount of annotated nematode genome data laid the foundation for target-based approaches in anthelmintic drug discovery and provided the hope to select for targets conserved among all nematodes of interest. Nevertheless, even “precedented” targets show species-specific characteristics that might influence drug effects. For instance, genetic variation in the molecular drug target has the potential to affect drug-binding affinity and influence the targets’ ability to interact with compounds (called druggability of a target). In other cases, depending on the species, the drug target could consist of different gene isoforms (Kashyap et al., 2019), direct one-to-one orthologues of the molecular target might be absent (Rufener et al., 2010), or gene duplications could appear, as shown for the two slo-1 genes found in Trichuris muris (Yilmaz et al., 2015) and the four H. contortus unc-29 gene copies (Duguet et al., 2016). For the latter, Duguet et al. (2016) elegantly demonstrated that all four H. contortus unc-29 gene copies acquired unique functional characteristics based on phenotypic rescue of transgenic C. elegans and heterologous expression in Xenopus laevis oocytes. Another layer of complexity is added when the actual drug target is composed of multiple subunits. Again, the well-studied genetic diversity of the LEV-sensitive nAChR subunits in different nematode species can be used as an example. In C. elegans, the LEV-sensitive nAChR consists of the five subunits UNC-38, UNC-63, UNC-29, LEV-1, and LEV-8 (Boulin et al., 2008). However, several genomes of closely related parasitic nematode species lack individual orthologs but still are sensitive to LEV (Duguet et al., 2016; Holden-Dye et al., 2013). Instead, different subunit compositions give rise to species-specific LEV-sensitive receptors with different pharmacological properties (Duguet et al., 2016; Holden-Dye et al., 2013). For H. contortus, a recent work by Blanchard and colleagues (Blanchard et al., 2018) provided functional evidence that the ACR-8 subunit is likely to replace LEV-8 in the LEV-sensitive receptor of this species. These findings make it difficult to assume that a common MoA applies to diverse nematode species.
Moreover, multi-subunit targets harbor the risk of species-specific target composition with potential consequences on drug potency and might therefore be considered more challenging to address using target-based approaches. In addition to the variation in druggability and target composition, the physiological role of a distinct protein might differ between species and thereby also affect drug performance. For instance GluCls, the physiologically relevant molecular target of ML, regulate pharyngeal pumping and motor activity in gastrointestinal nematodes as well as in C. elegans (Dent et al., 2000; Geary et al., 1993). However, although highly potent against distinct stages of filarial nematodes, ML fail to induce paralysis of the body wall or pharyngeal muscle when applied to the parasites in vitro at physiologically relevant concentrations (Geary et al., 1993). Indeed, the expression pattern of GluCl subunits shows co-localization with the excretory-secretory (E/S) apparatus of Brugia malayi (Moreno et al., 2010). This anatomical structure is considered as the main source of protein secretion in filarial nematodes, leading to the hypothesis that GluCls might play a role in the regulation of immunomodulatory processes (Moreno et al., 2010).
All these examples show that even a molecular target that is generally considered as validated can differ significantly between species and thereby reveal one of the major weaknesses of target-based screening approaches.
The power of C. elegans and its constantly growing molecular tool kit will play a crucial role in pinpointing species-specific target characteristics (see section 4) and thus support the selection of promising future drug targets. However, the output of this strategy to the future anthelmintic drug portfolio remains unknown. So far, no anthelmintic drug originating from target-based screening has made it to the market.
In summary, independent of the initial screening approach, the enormous diversity of nematodes makes the development of novel and safe anthelmintics challenging. Rather than focussing research to find the best model, multi-dimensional approaches should be chosen that combine target-based and whole-organism assays as well as multiple species early in the discovery phase. And finally, promising drug candidates will have to cross the boundaries from in vitro effects to deliver the proof of principle in an in vivo model, which is another significant hurdle.
3.2. Identification of resistance markers to improve current treatment strategies
As long as novel anthelmintic drug classes with resistance-breaking properties are missing, reliable resistance markers are urgently needed to improve current treatment strategies and to expand the lifetime of existing drugs. At least for some available drug classes, field-derived resistant parasite populations occurred before their MoA was fully understood (see section 2) (Prichard, 1990; van Wyk and Malan, 1988). Findings from anthelmintic drug target identification in C. elegans laboratory research were frequently used to search for resistance alleles in natural parasitic nematode populations (Kwa et al., 1994; Njue et al., 2004). Especially, molecular markers for BZ resistance, which are mainly based on single-nucleotide variants in beta-tubulin genes, are prime examples for the direct benefit of the C. elegans model (also see section 2). Another example for a direct translation of the C. elegans system to the field is the detection of MON resistance mutations in acr-23 (Bagnall et al., 2017). Nevertheless, resistance in natural nematode populations against drug classes like ML or nAChRs modulators can, at best, only be partially explained by mutations in major target genes (Kotze et al., 2014). Instead, anthelmintic resistance in natural nematode populations seems to be a complex trait that underlies multiple genetic mechanisms (Andersen et al., 2015; Doyle and Cotton, 2019; Ghosh et al., 2012; Hahnel et al., 2018; Niciura et al., 2019; Zamanian et al., 2018). These mechanisms may include differences in target gene expression (El-Abdellati et al., 2011; Kopp et al., 2009), alternative composition of heteromers using paralogs (Sarai et al., 2013, 2014), and/or alternative splicing of genes (Boulin et al., 2011; Fauvin et al., 2010; Kotze et al., 2014). Furthermore, increased drug efflux via transporters (Blackhall et al., 2008; James and Davey, 2009; Lespine et al., 2012; Peachey et al., 2017) or drug detoxification (Matouskova et al., 2016, 2018; Vokral et al., 2013; Yilmaz et al., 2017) might contribute to anthelmintic drug resistance. In general, it seems plausible that mutations that are tolerable under optimal lab conditions might be deleterious under natural selection, which could require more subtle adaptations (Geary and Thompson, 2001). Therefore, a focus on genetic variation in natural nematode populations appears to be more suitable to capture the complexity of resistance mechanisms in the field. With respect to C. elegans, new platforms and approaches facilitate the incorporation of natural genetic variation into anthelmintic research to elucidate resistance mechanisms (see section 4) (Hahnel et al., 2018; Zamanian et al., 2018). This trend is especially encouraging, as most classical C. elegans studies were performed on a single well-defined strain, N2, which represents not only a single genetic background but also one that has experienced adaptation to laboratory conditions for several decades (Sterken et al., 2015).
4. New perspectives for the C. elegans model in anthelmintic research
Encouragingly, one of the undeniable contributions of C. elegans as a model organism is to constantly push scientific innovations that have the potential to add new aspects to anthelmintic research and enlarge our repertoire of techniques. An outstanding example for this translation is RNA interference (RNAi) used to knock down the expression of specific genes, which was discovered in C. elegans (Fire et al., 1998) and has become a tool to characterize genes of interest in different parasite species (Dash et al., 2017; Maule et al., 2011; McCoy et al., 2015; Ward, 2015), including B. malayi (Aboobaker and Blaxter, 2003; Verma et al., 2017). Other aspects include the usage of the C. elegans genome as a reference for other nematode genome projects (International Helminth Genomes Consortium, 2019) and the large number of in silico resources and databases available (Antoshechkin and Sternberg, 2007; Cook et al., 2017; Grove et al., 2018; Howe et al., 2016; Kumar et al., 2012). Therefore, in the following section, we would like to focus on novel technical advances and innovations in (i) high-throughput screens, (ii) more precise phenotype assays, (iii) recent progress in genome-editing approaches such as CRISPR-Cas9, and (iv) the exploitation of natural C. elegans variation to elucidate genetic mechanisms of anthelmintic resistance (Table 3).
Table 3.
Examples of recent innovation in C. elegans with implications for anthelmintic research.
