Abstract
Although single cell RNA sequencing studies have begun providing compendia of cell expression profiles1–9, it has proven more difficult to systematically identify and localize all molecular types in individual organs to create a full molecular cell atlas. Here we describe droplet- and plate-based single cell RNA sequencing (scRNAseq) applied to ~75,000 human cells across all lung tissue compartments and circulating blood, combined with a multi-pronged cell annotation approach, which have allowed us to define the gene expression profiles and anatomical locations of 58 cell populations in the human lung, including 41 of 45 previously known cell types or subtypes and 14 new ones. This comprehensive molecular atlas elucidates the biochemical functions of lung cell types and the cell-selective transcription factors and optimal markers for making and monitoring them; defines the cell targets of circulating hormones and predicts local signaling interactions including sources and targets of chemokines in immune cell trafficking and expression changes on lung homing; and identifies the cell types directly affected by lung disease genes and respiratory viruses. Comparison to mouse identified 17 molecular types that appear to have been gained or lost during lung evolution and others whose expression profiles have been substantially altered, revealing extensive plasticity of cell types and cell-type-specific gene expression during organ evolution including expression switches between cell types. This atlas provides the molecular foundation for investigating how lung cell identities, functions, and interactions are achieved in development and tissue engineering and altered in disease and evolution.
58 molecular cell types of human lung
Since Malphigi10, dozens of lung cell types have been discovered by microscopy11, creating histological atlases that are the cellular foundation for pulmonary medicine. More recently, cell-type-specific markers12,13 were identified that provide molecular definitions and functions of the cell types14, reaching its apex in genome-wide expression profiling by scRNAseq15–19. We sought to create a comprehensive molecular cell atlas of adult human lung using scRNAseq, a substantial challenge because the 45 histological cell types have diverse structures, locations, and abundances varying over five orders of magnitude (Supplementary Table 1).
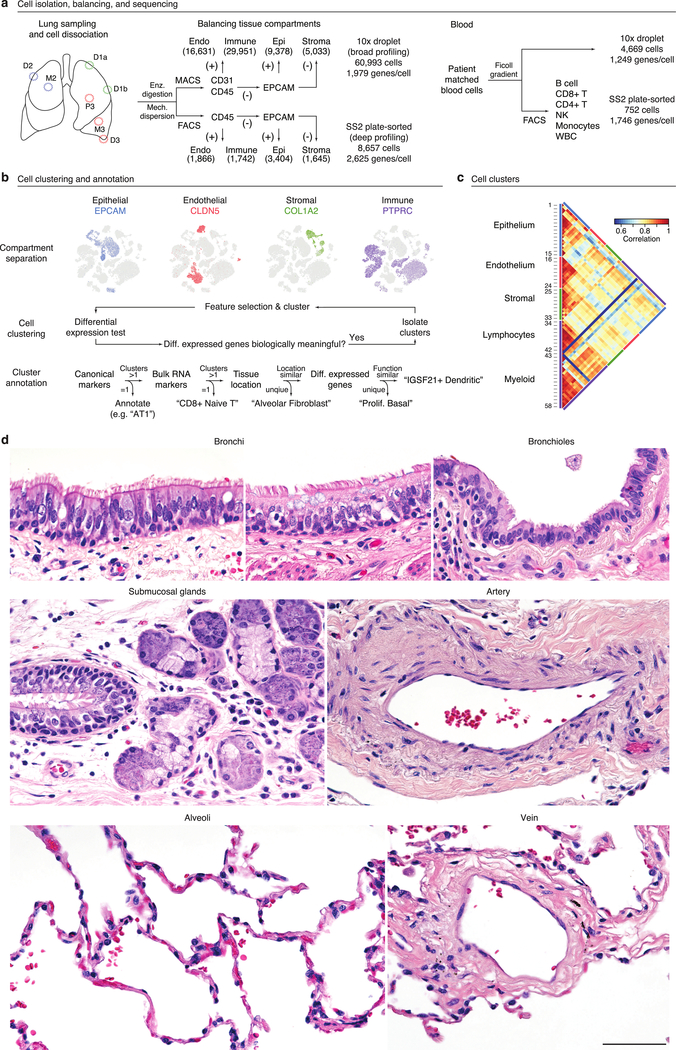
We acquired histologically normal lung tissue intraoperatively from bronchi (proximal), bronchiole (medial), and alveolar (distal) regions along with peripheral blood (Fig. ED1a,d). Lung samples were dissociated into cell suspensions, and each suspension was sorted into epithelial (EPCAM+), endothelial/immune (CD31+CD45+) and stromal (EPCAM−CD31−CD45−) populations (Supplementary Fig. 1a). This allowed us to balance tissue compartment representation for sequencing. We also sorted blood cells to balance immune lineages (Supplementary Fig. 1b). Sequencing libraries were prepared using 10x Chromium (10x) or SmartSeq2 (SS2)20. Higher throughput of 10x enabled discovery of rare cell types, whereas SS2 gave deeper transcriptomic information; there were also platform-specific idiosyncrasies in cell capture. We sequenced thousands of cells from each compartment for each subject (Supplementary Table 2) to directly compare cell types without batch correction, and did so for three subjects to address individual differences. High quality transcriptomes were obtained from ~75,000 cells (65,662 10x, 9,404 SS2).
We grouped cells based on expression of compartment-specific markers (Fig. ED1b), then iteratively clustered21 them for each subject to identify transcriptionally distinct cell populations. Populations between subjects were merged using cluster-specific marker genes for downstream analyses. Our approach identified 58 transcriptionally distinct cell populations (mean 51 per subject, Fig. ED1c, Supplementary Table 2), 37 more than a recent state-of-the-art study19.
Transcriptomes of canonical cell types
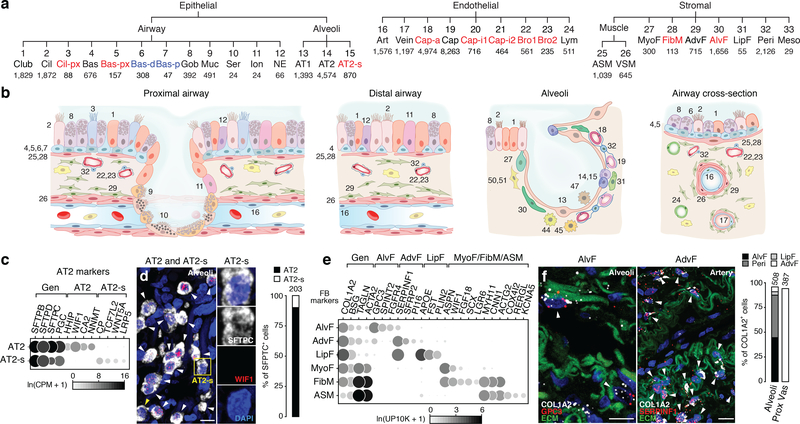
The 58 populations included 15 epithelial, 9 endothelial, 9 stromal, and 25 immune populations, greater than the number of classical cell types in each compartment (Supplementary Table 2). Using extant and newly identified (bronchial vessel) markers (Supplementary Table 1) and single molecule fluorescence in situ hybridization (smFISH), we identified clusters representing all but one classical lung cell type in epithelial, endothelial and stromal compartments (Fig. 1a,b).
Figure 1. Identities and locations of lung epithelial, endothelial, and stromal cell types.
a, Human lung molecular cell types identified after iterative clustering (each level of hierarchy is an iteration) of scRNAseq profiles of cells in indicated tissue compartments. Black, canonical types; blue, proliferating or differentiating subpopulations; red, novel populations. Number of cells shown below cluster name. b, Diagrams showing localization and morphology of each type (cell type numbering/names in (a) and Figure 2a). c, Dot plot of AT2 marker expression (10x dataset). UP10K, UMIs per 10,000. d, smFISH and quantification (n=203 cells scored, staining repeated in 2 subjects different from those profiled) for common AT2/AT2-s marker SFTPC (white) and specific AT2 marker WIF1 (red puncta, arrowheads). Bar, 10μm. AT2-s cells (SFTPCpos WIF1neg; box, enlarged at right, yellow arrowhead) is intermingled among AT2 cells (SFTPCpos WIF1pos, white arrowheads). e, Dot plot of stromal markers (10x dataset). f, smFISH and quantification for general fibroblast marker COL1A2 (white), alveolar fibroblast (AlvF) marker GPC3 (red, left) and adventitial fibroblast (AdvF) marker SERPINF1 (red, right). Blue, DAPI; Green, ECM (extracellular matrix, autofluorescence). Adventitial fibroblasts (arrowheads, right) localize around vessels (ECM). Graph, stromal cell type quantification in alveolar and proximal vascular regions (n=number of cells scored in each region, staining repeated in 2 subjects different from those profiled); pericyte, lipofibroblast markers in Figure ED4h,i). Bars, 10μm. For more details on statistics and reproducibility, please see Methods.
