Abstract
Background
Multiple organ dysfunction syndrome (MODS) occurs in the setting of a variety of pathologies including infection and trauma. Some patients decompensate and require Veno-Arterial extra corporeal membrane oxygenation (ECMO) as a palliating manoeuvre for recovery of cardiopulmonary function. The molecular mechanisms driving progression from MODS to cardiopulmonary collapse remain incompletely understood, and no biomarkers have been defined to identify those MODS patients at highest risk for progression to requiring ECMO support.
Methods
Whole blood RNA-seq profiling was performed for 23 MODS patients at three time points during their ICU stay (at diagnosis of MODS, 72 hours after, and 8 days later), as well as four healthy controls undergoing routine sedation. Of the 23 MODS patients, six required ECMO support (ECMO patients). The predictive power of conventional demographic and clinical features was quantified for differentiating the MODS and ECMO patients. We then compared the performance of markers derived from transcriptomic profiling including [1] transcriptomically imputed leukocyte subtype distribution, [2] relevant published gene signatures and [3] a novel differential gene expression signature computed from our data set. The predictive power of our novel gene expression signature was then validated using independently published datasets.
Finding
None of the five demographic characteristics and 14 clinical features, including The Paediatric Logistic Organ Dysfunction (PELOD) score, could predict deterioration of MODS to ECMO at baseline. From previously published sepsis signatures, only the signatures positively associated with patient's mortality could differentiate ECMO patients from MODS patients, when applied to our transcriptomic dataset (P-value ranges from 0.01 to 0.04, Student's test). Deconvolution of bulk RNA-Seq samples suggested that lower neutrophil counts were associated with increased risk of progression from MODS to ECMO (P-value = 0.03, logistic regression, OR=2.82 [95% CI 0.63 - 12.45]). A total of 30 genes were differentially expressed between ECMO and MODS patients at baseline (log2 fold change ≥ 1 or ≤ -1 with false discovery rate ≤ 0.01). These genes are involved in protein maintenance and epigenetic-related processes. Further univariate analysis of these 30 genes suggested a signature of seven DE genes associated with ECMO (OR > 3.0, P-value ≤ 0.05, logistic regression). Notably, this contains a set of histone marker genes, including H1F0, HIST2H3C, HIST1H2AI, HIST1H4, HIST1H2BL and HIST1H1B, that were highly expressed in ECMO. A risk score derived from expression of these genes differentiated ECMO and MODS patients in our dataset (AUC = 0.91, 95% CI 0.79-1.00, P-value = 7e-04, logistic regression) as well as validation dataset (AUC= 0.73, 95% CI 0.53-0.93, P-value = 2e-02, logistic regression).
Interpretation
This study demonstrates that transcriptomic features can serve as indicators of severity that could be superior to traditional methods of ascertaining acuity in MODS patients. Analysis of expression of signatures identified in this study could help clinicians in the diagnosis and prognostication of MODS patients after arrival to the Hospital.
Keywords: Multiple organ dysfunction syndrome, Veno-arterial extracorporeal membrane oxygenation, Paediatric intensive care unit, RNA-seq in paediatrics, Biomarker discovery
Abbreviation
- Multiple organ dysfunction syndrome
MODS
- Veno-Arterial Extracorporeal Membrane Oxygenation
ECMO
- patients did not develop MODS
no-MODS
- paediatric intensive care unit
PICU
- Differentially expressed
DE
- False discovery rate
FDR
- Area under curve
AUC
- principal component analysis
PCA
- Odds ratio
OR.
1. Introduction
Multiple organ dysfunction syndrome (MODS) is common in the paediatric intensive care unit (PICU), being diagnosed in the majority of patients with sepsis as well as many trauma patients [1]. MODS complicates a wide range of pathologies included severe hypoxemia and cardiorespiratory arrest [2]. Contemporary management of MODS is entirely supportive, and focused on addressing the underlying disease process.
The ICU course of MODS patients is very variable and not entirely dependent on the presenting symptoms. Some paediatric patients who develop MODS deteriorate and require intensive life support in the form of Veno-Arterial Extracorporeal Membrane Oxygenation (ECMO). It has been observed that paediatric patients requiring Veno-Arterial ECMO support (ECMO) have a 50-60% mortality rate [3]. Since no clinical scoring tool or molecular biomarker currently exists to identify the patients who may require advanced life support, the decision to initiate ECMO remains subjective based on the empirical experience of the multidisciplinary care team and criteria provided by the Extracorporeal Life Support Organization (ELSO) [4]. ELSO recommends ECMO only after less invasive measures such as high frequency ventilation, inhaled Nitric Oxide or prone positioning have failed [4]. In the paediatric population, there is a dire need for objective markers that predict the need for aggressive supportive measures such as ECMO that could simplify the decision-making process and enable earlier intervention for these patients. Therefore, developing biomarkers for identifying MODS patients at high risk of requiring ECMO support remains an unmet need.
Whole blood transcriptomic profiling has been evaluated to perform risk-stratification of sepsis patients, predict mortality in sepsis and better understand the pathogenesis of MODS [5]. A number of published gene expression signatures shed some light on the molecular mechanism of MODS [6,7]. However, none of the signatures were developed with a view towards identifying MODS patients that require ECMO support.
In this work, we present a cohort of healthy controls (CT) and MODS patients including a subset of whom progressed to requiring ECMO support (MODS vs ECMO). Here we use the term MODS to denote those MODS patients that did not require ECMO and ECMO for MODS patients deteriorated to needing Veno-Arterial ECMO support. These patients were assessed using a combination of conventional demographic and clinical markers, as well as whole blood transcriptomic profiling in an effort to identify diagnostic markers that can distinguish between the MODS and ECMO patient population.
2. Methods
2.1. Ethics
The IRB of this study (2016-062-SH/HDVCH) was approved by Spectrum Health, Grand Rapids, Michigan, on May 17, 2016. All the patients were minors and their parents were consented prior to recruitment into our study. After IRB approval, only deidentified data was used to adopt a short-term longitudinal design to assess the transcriptomic profiles of patients from the PICU at Helen DeVos Children's Hospital, Michigan.
2.2. Patients and blood sampling
Critically ill patients meeting criteria for MODS were determined by clinical observations that were first described by Proulx et al., 1996 [8] and recently used in the ABC PICU trial [9]. Once patients met criteria they were screened for eligibility and consented. Blood samples were collected at three time points: at recognition of MODS (0h), 72 hours after, and 8 days later (N=27). Samples were collected in PaxGene® tubes and stored at −80 °C. Healthy controls (N=4) were patients that presented for same day sedation. Samples from each control patient were obtained only once and were reported as 0h. Of the 23 MODS patients, 6 required Veno-Arterial ECMO support (“ECMO patients”). From admission to day 8, 47% of the MODS patients were discharged to home or out of the ICU to a medical floor. Patients who left the ICU did not have further blood draws. One patient from the ECMO group died during the study and two other MODS patients died six months later. Patients presenting with MODS are limited, so we consented and used all the MODS patients available in hospital during study period.
2.3. Sequencing
RNA samples were prepared using KAPA RNA HyperPrep Kit, and sequenced on an Illumina NextSeq500. Using ribosomal reduction RNAseq methodology, we were able to capture both cellular and acellular RNA signatures of all PICU patients.