| Implication for | Studies | Approach/tool | Evaluated in other nematodes |
|---|---|---|---|
| Whole-organism phenotype screening | Andersen et al. (2015) | Improving COPAS BIOSORT (Union Biometrica) platform for high-throughput C. elegans development and fecundity assays | – |
| Law et al. (2015) | Evaluation of C. elegans as heterologous expression platform of insect and nematode drug targets to screen for monoamineric agonists | – | |
| Burns et al. (2015) | Large anthelmintic screening approach using C. elegans. Promising candidates were validated in Cooperia onchophora and Haemonchus contortus. MoA elucidation in C. elegans mutant screens | – | |
| Partridge et al. (2018) | INVertebrate Automated Phenotyping Platform (INVAPP) - plate-based imager system for high-throughput screening for compounds with effect on motility and development of nematodes | in the same study INVAPP was also evaluated for H. contortus (L3), T. circumcincta (L3), and adult Trichuris muris | |
| Spensley et al. (2018) | Imager-based high-throughput assay system that captures acute drug effects and recovery over time | – | |
| Risi et al. (2019) | wMicroTracker (InVivo Biosystems) - Image-free, infrared-based motility assay device to evaluate time-dependent drug effects. | Also, suitable e.g. for larval stages of Ostertagia ostertagi, C. oncophora, H. contortus, T. circumcincta (Liu et al., 2019) | |
| novel phenotype assays | Lockery et al. (2012), Weeks et al. (2018) | Electrophysiological microfluidic chip system for the recording of nematode electropharyngeograms (EPG) | EPG recordings of A. ceylanicum L4 larval stages, A. suum L3 stages (Weeks et al., 2016) |
| Liu et al. (2013) | micro-electro-fluidic (MEF) approach for real-time monitoring of the locomotion of nematodes | – | |
| Ding et al. (2017) | Evaluation of a microfluidic feedback system control (FSC) to identify effective anthelmintic drug concentrations | – | |
| Phiri et al. (2017) | Dye-based detection of anthelmintic induced cuticle damage via a colorimetric read-out | – | |
| Banse et al. (2019) | Stress-Chip - Motility-based microfluidic chip assay measures behaviors of 600 animals in parallel | – | |
| CRISPR-Cas9 mediated drug target characterization | McDiarmid et al. (2018) | Development of CRISPR-Cas9 strategy for whole-gene replacement of human genes in C. elegans in combination with subsequent phenotype analyses for drug screens | – |
| Hahnel et al. (2018), Kitchen et al. (2019), Dilks et al. (2020) | Introduction and characterization of parasite resistance alleles in C. elegans using CRISPR-Cas9 | Successful CRISPR-Cas9 mediated gene knock out approaches in different Strongyloides species (Gang et al., 2017; Lok et al., 2017) | |
| Natural genetic variation | Cook et al. (2017) | C. elegans natural diversity resource (CeNDR) provides a large collection of C. elegans wild isolates as well as whole-genome sequence data. The CeNDR strain collection was successfully used to elucidate natural genetic variation in nematode anthelmintic drug responses (Zdraljevic et al., 2017; Hahnel et al., 2018) | – |
Whole-organism screens using parasitic nematodes are often limited by the amount of biological material that can be obtained, the limited access to relevant life-cycle stages, and the difficulty associated with laboratory culture of parasites. By contrast, C. elegans has an easy to maintain hermaphroditic life cycle and fast generation time, which allow cost- and time-efficient access to all stages in nearly unlimited quantities. Surprisingly, although included in pharmaceutical anthelmintic drug discovery programs, the results from most C. elegans high-throughput anthelmintic screens have not been published (Geary et al., 1999b). Nevertheless, automated high-throughput phenotypic read-out systems for C. elegans are under constant development. Significant progress has been made on automated assay systems that reduce human input in assay preparation and evaluation to increase throughput and to reduce individual bias. Many of these assay systems focus on motility (Buckingham et al., 2014; Partridge et al., 2018; Spensley et al., 2018) but also on fecundity (Andersen et al., 2015), animal growth (Andersen et al., 2015; Partridge et al., 2018; Zdraljevic et al., 2017), and survival (Lai et al., 2014). Applied assay systems range from large platforms like the COPAS BIOSORT (Union Biometrica) (Andersen et al., 2015) to infrared-based motility reader wMicroTracker (InVivoSystems) (Risi et al., 2019). Additionally, some studies took advantage of the genetic tool set of C. elegans by combining drug screens using wild-type and mutant strains (Burns et al., 2015; Mathew et al., 2016) or transgenic lines (Law et al., 2015). Burns and colleagues showed the power of C. elegans by screening over 67,000 small molecule compounds (Burns et al., 2015). They were able to show that compounds active against C. elegans are significantly more likely to be active against other clade V nematodes, including the parasites Cooperia oncophora and H. contortus. The approach was complemented by counter-screening against two different vertebrate cell lines and a resistance screen against C. elegans mutants (Burns et al., 2015). Another strategy was used in a recent study investigating the suitability of transgenic C. elegans strains as an anthelmintic screening platform (Law et al., 2015). The authors heterologously expressed monoamine receptors of parasitic nematodes in C. elegans null mutants to allow the expression of potential drug targets under physiological conditions and in the presence of nematode-specific accessory proteins and the nematode cuticle as a natural physiologically relevant barrier (Law et al., 2015). Encouragingly, some of these assays were already successfully evaluated for life cycle stages of parasitic nematodes (Liu et al., 2019; Partridge et al., 2018).
In addition to improvements in C. elegans high-throughput assays, current research has expanded to assay systems that allow measurement of more subtle drug effects. Compared to “classical” C. elegans assays that mostly focus on robust phenotypes like survival or motility inhibition of groups of animals, a focus on more subtle drug phenotypes has the potential to provide additional information on specific drug characteristics and increased detection sensitivity of drug effects on individual animals. For instance, progress has been made on microfluidic chip technologies to study anthelmintic responses on chemosensation (Aubry and Lu, 2017), motility (Carr et al., 2011; Ding et al., 2017; Liu et al., 2013) or feeding via electrophysiological recordings of the nematode pharynx (Lockery et al., 2012; Weeks et al., 2018). In general, microfluidic chip-based platforms provide microenvironments that can be tightly controlled (e.g. temperature) and manipulated (e.g. controlled buffer perfusion or change of buffer composition), which allows pre- and post-exposure measurements to define (i) baseline, (ii) onset of the maximum drug effect, and (iii) the drug effect over time. These advantages facilitate not only the collection of reproducible data but also the detection of phenotypes down to the individual animal level. Motility-based microfluidic systems like the High-Sensitivity Real-Time (HSRT) worm chip (Carr et al., 2011) and a micro-electro-fluidic approach (Liu et al., 2013) were designed to increase sensitivity in the detection of changes in motility of single worms after drug exposure. The latter approach is based on a microelectrode array that enables detection of nematode movement by measurement of local changes in electrical resistance and thus provides an imaging-free but precise recording system. The sensitivity of microfluidic motility and migration assays is further highlighted by a recent study, where Ding et al. (2017) were able to define effective drug combinations via an automated feedback control system. Other motility-based microfluidic chip systems like the Stress-Chip (Banse et al., 2019) were developed to increase throughput by measuring behaviors of 600 animals in parallel. Beside motility and migration recordings, microfluidic systems provide the possibility to assay a variety of other phenotypes in specific sub-tissues. One example is the eight-channel system that enables the electrophysiological recording of the nematode pharyngeal pumping (Lockery et al., 2012; Weeks et al., 2018). Weeks and colleagues not only confirmed inhibitory effects of ML on the nematode pharynx but were also able to provide evidence that different drug classes modulate pharynx activity in a class-specific manner (Weeks et al., 2018). Besides C. elegans, first steps to translate this approach to parasitic nematodes were made successfully (Weeks et al., 2016). Other assays allow the observation of more specific phenotypes like the measurement of ion fluxes in the excretory pore of C. elegans (Adlimoghaddam et al., 2014) or the detection of cuticular damage in nematodes (Phiri et al., 2017).
Indisputably, one of the success stories using C. elegans in anthelmintic research was based on its potential to serve as heterologous expression system for target genes from parasitic nematode species (Table 2). Recent progress in genome-editing approaches like CRISPR-Cas9 promise more precise expression of parasite target genes in a physiological nematode environment but also a direct transfer to parasitic species (Lok, 2019; Ward, 2015; Zamanian and Andersen, 2016). First developed in mammalian cells, CRISPR-Cas9 is well established in C. elegans and can be used to introduce specific mutations such as deletions, insertions, or point mutations into genomic loci of candidate genes to elucidate function of specific genetic variants (Farboud, 2017; Nance and Frokjaer-Jensen, 2019). The variety of modifications in combination with stable- and site-direct editing provides great advantages, especially for drug target characterization in the experimentally tractable and physiologically relevant C. elegans system. To mention only a few examples, two studies (Baskoylu et al., 2018; Wong et al., 2019) “humanized” the C. elegans orthologs of human neuronal expressed genes by introducing specific amino acid substitutions via CRISPR-Cas9 to investigate disease-related alleles in C. elegans mutants. Another recent study developed a CRISPR-Cas9 strategy for whole-gene replacement of human genes in C. elegans in combination with subsequent phenotype analyses for drug screens (McDiarmid et al., 2018). Although these examples and several other studies point toward the future of this technology, direct translation to anthelmintic research is still at its beginning. Among others, CRISPR-Cas9-mediated gene knock out was used to dissect C. elegans neuropeptide signaling involved in regulation of the dauer stage entry, including meta-analysis to identify corresponding parasite neuropeptide orthologs (Lee et al., 2017). Two other studies evaluated BZ resistance alleles of gastrointestinal nematodes in the C. elegans ben-1 gene via CRISPR-Cas9, to confirm that these alleles convey high levels of BZ resistance into the otherwise sensitive N2 strain (Hahnel et al., 2018; Kitchen et al., 2019). A more comprehensive approach was performed recently by Dilks et al. (2020). Multiple parasite BZ resistance alleles were introduced into the N2 background and quantitatively assessed for their effects on BZ resistance and organismal fitness in the presence or absence of the drug (Dilks et al., 2020). In addition, the first successful steps have been made to transfer the CRISPR-Cas9 technology from C. elegans to different Strongyloides species (Gang et al., 2017; Lok et al., 2017).