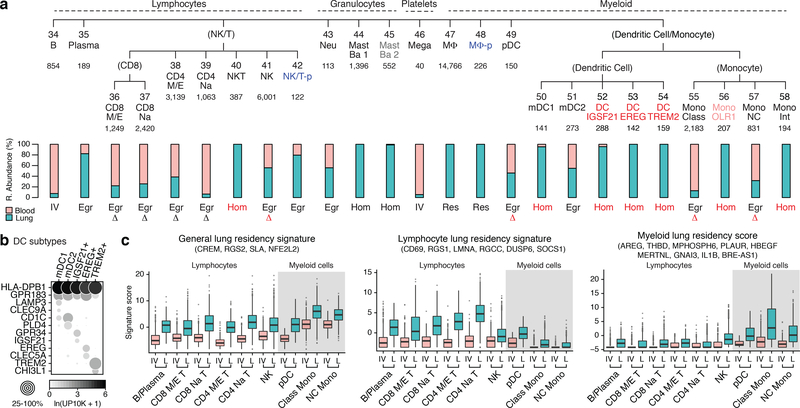
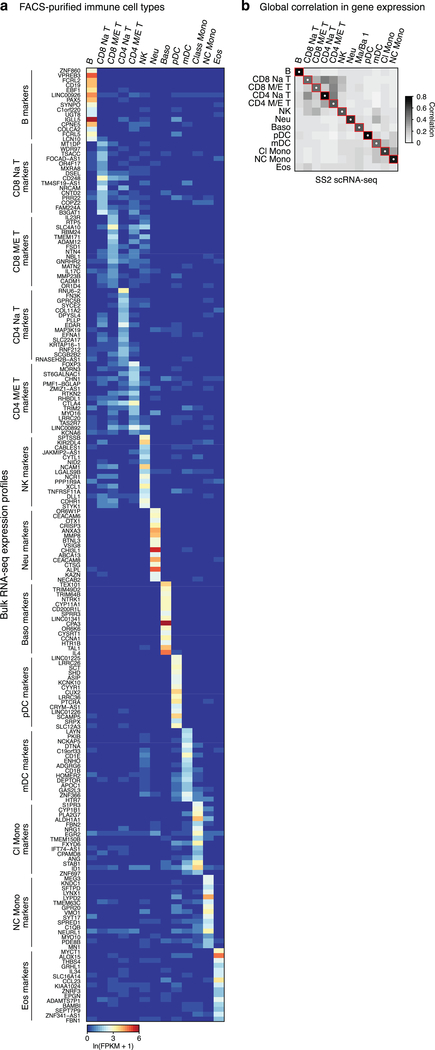
Immune cells were the most heterogeneous and included circulating, egressed, and lung resident cells. To aid identity assignment, we defined transcriptional profiles of circulating immune cells by by bulk RNA sequencing of 21 sorted, functionally-characterized classes of human blood cells (Fig. ED2a, Supplementary Table 3). We also obtained scRNAseq profiles of ~5,000 blood cells from two subjects whose lung cells we analyzed. Canonical immune markers and the ascertained panels of differentially-expressed genes were used to assign identities to 25 immune clusters from our lung and blood scRNAseq, including all but one previously known lung immune cell type (Figs. 2a,ED2b).
Figure 2. Identity and residency of lung immune cells.
a, Human lung immune molecular types clustered and annotated as in Figure 1a. Clusters 45 (grey) and 56 (light red) were found only in one subject. Bar graphs show relative abundance of each immune type in lung (blue) and blood (red) samples. Lung “resident” (Res) or “homing” (Hom) immune types, >90% enrichment in lung samples; “intravascular” (IV), >90% enrichment in blood; “egressed” (Egr), all other types (assignments are provisional because cell harvesting influences enrichment values). Red lettering, cells not previously known to home to (be enriched in) lung or change expression (delta symbol) following egression from blood. b, Dot plot showing expression (10x dataset) in dendritic cell clusters 50–54 of, from top row to bottom: two canonical dendritic markers, four myeloid dendritic (mDC1, mDC2) markers, and six markers for three novel dendritic populations (IGSF21+, EREG+, and TREM2+). c, Box-and-whisker plots of general, lymphocyte-specific, and myeloid-specific lung residency (egression) signature scores (of cells in panel a) based on expression of indicated genes in 10x profiles of indicated immune types isolated from blood (IV) or lung (L). Many previously known lymphocyte residency genes (e.g. S1PR1, RUNX3, RBPJ, HOBIT) were lowly expressed and only uncovered in SS2 profiles. Gray shading, myeloid cells. n, cells in each box-and-whisker from left to right are 725; 187; 419; 771; 631; 1,411; 594; 2,419; 644; 288; 519; 4,250; 21; 116; 1,064; 1,013; 200; and 604. For more details on statistics and reproducibility, please see Methods.
Our approach defined genome-wide expression profiles for nearly all classical lung cell types (41 of 45, 91%), from most abundant (capillaries, ~23% of lung cells) to exceedingly rare (ionocytes, 0.01%) (Supplementary Table 1). One-quarter (11 of 45) previously lacked high quality single cell transcriptomes. The only classical types not captured are extremely rare (neurons, glia), primarily found in disease (tuft cells)22, or require special isolation methods (eosinophils).
New lung cell types, subtypes and states
Many canonical types were represented by more than one cluster, so the specific identities of 25 clusters remained uncertain. All but one were found in multiple subjects so were unlikely to be subject-specific (Supplementary Table 2). This suggested the distinct expression profiles uncovered represented discrete molecular states or novel cell types or subtypes. To distinguish these possibilities, we analyzed the differentially-expressed genes and examined cell structure and location.
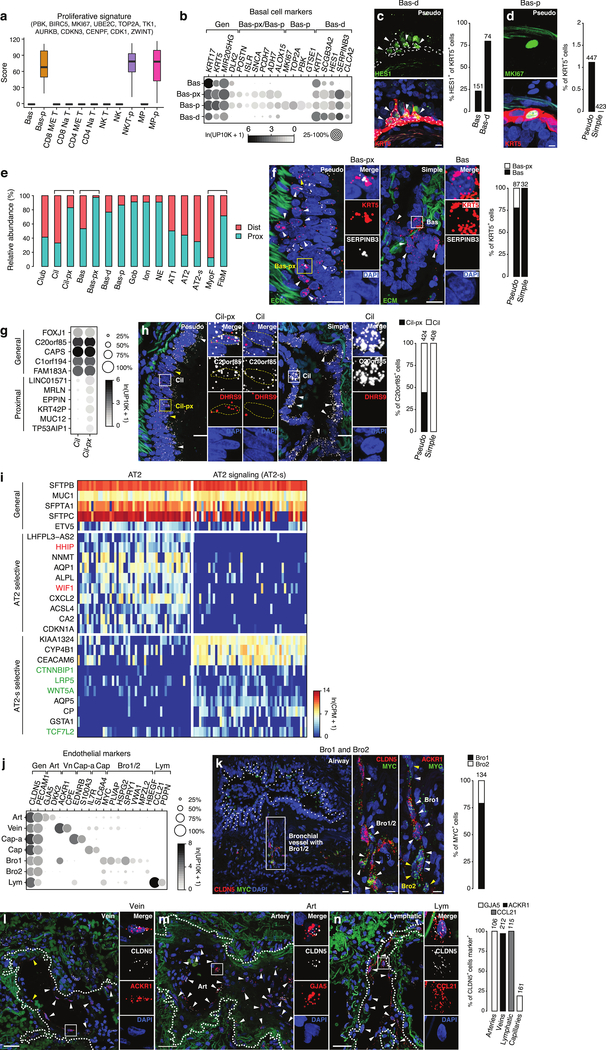
We first identified clusters representing common cell states. Three clusters (Bas-p, NK/T-p, MP-p) were enriched in expression of cell cycle genes, indicating they represent proliferative states of basal, Natural Killer, T cells, and macrophages, respectively, and are the most proliferative lung cell types (Fig. ED3a). Another cluster (Bas-d) had reduced KRT5 and increased HES1, KRT7, and SCGB3A2 expression, indicating active differentiation to other epithelial fates23,24, consistent with their transitional morphology (Fig. ED3b,c). Proliferating and differentiating basal cells derived mostly from proximal lung samples (Fig. ED3d,e), suggesting one-third of proximal basal cells are active.
The other basal cell clusters were quiescent and localized to proximal (large, pseudostratified) airways (Bas-px), or both proximal and distal (small, simple) airways (Bas) (Fig. ED3e,f). Bas-px and Bas are distinguished by hundreds of genes, suggesting they are molecularly distinct cell types differing in hormone production (ALOX15,ADH7,SNCA) and adhesion (POSTN,ISLR,PCDH7) (Fig. ED3b). There were also distinct clusters of ciliated cells along the proximal-distal axis (Fig. ED3g,h).
We uncovered two clusters of alveolar type 2 (AT2) cells (Fig. 1c), which produce surfactant that prevents alveolar collapse. These are intermingled throughout the alveolar epithelium (Fig. 1d). One (WIF1+HHIP+CA2+) expressed higher levels of some canonical AT2 markers (SFTPA1,SFTPC,ETV5) and selectively expressed inhibitors of Wnt (WIF1) and Hedgehog (HHIP) signaling and the cell cycle (CDKN1A), implying they are quiescent (Fig. ED3i, left). The other, 10-fold less abundant cluster (AT2-signaling, AT2-s) selectively expressed Wnt signaling (WNT5A,LRP5,CTNNBIP,TCF4/TCF7L2) and detoxification (CP, GSTA1, CYP4B1) genes (Fig. ED3i, right). AT2-s could be alveolar stem cells, homologous to the rare, Wnt-active subpopulation of mouse AT2 cells (AT2stem)25,26. However, homology between human AT2-s and mouse AT2stem is provisional because although both show elevated Wnt signaling or components, the many other expression differences between human AT2-s and “bulk” AT2 are not shared by mouse AT2stem.