2.4. Validation datasets
For validation, we were unable to identify any analogous publicly available gene expression datasets that included paediatric MODS patients with measurements at multiple timepoints. There were many datasets of paediatric patients with sepsis but none that MODS patients that progressed onto ECMO. We therefore chose a dataset describing an adult cohort (23-63 years) that developed MODS in the hyperacute phase of trauma [10]. This dataset was used as an independent cohort to validate our signature genes. The MODS patients in this validation dataset were those patients, who require intensive care support (ICU) for their survival (similar to our ECMO patients) and those do not need ICU support were labelled as “noMODS” (similar to our MODS patients) as described in patient demographics [10]. In addition, a single cell RNA-Seq dataset was also available for adult ECMO patients [11]. We used the immune cell markers from this dataset to validate our immune response analysis.
2.5. Bioinformatics analysis
2.5.1. RNA-Seq data analysis
All the sequencing reads were mapped on Hg38 transcriptome using the ENSEMBL GRCh38.p3 annotation with the STAR aligner [12]. The edgeR package [13] was used for quantification of differentially expressed (DE) genes with criteria: log2 fold change ≥ 1 or ≤ -1 with adjusted P-value (False Discover Rate) < 0.01. DE genes were identified between the two groups in all the three-time points separately. The DE genes were used for co-expression network analysis using CEMiTools package [14]. The gene ontology (GO) enrichment of DE genes was performed using the clusterProfiler R package [15]. Biological processes with P-value ≤ 0.001 were considered as significantly enriched. Dotplot function provided in clusterProfiler was used to visualize enriched pathways. In addition, gene interaction network was visualized using STRING: functional protein association network (https://string-db.org/).
2.5.2. Immune cell deconvolution
CIBERSORT was used to estimate the relative composition of immune cells in bulk RNA-Seq samples [16] using a machine learning model named as nu–support vector regression (ν-SVR) [17]. For each patient, a complete blood count (CBC) was obtained upon presentation as part of their standard of care clinical evaluation. We were therefore able to calculate estimated absolute counts for each leukocyte subpopulation. This was done by multiplying the proportion for each subpopulation as determined by CIBERSORT to the total white blood cell count from the CBC. This analysis was validated by comparing the absolute neutrophil counts (ANC) as estimated by CIBERSORT with the ANC reported by the clinical laboratory. In addition, we obtained gene markers specific to different immune cells and pathways important for MODS patients described previously [18]. Genes specific to immune cells and pathways were used for gene set enrichment analysis (ssGSEA) in each sample using GSVA package [19]. The ssGSEA score for each cell represents the enrichment of that cell markers in each sample.
2.5.3. Statistical analyses
All plots and statistical analyses were carried out using R programming language (v3.5.1) (https://www.r-project.org/). By default, two-sided student's t-test was performed to compute the significance between two groups (either Control vs MODS or MODS vs ECMO). For categorical variables, Fisher's exact test was used to compute association and Pearson correlation method was used to compute the correlation between continuous variables. The generalized linear model function (glm) was used to calculate odds ratio (OR, 95% CI, MODS as the reference). Principal component analysis (PCA) of gene expression profiles was performed using the prcomp function. The risk score was estimated using the signature gene expression for each patient based on the geometric mean [5,[20], [21], [22]]. The geometric mean for x1, x2, ..., xn was calculated as follows:
A risk score was further used to re-classify patients into two groups and receiver operating characteristic (ROC) and area under curve (AUC) were adopted to assess the performance using the pROC package [23].
3. Results
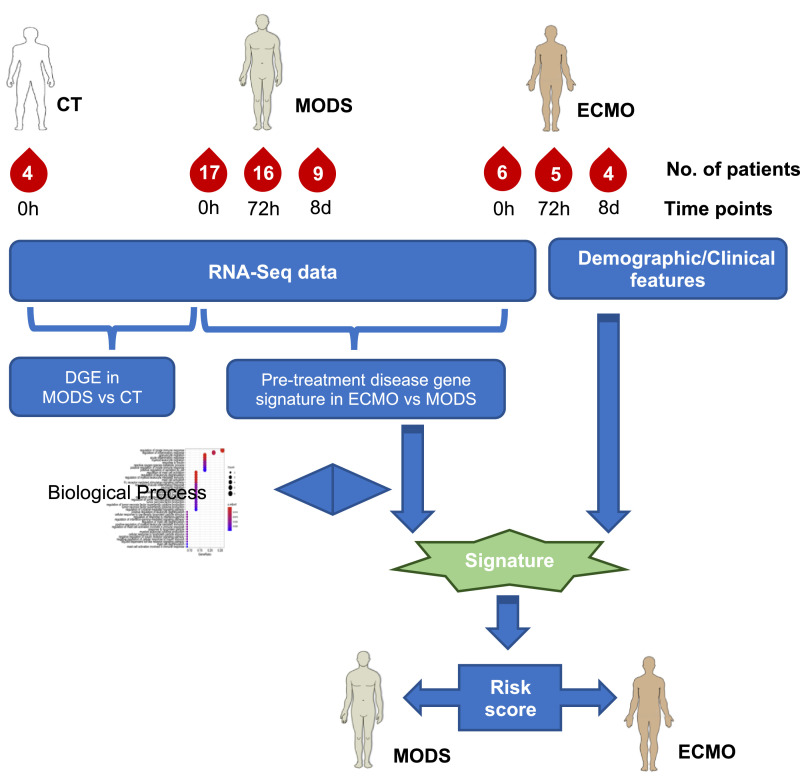
The workflow of the study is summarized in Fig. 1. Patient demographics and baseline clinical parameters are provided in Table 1, with 72h and 8 days values presented in supplemental Table S1. In total, five demographic characteristics (i.e., age, gender, BMI, weight, height) and 14 different clinical features were examined for all patients. There was high variation between MODS and ECMO for many clinical parameters (e.g., platelet count), diluting the predictive power of these measures.
Fig. 1.
An overview of the analysis. DGE: Differential gene expression.
Table 1.
Patient demographics at baseline (Pre-ECMO,0h) time point.
| RNA-Seq cohort | ||||
| Demographics | Control | MODS | ECMO | P-value |
| Time | 0h | 0h | 0h | - |
| Number | 4 | 17 | 6 | - |
| Age (months) | 84.75(28-122) | 90(0.14-202) | 63.25(0.5-202) | 0.54 |
| Male | 2 | 10 | 5 | 0.36 |
| Female | 2 | 7 | 1 | |
| BMI | 17(14-21) | 20.3(13-38.5) | 19(14-32.4) | 0.74 |
| Weight | 26.5(12-35) | 42.85(3.5-178) | 25.87(3.9-81) | 0.35 |
| Height | 122(80-142) | 103(51-157) | 90(53-160) | 0.59 |
| Mortality | - | 2 | 1 | |
| Clinical Features | ||||
| Liver Failure (%) | - | 30 | 50 | - |
| Bilirubin | - | 0.92(0.1-5.6) | 0.51(0.1-1.1) | 0.28 |
| AST | - | 258.88(13-3296) | 215.67(7-726) | 0.85 |
| Albumin | - | 2.35(1.6-3.5) | 2.35(1.9-2.8) | 0.96 |
| CRP | - | 83.75(0.3-234) | 75.75(2.8-211) | 0.87 |
| Renal Failure (%) | - | 89 | 100 | - |
| Creatinine | - | 0.65(0.13-0.29) | 0.77(0.22-0.29) | 0.71 |
| Lactate | - | 2.2(0.9-4.6) | 6.05(0.6-14.5) | 0.19 |
| WBC | - | 14.9(3.95-62.6) | 12.67(4.6-20) | 0.56 |
| platelet | - | 232(37-718) | 208(92-378) | 0.68 |
| PELOD Score | - | 14.37(1-32) | 12.5(10-20) | 0.21 |
| Bacterial infection (%) | - | 35 | 33 | |
| Viral infection (%) | - | 52 | 50 | |
| Inotrope usage | 88 | 100 | ||
| Respiratory failure (%) | - | 100 | 100 | |
| Neurological (%) | - | 23 | 33 | |
Where relevant, mean(range). T-test and fisher's exact test was used to compute P-value between MODS and ECMO.