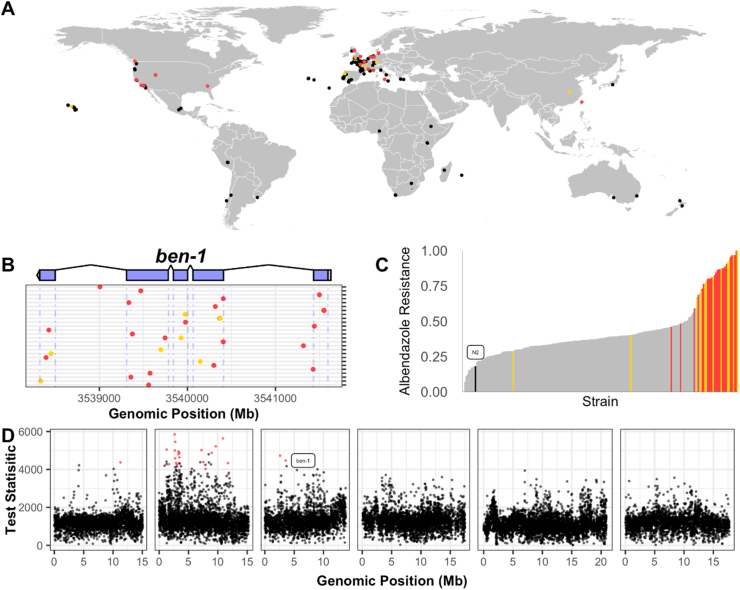
As mentioned previously, most anthelmintic research in C. elegans was performed with the laboratory strain N2, which represents a single genetic background. The applicability of C. elegans results to parasitic nematode species is limited by this lack of genetic diversity. For example, C. elegans wild strains differ significantly in responses to BZ (Hahnel et al., 2018). The tremendous progress, including the discovery of beta-tubulin as a target of BZ, would have been impossible if any of nearly a third of the other strains in the C. elegans species were chosen as the representative wild-type strain. Importantly, parasitic nematode species have some of the highest levels of diversity in the animal kingdom (De Ley, 2006), so investigators should take advantage of natural diversity in the free-living models as well. To meet this need, the laboratory strain does not reflect the natural genetic variation present in natural nematode populations. The C. elegans Natural Diversity Resource (CeNDR) provides tools and methods to leverage the natural variation for this species (Fig. 2) (Cook et al., 2017). First, wild C. elegans strains isolated by researchers and citizen-scientists are archived and sent to investigators upon request. These wild strains can be assayed for responses to anthelmintic compounds. Second, whole-genome sequence data and identified variants are available to the community. These data can be queried to identify orthologous genes between C. elegans and parasites of interest. These orthologous genes might harbor variation in specific C. elegans strains, and these strains can be assayed for anthelmintic responses. Third, if wild strains are measured for anthelmintic responses, genome-wide association studies can connect genomic variation to drug response differences (Hahnel et al., 2018; Zamanian et al., 2018). This unbiased approach enables future discoveries of drug resistance mechanisms.
Fig. 2.
The Caenorhabditis elegans natural diversity resource (CeNDR) provides insights into genetic and phenotypic variation of natural C. elegans population.
(A) The map shows global origin of C. elegans strains that are available to the community through CeNDR. Overall, 249 genetically diverse strains were collected over the past 50 years by researchers and citizen-scientists from different regions and substrates all over the world. All isolated strains were cryopreserved and genome sequenced. (B) Genomic variation in anthelmintic drug targets can be queried with the CeNDR Variant Browser tool (https://www.elegansvariation.org/data/browser/). As an example, the genetic variation in the beta-tubulin gene ben-1, a major resistance gene for benzimidazoles, is shown. Genetic variants with predicted moderate (e.g. missense variants) or high effects (e.g. frame shift variants) are displayed in yellow or red, respectively. (C) CeNDR strains vary in their phenotypic response to anthelmintic drugs. As an example, the relative resistance to albendazole (ABZ) is shown (Hahnel et al., 2018). Each bar represents a single CeNDR strain, included in an ABZ exposure assay, sorted by their relative ABZ resistance. Strains that have ben-1 variants with predicted moderate or high effects are colored in yellow and red, respectively. Strains similar to the N2 reference genome with respect to the ben-1 locus are shown in grey. Distribution of strains with a ben-1 variant indicate a correlation between ben-1 and ABZ resistance in C. elegans wild isolates. (D) Phenotype and genotype data of wild isolates can be used to identify genomic regions or genes that correlate with an observed resistance phenotype. The example shows the Manhattan plot of a genome-wide association study performed to identify genetic determinants of BZ resistance in CeNDR strains (Hahnel et al., 2018). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
5. Conclusions
The C. elegans laboratory model has made broad impacts in a variety of biological disciplines over more than four decades. Beside its groundbreaking contributions to our understanding of different aspects of general biology, the experimentally tractable C. elegans model was successfully used for identifying and characterizing molecular drug targets of all major anthelmintic classes and provided the first validation of selected resistance alleles. However, in order to face the global threat of parasitic nematodes, novel anthelmintic drugs need to be discovered and we need to understand how resistance arises in diverse parasitic nematode species to improve current treatment strategies. A growing number of studies provides insights into the enormous diversity within the phylum, suggesting that a focus on a single species alone is not sufficient to address these ambitious goals.
With C. elegans a powerful system is in place being an enormous and constant source for both incremental as well as disruptive scientific innovation. Steady progress on assay platforms, heterologous expression, or wild isolates will help to evaluate drug candidates, characterize drug targets and provide insights into the natural genetic variation in nematodes. To leverage these advantages in the most meaningful way, an essential task will be to integrate the power of the C. elegans model into cross-species approaches, including parasite species from different clades, to allow robust assumptions with broad implications for anthelmintic research.
Declaration of competing interest
Steffen R. Hahnel (SRH), Iring Heisler (IH) and Daniel Kulke (DK) are employees of Elanco Animal Health. Elanco Animal Health develops and sells veterinary pharmaceuticals including dewormers. Except of SRH, IH and DK, Elanco Animal Health was not involved in the preparation of the manuscript. The decision to publish the manuscript was jointly taken.
Acknowledgements
We thank the Bayer Life Sciences Collaboration (LSC) (Germany) for funding SRH, the Pew Foundation (United States) for funding ECA, and the NIH Biotechnology Training Grant (T32 GM008449) (United States) for funding CMD.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.09.005.
Contributor Information
Steffen R. Hahnel, Email: steffen.hahnel@elancoah.com.
Clayton M. Dilks, Email: claytondilks2923@u.northwestern.edu.
Iring Heisler, Email: iring.heisler@elancoah.com.
Erik C. Andersen, Email: andersen@northwestern.edu.