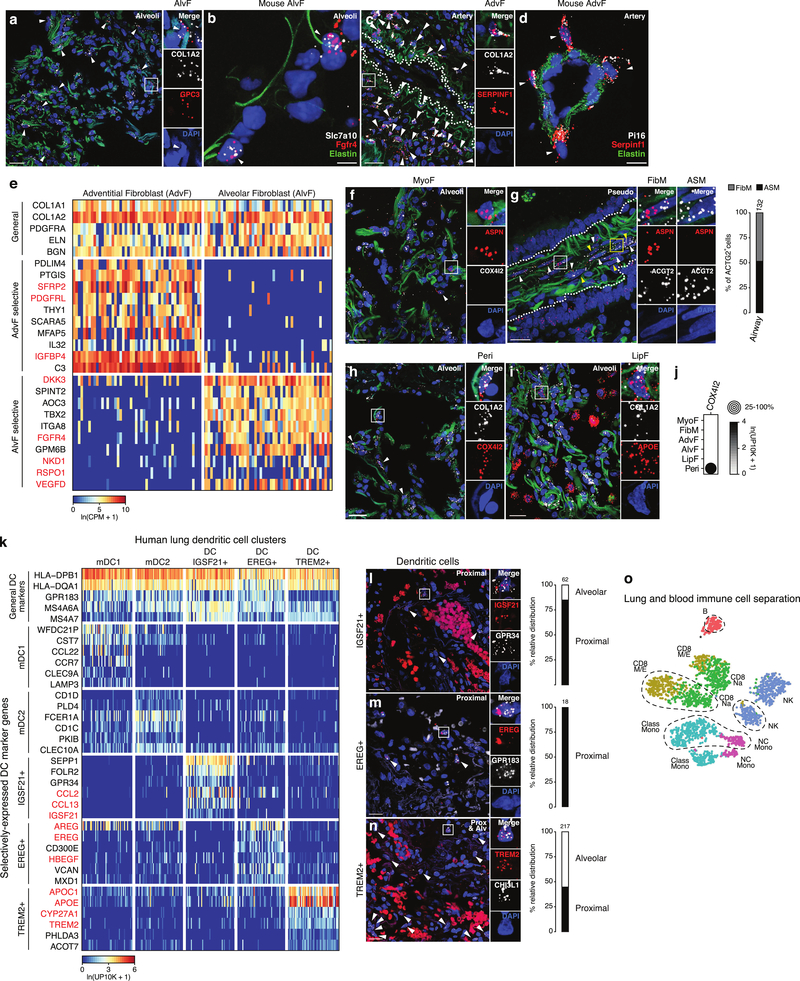
We found unexpected molecular diversity in the endothelial compartment (Fig. ED3j). Two populations were identified as bronchial (Bro1, Bro2) by their localization around bronchi (Fig. ED3k). Thus bronchial endothelial cells are distinct from their counterparts in the pulmonary circulation, distinguished by matrix (VWA1,HSPG2), fenestrated morphology27 (PLVAP) and cell cycle associated (MYC,HBEGF) genes. Four clusters of endothelial cells in the pulmonary circulation expressed capillary markers. Two (Cap-a, Cap) are intermingled alveolar capillary cell types (Gillich et al, accompanying paper); the others are rare capillary types showing mixed Cap-a and Cap features (capillary “intermediates” Cap-i1, Cap-i2).
We identified new types in the stroma, the least characterized compartment. Two clusters expressed classical fibroblast markers (BSG,COL1A2) (Fig. 1e) but one (SPINT2+FGFR4+GPC3+) localized to alveoli (“alveolar fibroblasts”) and the other (SFRP2+PI16+SERPINF1+) to vascular adventitia and nearby airways (“adventitial fibroblasts”) (Figs. 1f, ED4a–d). Both expressed genes involved in canonical fibroblast functions (matrix biosynthesis, adhesion, signaling regulators) but the specific genes often differed (Fig. ED4e). Each cluster also has distinct functions: expression of voltage-gated sodium channel SCN7A and glutamate receptor GRIA1 suggest alveolar fibroblasts are excitable cells with glutamatergic input (Supplementary Table 4). Their profiles also suggest novel, shared functions including immune cell recruitment (IL1RL1,IL32,CXCL2,MHCII) and the complement system (C2,C3,C7,CFI,CFD,CFH,CFB).
Two stromal clusters were enriched for ACTA2, a canonical marker of myofibroblasts (Fig. 1e), which help form alveoli and can act inappropriately in disease. One (WIF1+FGF18+ASPN+) is classical myofibroblasts and localized to alveolar ducts (Fig. ED4f). The other (named “fibromyocytes”) showed higher expression of contractile genes (MYH11,CNN1,TAGLN), was preferentially isolated from proximal lung samples, and was found both intermingled with airway smooth muscle and in alveoli (Figs. ED3e,ED4g). Both populations shared expression of genes for canonical fibroblast functions, though the specific genes differed from alveolar and adventitial fibroblasts (Supplementary Table 4).
Lung immune cell residency signatures
To distinguish between lung resident, egressed, and circulating immune cells, we compared the relative abundance of each immune population in lung and peripheral blood samples from the same subject (Fig. 2a). Eleven clusters (including alveolar macrophages, as expected28) were comprised of cells only from lung samples, with no or rare exception, indicating they are lung resident or greatly enriched. This included three novel lung dendritic populations: IGSF21+ and rare EREG+ dendritic cells express asthma genes (CCL2,CCL13,IGSF21) and developmental signals (EREG,VEGFA,AREG), respectively, and both localize to proximal vessels; TREM2+ dendritic cells localize to vessels and alveoli and express lipid machinery (APOC1,APOE,CYP27A1) (Figs. 2b,ED4k–n).
The other immune cell types were found in both lung and blood samples. For some types, every cell—whether from lung or blood — clustered together. However, for others cells from lung formed a separate cluster (Fig. ED4o). Some of the differentially-expressed genes may be due to technical differences (e.g. collagenase treatment of lung29, circulating RNA in blood30), but others such as upregulation in lung cells of lymphocyte-residence gene CD69 likely represent genes induced following egression31. We identified a core transcriptional signature for all human lung resident lymphocytes (Fig. 2c), which overlaps a residence signature found by bulk RNAseq of CD8+ T cells in mouse spleen, gut and liver32. We also found a residency signature for lung myeloid cells that overlaps the lymphocyte signature, supporting a core residency program for immune cells plus specific subprograms for myeloid cells and lymphocytes.
Cell markers, regulators, interactions
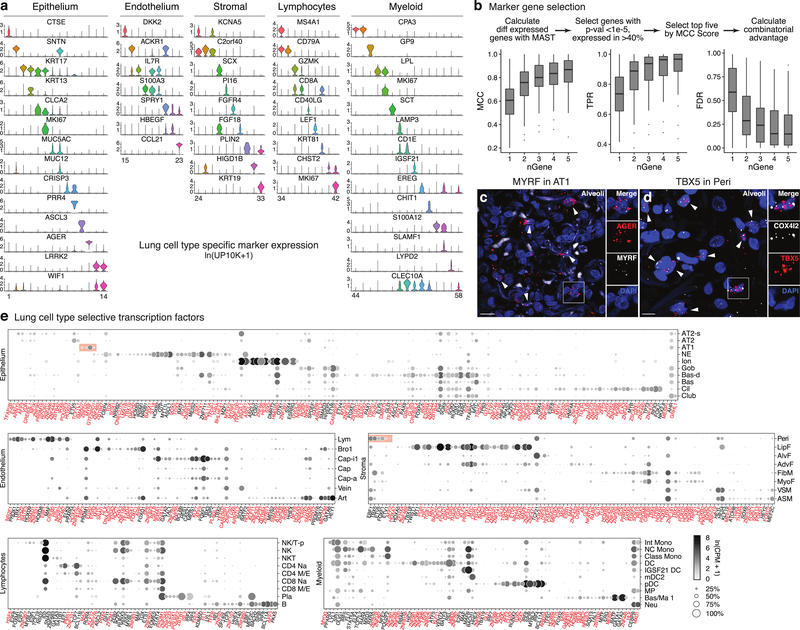
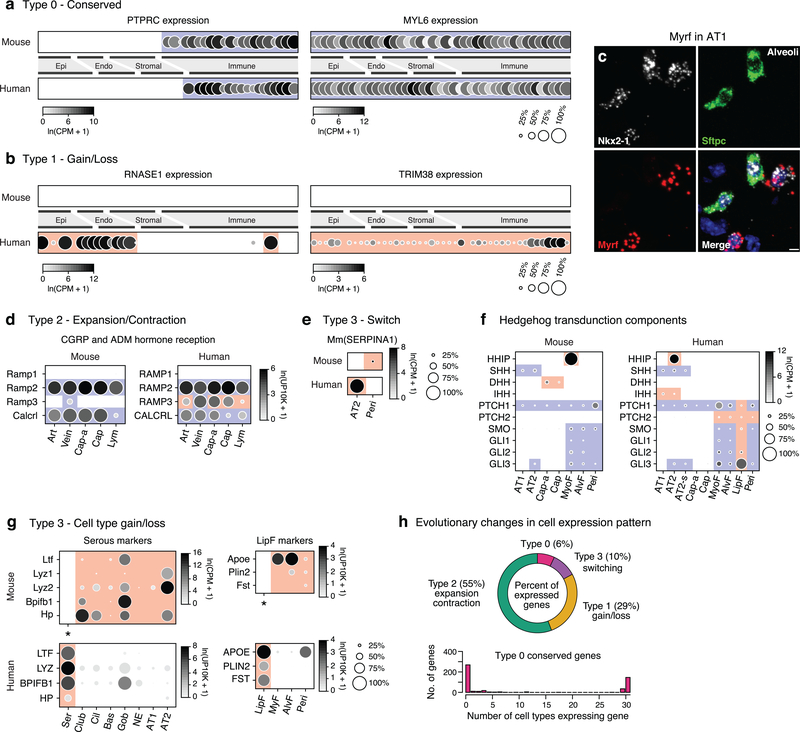
We identified optimal markers for each previously known and newly identified lung cell type (Fig. ED5a, Supplementary Table 4). A battery of ~200 markers can distinguish virtually all types (Fig. ED5b), so could be used with multiplexed smFISH33–35 to simultaneously detect in clinical specimens alterations in their numbers and relationships. A similar compendium of membrane protein markers (Supplementary Table 4) could be used to purify or therapeutically target specific lung cell types. We also identified ~400 transcription factors enriched in each cell type (Fig. ED5e, Supplementary Table 4), putative “master regulators” that could help create all lung cell types by cellular reprogramming. These include what may be long-sought master regulators of AT1 cells (e.g. MYRF), which comprise the gas exchange surface, and of pericytes (TBX5) (Fig. ED5c,d).