Some outcomes differed significantly between MODS and ECMO, specifically the renal failure rate (89% in MODS and 100% in ECMO) and liver failure rate (30% in MODS and 50% in ECMO). However, no baseline demographic or clinical parameter, including PELOD score, was predictive of progression from MODS to ECMO. This observation highlighted the need to explore molecular features for identifying risk markers.
3.1. Immune cells deconvolution and transcriptome analysis
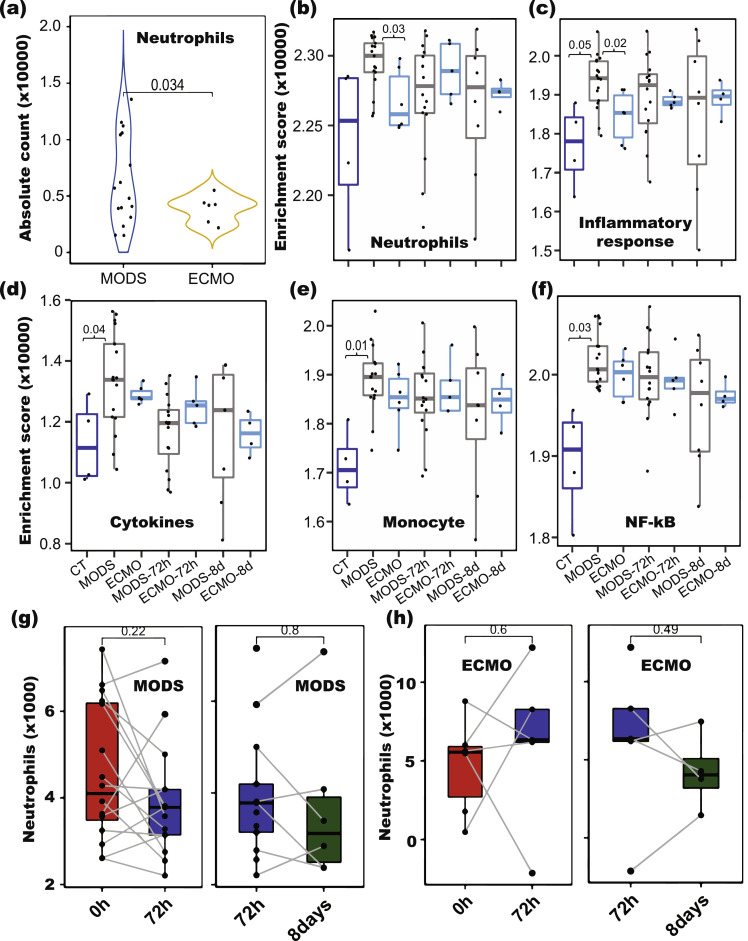
Immune responses were examined for individual patients and compared to elucidate their role. The relative proportions of immune cell subtypes were estimated using CIBERSORT based on bulk RNA-Seq data. WBC counts obtained upon arrival in the emergency department were used to quantify the absolute abundance of immune cell subtypes. The ANC as determined by the clinical laboratory and the ANC derived from CIBERSORT were high correlated (correlation value 0.85) (Figure S1), suggesting the high fidelity of the inferred leukocyte subtype composition. Comparison of neutrophils between ECMO and MODS showed decreased level in ECMO (P-value = 0.03, OR=2.82 [95% CI 0.63 – 12.45], logistic regression) as compared to MODS (Fig. 2a). Interestingly, the two lowest neutrophil counts were among MODS. Clinical data of these two patients revealed that one patient did not survive and another had the PELOD score of 32, the highest score among all patients, suggesting that these patients had a risk profile similar to the ECMO patients despite not being started on ECMO.
Fig. 2.
Immune cell composition analyses in ECMO and MODS patients. (a) Neutrophils counts computed from CIBERSORT decreased in ECMO (P = 0.034, Student's T-test) compared to MODS at baseline. (b-f) Enrichment of genes involved in various immune responses (Monocytes, Cytokines, NF-kB, Neutrophils and Inflammation) in CT, MODS and ECMO at different time points (0h, 72h and 8d). Abundance of neutrophils in MODS (g) and ECMO (h) patients at different time points (0h, 72h and 8days). Blue colour - control (CT), grey colour - MODS patients and cyan colour - ECMO patients, filled dark red colour-0h, filled blue colour-72h and filled green colour-8 days. Student's T-test was used to compute p values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We then examined the expression of marker genes of neutrophils (from CIBERSORT), monocytes, cytokines and genes involved in NF-kB and inflammatory response from Hall et al., 2007 [18]. All the marker genes were down-regulated in ECMO compared to MODS (Figure S2-S6). In addition to CIBERSORT, ssGSEA was performed on cell-type specific biomarker genes for each cell type and pathways in order to confirm the findings. Neutrophil cells (ssGSEA score) and inflammatory response pathway displayed significantly (P-value < 0.03, Student's T-test) decreased in ECMO compared to MODS (Fig. 2b and 2c). This observation is aligned to CIBERSORT results showing the decrease level of neutrophils. In contrast, marker genes pertaining to monocytes, cytokines, and NF-kB displayed a significant higher enrichment in MODS compared to CT (P-value < 0.04, Student's T-test) (Fig. 2d-2f).
The finding of changes in the neutrophil count was independently validated using additional single cell RNA-seq data of ECMO adult patients data [11], where we observed decrease of expression of neutrophil gene markers and genes involved in inflammatory response in deceased ECMO patients compared to patients that survived (Figure S7 and S8). Further, paired comparison of neutrophil levels for each patient showed no significant change across different time points (Fig. 2g and 2h).