Daniel Kulke, Email: daniel.kulke@elancoah.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aboobaker A.A., Blaxter M.L. Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Mol. Biochem. Parasitol. 2003;129:41–51. doi: 10.1016/s0166-6851(03)00092-6. [DOI] [PubMed] [Google Scholar]
- Aceves J., Erlij D., Martinez-Maranon R. The mechanism of the paralysing action of tetramisole on Ascaris somatic muscle. Br. J. Pharmacol. 1970;38:602–607. doi: 10.1111/j.1476-5381.1970.tb10601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlimoghaddam A., Weihrauch D., O'Donnell M.J. Localization of K(+), H(+), Na(+) and Ca(2)(+) fluxes to the excretory pore in Caenorhabditis elegans: application of scanning ion-selective microelectrodes. J. Exp. Biol. 2014;217:4119–4122. doi: 10.1242/jeb.112441. [DOI] [PubMed] [Google Scholar]
- Altreuther G., Buch J., Charles S.D., Davis W.L., Krieger K.J., Radeloff I. Field evaluation of the efficacy and safety of emodepside/praziquantel spot-on solution against naturally acquired nematode and cestode infections in domestic cats. Parasitol. Res. 2005;97(Suppl. 1):S58–s64. doi: 10.1007/s00436-005-1445-0. [DOI] [PubMed] [Google Scholar]
- Andersen E.C., Shimko T.C., Crissman J.R., Ghosh R., Bloom J.S., Seidel H.S., Gerke J.P., Kruglyak L. A powerful new quantitative genetics platform, combining Caenorhabditis elegans high-throughput fitness assays with a large collection of recombinant strains. G3 (Bethesda, Md.) 2015;5:911–920. doi: 10.1534/g3.115.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin I., Sternberg P.W. The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nat. Rev. Genet. 2007;8:518–532. doi: 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- Aubry G., Lu H. Droplet array for screening acute behaviour response to chemicals in Caenorhabditis elegans. Lab Chip. 2017;17:4303–4311. doi: 10.1039/c7lc00945c. [DOI] [PubMed] [Google Scholar]
- Aubry M.L., Cowell P., Davey M.J., Shevde S. Aspects of the pharmacology of a new anthelmintic: pyrantel. Br. J. Pharmacol. 1970;38:332–344. doi: 10.1111/j.1476-5381.1970.tb08521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall N.H., Ruffell A., Raza A., Elliott T.P., Lamb J., Hunt P.W., Kotze A.C. Mutations in the Hco-mptl-1 gene in a field-derived monepantel-resistant isolate of Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2017;7:236–240. doi: 10.1016/j.ijpddr.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse S.A., Blue W.B., Robinson K.J., Jarrett C.M., Phillips P.C. The Stress-Chip: A microfluidic platform for stress analysis in Caenorhabditis elegans. PLoS One. 2019;14(5):e0216283. doi: 10.1371/journal.pone.0216283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman M.M., Marriner S.E., Bogan J.A. The binding and subsequent inhibition of tubulin polymerization in Ascaris suum (in vitro) by benzimidazole anthelmintics. Biochem. Pharmacol. 1984;33:3037–3040. doi: 10.1016/0006-2952(84)90605-1. [DOI] [PubMed] [Google Scholar]
- Baskoylu S.N., Yersak J., O'Hern P., Grosser S. Vol. 14. 2018. (Single copy/knock-in models of ALS SOD1 in C. elegans suggest loss and gain of function have different contributions to cholinergic and glutamatergic neurodegeneration). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur R., Beech R., Sigel E., Rufener L. Monepantel irreversibly binds to and opens Haemonchus contortus MPTL-1 and Caenorhabditis elegans ACR-20 receptors. Mol. Pharmacol. 2015;87:96–102. doi: 10.1124/mol.114.095653. [DOI] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. P-glycoprotein selection in strains of Haemonchus contortus resistant to benzimidazoles. Vet. Parasitol. 2008;152:101–107. doi: 10.1016/j.vetpar.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Blanchard A., Fabrice Guégnard F., Charvet C.L., Crisford A., Courtot E., Sauvé C., Harmache A., Duguet T., O’Connor V., Castagnone-Sereno P., Reaves B., Wolstenholme A.J., Beech R.N., Holden-Dye L., Neveu C. Deciphering the molecular determinants of cholinergic anthelmintic sensitivity in nematodes: when novel functional validation approaches highlight major differences between the model Caenorhabditis elegans and parasitic species. PLoS Pathog. 2018;14(5) doi: 10.1371/journal.ppat.1006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M.L. Nematoda: genes, genomes and the evolution of parasitism. Adv. Parasitol. 2003;54:101–195. doi: 10.1016/s0065-308x(03)54003-9. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Koutsovoulos G. The evolution of parasitism in Nematoda. Parasitology. 2015;142(Suppl. 1):S26–S39. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T., Gielen M., Richmond J.E., Williams D.C., Paoletti P., Bessereau J.L. Eight genes are required for functional reconstitution of the Caenorhabditis elegans levamisole-sensitive acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18590–18595. doi: 10.1073/pnas.0806933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T., Fauvin A., Charvet C.L., Cortet J., Cabaret J., Bessereau J.L., Neveu C. Functional reconstitution of Haemonchus contortus acetylcholine receptors in Xenopus oocytes provides mechanistic insights into levamisole resistance. Br. J. Pharmacol. 2011;164:1421–1432. doi: 10.1111/j.1476-5381.2011.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T., Rapti G., Briseno-Roa L., Stigloher C., Richmond J.E., Paoletti P., Bessereau J.L. Positive modulation of a Cys-loop acetylcholine receptor by an auxiliary transmembrane subunit. Nat. Neurosci. 2012;15:1374–1381. doi: 10.1038/nn.3197. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H.D., Matzuk A.R., Ilves R., Peterson L.H., Harris S.A., Sarett L.H., Egerton J.R., Yakstis J.J., Campbell W.C., Cuckler A.C. Antiparasitic drugs. IV. 2-(4’-THIAZOLYL)-BENZIMIDAZOLE, a new anthelmintic. J. Am. Chem. Soc. 1961;83(7):1764–1765. [Google Scholar]
- Brown L.A., Jones A.K., Buckingham S.D., Mee C.J., Sattelle D.B. Contributions from Caenorhabditis elegans functional genetics to antiparasitic drug target identification and validation: nicotinic acetylcholine receptors, a case study. Int. J. Parasitol. 2006;36:617–624. doi: 10.1016/j.ijpara.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Buckingham S.D., Partridge F.A., Sattelle D.B. Automated, high-throughput, motility analysis in Caenorhabditis elegans and parasitic nematodes: applications in the search for new anthelmintics. Int. J. Parasitol. Drugs Drug Resist. 2014;4:226–232. doi: 10.1016/j.ijpddr.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull K., Cook A., Hopper N.A., Harder A., Holden-Dye L., Walker R.J. Effects of the novel anthelmintic emodepside on the locomotion, egg-laying behaviour and development of Caenorhabditis elegans. Int. J. Parasitol. 2007;37:627–636. doi: 10.1016/j.ijpara.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Burns A.R., Luciani G.M., Musso G., Bagg R., Yeo M., Zhang Y., Rajendran L., Glavin J., Hunter R., Redman E., Stasiuk S., Schertzberg M., Angus McQuibban G., Caffrey C.R., Cutler S.R., Tyers M., Giaever G., Nislow C., Fraser A.G., MacRae C.A., Gilleard J., Roy P.J. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat. Commun. 2015;6:7485. doi: 10.1038/ncomms8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W.C. Benzimidazoles: veterinary uses. Parasitol. today. 1990;6:130–133. doi: 10.1016/0169-4758(90)90231-r. [DOI] [PubMed] [Google Scholar]
- Campbell W.C. History of avermectin and ivermectin, with notes on the history of other macrocyclic lactone antiparasitic agents. Curr. Pharmaceut. Biotechnol. 2012;13:853–865. doi: 10.2174/138920112800399095. [DOI] [PubMed] [Google Scholar]
- Campbell W.C., Fisher M.H., Stapley E.O., Albers-Schonberg G., Jacob T.A. Ivermectin: a potent new antiparasitic agent. Science. 1983;221:823–828. doi: 10.1126/science.6308762. [DOI] [PubMed] [Google Scholar]
- Carr J.A., Parashar A., Gibson R., Robertson A.P., Martin R.J., Pandey S. A microfluidic platform for high-sensitivity, real-time drug screening on C. elegans and parasitic nematodes. Lab Chip. 2011;11:2385–2396. doi: 10.1039/c1lc20170k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier J., van der Voort M., Kenyon F., Skuce P., Vercruysse J. Chasing helminths and their economic impact on farmed ruminants. Trends Parasitol. 2014;30:361–367. doi: 10.1016/j.pt.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Cook A., Aptel N., Portillo V., Siney E., Sihota R., Holden-Dye L., Wolstenholme A. Caenorhabditis elegans ivermectin receptors regulate locomotor behaviour and are functional orthologues of Haemonchus contortus receptors. Mol. Biochem. Parasitol. 2006;147:118–125. doi: 10.1016/j.molbiopara.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Cook D.E., Zdraljevic S., Roberts J.P., Andersen E.C. Vol. 45. 2017. pp. D650–d657. (CeNDR, the Caenorhabditis elegans natural diversity resource). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi A.K., Wightman B., Chalfie M. A Transparent window into biology: a primer on Caenorhabditis elegans. Worm : the online review of C. elegans biology. 2015:1–31. doi: 10.1895/wormbook.1.177.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot E., Charvet C.L., Beech R.N., Harmache A., Wolstenholme A.J., Holden-Dye L., O'Connor V., Peineau N., Woods D.J., Neveu C. Functional characterization of a novel class of morantel-sensitive acetylcholine receptors in nematodes. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisford A., Murray C., O'Connor V., Edwards R.J., Kruger N., Welz C., von Samson-Himmelstjerna G., Harder A., Walker R.J., Holden-Dye L. Selective toxicity of the anthelmintic emodepside revealed by heterologous expression of human KCNMA1 in Caenorhabditis elegans. Mol. Pharmacol. 2011;79:1031–1043. doi: 10.1124/mol.111.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culetto E., Baylis H.A., Richmond J.E., Jones A.K., Fleming J.T., Squire M.D., Lewis J.A., Sattelle D.B. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J. Biol. Chem. 2004;279(41):42476–42483. doi: 10.1074/jbc.M404370200. [DOI] [PubMed] [Google Scholar]