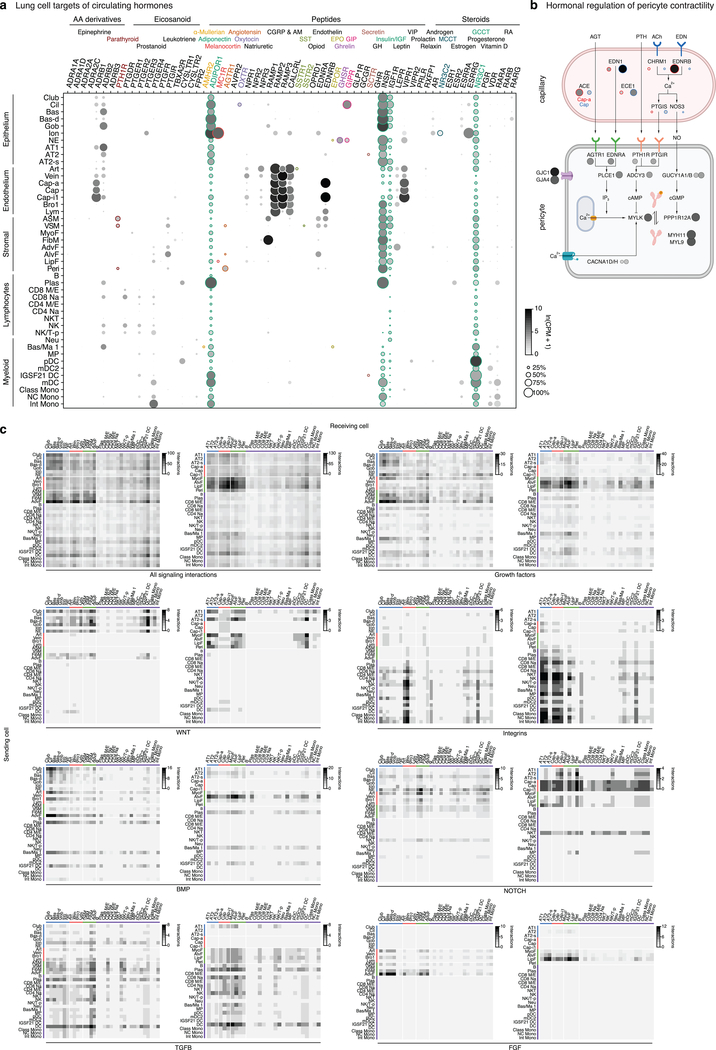
The atlas allowed us to map the cell targets of circulating hormones, based on expression of their cognate receptors. Receptors for some hormones are broadly expressed, implying direct action throughout the lung (Fig. ED6a). Other hormones have specific and unexpected targets, such as somatostatin (SSTR1, arteries), melanocortin (MC1R, ionocytes), and oxytocin (OXTR, ciliated cells). Pericytes are predicted targets of multiple hormones, which could impact their contractile machinery to regulate alveolar perfusion (Fig. ED6b). Receptors for half the hormones were not detectably expressed so may not directly influence lung physiology. We also mapped local signaling interactions by examining expression of ligands and receptors, which predicts up to hundreds of interactions among neighboring cell types (Fig. ED6c, Supplementary Table 5).
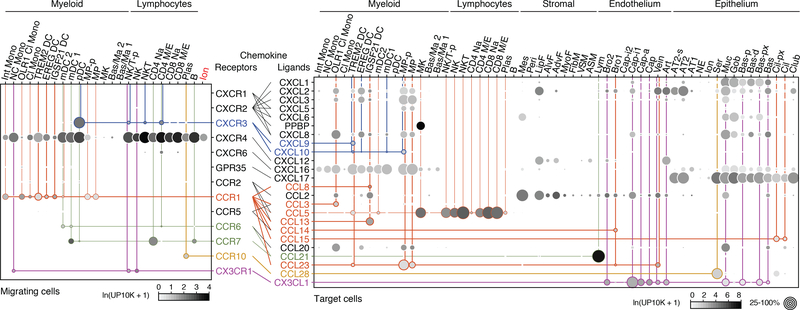
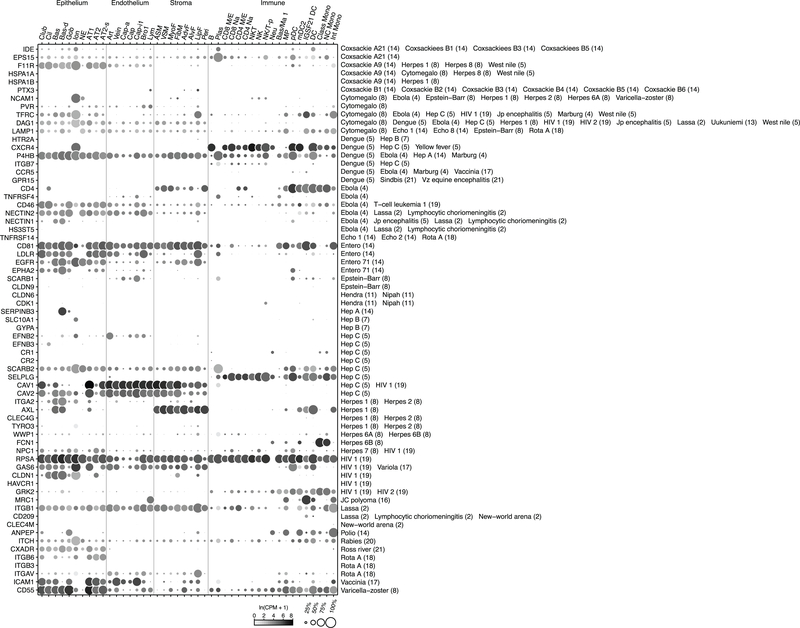
Expression of chemokine receptors illuminated immune cell homing (Fig. 3). Our data confirmed canonical homing interactions such as CD4+ T cells to lymphatic vessels, and provides specificity for others such as plasma cell homing to epithelial mucosa through CCL28 from serous cells. It also predicts new interactions such as CX3CR1-mediated homing of nonclassical monocytes to CX3CL1-expressing endothelial and airway epithelial cells. All three novel dendritic populations express CCR1, which could mediate their attraction to veins (CCL23), bronchial vessels (CCL14), ciliated cells (CCL15), and lymphocytes (CCL5). Ionocytes are the only non-immune cell to express appreciable levels of any chemokine receptor (CXCR4).
Figure 3. Chemokine signaling predicts immune cell homing in lung.
Dot plots showing expression of chemokine receptors (left) and ligands (right) in human lung cells (10x dataset); only cell types and chemokines with detected expression are shown. Colored lines connect ligand sources (target cells) with migrating immune cell types and ionocytes (Ion, red) expressing cognate receptor; thicker lines indicate previously unknown interactions. For more details on statistics and reproducibility, please see Methods.
Mapping cellular focus of lung diseases
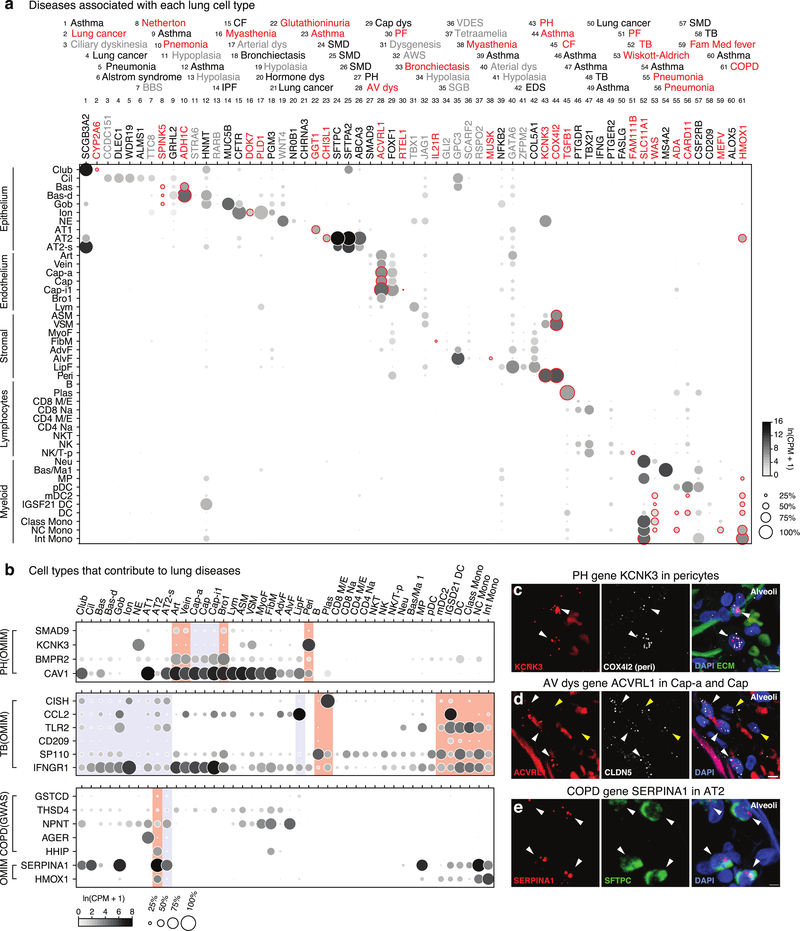
We determined expression of 233 extant lung disease genes (Fig. ED7). Disease genes with cell-type-specific expression (Fig. ED8a) and cell types expressing many genes for a specific disease (Fig. ED8b) are of special interest because they can pinpoint the cellular origin of disease. This supported known or suspected ‘culprit’ cells for 27 genes involved in 12 diseases, and identified potential novel culprits for 21 genes implicated in 15 diseases including pericytes in pulmonary hypertension, capillaries in atrioventricular dysplasia, and AT2 cells in COPD. We confirmed pericyte, capillary, and AT2 expression of disease genes by smFISH (Fig. ED8c–e).