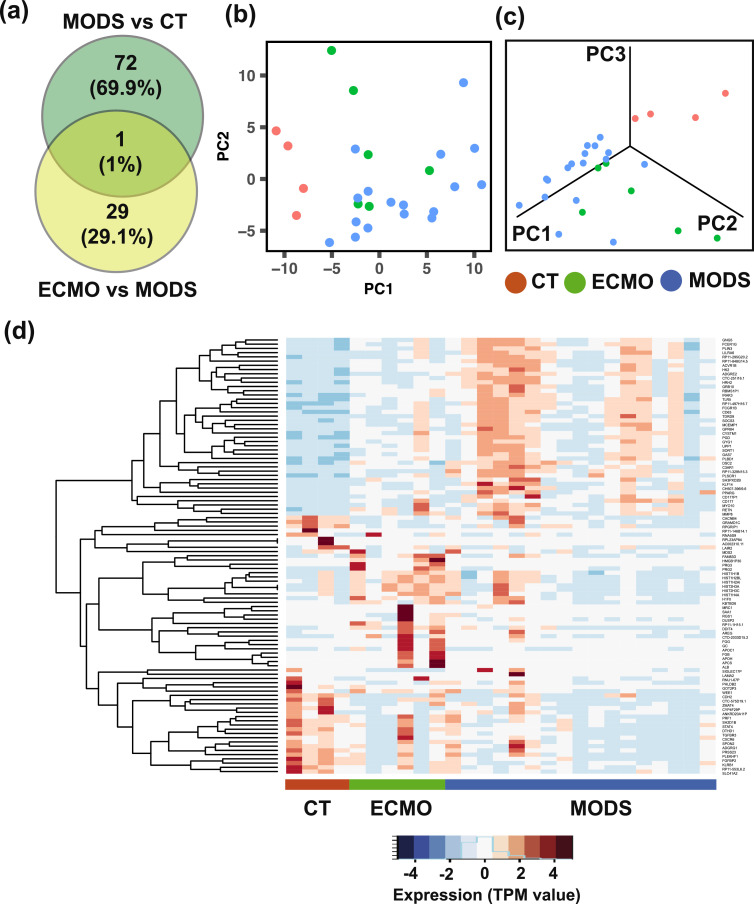
Furthermore, differential expression (DE) analysis between MODS and control (CT) as well as between ECMO and MODS was performed at baseline (0h). A total of 73 DE genes (log2 fold change ≥ 1 or ≤ -1 with FDR < 0.01) between MODS and CT, and 30 DE genes between ECMO and MODS were identified at baseline (Fig. 3a). Comparison of DE genes from these two groups showed only one pseudogene (RNU1-67P) common to these two DE lists. As expected, these DE genes clearly separate CT, MODS and ECMO patients (Fig. 3b and 3c) in reduced dimensional space. Heatmap visualization of these genes highlights their differential patterns of expression between groups (Fig. 3d). In addition, 56 and 33 DE genes between MODS and CT were identified at 72h and 8d time points, respectively (Figure S9a), while 7 and 2 DE genes were identified between ECMO and MODS at 72h and 8d time points, respectively (Figure S9b). Only one gene (pseudogene- RNU1-67P) was common among all the three time points (0h, 72h and 8d) in both comparisons.
Fig. 3.
Differential gene expression analyses at baseline (0h). (a) Comparison of differentially expressed (DE) genes between MODS vs. control (CT), and ECMO vs. MODS at baseline (0h). (b) First two principal component analysis and (c) first three principal component analysis, using the union of DE genes obtained from the comparison between MODS and CT, and that those between ECMO and MODS at baseline. Patients are clustered by their pathology group (CT, MODS and ECMO). (d) Expression of the DE genes.
3.2. Biological processes and co-expression networks regulated by DE genes
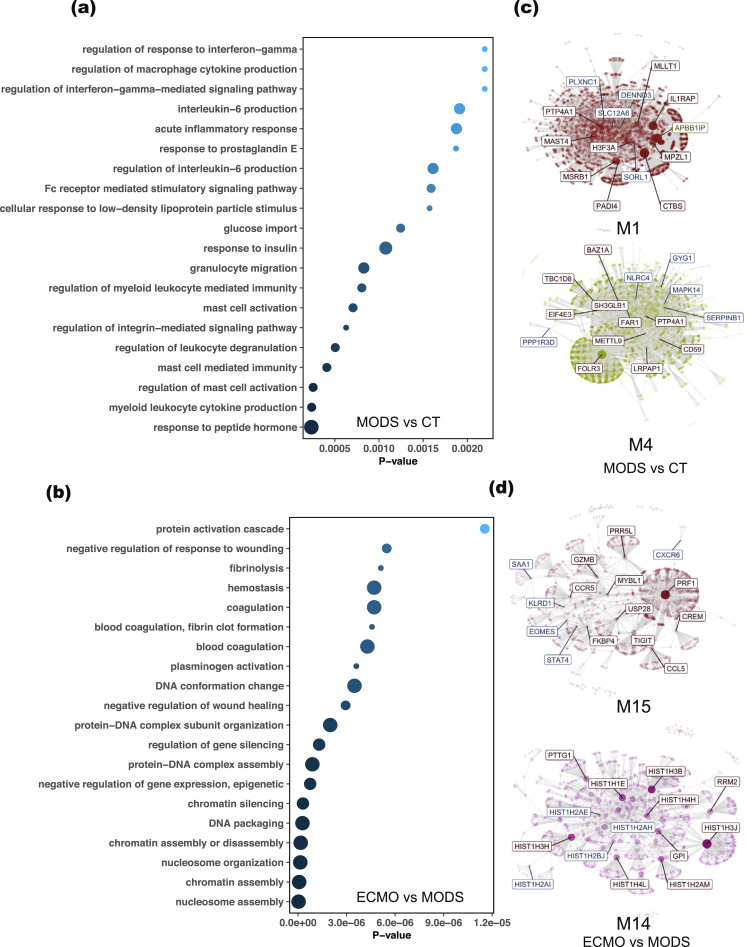
Gene ontology (GO) enrichment analysis of total DE genes from MODS to CT comparison revealed that immune-related (regulation of leukocytes degranulation, mast cell activation, acute inflammatory response, interleukin-6 production, and cytokine production) and glucose import pathways are enriched in MODS compared to CT (Fig. 4a). Notable genes included in immune responses are ADGRE2, C3AR1, CD177, FCER1G, IRAK3, MMP8, PLSCR1, PPARG, SOCS3, and TLR5. Similar pathways were also observed in a previous analysis between MODS and CT [10]. In addition, gene expression related to epigenetic processes (e.g., regulation of gene silence, DNA packaging, chromatin assembly) was activated in ECMO compared to MODS (Fig. 4b).
Fig. 4.
Gene enrichment and co-expression network analysis of DE genes in MODS and CT, and in ECMO and MODS. (a) Gene ontology (GO) enrichment of DE genes from MODS to CT showed their involvement in immune responses. (b) However, GO enrichment of DE genes from ECMO to MODS displayed enrichment in epigenetic regulations. (c) The DE genes obtained from the comparison of MODS and CT were clustered into two separate groups. (d) Similarly, two co-expression networks were created after mapping the DE genes in ECMO and MODS. The highlighted genes in co-expressed networks are hub genes. Notably, many DE genes from both comparisons were shared in module 15 (M15), suggesting phase transition. Size of circles in GO represents the number of mapped genes.
Further, co-expression analysis was performed to delineate the relationships between gene expression and their regulated pathways. The transcripts per million (TPM) count of all the genes for baseline patients was used to create co-expressed network modules. The DE genes from MODS to CT and ECMO to MODS were mapped on these modules and identified the corresponding modules. Two modules were identified in each comparison (Fig. 4c and 4d). Notably, some of the DE genes from MODS to CT were mapped on module M15 of ECMO to MODS, deciphering the phase transition of MODS to ECMO support. Module M14 was specific to the comparison of ECMO and MODS, whereas modules M1 and M4 were specific to the comparison of MODS and CT.