- Cully D.F., Vassilatis D.K., Liu K.K., Paress P.S., Van der Ploeg L.H., Schaeffer J.M., Arena J.P. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature. 1994;371:707–711. doi: 10.1038/371707a0. [DOI] [PubMed] [Google Scholar]
- Dash M., Dutta T.K., Phani V., Papolu P.K., Shivakumara T.N., Rao U. RNAi-mediated disruption of neuropeptide genes, nlp-3 and nlp-12, cause multiple behavioral defects in Meloidogyne incognita. Biochem. Biophys. Res. Commun. 2017;490:933–940. doi: 10.1016/j.bbrc.2017.06.143. [DOI] [PubMed] [Google Scholar]
- Dawson P.J., Gutteridge W.E., Gull K. A comparison of the interaction of anthelmintic benzimidazoles with tubulin isolated from mammalian tissue and the parasitic nematode Ascaridia galli. Biochem. Pharmacol. 1984;33:1069–1074. doi: 10.1016/0006-2952(84)90515-x. [DOI] [PubMed] [Google Scholar]
- De Ley P. A quick tour of nematode diversity and the backbone of nematode phylogeny. Worm : the online review of C. elegans biology. 2006:1–8. doi: 10.1895/wormbook.1.41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J.A., Smith M.M., Vassilatis D.K., Avery L. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2674–2679. doi: 10.1073/pnas.97.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A., Halpenny C.M., Churcher T.S., Mwandawiro C., Kihara J., Kaplan R.M., Streit T.G., Idaghdour Y., Scott M.E., Basanez M.G., Prichard R.K. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS Neglected Trop. Dis. 2013;7:e2247. doi: 10.1371/journal.pntd.0002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks C.M., Hahnel S.R., Sheng Q., Long L., McGrath P.T., Andersen E.C. Quantitative benzimidazole resistance and fitness effects of parasitic nematode beta-tubulin alleles. International journal for parasitology. Drugs and drug resistance. 2020;14:28–36. doi: 10.1016/j.ijpddr.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Njus Z., Kong T., Su W., Ho C.M., Pandey S. Effective drug combination for Caenorhabditis elegans nematodes discovered by output-driven feedback system control technique. Sci Adv. 2017;3(10):eaao1254. doi: 10.1126/sciadv.aao1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris M., De Ley P., Blaxter M.L. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol. today. 1999;15:188–193. doi: 10.1016/s0169-4758(99)01439-8. [DOI] [PubMed] [Google Scholar]
- Doyle S.R., Cotton J.A. Genome-wide approaches to investigate anthelmintic resistance. Trends Parasitol. 2019;35:289–301. doi: 10.1016/j.pt.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 1989;109:2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet T.B., Charvet C.L., Forrester S.G., Wever C.M., Dent J.A., Neveu C., Beech R.N. Recent duplication and functional divergence in parasitic nematode levamisole-sensitive acetylcholine receptors. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert J. Alternatives to animal experimentation in parasitology. Vet. Parasitol. 1997;71:99–120. doi: 10.1016/s0304-4017(97)00027-7. [DOI] [PubMed] [Google Scholar]
- El-Abdellati A., De Graef J., Van Zeveren A., Donnan A., Skuce P., Walsh T., Wolstenholme A., Tait A., Vercruysse J., Claerebout E., Geldhof P. Altered avr-14B gene transcription patterns in ivermectin-resistant isolates of the cattle parasites, Cooperia oncophora and Ostertagia ostertagi. Int. J. Parasitol. 2011;41:951–957. doi: 10.1016/j.ijpara.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Elfawal M.A., Savinov S.N., Aroian R.V. Drug screening for discovery of broad-spectrum agents for soil-transmitted nematodes. Sci. Rep. 2019;9:12347. doi: 10.1038/s41598-019-48720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B. Targeted genome editing in Caenorhabditis elegans using CRISPR/Cas9. Wiley interdisciplinary reviews. Dev. Biol. 2017;6 doi: 10.1002/wdev.287. [DOI] [PubMed] [Google Scholar]
- Fauvin A., Charvet C., Issouf M., Cortet J., Cabaret J., Neveu C. cDNA-AFLP analysis in levamisole-resistant Haemonchus contortus reveals alternative splicing in a nicotinic acetylcholine receptor subunit. Mol. Biochem. Parasitol. 2010;170:105–107. doi: 10.1016/j.molbiopara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Flecknell P. Replacement, reduction and refinement. ALTEX. 2002;19:73–78. [PubMed] [Google Scholar]
- Fleming J.T., Squire M.D., Barnes T.M., Tornoe C., Matsuda K., Ahnn J., Fire A., Sulston J.E., Barnard E.A., Sattelle D.B., Lewis J.A. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. Off. J. Soc. Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks C.J., Holden-Dye L., Williams R.G., Pang F.Y., Walker R.J. A nematode FMRFamide-like peptide, SDPNFLRFamide (PF1), relaxes the dorsal muscle strip preparation of Ascaris suum. Parasitology. 1994;108(Pt 2):229–236. doi: 10.1017/s0031182000068335. [DOI] [PubMed] [Google Scholar]
- Gang S.S., Castelletto M.L., Bryant A.S. Vol. 13. 2017. (Targeted Mutagenesis in a Human-Parasitic Nematode). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary T.G., Thompson D.P. Caenorhabditis elegans: how good a model for veterinary parasites? Vet. Parasitol. 2001;101:371–386. doi: 10.1016/s0304-4017(01)00562-3. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Sims S.M., Thomas E.M., Vanover L., Davis J.P., Winterrowd C.A., Klein R.D., Ho N.F., Thompson D.P. Haemonchus contortus: ivermectin-induced paralysis of the pharynx. Exp. Parasitol. 1993;77:88–96. doi: 10.1006/expr.1993.1064. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Bowman J.W., Friedman A.R., Maule A.G., Davis J.P., Winterrowd C.A., Klein R.D., Thompson D.P. The pharmacology of FMRFamide-related neuropeptides in nematodes: new opportunities for rational anthelmintic discovery? Int. J. Parasitol. 1995;25:1273–1280. doi: 10.1016/0020-7519(95)00064-9. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Sangster N.C., Thompson D.P. Frontiers in anthelmintic pharmacology. Vet. Parasitol. 1999;84:275–295. doi: 10.1016/s0304-4017(99)00042-4. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Thompson D.P., Klein R.D. Mechanism-based screening: discovery of the next generation of anthelmintics depends upon more basic research. Int. J. Parasitol. 1999;29:105–112. doi: 10.1016/s0020-7519(98)00170-2. discussion 113-104. [DOI] [PubMed] [Google Scholar]
- Ghosh R., Andersen E.C., Shapiro J.A., Gerke J.P., Kruglyak L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 2012;335:574–578. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J.S. The use of Caenorhabditis elegans in parasitic nematode research. Parasitology. 2004;128(Suppl. 1):S49–S70. doi: 10.1017/S003118200400647X. [DOI] [PubMed] [Google Scholar]
- Glendinning S.K., Buckingham S.D., Sattelle D.B., Wonnacott S., Wolstenholme A.J. Glutamate-gated chloride channels of Haemonchus contortus restore drug sensitivity to ivermectin resistant Caenorhabditis elegans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon H.M. Thiabendazole: a highly effective anthelmintic for sheep. Nature. 1961;191:1409–1410. doi: 10.1038/1911409a0. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Almedom R.B., Schedletzky T., Anderson S.D., Yates J.R., 3rd, Schafer W.R. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove C., Cain S., Chen W.J., Davis P., Harris T., Howe K.L., Kishore R., Lee R., Paulini M., Raciti D., Tuli M.A., Van Auken K., Williams G. Using WormBase: a genome biology resource for Caenorhabditis elegans and related nematodes. Methods Mol. Biol. 2018;1757:399–470. doi: 10.1007/978-1-4939-7737-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest M., Bull K., Walker R.J., Amliwala K., O'Connor V., Harder A., Holden-Dye L., Hopper N.A. The calcium-activated potassium channel, SLO-1, is required for the action of the novel cyclo-octadepsipeptide anthelmintic, emodepside, in Caenorhabditis elegans. Int. J. Parasitol. 2007;37:1577–1588. doi: 10.1016/j.ijpara.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Hahnel S.R., Zdraljevic S., Rodriguez B.C., Zhao Y., McGrath P.T., Andersen E.C. Vol. 14. 2018. (Extreme Allelic Heterogeneity at a Caenorhabditis elegans Beta-Tubulin Locus Explains Natural Resistance to Benzimidazoles). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder A. The biochemistry of Haemonchus contortus and other parasitic nematodes. Adv. Parasitol. 2016;93:69–94. doi: 10.1016/bs.apar.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Harder A., von Samson-Himmelstjerna G. Cyclooctadepsipeptides--a new class of anthelmintically active compounds. Parasitol. Res. 2002;88:481–488. doi: 10.1007/s00436-002-0619-2. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L., Walker R.J. Avermectin and avermectin derivatives are antagonists at the 4-aminobutyric acid (GABA) receptor on the somatic muscle cells of Ascaris; is this the site of anthelmintic action? Parasitology. 1990;101(Pt 2):265–271. doi: 10.1017/s0031182000063320. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L., Walker R.J. 2014. Anthelmintic Drugs and Nematicides: studies in Caenorhabditis elegans. Worm Online Review of C. elegans Biology; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden-Dye L., Joyner M., O'Connor V., Walker R.J. Nicotinic acetylcholine receptors: a comparison of the nAChRs of Caenorhabditis elegans and parasitic nematodes. Parasitol. Int. 2013;62:606–615. doi: 10.1016/j.parint.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Alvarado M., Basanez M.G., Bolliger I., Bourne R., Boussinesq M., Brooker S.J., Brown A.S., Buckle G., Budke C.M., Carabin H., Coffeng L.E., Fevre E.M., Furst T., Halasa Y.A., Jasrasaria R., Johns N.E., Keiser J., King C.H., Lozano R., Murdoch M.E., O'Hanlon S., Pion S.D., Pullan R.L., Ramaiah K.D., Roberts T., Shepard D.S., Smith J.L., Stolk W.A., Undurraga E.A., Utzinger J., Wang M., Murray C.J., Naghavi M. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., Harris T.W., Kishore R., Lee R., Lomax J., Li Y., Muller H.M., Nakamura C., Nuin P., Paulini M., Raciti D., Schindelman G., Stanley E., Tuli M.A., Van Auken K., Wang D., Wang X., Williams G., Wright A., Yook K., Berriman M., Kersey P., Schedl T., Stein L., Sternberg P.W. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44(D1):D774–80. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Ellis B.L., Yiu Y.Y., Miller M.M., Urban J.F., Shi L.Z., Aroian R.V. An extensive comparison of the effect of anthelmintic classes on diverse nematodes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt V.L., Tsai I.J., Coghlan A., Reid A.J., Holroyd N., Foth B.J., Tracey A., Cotton J.A., Stanley E.J., Beasley H., Bennett H.M., Brooks K., Harsha B., Kajitani R., Kulkarni A., Harbecke D., Nagayasu E., Nichol S., Ogura Y., Quail M.A., Randle N., Xia D., Brattig N.W., Soblik H., Ribeiro D.M., Sanchez-Flores A., Hayashi T., Itoh T., Denver D.R., Grant W., Stoltzfus J.D., Lok J.B., Murayama H., Wastling J., Streit A., Kikuchi T., Viney M., Berriman M. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 2016;48:299–307. doi: 10.1038/ng.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium Comparative genomics of the major parasitic worms. Nat. Genet. 2019;51:163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Janssen P.A. The levamisole story. Prog. Drug Res. 1976;20:347–383. doi: 10.1007/978-3-0348-7094-8_11. [DOI] [PubMed] [Google Scholar]
- Jex A.R., Liu S., Li B., Young N.D., Hall R.S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G.A., Harris T.W., Campbell B.E., Vlaminck J., Wang T., Cantacessi C., Schwarz E.M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P.W., Yang H., Wang J., Wang J., Gasser R.B. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Ducray P., Jung M., Clover R., Rufener L., Bouvier J., Weber S.S., Wenger A., Wieland-Berghausen S., Goebel T., Gauvry N., Pautrat F., Skripsky T., Froelich O., Komoin-Oka C., Westlund B., Sluder A., Maser P. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Gauvry N., Schorderet Weber S., Skripsky T., Bouvier J., Wenger A., Schroeder F., Desaules Y., Hotz R., Goebel T., Hosking B.C., Pautrat F., Wieland-Berghausen S., Ducray P. Identification of the amino-acetonitrile derivative monepantel (AAD 1566) as a new anthelmintic drug development candidate. Parasitol. Res. 2008;103:931–939. doi: 10.1007/s00436-008-1080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kashyap S.S., Verma S., Voronin D., Lustigman S., Kulke D., Robertson A.P., Martin R.J. Emodepside has sex-dependent immobilizing effects on adult Brugia malayi due to a differentially spliced binding pocket in the RCK1 region of the SLO-1 K channel. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen S., Ratnappan R., Han S., Leasure C., Grill E., Iqbal Z., Granger O., O’Halloran D.M., Hawdon J.M. Isolation and characterization of a naturally occurring multidrug-resistant strain of the canine hookworm, Ancylostoma caninum. Int. J. Parasitol. 2019;49(5):397–406. doi: 10.1016/j.ijpara.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp S.R., Coleman G.T., Traub R.J., McCarthy J.S., Kotze A.C. Acetylcholine receptor subunit genes from Ancylostoma caninum: altered transcription patterns associated with pyrantel resistance. Int. J. Parasitol. 2009;39:435–441. doi: 10.1016/j.ijpara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Hunt P.W., Skuce P., von Samson-Himmelstjerna G., Martin R.J., Sager H., Krücken J., Hodgkinson J., Lespine A., Jex A.R., Gilleard J.S., Beech R.N., Wolstenholme A.J., Demeler J., Robertson A.P., Charvet C.L., Neveu C., Kaminsky R., Rufener L., Alberich M., Menez C., Prichard R.K. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int. J. Parasitol. Drugs Drug Resist. 2014;4:164–184. doi: 10.1016/j.ijpddr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krücken J., Harder A., Jeschke P., Holden-Dye L., O'Connor V., Welz C., von Samson-Himmelstjerna G. Anthelmintic cyclcooctadepsipeptides: complex in structure and mode of action. Trends Parasitol. 2012;28:385–394. doi: 10.1016/j.pt.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Krücken J., Fraundorfer K., Mugisha J.C., Ramunke S., Sifft K.C., Geus D., Habarugira F., Ndoli J., Sendegeya A., Mukampunga C., Bayingana C., Aebischer T., Demeler J., Gahutu J.B., Mockenhaupt F.P., von Samson-Himmelstjerna G. Reduced efficacy of albendazole against Ascaris lumbricoides in Rwandan schoolchildren. Int. J. Parasitol. Drugs Drug Resist. 2017;7:262–271. doi: 10.1016/j.ijpddr.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulke D., von Samson-Himmelstjerna G., Miltsch S.M., Wolstenholme A.J., Jex A.R., Gasser R.B., Ballesteros C., Geary T.G., Keiser J., Townson S., Harder A., Krücken J. Characterization of the Ca2+-gated and voltage-dependent K+-channel Slo-1 of nematodes and its interaction with emodepside. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Schiffer P.H., Blaxter M. 959 Nematode Genomes: a semantic wiki for coordinating sequencing projects. Nucleic Acids Res. 2012;40:D1295–D1300. doi: 10.1093/nar/gkr826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Lacey E. The role of the cytoskeletal protein, tubulin, in the mode of action and mechanism of drug resistance to benzimidazoles. Int. J. Parasitol. 1988;18:885–936. doi: 10.1016/0020-7519(88)90175-0. [DOI] [PubMed] [Google Scholar]
- Lai Y., Xiang M., Liu S., Li E., Che Y., Liu X. A novel high-throughput nematicidal assay using embryo cells and larvae of Caenorhabditis elegans. Exp. Parasitol. 2014;139:33–41. doi: 10.1016/j.exppara.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Law W., Wuescher L.M., Ortega A., Hapiak V.M., Komuniecki P.R., Komuniecki R. Heterologous expression in remodeled C. elegans: a platform for monoaminergic agonist identification and anthelmintic screening. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecova L., Stuchlikova L., Prchal L., Skalova L. Monepantel: the most studied new anthelmintic drug of recent years. Parasitology. 2014;141:1686–1698. doi: 10.1017/S0031182014001401. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Shih P.Y., Schaedel O.N., Quintero-Cadena P., Rogers A.K., PaulSternberg WP.W. FMRFamide-like peptides expand the behavioral repertoire of a densely connected nervous system. Proc Natl Acad Sci U S A. 2017;114(50):E10726–E10735. doi: 10.1073/pnas.1710374114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespine A., Menez C., Bourguinat C., Prichard R.K. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: prospects for reversing transport-dependent anthelmintic resistance. Int. J. Parasitol. Drugs Drug Resist. 2012;2:58–75. doi: 10.1016/j.ijpddr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.A., Wu C.H., Berg H., Levine J.H. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Martin R.J., Dong L. Micro-electro-fluidic grids for nematodes: a lens-less, image-sensor-less approach for on-chip tracking of nematode locomotion. Lab Chip. 2013;13:650–661. doi: 10.1039/c2lc41174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Landuyt B., Klaassen H., Geldhof P., Luyten W. Screening of a drug repurposing library with a nematode motility assay identifies promising anthelmintic hits against Cooperia oncophora and other ruminant parasites. Vet. Parasitol. 2019;265:15–18. doi: 10.1016/j.vetpar.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Lockery S.R., Hulme S.E., Roberts W.M., Robinson K.J., Laromaine A., Lindsay T.H., Whitesides G.M., Weeks J.C. A microfluidic device for whole-animal drug screening using electrophysiological measures in the nematode C. elegans. Lab Chip. 2012;12:2211–2220. doi: 10.1039/c2lc00001f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok J.B. CRISPR/Cas9 mutagenesis and expression of dominant mutant transgenes as functional genomic approaches in parasitic nematodes. Front. Genet. 2019;10:656. doi: 10.3389/fgene.2019.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok J.B., Shao H., Massey H.C., Li X. Transgenesis in Strongyloides and related parasitic nematodes: historical perspectives, current functional genomic applications and progress towards gene disruption and editing. Parasitology. 2017;144:327–342. doi: 10.1017/S0031182016000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubega G.W., Prichard R.K. Specific interaction of benzimidazole anthelmintics with tubulin: high-affinity binding and benzimidazole resistance in Haemonchus contortus. Mol. Biochem. Parasitol. 1990;38:221–232. doi: 10.1016/0166-6851(90)90025-h. [DOI] [PubMed] [Google Scholar]