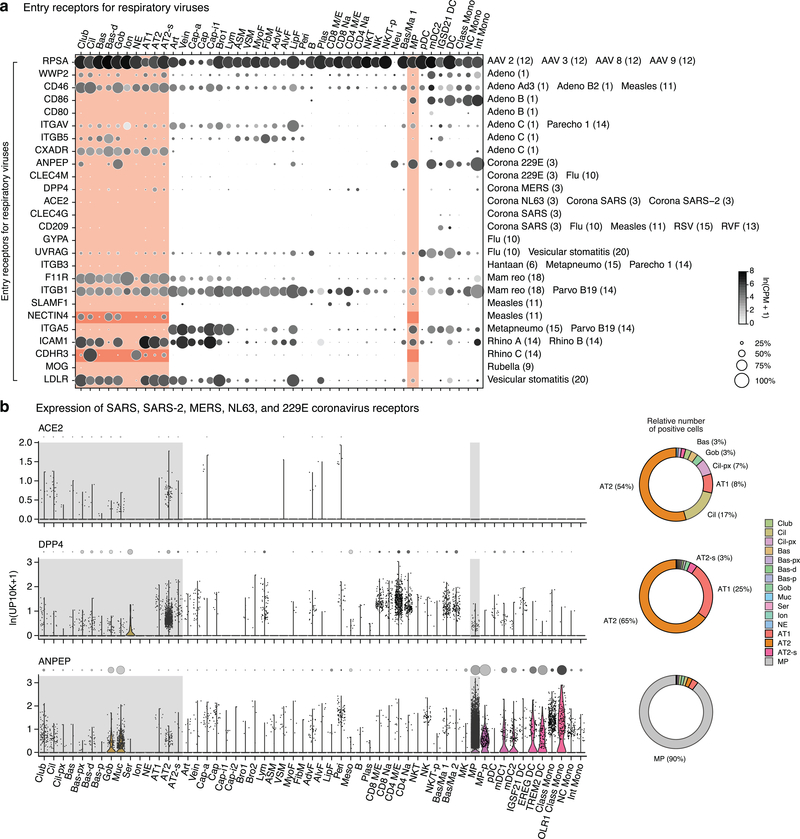
We mapped expression of 80 genes encoding virus receptors, including 26 used by respiratory viruses (Figs. ED9a,ED10). NECTIN4 (measles virus receptor) was enriched in club, ciliated, differentiating basal, and goblet cells, and CDHR3 (“common cold” Rhinovirus C) was enriched in ciliated and neuroendocrine cells, implying infections initiate in those bronchial types. By contrast, ACE2 (SARS, COVID-19 coronaviruses) and DPP4 (MERS coronavirus) were both detected in AT2 cells (Fig. ED9b), consistent with severe alveolar pathology36.
Evolution of cell types and expression
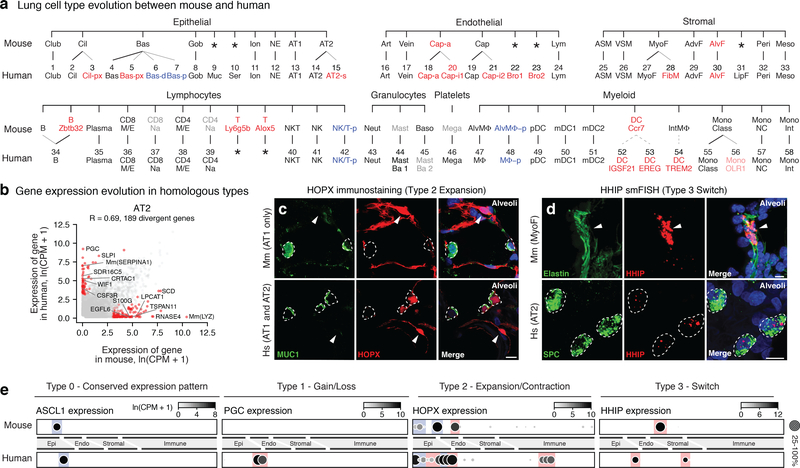
Construction of a mouse lung atlas2 plus additional cells annotated as above for human (Supplementary Table 6) allowed analysis of evolutionary conservation of lung cell types and their transcriptomes. Homologous cell types were assigned by conserved expression of cell-type markers (Fig. 4a). Remarkably, mice appear to lack 17 (29%) of the 58 human lung types including 12 of the 14 (86%) newly identified types. Some missing mouse populations might be rare, transient, unstable, or too diverged to relate transcriptionally so may be uncovered by further studies. By contrast, just five mouse cell populations, all immune, were not found in human. This suggests substantial diversification of lung cell types during mammalian evolution.
Figure 4. Evolutionary divergence of lung cell types and expression patterns.
a, Mouse (top) lung molecular cell types (profiled and identified as for human, see Methods) aligned with homologous human types (bottom, Figs. 2a, 3a) by expression of classical markers in Supplementary Table 6. Thin lines, evolutionary expansions; dashed lines, potential expansions of functionally-related types. Red text, newly identified populations (light red, identified in only one subject); blue, cell states more abundant in human; gray, extant mouse cell types not captured in our data or found in only one patient in human; *, missing cell types. b, Scatter plot comparing average expression levels (dots) in AT2 cells of each expressed human gene and mouse ortholog (SS2 datasets; n, 3,404 human and 318 mouse AT2 cells). R, Pearson correlation coefficient. Red dots, divergent genes (selected ones indicated) expressed 20-fold higher in either species, p<0.05 (‘MAST’ differential gene expression test). Scale, ln(CPM+1). c, Alveolar sections from mouse (top, Mm) and human (bottom, Hs) immunostained for HOPX (red) and AT2 marker MUC1 (green), and DAPI (blue). HOPX is expressed selectively in AT1 cells (arrowheads) in mouse but in human expression has expanded to AT2 and AT2-s cells (dashed circles). Bars, 10μm. Staining repreated on 3 subjects and mice. d, Alveolar sections from mouse (top) and human (bottom) probed by smFISH for Hhip and HHIP (red) and hydrazide staining for myofibroblast marker elastin (green) in mouse and smFISH for AT2 marker SFTPC (green) in human. Note HHIP expression switch from myofibroblast (mouse, arrowhead) to AT2 cells (human, dashed circles). Bars, 10μm. Staining repreated on 3 human subjects and mice. e, Dot plots of expression (SS2 datasets) of homologous genes indicated in mouse and human lung cell types (ordered as in panel a) exemplifying the four observed scenarios (Type 0,1,2,3) for evolution of cellular expression pattern. Colors highlight cell types with conserved (blue) and diverged (red) expression. For more details on statistics and reproducibility, please see Methods.
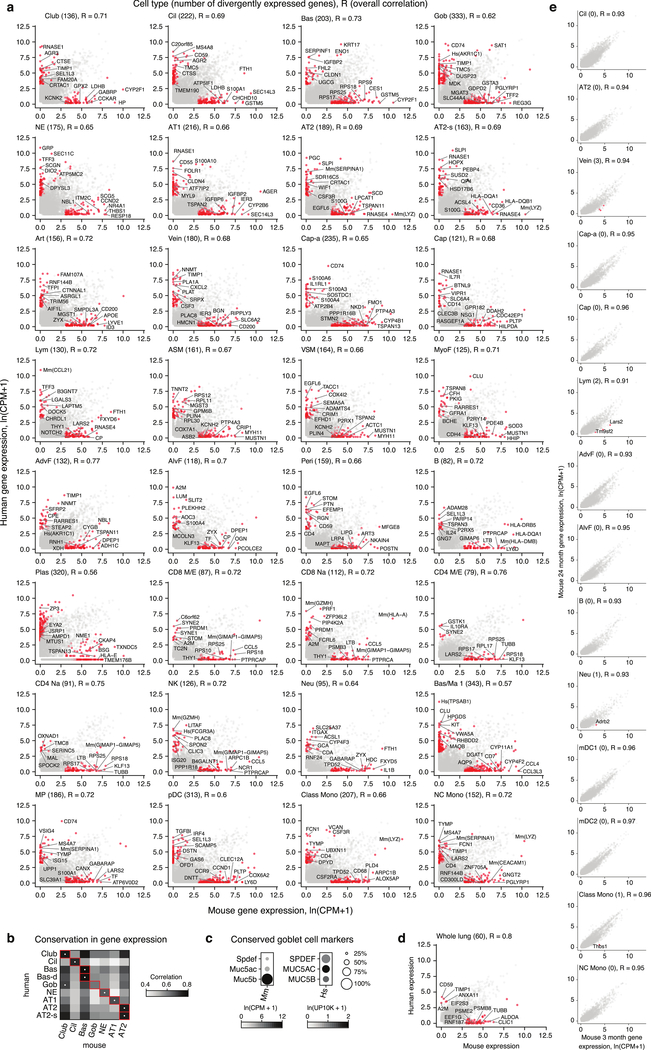
We compared expression levels of all active genes in each human cell type with those of the orthologous genes in the corresponding mouse type (Fig. ED11a, Supplementary Table 7). Most cell types correlated best with their counterparts across species, but surprisingly one human type (goblet) showed greater correlation with another mouse type (club, R=0.68 versus 0.63) (Fig. ED11b)—despite conserved expression of canonical markers and master regulator SPDEF (Fig. ED11c). Corresponding cell types diverged in expression (≥20-fold change, p<0.05) of hundreds of genes, such as SERPINA1, PGC, WIF1, and LYZ in AT2 cells (Fig. 4b). Lung as a whole had fewer diverged genes than any cell type, suggesting expression lost in one type is gained in another (Fig. ED11d). Diverged genes varied above age-related expression changes in mice (Fig. ED11e) and included canonical cell type markers, transcription factors, signaling molecules, and disease genes.