Pathways analysis of each module showed that genes in module M1 were involved in immune responses (Figure S10a) and genes in module M4 were involved in glucose metabolisms and glycogen breakdown (Figure S10b). However, module M15 (shared by both comparisons) showed enrichment of signalling pathways and proteins maintenance (Figure S10c). Module M14 belonging to genes that differed between ECMO and MODS was enriched with genes related to DNA damage, DNA maintenance and histone acetylation (Figure S10d). Together, DE analysis showed enrichment of immune related and glycogenolysis pathways in MODS, while protein maintenance and epigenetic-related pathways were enriched in ECMO. The protein-protein interaction network of the DE genes also revealed two distinct clusters: histone activation and blood coagulation were uniquely enriched in ECMO (Figure S11).
The GO enrichment analysis and co-expression analysis of DE genes expressed at 72h and 8d did not show any significantly enriched pathway in any of the comparisons. This observation may suggest that the MODS and ECMO patients have important physiological differences at baseline, but other processes obfuscate these differences as diverse disease processes and therapeutic interventions unfold. Such baseline differences could be exploited for prognostic and potentially diagnostic purposes.
3.3. Identification of molecular signatures associated with ECMO
In 2018, Sweeney et al. [5] evaluated four prognostic biomarker signatures consisting of genes positively or negatively correlated with mortality in sepsis. We computed the geometric mean of the expression of these signature genes and investigated whether these values could be used as risk scores for MODS to ECMO progression. We observed that the risk scores derived from the signature genes that are positively correlated with mortality among sepsis patients could differentiate ECMO and MODS (P-value ranges from 0.04 to 0.01, Student's T-test) (Figure S12).
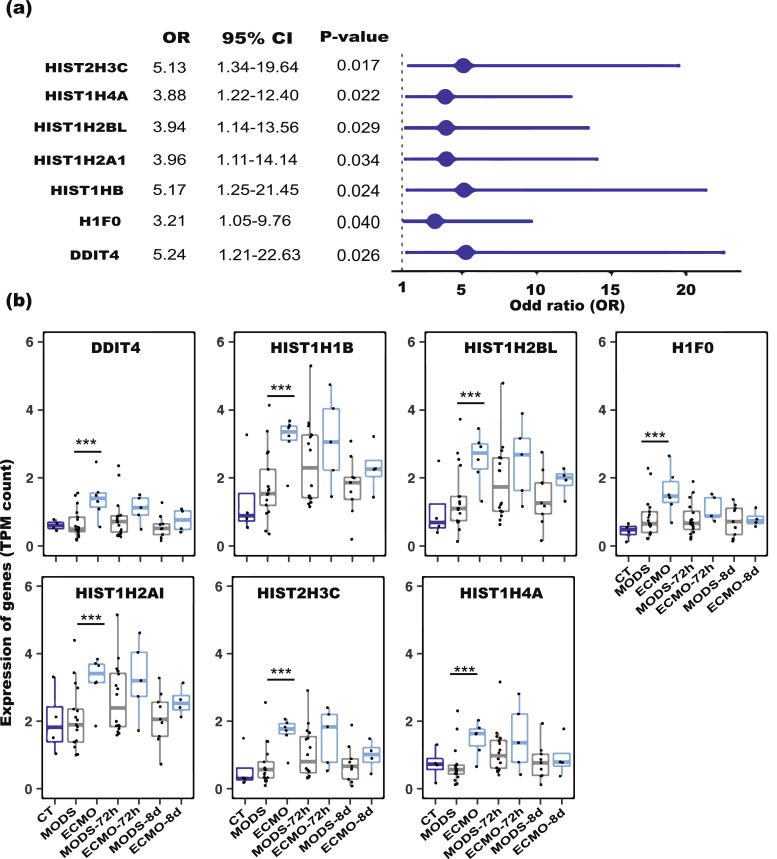
We next sought to derive the predictive power of the differentially expressed genes identified between ECMO and MODS. Seven genes from our differential gene expression analysis demonstrated a very strong association with MODS for their progression to ECMO (P-value < 0.04, logistic regression) (Fig. 5a) and these were used to create a signature for ECMO prediction. Most of these genes belong to the histone family (HIST2H3C, HIST1H4A, HIST1H2AI, HIST1H1B, HISTH2BL, and H1F0, Table 2) and these were expressed significantly higher in ECMO than MODS (P-value < 3.5e-6, Student's T-test) (Fig. 5b). In addition, the Human Protein Atlas dataset showed the enhanced expression of some genes in neutrophils (Figure S14).
Fig. 5.
Univariate analysis of differentially expressed (DE) genes in ECMO and MODS. (a) Odds ratio of the DE genes between ECMO and MODS (reference). A total of 7 genes from 30 DE genes are significant (OR > 1 and P value < 0.05, logistic regression). (b) Expression of the DE genes in CT, ECMO and MODS patients at different time points. The higher expression of the genes in ECMO than in MODS at three time points (0h, 72h and 8d) suggests their strong association with the deterioration from MODS to ECMO. Blue colour displayed- control (CT), grey colour displayed- MODS patients and cyan colour displayed- ECMO patients. *** P-value < 1E-06 (Student's T-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
List of signature genes strongly associated with ECMO.
| Gene | Ensembl | Log2 fold change | Log CPM | P-value | Adj. P-value | Protein coding | Function |
|---|---|---|---|---|---|---|---|
| DDIT4 | ENSG00000168209 | 2.02 | 3.74 | 4.36E-07 | 0.0015 | Y | DNA-damage-inducible transcript 4 |
| HIST1H1B | ENSG00000184357 | 1.92 | 4.23 | 2.43E-06 | 0.0055 | Y | histone cluster 1, H1b |
| HIST1H2BL | ENSG00000185130 | 1.94 | 2.93 | 4.84E-06 | 0.0091 | Y | histone cluster 1, H2bl |
| H1F0 | ENSG00000189060 | 1.72 | 4.5 | 3.50E-06 | 0.0073 | Y | H1 histone family, member 0 |
| HIST1H2AI | ENSG00000196747 | 1.61 | 3.85 | 3.43E-06 | 0.0073 | Y | histone cluster 1, H2ai |
| HIST2H3C | ENSG00000203811 | 2.11 | 3.87 | 9.08E-07 | 0.0026 | Y | histone cluster 2, H3c |
| HIST1H4A | ENSG00000278637 | 1.85 | 0.24 | 1.02E-06 | 0.0028 | Y | histone cluster 1, H4a |
3.4. Re-classification of patients and signature-based risk estimation
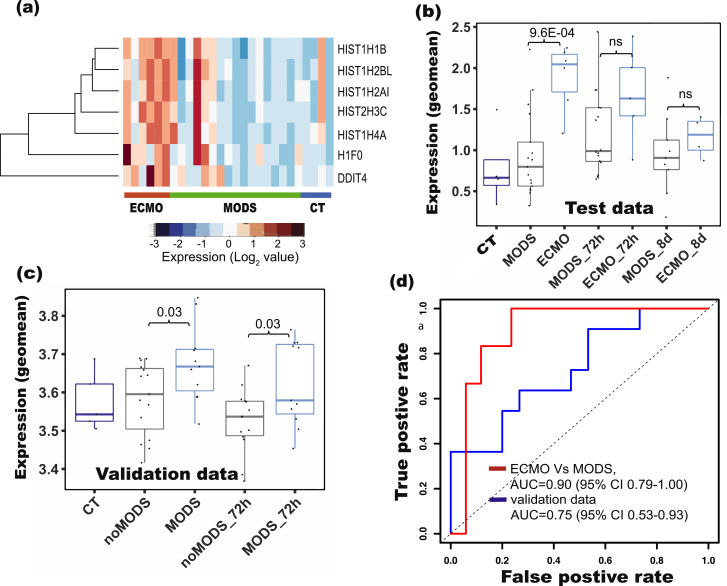
Expression of the genes in our 7 gene risk signatures was higher in ECMO than MODS (Fig. 6a). Interestingly, when the additional time points (72h and 8d) were added, these signature genes were not different in MODS and ECMO and could also be confirmed by the overlap of patients (Figure S15a and S15b). The risk scores derived from these genes were significantly different between ECMO and MODS (95%CI 1.54-42.91, P-value=0.001, logistic regression; P-value = 9.6E-04, Student's T-test, Fig. 6b) at baseline. In contrast, risk scores of MODS patients at 72h and 8d are close to those of ECMO patients at 72h and 8d (Fig. 6b).