- Lustigman S., Prichard R.K., Gazzinelli A., Grant W.N., Boatin B.A., McCarthy J.S., Basanez M.G. A research agenda for helminth diseases of humans: the problem of helminthiases. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.J., Verma S., Levandoski M., Clark C.L., Qian H., Stewart M., Robertson A.P. Drug resistance and neurotransmitter receptors of nematodes: recent studies on the mode of action of levamisole. Parasitology. 2005;131(Suppl. l):S71–S84. doi: 10.1017/S0031182005008668. [DOI] [PubMed] [Google Scholar]
- Mathew M.D., Mathew N.D., Miller A., Simpson M., Au V., Garland S., Gestin M., Edgley M.L., Flibotte S., Balgi A., Chiang J., Giaever G., Dean P., Tung A., Roberge M., Roskelley C., Forge T., Nislow C., Moerman D. Using C. elegans forward and reverse genetics to identify new compounds with anthelmintic activity. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova P., Vokral I., Lamka J., Skalova L. The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 2016;32:481–491. doi: 10.1016/j.pt.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Matouskova P., Lecova L., Laing R., Dimunova D., Vogel H., Raisova Stuchlikova L., Nguyen L.T., Kellerova P., Vokral I., Lamka J., Szotakova B., Varady M., Skalova L. UDP-glycosyltransferase family in Haemonchus contortus: phylogenetic analysis, constitutive expression, sex-differences and resistance-related differences. Int. J. Parasitol. Drugs Drug Resist. 2018;8:420–429. doi: 10.1016/j.ijpddr.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A.G., Geary T.G., Bowman J.W., Marks N.J., Blair K.L., Halton D.W., Shaw C., Thompson D.P. Inhibitory effects of nematode FMRFamide-related peptides (FaRPs) on muscle strips from Ascaris suum. Invertebr. Neurosci. 1995;1:255–265. doi: 10.1007/BF02211027. [DOI] [PubMed] [Google Scholar]
- Maule A.G., McVeigh P., Dalzell J.J., Atkinson L., Mousley A., Marks N.J. An eye on RNAi in nematode parasites. Trends Parasitol. 2011;27:505–513. doi: 10.1016/j.pt.2011.07.004. [DOI] [PubMed] [Google Scholar]
- McCoy C.J., Warnock N.D., Atkinson L.E., Atcheson E., Martin R.J., Robertson A.P., Maule A.G., Marks N.J., Mousley A. RNA interference in adult Ascaris suum--an opportunity for the development of a functional genomics platform that supports organism-, tissue- and cell-based biology in a nematode parasite. Int. J. Parasitol. 2015;45:673–678. doi: 10.1016/j.ijpara.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDiarmid T.A., Au V., Loewen A.D., Liang J., Mizumoto K., Moerman D.G., Catharine H Rankin C.H. CRISPR-Cas9 human gene replacement and phenomic characterization in Caenorhabditis elegans to understand the functional conservation of human genes and decipher variants of uncertain significance. Dis Model Mech. 2018;11(12) doi: 10.1242/dmm.036517. dmm036517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVeigh P., Atkinson L., Marks N.J., Mousley A., Dalzell J.J., Sluder A., Hammerland L., Maule A.G. Parasite neuropeptide biology: seeding rational drug target selection? Int. J. Parasitol. Drugs Drug Resist. 2012;2:76–91. doi: 10.1016/j.ijpddr.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani H., Komai T., Mizuno D. Antitumor and antiviral effect of riboside of benzimidazole derivatives. Jpn. J. Med. Sci. Biol. 1960;13:147–153. doi: 10.7883/yoken1952.13.147. [DOI] [PubMed] [Google Scholar]
- Moreno Y., Nabhan J.F., Solomon J., Mackenzie C.D., Geary T.G. Ivermectin disrupts the function of the excretory-secretory apparatus in microfilariae of Brugia malayi. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20120–20125. doi: 10.1073/pnas.1011983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Frokjaer-Jensen C. Vol. 212. 2019. pp. 959–990. (The Caenorhabditis elegans Transgenic Toolbox). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niciura S.C.M., Tizioto P.C., Moraes C.V., Cruvinel G.G., de Albuquerque A.C.A., Santana R.C.M., Chagas A.C.S., Esteves S.N., Benavides M.V., do Amarante A.F.T. Extreme-QTL mapping of monepantel resistance in Haemonchus contortus. Parasites Vectors. 2019;12:403. doi: 10.1186/s13071-019-3663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet A.J., Zarlenga D.S., Knox D.P., Meikle L.I., Wildblood L.A., Matthews J.B. A calcium-activated apyrase from Teladorsagia circumcincta: an excretory/secretory antigen capable of modulating host immune responses? Parasite Immunol. 2011;33:236–243. doi: 10.1111/j.1365-3024.2011.01278.x. [DOI] [PubMed] [Google Scholar]
- Njue A.I., Hayashi J., Kinne L., Feng X.P., Prichard R.K. Mutations in the extracellular domains of glutamate-gated chloride channel alpha3 and beta subunits from ivermectin-resistant Cooperia oncophora affect agonist sensitivity. J. Neurochem. 2004;89:1137–1147. doi: 10.1111/j.1471-4159.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- Partridge F.A., Brown A.E., Buckingham S.D., Willis N.J., Wynne G.M., Forman R., Else K.J., Morrison A.A., Matthews J.B., Russell A.J., Lomas D.A., Sattelle D.B. An automated high-throughput system for phenotypic screening of chemical libraries on C. elegans and parasitic nematodes. Int. J. Parasitol. Drugs Drug Resist. 2018;8:8–21. doi: 10.1016/j.ijpddr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peachey L.E., Pinchbeck G.L., Matthews J.B., Burden F.A., Lespine A., von Samson-Himmelstjerna G., Krücken J., Hodgkinson J.E. P-glycoproteins play a role in ivermectin resistance in cyathostomins. Int. J. Parasitol. Drugs Drug Resist. 2017;7:388–398. doi: 10.1016/j.ijpddr.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phiri A.M., Di D.E.P., Buttle D.J., Behnke J.M. A novel assay for the detection of anthelmintic activity mediated by cuticular damage to nematodes: validation on Caenorhabditis elegans exposed to cysteine proteinases. Parasitology. 2017;144:583–593. doi: 10.1017/S0031182016002353. [DOI] [PubMed] [Google Scholar]
- Prichard R.K. Anthelmintic resistance in nematodes: extent, recent understanding and future directions for control and research. Int. J. Parasitol. 1990;20:515–523. doi: 10.1016/0020-7519(90)90199-w. [DOI] [PubMed] [Google Scholar]
- Prichard R.K., Basáñez M.G., Boatin B.A., McCarthy J.S., García H.H., Yang G.J., Sripa B., Lustigman S. A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS Neglected Trop. Dis. 2012;6:e1549. doi: 10.1371/journal.pntd.0001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quist C.W., Smant G., Helder J. Evolution of plant parasitism in the phylum Nematoda. Annu. Rev. Phytopathol. 2015;53:289–310. doi: 10.1146/annurev-phyto-080614-120057. [DOI] [PubMed] [Google Scholar]
- Risi G., Aguilera E., Lados E., Suarez G. Vol. 6. 2019. (Caenorhabditis elegans Infrared-Based Motility Assay Identified New Hits for Nematicide Drug Development). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener L., Keiser J., Kaminsky R., Maser P., Nilsson D. Phylogenomics of ligand-gated ion channels predicts monepantel effect. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lancheros E., Viau C., Walter T.N., Francis A., Geary T.G. Activity of novel nicotinic anthelmintics in cut preparations of Caenorhabditis elegans. Int. J. Parasitol. 2011;41:455–461. doi: 10.1016/j.ijpara.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Salinas G., Risi G. Caenorhabditis elegans: nature and nurture gift to nematode parasitologists. Parasitology. 2018;145:979–987. doi: 10.1017/S0031182017002165. [DOI] [PubMed] [Google Scholar]
- Sarai R.S., Kopp S.R., Coleman G.T., Kotze A.C. Acetylcholine receptor subunit and P-glycoprotein transcription patterns in levamisole-susceptible and -resistant Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2013;3:51–58. doi: 10.1016/j.ijpddr.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai R.S., Kopp S.R., Coleman G.T., Kotze A.C. Drug-efflux and target-site gene expression patterns in Haemonchus contortus larvae able to survive increasing concentrations of levamisole in vitro. Int. J. Parasitol. Drugs Drug Resist. 2014;4:77–84. doi: 10.1016/j.ijpddr.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Takagi M., Yaguchi T., Miyadoh S., Okada T., Koyama M. A new anthelmintic cyclodepsipeptide, PF1022A. J. Antibiot. (Tokyo) 1992;45:692–697. doi: 10.7164/antibiotics.45.692. [DOI] [PubMed] [Google Scholar]
- Schwab A.E., Boakye D.A., Kyelem D., Prichard R.K. Detection of benzimidazole resistance-associated mutations in the filarial nematode Wuchereria bancrofti and evidence for selection by albendazole and ivermectin combination treatment. Am. J. Trop. Med. Hyg. 2005;73:234–238. [PubMed] [Google Scholar]
- Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A., Hall R.S., Mondal A., Howe A.C., Pell J., Hofmann A., Boag P.R., Zhu X.Q., Gregory T., Loukas A., Williams B.A., Antoshechkin I., Brown C., Sternberg P.W., Gasser R.B. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoop W.L., Egerton J.R., Eary C.H., Suhayda D. Anthelmintic activity of paraherquamide in sheep. J. Parasitol. 1990;76:349–351. [PubMed] [Google Scholar]
- Simpkin K.G., Coles G.C. The use of Caenorhabditis elegans for anthelmintic screening. J. Chem. Technol. Biotechnol. 1981;31:66–69. [Google Scholar]
- Sloan M.A., Reaves B.J., Maclean M.J., Storey B.E., Wolstenholme A.J. Expression of nicotinic acetylcholine receptor subunits from parasitic nematodes in Caenorhabditis elegans. Mol. Biochem. Parasitol. 2015;204:44–50. doi: 10.1016/j.molbiopara.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spensley M., Del Borrello S., Pajkic D., Fraser A.G. Acute Effects of Drugs on Caenorhabditis elegans Movement Reveal Complex Responses and Plasticity. G3 (Bethesda) 2018;8(9):2941–2952. doi: 10.1534/g3.118.200374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken M.G., Snoek L.B., Kammenga J.E., Andersen E.C. The laboratory domestication of Caenorhabditis elegans. Trends Genet. TIG (Trends Genet.) 2015;31:224–231. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambo E., Khater E.I., Chen J.H., Bergquist R., Zhou X.N. Nobel prize for the artemisinin and ivermectin discoveries: a great boost towards elimination of the global infectious diseases of poverty. Infect. Dis. Poverty. 2015;4:58. doi: 10.1186/s40249-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I., Folkers K., Horsfall F.L., Jr. Inhibition of influenza virus multiplication by alkyl derivatives of benzimidazole. I. Kinetic aspects of inhibition by 2,5-dimethylbenzimidazole as measured by infectivity titrations. J. Exp. Med. 1953;98:219–227. doi: 10.1084/jem.98.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. The efficacy of amidantel, a new anthelmintic, on hookworms and ascarids in dogs. Tropenmed. Parasitol. 1979;30:404–408. [PubMed] [Google Scholar]
- Thomas J.H., Neff N.F., Botstein D. Isolation and characterization of mutations in the beta-tubulin gene of Saccharomyces cerevisiae. Genetics. 1985;111:715–734. doi: 10.1093/genetics/111.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto B., Cortada L., Murungi L.K., Haukeland S., Coyne D.L. Management of cyst and root knot nematodes: a chemical ecology perspective. J. Agric. Food Chem. 2018;66:8672–8678. doi: 10.1021/acs.jafc.8b01940. [DOI] [PubMed] [Google Scholar]
- Towers P.R., Edwards B., Richmond J.E., Sattelle D.B. The Caenorhabditis elegans lev-8 gene encodes a novel type of nicotinic acetylcholine receptor alpha subunit. J. Neurochem. 2005;93(1):1–9. doi: 10.1111/j.1471-4159.2004.02951.x. [DOI] [PubMed] [Google Scholar]
- Trap C., Fu B., Le Guerhier F., Liu M., Le Rhun D., Romand T., Perret C., Blaga R., Boireau P. Cloning and analysis of a cDNA encoding a putative serine protease comprising two trypsin-like domains of Trichinella spiralis. Parasitol. Res. 2006;98:288–294. doi: 10.1007/s00436-005-0075-x. [DOI] [PubMed] [Google Scholar]
- Trudgill D.L., Blok V.C. Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 2001;39:53–77. doi: 10.1146/annurev.phyto.39.1.53. [DOI] [PubMed] [Google Scholar]
- Turner H.C., Bettis A.A., Chu B.K., McFarland D.A., Hooper P.J., Ottesen E.A., Bradley M.H. The health and economic benefits of the global programme to eliminate lymphatic filariasis (2000-2014) Infect. Dis. Poverty. 2016;5:54. doi: 10.1186/s40249-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk J.A., Malan F.S. Resistance of field strains of Haemonchus contortus to ivermectin, closantel, rafoxanide and the benzimidazoles in South Africa. Vet. Rec. 1988;123:226–228. doi: 10.1136/vr.123.9.226. [DOI] [PubMed] [Google Scholar]
- Verma S., Kashyap S.S., Robertson A.P., Martin R.J. Functional genomics in Brugia malayi reveal diverse muscle nAChRs and differences between cholinergic anthelmintics. Proc. Natl. Acad. Sci. U.S.A. 2017;114:5539–5544. doi: 10.1073/pnas.1619820114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney M. The genomic basis of nematode parasitism. Brief Funct Genomics. 2018;17:8–14. doi: 10.1093/bfgp/elx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokral I., Jirasko R., Stuchlikova L., Bartikova H., Szotakova B., Lamka J., Varady M., Skalova L. Biotransformation of albendazole and activities of selected detoxification enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Vet. Parasitol. 2013;196:373–381. doi: 10.1016/j.vetpar.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Ward J.D. Rendering the intractable more tractable: tools from Caenorhabditis elegans ripe for import into parasitic nematodes. Genetics. 2015;201:1279–1294. doi: 10.1534/genetics.115.182717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J.C., Roberts W.M., Robinson K.J., Keaney M., Vermeire J.J., Urban J.F., Jr., Lockery S.R., Hawdon J.M. Microfluidic platform for electrophysiological recordings from host-stage hookworm and Ascaris suum larvae: a new tool for anthelmintic research. Int. J. Parasitol. Drugs Drug Resist. 2016;6:314–328. doi: 10.1016/j.ijpddr.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J.C., Robinson K.J., Lockery S.R., Roberts W.M. Anthelmintic drug actions in resistant and susceptible C. elegans revealed by electrophysiological recordings in a multichannel microfluidic device. Int. J. Parasitol. Drugs Drug Resist. 2018;8:607–628. doi: 10.1016/j.ijpddr.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz C., Krüger N., Schniederjans M., Miltsch S.M., Krücken J., Guest M., Holden-Dye L., Harder A., von Samson-Himmelstjerna G. SLO-1-channels of parasitic nematodes reconstitute locomotor behaviour and emodepside sensitivity in Caenorhabditis elegans slo-1 loss of function mutants. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker J.H., Carlson S.A., Jones D.E., Brewer M.T. Molecular mechanisms for anthelmintic resistance in strongyle nematode parasites of veterinary importance. J. Vet. Pharmacol. Therapeut. 2017;40:105–115. doi: 10.1111/jvp.12330. [DOI] [PubMed] [Google Scholar]
- Williamson A.L., Brindley P.J., Knox D.P., Hotez P.J., Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;19:417–423. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Williamson A.L., Lustigman S., Oksov Y., Deumic V., Plieskatt J., Mendez S., Zhan B., Bottazzi M.E., Hotez P.J., Loukas A. Ancylostoma caninum MTP-1, an astacin-like metalloprotease secreted by infective hookworm larvae, is involved in tissue migration. Infect. Immun. 2006;74:961–967. doi: 10.1128/IAI.74.2.961-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W.R., Brugman K.I., Maher S., Oh J.Y., Howe K., Kato M., Sternberg P.W. Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum. Mol. Genet. 2019;28:2271–2281. doi: 10.1093/hmg/ddz051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.J., Lauret C., Geary T. Anthelmintic discovery and development in the animal health industry. Expet Opin. Drug Discov. 2007;2:S25–S33. doi: 10.1517/17460441.2.S1.S25. [DOI] [PubMed] [Google Scholar]
- Wright J.B. The chemistry of the benzimidazoles. Chem. Rev. 1951;48:397–541. [PubMed] [Google Scholar]
- Xiao S.H., Hui-Ming W., Tanner M., Utzinger J., Chong W. Tribendimidine: a promising, safe and broad-spectrum anthelmintic agent from China. Acta Trop. 2005;94:1–14. doi: 10.1016/j.actatropica.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Xiong H., Pears C., Woollard A. An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 2017;7:9839. doi: 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz E., Kulke D., von Samson-Himmelstjerna G., Krücken J. Identification of novel splice variants of the voltage- and Ca(2)(+)-dependent K(+)-channel SLO-1 of Trichuris muris. Mol. Biochem. Parasitol. 2015;199:5–8. doi: 10.1016/j.molbiopara.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Yilmaz E., Ramunke S., Demeler J., Krücken J. Comparison of constitutive and thiabendazole-induced expression of five cytochrome P450 genes in fourth-stage larvae of Haemonchus contortus isolates with different drug susceptibility identifies one gene with high constitutive expression in a multi-resistant isolate. Int. J. Parasitol. Drugs Drug Resist. 2017;7:362–369. doi: 10.1016/j.ijpddr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Andersen E.C. Prospects and challenges of CRISPR/Cas genome editing for the study and control of neglected vector-borne nematode diseases. FEBS J. 2016;283:3204–3221. doi: 10.1111/febs.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Cook D.E., Zdraljevic S., Brady S.C., Lee D., Lee J., Andersen E.C. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Strand C., Seidel H.S., Cook D.E., Doench J.G., Andersen E.C. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 2017;13(7) doi: 10.1371/journal.pgen.1006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E.W., Wolf M.L., Alexander-Bowman S.J., Thomas E.M., Davis J.P., Groppi V.E., Lee B.H., Thompson D.P., Geary T.G. Anthelmintic paraherquamides are cholinergic antagonists in gastrointestinal nematodes and mammals. J. Vet. Pharmacol. Therapeut. 2002;25:241–250. doi: 10.1046/j.1365-2885.2002.00423.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.