Evolutionary changes in expression grouped into four types (Supplementary Table 7). Type 0 (“conserved”) genes are expressed in the same cell types in mouse and human (Figs. 4e,ED13a). Type 1 (“expression gain/loss”) genes show simple gain (or loss) of expression between species, which involved a single cell type (Type 1a, PGC, Fig. 4e), multiple types (Type 1b, RNASE1, Fig. ED12b), or entire lung (Type 1c, TRIM38, Fig. ED12b). Type 2 (“expression expansion/contraction”) changes involved gain (or loss) of expression in additional lung cell types, expanding (or contracting) expression of the gene during evolution. For example, Hopx, the canonical AT1 transcription factor in mouse, is expressed in both AT1 and AT2 cells in human (Fig. 4c,e), implying existence of other AT1 transcription factors such as MYRF, which is AT1-selective in both species (Fig. ED12c). Expanded expression of RAMP3, co-receptor for vasodilators CGRP and adrenomedullin, presumably alters pulmonary vascular response to these hormones (Fig. ED12d).
Type 3 (“expression switch”) changes involve a switch in expression from one cell type to another. Two medically important examples are COPD/emphysema genes SERPINA1 and HHIP, both selectively expressed in AT2 cells in human but alveolar stromal cells in mice (Figs. 4d,e;ED12e); other hedgehog pathway components were mostly conserved (Fig. ED12f). Extreme examples occurred during evolution of species-specific cell types, such as consolidation in expression of anti-bacterial enzymes (LTF,LYZ,BPIFB1) from multiple mouse airway cells into human-specific serous cells, and consolidation of broadly expressed lipid-handling genes (PLIN2,APOE) from mouse alveolar fibroblasts (which can contain lipid droplets) and myofibroblasts to human-specific lipofibroblasts (Fig. ED12g).
Despite general conservation of cell type expression patterns noted above, just ~6% of expressed genes showed fully conserved patterns (Type 0), most extremely specific or broadly expressed (Fig. ED12h, Supplementary Table 8). Thus, expression patterns of nearly all genes are evolutionarily labile, most undergoing broadening (~55%, Type 2) or simple gain/loss (29%, Type 1) and rarely cell type switching (10%, Type 3) (Supplementary Table 9).
Discussion
We constructed a comprehensive expression atlas of human lung comprising 58 molecular types and their locations (Fig. 1b) including 41 of 45 previously known cell types, all but the exceedingly rare. We identified 14 novel populations across all four compartments that are as distinct molecularly as the canonical cell types; each must be thoroughly characterized, as done for new capillary types (Gillich et al., accompanying paper). If there are other lung cell types, they must be exceedingly rare, fragile, region- or stage-specific, or so similar to the 58 they are not resolved by current methods.
The atlas has broad implications for physiology and medicine, providing insight into the functions, regulation, and interactions of the known and new cell types. It identifies those directly affected by hormones, viruses, and extant lung disease genes, and distinguishes lung resident and homing immune cell types and infers their expression changes following egression from circulation and the cellular sources of homing signals. The atlas defines type-selective transcription factors for creating cells to engineer a lung, and provides optimal markers and a benchmark for monitoring all types and how they change during development, aging, disease, and evolution.
Mice appear to lack 17 of the 58 human lung types, including most (12 of 14) of the newly discovered types. This suggests a significant expansion of cell types in the human lineage, perhaps for new functions, durability, or regenerative capacity of our 6000-fold larger lungs and 30-times longer lifespan37,38. Even homologous cell types diverged in expression of hundreds of genes. Indeed, just 6% of expressed genes had fully conserved expression patterns across the lung, implying widespread gain, loss, or conversion of cell-type-specific transcriptional enhancers during mammalian evolution. It will be important to unravel the genetic mechanisms and functional consequences of these changes, and to elucidate the selective forces operative for genes with fully conserved expression. The evolutionary cell type and expression changes predict where mouse will fail to model human lung physiology and disease.
The success of our atlas relied on: procuring fresh tissue across the organ plus matched blood; balancing tissue compartments to ensure broad cell representation; extensive profiling of each subject using broad cell capture and deep gene coverage scRNAseq strategies; clustering subject and compartment data separately and iteratively; assigning cell identies using extant markers, functions of selectively-expressed genes, and tissue localization. Applying the approach to other organs could create a comprehensive human molecular cell atlas.
Methods
Human lung tissue and peripheral blood
Freshly resected lung tissue was procured intraoperatively from patients undergoing lobectomy for focal lung tumors. Normal lung tissues (~5 cm3) were obtained from uninvolved regions and annotated for the specific lung lobe and location along the airway or periphery. Pathological evaluation (by G.B.) confirmed normal histology of the profiled regions, except for areas of very mild emphysema in Patient 1. Patient 1 was a 75 year-old male with a remote history of smoking, diagnosed with early stage adenocarcinoma who underwent left upper lobe (LUL) lobectomy; two blocks of normal tissue were obtained from lung periphery (“Distal 1a and 1b”). Patient 2 was a 46 year-old male, non-smoker with a right middle lobe (RML) endobronchial carcinoid, who underwent surgical resection of the right upper and middle lobes; two blocks of tissue were selected from mid-bronchial region (“Medial 2”) and periphery (“Distal 2”) of right upper lobe (RUL). Patient 3 was a 51 year-old female, non-smoker with a LLL endobronchial typical carcinoid, who underwent LLL lobectomy; three tissue blocks were resected from the bronchus (“Proximal 3”), mid-bronchial (“Medial 2”), and periphery (“Distal 3”) of the LLL. All tissues were received and immediately placed in cold phosphate buffered saline (PBS) and transported on ice directly to the research lab for single cell dissociation procedures. Peripheral blood was collected from patients 1 and 3 in EDTA tubes. For bulk RNAseq of canonical immune populations, whole blood from healthy human donors was obtained from AllCells Inc in EDTA tubes. Patient tissues were obtained under a protocol approved by Stanford University’s Human Subjects Research Compliance Office (IRB 15166) and informed consent was obtained from each patient prior to surgery. All experiments followed applicable regulations and guidelines.
Mouse lung tissue
Lung tissue for Tabula Muris Senis39 was obtained as previously described. We obtained additional tissue from two mice expressing Cre recombinase and two expressing estrogen-inducible Cre recombinase (Cre-ERT2) for conditional cell-specific labeling in vivo with the gene-targeted alleles FVB-Tbx4-LME-Cre40,41 (lung stroma) and B6.129-Axin2-Cre-ERT240, respectively. Cre-dependent reporter alleles Rosa26ZsGreen1, which expresses cytosolic ZsGreen1 following Cre-mediated recombination, and Rosa26mTmG, which expresses membrane-targeted GFP (mGFP) following recombination and membrane-targeted tdTomato (mTomato) in all other tissues, were used to label cells expressing Tbx4 and Axin2, respectively42,43. Induction of the Axin2-Cre-ERT2 allele was done by intraperitoneal injection of tamoxifen (3 mg) once a day for three days as described25. All mouse experiments followed applicable regulations and guidelines and were approved by the Institutional Animal Care and Use Committee at Stanford University (Protocol 9780).
Isolation of lung and blood cells
Individual human lung samples were dissected, minced, and placed in digestion media (400 μg/ml Liberase DL (Sigma 5466202001) and 100 μg/ml elastase (Worthington LS006365) in RPMI (Gibco 72400120) in a gentleMACS c-tube (Miltenyi 130–096-334). Samples were partially dissociated by running ‘m_lung_01’ on a gentleMACS Dissociator (Miltenyi 130–093-235), incubated on a Nutator at 37°C for 30 minutes, and then dispersed to a single cell suspension by running ‘m_lung_02’. Processing buffer (5% fetal bovine serum in PBS) and DNAse I (100 μg/ml, Worthington LS006344) were then added and the samples rocked at 37°C for 5 minutes. Samples were then placed at 4oC for the remainder of the protocol. Cells were filtered through a 100 μm filter, pelleted (300 x g, 5 minutes, 4°C), and resuspended in ACK red blood cell lysis buffer (Gibco A1049201) for 3 minutes, after which the buffer was inactivated by adding excess processing buffer. Cells were then filtered through a 70 μm strainer (Fisherbrand 22363548), pelleted again (300 x g, 5 minutes, 4°C), and resuspended in magnetic activated cell sorting (MACS) buffer (0.5% BSA, 2 mM EDTA in PBS) with Human FcR Blocking Reagent (Miltenyi 130–059-901) to block non-specific binding of antibodies (see below).
Immune cells, including granulocytes, were isolated from peripheral blood using a high density ficoll gradient44. Briefly, peripheral blood was diluted 10-fold with FACS buffer (2% FBS in PBS), carefully layered on an RT Ficoll gradient (Sigma HISTOPAQUE®−1119), and centrifuged at 400 x g for 30 minutes at room temperature. The buffy coat was carefully removed, diluted 5-fold with FACS buffer, pelleted (300 x g, 5 minutes, 4°C), and incubated in ice cold FACS buffer containing DNAse I (Worthington LS006344) for 10 minutes at 4°C. Clumps were separated by gentle pipetting to create a single cell suspension.
Mouse lung samples were processed into single cell suspensions as previously described2. Briefly, each lung was dissected, minced, and placed in gentleMACS c-tubes (Miltenyi 130–096-334) with digestion buffer (400 μg/ml Liberase DL (Sigma 5466202001) in RPMI (Gibco 72400120)). The minced tissue was partially dissociated by running ‘m_lung_01’ on a gentleMACS Dissociator (Miltenyi 130–093-235), incubated at 37°C on a nutator for 30 minutes, completely dissociated on a gentleMACS by running ‘m_lung_02’, and kept at 4oC or on ice for the remainder of the protocol. Cells were washed with 5% FBS in PBS, centrifuged at 300 x g for 5 minutes, resuspended in 5% FBS in PBS, filtered through a 70 μm strainer (Fisherbrand 22363548), and centrifuged again and resuspended in FACS buffer (2% FBS in PBS).
Magnetic separation of lung tissue compartments
Immune and endothelial cells were overrepresented in our previous mouse single cell suspensions. To partially deplete these populations in our human samples, we stained cells isolated from lung with MACS microbeads conjugated to CD31 and CD45 (Miltenyi 130–045-801, 130–091-935) then passed them through an LS MACS column (Miltenyi, 130–042-401) on a MidiMACS Separator magnet (Miltenyi, 130–042-302). Cells retained on the column were designated “immune and endothelial enriched.” The flow through cells were then split, with 80% immunostained for FACS (see below) and the remaining 20% stained with EPCAM microbeads (Miltenyi 130–061-101). EPCAM stained cells were passed through another LS column. Cells retained on the column were labeled “epithelial enriched”, and cells that flowed through were designated “stromal”.