Fig. 6.
Signature based re-classification of patients in the test (CT, MODS and ECMO) dataset and validation dataset. (a) Heatmaps showed the clustering of signature genes in ECMO patients compared to control (CT) and MODS patients. Risk scores derived from the signature genes showed difference in (b) ECMO and MODS in our data, and in (c) MODS (require ICU support similar to our ECMO patients) and noMODS (do not require ICU support similar to our MODS patients) in the validation data (Cabrera et at., 2017) [10]. (d) Receiver operating characteristics (ROC) of the classification using our data and the validation data. A risk score for each patient was computed based on the geometric mean of the signature gene expression. Risk scores were strongly associated with ECMO and can be helpful to predict the probability of the MODS patients who require ECMO support.
Due to the lack of an appropriate paediatric cohort, we used previously published microarray data of adult patients that developed MODS after a major trauma as validation data. The authors had categorized the patients into two groups, those that developed MODS (require ICU support similar to our ECMO patients) and those that did not (noMODS) (do not require ICU support similar to our MODS patients); however these were more sick compared to controls [10]. In their cohort, the risk score derived from our signature was significantly higher (95%CI 1.02-10.35, P-value=0.04, logistic regression; P-value = 0.027, Student's T-test) in MODS than noMODS (Fig. 6c).
We further found that our signature genes can also classify patients (noMODS, and MODS) in the validation cohort at 0h (Figure S17a) as well as 72h timepoint (Figure S17b). Using logistic regression to train the risk scores led to a remarkable separation (AUC of 0.90 [95%CI 0.79-1.00] for ECMO and MODS patients at baseline in our data and AUC of 0.75 [95%CI 0.53-0.93] in the validation set) of two group of patients from our data as well as validation data, indicating a strong association of risk scores with MODS deterioration (Fig. 6d).
4. Discussion
The decision to initiate ECMO is often subjective, determined by the clinical judgement of the multidisciplinary care team in a very stressful and dynamic setting as opposed to quantitative measures of pathophysiology. ELSO does provide criteria that recommends the initiation of ECMO in paediatric patients using evidence that have evolved over time to predict mortality [4]. ELSO recommends ECMO only after less invasive measures such as high frequency ventilation, inhaled Nitric Oxide or prone positioning have failed. This process is time consuming and still involves some trial and error and the presence of some more objective data could enhance clinical decision making [4]. Biological sampling of the exact failing organs is impractical if not impossible but circulating white blood cells may serve as a proxy readout of the stress being experienced by multiple organ systems. We employed transcriptomics of peripheral white cells in an effort to improve our understanding of the response of circulating cells to multi-organ failure and its progression to either recovery or cardiopulmonary collapse culminating in the need for extra corporeal life support.
White blood cells are uniquely suited for this because aside from a few exceptions (e.g., memory T cells, some tissue macrophages), most of the mature blood cell types are mitotically inactive, metabolically active and relatively short-lived with half-lives ranging over hours to a few days. Thus, they are reflective of the environment they course through [24]. We found the gene AREG which regulates Amphiregulin a mediator for macrophage activity were preferentially activated in patients prior to ECMO [25,26]. Amphiregulin has been shown to an essential cardioprotective mediator produced by cardiac Ly6C macrophages in response to fluid overload, which is very common in MODS [27].
The activation of immune response and glycogenolysis in MODS compared to CT showed that patients in MODS need excessive energy for cellular homeostasis and activation of immune response against the initial infections. However, during the transition from MODS to ECMO, various signalling and protein maintenance pathways also got activated. Notably, DNA repair, DNA methylation and other epigenetic changes were activated in the patients who deceased further and needed ECMO support. This aligns with the decompensated state that patients needing ECMO often experience from hypoxia, inadequate circulation and cardiorespiratory arrest. This leads to the activation of various oxidative stress and inflammatory responses resulting in DNA methylation and repair as observed in various disease and cancer [28,29].
One of the key observations is the enrichment and strong association of histone genes with ECMO. The histone octamer HIST2H3C, HIST1H2AI, HIST1H4, and HIST1H1B, are genes that increase the availability of histones. Among these histones, HIST2H3C, HIST1H2AI, and HIST1H4A are highly expressed in neutrophils (Figure S14). Histones are a protein class, containing histone H1 and the core histones H2A, H2B, H3, and H4 [30] that are involved in numerous biological processes, largely through repressing transcription [31,32]. These are important due to their capability to determine if DNA is accessible for transcription and they have a major impact on gene expression, too [33]. However, to allow processes like transcription or replication, this structure needs to change dynamically from a condensed state to an open one.
Genes that are associated with the histone cluster were found to be elevated. Increases in serum histones have previously been shown to be elevated in patients with sepsis and heart failure [34,35]. In addition, higher concentrations of circulatory histones are associated with poor survival in patients undergoing ECMO [36]. The increased availability of histones in pathologies that concur with a prolonged inflammatory response as is the case of sepsis. This is not only due to tissue damage but also to a second source: activated neutrophils generate neutrophil extracellular traps (NETs), structures made of cellular components which include specifically modified histones [37]. Generation of circulating histones from NETs or from necrotic neutrophils implies the release of a high concentration of histones to the bloodstream. Both processes, NET and apoptosis and necrosis of neutrophils and other immune cells, contribute to the pathogenesis of sepsis. NET however has been linked to organ failure [38,39,40]. In this study we showed that these processes are active enough to be uncovered by gene-expression. There is now evidence being accumulated that an aptamer-based therapeutic approach directed specifically against histones could potentially reverse some of the clinical findings seen in histone mediated diseases [41]. Recently, molecular medications such as nuclease-resistant RNA aptamers have been used in experimental MODS to bind with high affinity and specificity to human histones H3 and H4 implicated in MODS [41,42].