Flow cytometry and cell sorting
Lysis plates for single cell mRNA sequencing were prepared as previous described2. 96-well lysis plates were used for cells from the blood and mouse samples and contained 4 μL of lysis buffer instead of 0.4 μL.
Following negative selection against immune and endothelial cells by MACS, the remaining human lung cells were incubated with FcR Block (Becton Dickinson (BD) 564219) for 5 minutes and stained with directly conjugated anti-human CD45 (Biolegend 304006) and EPCAM (eBioscience 25–9326-42) antibodies on a Nutator for 30 minutes at the manufacturer’s recommended concentration. Cells were then pelleted (300 x g, 5 minutes, 4°C), washed with FACS buffer three times, then incubated with cell viability marker Sytox blue (1:3000, ThermoFisher S34857) and loaded onto a Sony SH800S cell sorter. Living single cells (Sytox blue-negative) were sorted into lysis plates based on three gates: EPCAM+CD45− (designated “epithelial”), EPCAM−CD45+ (designated “immune”), and EPCAM−CD45− (designated “endothelial or stromal”).
Immune cells from subject matched blood were incubated with FcR Block and Brilliant Violet buffer (BD 563794) for 20 minutes and then stained with directly conjugated anti-human CD3 (BD 563548), CD4 (BD 340443), CD8 (BD 340692), CD14 (BD 557831), CD19 (Biolegend 302234), CD47 (BD 563761), CD56 (BD 555516), and CD235a (BD 559944) antibodies for 30 minutes at the manufacturer’s recommended concentration. Cells were pelleted (300 x g, 5 minutes, 4°C), washed with FACS buffer twice, and then incubated with the viability marker propidium iodide and loaded onto a BD FACSAria II cell sorter. Living (propidium iodide-negative) single, non-red blood (CD235a−) cells were sorted into lysis plates along with specific immune populations: B cells (CD19+CD3−), CD8+ T cells (CD8+), CD4+ T cells (CD4+), NK cells (CD19−CD3−CD56+CD14−), classical monocytes (CD19− CD3−CD56−CD14+). After sorting, plates were quickly sealed, vortexed, spun down for 1 minute at 1000 x g, snap frozen on dry ice, and stored at −80 until cDNA synthesis.
Mouse cells were incubated with the viability marker DAPI and loaded onto a BD Influx cell sorter. Living (DAPI-negative) single cells were sorted into lysis plates based on presence or absence of the fluorescent lineage label (mEGFP for Axin2-Cre-ERT2, ZsGreen1 for Tbx4-LME-Cre).
Immune cells for bulk mRNA sequencing were incubated with FcR Block for 20 minutes and then stained with one of six panels of directly conjugated antibodies for 30 minutes at the manufacturers recommended concentration: anti-human CD16 (BD 558122), CD123 (BD 560826), CCR3 (R&D FAB155F), ITGB7 (BD 551082), CD3 (BD 555341), CD14 (Invitrogen MHCD1406), CD19 (BD 555414), and CD56 (BD 555517) (“basophils, neutrophils and eosinophils”); anti-human CD16 (BD 558122), CD14 (BD 347497), CD4 (BD 340443), CD3 (BD 555341), CD8 (BD 555368), CD19 (BD 555414), and CD56 (BD 555517) (“classical and nonclassical monocytes”); anti-human CD16 (BD 558122), CD1c (Miltenyi Biotec 130–098-007), CD11c (BD 340544), CCR3 (R&D FAB155F), CD123 (BD 560826), HLA-DR (BD 335796), CD3 (BD 555341), CD4 (BD 555348), CD8 (BD 555368), CD14 (Invitrogen MHCD1406), CD19 (BD 555414), and CD56 (BD 555517) (“pDCs, mDCs, CD16+ DCs”); anti-human IgM/IgD (BD 555778), CD19 (BD 557835), CD27 (BD 558664), CD20 (BD 335794), CD3 (BD 555341), CD4 (BD 555348), CD14 (Invitrogen MHCD1406), and CD56 (BD 555517) (“B cells”); anti-human CD16 (BD 558122), CD57 (BD 347393), CD56 (BD 557747), CD3 (BD 555341), CD4 (BD 555348), CD14 (Invitrogen MHCD1406), and CD19 (BD 555414) (“NK cells”); and anti-human CD45RA (Biolegend 304118), CCR7 (R&D FAB197F), CD62L (BD 555544), CD45RO (BD Pharmingen 560608), CD4 (BD 340443), CD8 (BD 340584), CD11b (BD 555389), CD14 (Invitrogen MHCD1406), CD19 (BD 555414), CD56 (BD 555517) (“T cells”). Cells were washed with FACS buffer twice, incubated with the viability marker propidium iodide and loaded onto a BD FACSAria II cell sorter. 40,000 cells from 21 canonical immune populations (Supplementary Table 3) were sorted in duplicate into Trizol LS (Invitrogen 10296010).
After sorting, all plates and samples were quickly sealed, vortexed, spun down for 1 minute at 1000 x g and then snap frozen on dry ice and stored at −80 until cDNA synthesis.
Single cell mRNA sequencing
mRNA from single cells sorted from human and mouse lungs and human blood into lysis plates was reverse transcribed to complementary DNA (cDNA) and amplified as previously described2. Illumina sequencing libraries for cDNA from single cells were prepared as previously described2. Briefly, cDNA libraries were prepared using the Nextera XT Library Sample Preparation kit (Illumina, FC-131–1096). Nextera tagmentation DNA buffer (Illumina) and Tn5 enzyme (Illumina) were added, and the sample was incubated at 55°C for 10 minutes. The reaction was neutralized by adding “Neutralize Tagment Buffer” (Illumina) and centrifuging at room temperature at 3,220 x g for 5 minutes. Mouse samples were then indexed via PCR by adding i5 indexing primer, i7 indexing primer, and Nextera NPM mix (Illumina). Human samples were similarly indexed via PCR using custom, dual-unique indexing primers (IDT)2.
Following library preparation, wells of each library plate were pooled using a Mosquito liquid handler (TTP Labtech), then purified twice using 0.7x AMPure beads (Fisher A63881). Library pool quality was assessed by capillary electrophoresis on a Tapestation system (Agilent) with either a high sensitivity or normal D5000 ScreenTape assay kit (Agilent) or Fragment analyzer (AATI), and library cDNA concentrations were quantified by qPCR (Kapa Biosystems KK4923) on a CFX96 Touch Real-Time PCR Detection System (Biorad). Plate pools were normalized and combined equally to make each sequencing sample pool. A PhiX control library was spiked in at 1% before sequencing. Human libraries were sequenced on a NovaSeq 6000 (Illumina) and mouse libraries on a NextSeq 500 (Illumina).
Cells isolated from each compartment (“immune and endothelial enriched”, “epithelial enriched”, “stromal”) and subject blood were captured in droplet emulsions using a Chromium Single-Cell instrument (10x Genomics) and libraries were prepared using the 10x Genomics 3’ Single Cell V2 protocol as previously described2. All 10x libraries were pooled and sequenced on a NovaSeq 6000 (Illumina).
Immune cell bulk mRNA sequencing
Total RNA from bulk-sorted canonical immune populations was reverse transcribed to cDNA, amplified, and prepared as sequencing libraries as previously described44. Libraries were sequenced on a NextSeq 500 (Illumina).
Immunohistochemistry
Mouse and human lungs were collected as previously described25,45. After inflation, lungs were removed en bloc, fixed in 4% paraformaldehyde (PFA) overnight at 4°C with gentle rocking, then cryo-embedded in Optimal Cutting Temperature compound (OCT, Sakura) and sectioned using a cryostat (Leica) onto Superfrost Plus Microscope Slides (Fisherbrand). Immunohistochemistry was performed using primary antibodies raised against the following antigens and used at the indicated dilutions to stain slides overnight at 4°C: anti-proSP-C (rabbit, Chemicon AB3786, 1:250 dilution), HES1 (rabbit, Cell Signaling 11988S clone D6P2U, 1:100), MUC-1 (hamster, Thermo Scientific HM1630, clone MH1, 1:250), Ki67 (rat, DAKO M7249 clone MIB-1, 1:100), and Keratin-5 (chicken, Biolegend 905901, 1:100). Primary antibodies were detected with Alexa Fluor-conjugated secondary antibodies (Jackson ImmunoResearch) unless otherwise noted, then mounted in Vectashield containing DAPI (5 ug/ml, Vector labs). Images were acquired with a laser scanning confocal fluorescence microscope (Zeiss LSM780) and processed with Fiji (version 2.0) and Imaris (version 9.2.0, Oxford Instruments). Immunostaining experiments were performed on at least 2 human or mouse subjects distinct from the donors used for sequencing, and quantifications were based on at least 10 fields of view in each.