This study shows that serial whole-blood transcriptomic profiling holds a great promise to predict which MODS patients may need ECMO support. Several published gene signatures developed to predict mortality showed a significance in predicting ECMO, but none of them suffice as a marker in our case. Our new signature genes could remarkably differentiate MODS and ECMO. Their association with ECMO is considerably strong and is also able to distinguish the severe and moderate adult MODS patients in the validation cohort. This showed the broad uses of this signature for diagnosing the patients needed ICU support. The risk score derived from the signature genes for each patient can be used to classify patients into two groups (ECMO and MODS) in our cohort. This is important because in spite of the limited sample size, using paediatric ECMO samples, the multiple time points and validation datasets increase the robustness of our findings. Furthermore, the study included patients, where sepsis was not the primary cause of MODS indicating that histone signatures that occur in patients with MODS do so regardless of the initial insult. The signature genes need further evaluation by prospective studies in paediatric MODS/ECMO patients as these pathways are being currently explored as therapeutic targets in multiple diseases [43,44]. Nevertheless, this study is one of the first to demonstrate that there exists the potential for using clinical and transcriptomic features in identifying MODS patients from those requiring ECMO. The earliest identification of the expression factors in patients with MODS could be used by the clinical team to predict which patient may need aggressive life support measures such as ECMO.
This study has a few limitations. For the diseases like MODS, patients and research resources are scant and no reliable preclinical models are readily available, so starting with a relatively small cohort is one effective way, if not the only, to derive the findings that are critical to drive and design a subsequent larger cohort study. In addition, due to the limitation of patients and resources, finding a validation dataset is also quite difficult, thus adult patients with similar clinical conditions were used as a surrogate. Lastly, as patients in this study presented very diverse clinical and biological characteristics, finding matched control samples was also challenging. The ECMO cases are also less than 50% of the ones with only MODS and the transcriptomic analysis still revealed some robust biomarkers. This work may be of some help to guide the diagnosis of infected patients (irrespective of pathogenesis) at a higher risk for progression to requiring ECMO and the findings warrant the investigation of these biomarkers in a larger patient cohort in the future.
5. Data and code availability
The codes used in these analyses are available at https://github.com/Bin-Chen-Lab/MODS. The processed data used in this study is available through NCBI GEO accession GSE144406.
6. Funding sources
The trial was designed and run by the investigative team, who received financial support from the Spectrum Health Office of Research Administration (SHORA). Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM134307. The funders had no role in data collection, data analysis, data interpretation, or writing of the manuscript. RS, MLL, PAN, KL, JX, DN, EJK, JWP, GZ, ASB, BC and SR had access to all study data and had final responsibility for the decision to submit for publication.
7. Author contributions
Conceived and designed the experiments: BC, SR and RS. Performed the experiments: RS, ML and DM. Analysed the data: RS and PN. Contributed material/analysis tools: KL, JX, EK, JWP, GZ, ASB. Wrote the paper: RS, BC, EK, SR and ML. Supervised the study: BC and SR. All authors read and approved the final version of manuscript.
8. Funding
Spectrum Health Office of Research Administration (SHORA), and R01GM134307, National Institute of General Medical Sciences, National Institutes of Health
9. Research in context
9.1. Evidence before this study
MODS is common in paediatrics patients in the intensive care unit. Some MODS patients need ECMO support as manoeuvre for survival. No clinical scoring tool or molecular biomarker currently exists to identify the patients who may require ECMO support, the decision to initiate ECMO remains subjective based on the empirical experience of the multidisciplinary care team. Whole blood transcriptomic profiling has been widely explored to develop biomarkers for diagnosis or prognosis in many diseases.
9.2. Added value of this study
We performed transcriptome analysis of whole blood samples taken from MODS and ECMO (MODS patients needed ECMO support) patients. Bioinformatics analysis of differentially expressed genes suggested that various epigenetic related pathways and DNA methylation were activated in ECMO patients as compared to MODS. Cellular deconvolution analysis revealed that neutrophil level decreased in ECMO. In addition, we developed a signature of seven genes, presenting diagnostic potential for ECMO patients at base line.
9.3. Implications of all the available evidence
The decreased level of neutrophils reflect that immune responses would have been compromised in ECMO patients. In addition, a large number of mechanisms including inflammation and oxidative stress might have activated in the ECMO group due to hypoxia and cardiorespiratory arrest, resulting in the activation of DNA methylation and DNA repair processes. The earliest identification of the expression patterns in patients with MODS could be used by the clinical team to predict which patient may need aggressive life support measures such as ECMO.
Declaration of competing interest
AB is president and cofounder of Hibiskus Biopharma, Inc, and declares that none of the work in this manuscript is related to the interests of the company. All other authors declare no competing interests.
Acknowledgment
The authors gratefully acknowledge to the participants parents and fellow staff member for their help and cooperation during the study. We would also like to thank Dr. Hui Shen for critical comments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103122.
Contributor Information
Rama Shankar, Email: ramashan@msu.edu.
Mara L. Leimanis, Email: Mara.Leimanis@spectrumhealth.org.
Ke Liu, Email: liuke2@msu.edu.
Jing Xing, Email: xingjin1@msu.edu.
Eric J. Kort, Email: eric.kort@helendevoschildrens.org.
Jeremy W Prokop, Email: prokopje@msu.edu.
Guoli Zhou, Email: zhoug@msu.edu.
André S Bachmann, Email: bachma26@msu.edu.
Bin Chen, Email: chenbi12@msu.edu.
Surender Rajasekaran, Email: surender.rajasekaran@spectrumhealth.org.
Appendix. Supplementary materials
References
- 1.Weiss SL, Fitzgerald JC, Pappachan J, Wheeler D, Jaramillo-Bustamante JC, Salloo A. Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am J Respir Crit Care Med. 2015;191(10):1147–1157. doi: 10.1164/rccm.201412-2323OC. May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bestati N, Leteurtre S, Duhamel A, Proulx F, Grandbastien B, Lacroix J. Differences in organ dysfunctions between neonates and older children: a prospective, observational, multicenter study. Crit Care. 2010;14(6):R202. doi: 10.1186/cc9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenks CL, Raman L, Dalton HJ. Pediatric extracorporeal membrane oxygenation. Crit Care Clin. 2017;33(4):825–841. doi: 10.1016/j.ccc.2017.06.005. Oct. [DOI] [PubMed] [Google Scholar]
- 4.Brain MJ, Butt WW, MacLaren G. Physiology of extracorporeal life support (ECLS) In: Schmidt GA, editor. Extracorporeal Life Support for Adults [Internet] Springer New York; New York, NY: 2016. pp. 1–60. editor. Available from: https://doi.org/10.1007/978-1-4939-3005-0_1. [Google Scholar]
- 5.Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. 2018;9(1):694. doi: 10.1038/s41467-018-03078-2. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7(287) doi: 10.1126/scitranslmed.aaa5993. May 13287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018;46(6):915–925. doi: 10.1097/CCM.0000000000003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proulx F, Fayon M, Farrell CA, Lacroix J, Gauthier M. Epidemiology of sepsis and multiple organ dysfunction syndrome in children. Chest. 1996;109(4):1033–1037. doi: 10.1378/chest.109.4.1033. Apr. [DOI] [PubMed] [Google Scholar]
- 9.Spinella PC, Tucci M, Fergusson DA, Lacroix J, Hébert PC, Leteurtre S. Effect of fresh vs standard-issue red blood cell transfusions on multiple organ dysfunction syndrome in critically ill pediatric patients: a randomized clinical trial. JAMA. 2019;322(22):2179–2190. doi: 10.1001/jama.2019.17478. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrera CP, Manson J, Shepherd JM, Torrance HD, Watson D, Longhi MP. Signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: a prospective cohort study. Schreiber M, editor. PLoS Med [Internet] 2017;14(7) doi: 10.1371/journal.pmed.1002352. Jul 17 [cited 2020 Feb 10]Available from: https://dx.plos.org/10.1371/journal.pmed.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kort EJ, Weiland M, Grins E, Eugster E, Milliron H, Kelty C. Single cell transcriptomics is a robust approach to defining disease biology in complex clinical settings [Internet] Genomics. 2019 Mar [cited 2020 Feb 10]. Available from: http://biorxiv.org/lookup/doi/10.1101/568659. [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo PST, Ferreira GR, Cardozo LE, Bürger MC, Arias-Carrasco R, Maruyama SR. CEMiTool: a bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinformatics. 2018;19(1):56. doi: 10.1186/s12859-018-2053-1. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schölkopf B, Smola AJ, Williamson RC, Bartlett PL. New support vector algorithms. Neural Computation [Internet] 2000;12(5):1207–1245. doi: 10.1162/089976600300015565. May [cited 2020 Feb 10]Available from: http://www.mitpressjournals.org/doi/10.1162/089976600300015565. [DOI] [PubMed] [Google Scholar]
- 18.Hall MW, Gavrilin MA, Knatz NL, Duncan MD, Fernandez SA, Wewers MD. Monocyte mRNA phenotype and adverse outcomes from pediatric multiple organ dysfunction syndrome. Pediatr Res. 2007;62(5):597–603. doi: 10.1203/PDR.0b013e3181559774. [DOI] [PubMed] [Google Scholar]
- 19.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics [Internet]. 2013 [cited 2020 May 13];14(1):7. Available from: http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed]
- 20.Goymer P. Which mean do you mean? Nat Rev Genet [Internet] 2005;6(12):877. Dec [cited 2020 Aug 30]Available from: http://www.nature.com/articles/nrg1758. [Google Scholar]
- 21.Bengtsson M, Ståhlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15(10):1388–1392. doi: 10.1101/gr.3820805. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, et al. Determination of the optimal number of control genes for normalization. 2011; Available from: https://figshare.com/articles/figure/Determination_of_the_optimal_number_of_control_genes_for_normalization/607
- 23.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. Mar 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boettcher S, Manz MG. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 2017;38(5):345–357. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Meng C, Liu G, Mu H, Zhou M, Zhang S, Xu Y. Amphiregulin may be a new biomarker of classically activated macrophages. Biochem Biophys Res Commun. 2015;466(3):393–399. doi: 10.1016/j.bbrc.2015.09.037. Oct 23. [DOI] [PubMed] [Google Scholar]
- 26.Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42(2):216–226. doi: 10.1016/j.immuni.2015.01.020. Feb 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiu K, Shibata M, Nakayama Y, Ogata F, Matsumoto S, Noshita K. A heart-brain-kidney network controls adaptation to cardiac stress through tissue macrophage activation. Nat Med. 2017;23(5):611–622. doi: 10.1038/nm.4326. May. [DOI] [PubMed] [Google Scholar]
- 28.Donkena KV, Young CYF, Tindall DJ. Oxidative stress and DNA methylation in prostate cancer. Obstetr Gynecol Int. 2010 doi: 10.1155/2010/302051. [Internet]. 2010 [cited 2020 Aug 31]1–14. Available from: http://www.hindawi.com/journals/ogi/2010/302051/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida N, Kudo M. Oxidative stress and epigenetic instability in human hepatocarcinogenesis. Dig Dis [Internet]. 2013 [cited 2020 Aug 31];31(5–6):447–53. Available from: https://www.karger.com/Article/FullText/355243 [DOI] [PubMed]
- 30.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 31.Grant PA. A tale of histone modifications. Genome Biol. 2001;2(4) doi: 10.1186/gb-2001-2-4-reviews0003. REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. Aug 10. [DOI] [PubMed] [Google Scholar]
- 33.Falvo JV, Jasenosky LD, Kruidenier L, Goldfeld AE. Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv Immunol. 2013;118:37–128. doi: 10.1016/B978-0-12-407708-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alhamdi Y, Abrams ST, Cheng Z, Jing S, Su D, Liu Z. Circulating histones are major mediators of cardiac injury in patients with sepsis. Crit Care Med. 2015;43(10):2094–2103. doi: 10.1097/CCM.0000000000001162. Oct. [DOI] [PubMed] [Google Scholar]
- 35.Ekaney ML, Otto GP, Sossdorf M, Sponholz C, Boehringer M, Loesche W. Impact of plasma histones in human sepsis and their contribution to cellular injury and inflammation. Crit Care. 2014;18(5):543. doi: 10.1186/s13054-014-0543-8. Sep 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen Z, Jin Y, Jiang X, Sun M, Arman N, Wen T. Extracellular histones indicate the prognosis in patients undergoing extracorporeal membrane oxygenation therapy. Perfusion. 2019;34(3):211–216. doi: 10.1177/0267659118809557. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15(11):1318–1321. doi: 10.1038/nm.2053. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czaikoski PG, Mota JMSC, Nascimento DC, Sônego F, Castanheira FVe S, Melo PH. Neutrophil extracellular traps induce organ damage during experimental and clinical sepsis. PLoS ONE. 2016;11(2) doi: 10.1371/journal.pone.0148142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li RHL, Tablin F. A comparative review of neutrophil extracellular traps in sepsis. Front Vet Sci. 2018;5:291. doi: 10.3389/fvets.2018.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakazawa D, Kumar SV, Marschner J, Desai J, Holderied A, Rath L. Histones and neutrophil extracellular traps enhance tubular necrosis and remote organ injury in ischemic AKI. J Am Soc Nephrol. 2017;28(6):1753–1768. doi: 10.1681/ASN.2016080925. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urak KT, Blanco GN, Shubham S, Lin L-H, Dassie JP, Thiel WH. RNA inhibitors of nuclear proteins responsible for multiple organ dysfunction syndrome. Nat Commun. 2019 10;10(1):116. doi: 10.1038/s41467-018-08030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samanta S, Rajasingh S, Cao T, Dawn B, Rajasingh J. Epigenetic dysfunctional diseases and therapy for infection and inflammation. Biochim Biophys Acta Mol Basis Dis. 2017;1863(2):518–528. doi: 10.1016/j.bbadis.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thangavel J, Samanta S, Rajasingh S, Barani B, Xuan Y-T, Dawn B. Epigenetic modifiers reduce inflammation and modulate macrophage phenotype during endotoxemia-induced acute lung injury. J Cell Sci. 2015;128(16):3094–3105. doi: 10.1242/jcs.170258. Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The codes used in these analyses are available at https://github.com/Bin-Chen-Lab/MODS. The processed data used in this study is available through NCBI GEO accession GSE144406.