Single molecule in situ hybridization
Samples were fixed in either 10% neutral buffered formalin, dehydrated with ethanol and embedded in paraffin wax or fixed in 4% paraformaldehyde and embedded in OCT compound. Sections from paraffin (5 μm) and OCT (20 μm) blocks were processed using standard pre-treatment conditions for each per the RNAscope multiplex fluorescent reagent kit version 2 (Advanced Cell Diagnostics) assay protocol. TSA-plus fluorescein, Cy3 and Cy5 fluorophores were used at 1:500 dilution. Micrographs were acquired with a laser scanning confocal fluorescence microscope (Zeiss LSM780) and processed with ImageJ and Imaris (version 9.2.0, Oxford Instruments). smFISH experiments were performed on at least 2 human or mouse subjects distinct from the donors used for sequencing, and quantifications were based on at least 10 fields of view in each. For smFISH, fields of view were scored manually, calling a cell positive for each gene probed if its nucleus had >3 associated expression puncta. Proprietary (Advanced Cell Diagnostics) probes used were: KRT5 (547901-C2), SERPINB3 (828601-C3), SFTPC (452561-C2), WIF1 (429391), CLDN5 (517141-C2, 517141-C3), MYC (311761-C3), ACKR1 (525131, 525131-C2), COL1A2 (432721), GPC3 (418091-C2), SERPINF1 (564391-C3), C20rf85 (560841-C3), DHRS9 (467261), GJA5 (471431), CCL21 (474371-C2), COX4I2 (570351-C3), APOE (433091-C2), ACGT2 (828611-C2), ASPN (404481), IGSF21 (572181-C3), GPR34 (521021), EREG (313081), GPR183 (458801-C2), TREM2 (420491-C3), CHI3L1 (408121), MYRF (499261), AGER (470121-C3), TBX5 (564041), KCNK3 (536851), ACVRL1 (559221), SERPINA1 (435441), HHIP (464811), Slc7a10 (497081-C2), Fgfr4 (443511), Pi16 (451311-C2), Serpinf1 (310731), Hhip (448441-C3), Sftpc (314101-C2), Nkx2–1 (434721-C3), and Myrf (524061).
Bioinformatic Methods
Read alignments and quality control
Reads from single cells isolated using 10x chromium were demultiplexed and then aligned to the GRCh38.p12 human reference (from 10x Genomics) using Cell Ranger (version 2.0, 10x Genomics). Cells with fewer than 500 genes detected or 1000 unique molecular identifiers (UMIs) were excluded from further analyses.
Reads from single cells isolated by flow cytometry were demultiplexed using bcl2fastq (version 2.19.0.316, Illumina), pruned for low nucleotide quality scores and adapter sequences using skewer (version 0.2.2), and aligned to either (depending on organism) the GRCh38.p12 human reference genome with both the gencode-vH29 and NCBI-108 annotations or the GRCm38.p6 mouse reference genome with the NCBI-106 annotation (with fluorescent genes mEGFP, tdTomato, and ZsGreen1 supplemented) using STAR (version 2.6.1d) in two-pass mapping mode, in which the first pass identifies novel splice junctions and the second pass aligns reads after rebuilding the genome index with the novel junctions. The number of reads mapping to each annotated gene were calculated by STAR during the second pass alignment, and cells with fewer than 500 genes detected or 50,000 mapped reads were excluded from later analyses. Reads from mRNA sequencing of canonical immune populations were demultiplexed, aligned, and quantified using the same pipeline.
Cell clustering, doublet calling, and annotation
Expression profiles of cells from different subjects and different capture approaches (10x and SS2) were clustered separately using the R software package Seurat (version 2.3)46. Briefly, counts (SS2) and UMIs (10x) were normalized across cells, scaled per million (SS2) or per 10,000 (10x), and converted to log scale using the ‘NormalizeData’ function. These values were converted to z-scores using the ‘ScaleData’ command and highly variable genes were selected with the ‘FindVariableGenes’ function with a dispersion cutoff of 0.5. Principle components were calculated for these selected genes and then projected onto all other genes with the ‘RunPCA’ and ‘ProjectPCA’ commands. Clusters of similar cells were detected using the Louvain method for community detection including only biologically meaningful principle components (see below) to construct the shared nearest neighbor map and an empirically set resolution, as implemented in the ‘FindClusters’ function.
When clustering all cells from a single subject at once, we found that the first principal components defining heterogeneity represented differences in tissue compartment, but some cell types within a compartment (e.g., basal, goblet club, neuroendocrine and ionocyte) had a tendency to co-cluster. Clusters were therefore grouped based on expression of tissue compartment markers (e.g. EPCAM, CLDN5, COL1A2, and PTPRC) using the ‘SubsetData’ command and the same procedure (from ‘ScaleData’ onwards) was applied iteratively to each tissue compartment until the markers enriched in identified clusters, identified using the ‘MAST’ statistical framework47 implemented in the ‘FindMarkers’ command, were no longer biologically meaningful (e.g. clusters distinguished by dissociation-induced genes29, ribosomal genes, mitochondrial genes, or ambient RNA released by abundant cells such as RBCs30). Doublets were identified by searching for cells with substantial and coherent expression profiles from two or more tissue compartments and/or cell types.
To assign clusters identities, we first compiled a list of all established lung cell types, their abundances, their classical markers, and any RNA markers (when available) (Supplementary Table 1). RNA markers for canonical immune populations (Supplementary Table 3) were obtained from bulk mRNA sequencing by correlating the average expression (each captured in duplicate) with a test vector where the target population position equaled 10 and all others equaled 0 (see GitHub for details). Clusters were assigned a canonical identity based on enriched expression of these marker genes. Pearson correlations were calculated between the average expression profiles from each immune cluster for all cells in the SS2 with the average bulk profiles using the ‘cor’ function in R. There were no clusters that lacked expression of canonical marker genes. When two or more clusters were assigned the same identity, we first determined whether their tissue locations differed substantially (e.g. proximal versus distal, alveolar versus adventitial) and prepended these locations when applicable. When both clusters localized to the same tissue region (e.g. capillary endothelial cells or AT2 cells), we next compared their differentially expressed genes head-to-head to identify differences in molecular functions. These functional differences were also prepended, when applicable (e.g. Signaling AT2 versus AT2, Proliferative Basal versus Basal). If the clusters could not be resolved by location or function, we prepended a representative marker gene to their “canonical” identity (e.g. IGSF21+ Dendritic, EREG+ Dendritic, and TREM2+ Dendritic). Cells from different subjects with the same annotation were merged into a single group for all downstream analyses.
Approximately 35,000 mouse lung and blood cell expression profiles by SS2 and 10x from Tabula Muris Senis2 were combined with 522 cells isolated from Axin2-Cre-ERT2 > Rosa26mTmG (A.N.N.) and Tbx4-LME-Cre > Rosa26ZsGreen1 (K.J.T.) mice and amplified by SS2. Cells were stratified by technology (10x versus SS2), re-clustered and re-annotated using the strategy described above for human lung cells.
Re-annotation of existing human lung single cell RNA sequencing datasets
UMI tables were obtained from the Gene Expression Omnibus (GSE122960 for Reyfman et al, GSE130148 for Braga et al), clustered, and annotated using the strategy described above. New annotations for each cell are available on GitHub (see below).
Cell type pairwise correlations
We obtained average expression profiles for each cell type from all cells in the 10x dataset, supplemented with the average expression profile from neutrophils in the SS2 dataset, and calculated pairwise Pearson correlation coefficients using the ‘cor’ function in R.
Identification of proliferation signature
Expression profiles from matched proliferating and quiescent cell types were compared head-to-head using the ‘MAST’ statistical framework implemented in the ‘FindMarkers’ command in Seurat. Differentially-expressed genes common in each proliferating cell type were converted to z-scores using the ‘ScaleData’ command in Seurat, and summed to create a “proliferation score” for each cell in the 10x dataset.
Identification of immune egression signatures
Blood and tissue expression profiles for each immune cell type were compared head-to-head using the ‘MAST’ statistical framework implemented in the ‘FindMarkers’ command in Seurat. Differentially-expressed genes common in each subject were screened for dissociation artifact and contamination by red blood cells. Genes specific to tissue immune cells were binned based on their breadth of expression (lymphocyte, myeloid, or both), converted to z-scores using the ‘ScaleData’ command in Seurat, and summed to create an “egression score” for each cell in the 10x dataset.
Identification of enriched marker genes, transcription factors, and disease genes
Differentially-expressed genes for each annotated cell type relative to the other cells within its tissue compartment were identified using the ‘FindMarkers’ command in Seurat with the ‘MAST’ statistical framework after downsampling each cell type to 100 (SS2) or 500 (10x) cells. To obtain the most sensitive and specific markers for each cell type, we ranked enriched genes, with a p-value less than 10−5 and a sensitivity greater than 0.4, by their Mathews Correlation Coefficients (MCCs) calculated for each cell type from all cells in the 10x dataset (numbers available in Supplementary Table 2). To measure the utility of using multiple markers in assigning cell identities, we calculated MCC scores for all possible combinations of each cell type’s top five marker genes.
Enriched genes were annotated as transcription factors or genes associated with pulmonary pathology based on lists compiled from The Animal Transcription Factor Database48, The Online Mendelian Inheritance in Man Catalog (OMIM)49, and Genome Wide Association Studies (GWAS) obtained from the EMBL-EBI Catalog50 (EFO IDs 0000270, 0000341, 0000464, 0000571, 0000702, 0000707, 0000708, 0000768, 0001071, 0003060, 0003106, 0004244, 0004312, 0004313, 0004314, 0004647, 0004713, 0004806, 0004829, 0005220, 0005297, 0006505, 0006953, 0007627, 0007744, 0007944, 0008431, 0009369, 0009370; GO IDs 0031427, 0097366; Orphanet IDs 586 182098; log(p-value) < −20, statistical tests vary in indicated studies). Viral entry genes were obtained from Gene Ontology (GO:0046718) and then curated and associated with their cognate virus(es) based on literature citations available in our GitHub repository.
Cellular interaction and hormone target mapping
Interactions between cell types were predicted using CellPhoneDB (‘statistical_analysis’ method) with all cells in the SS2 dataset, as previously described6. For our targeted analyses, we curated the chemokine receptor-ligand interaction map and list of hormone receptors from an extensive literature search (available on GitHub, see below).
Human and mouse gene alignment, cell type correlation, and gene expression comparisons
The gene expression matrices from our human SS2 cells and the Tabula Muris Senis SS2 cells, supplemented with the 522 mouse cells from Axin2-CreER > mTmG and Tbx4-Cre > ZsGreen1 described above, were collapsed to HomologyIDs obtained from the Mouse Genome Informatics database to enable direct comparison. We obtained mean expression profiles for each cell type from all cells in the SS2 dataset and calculated pairwise Pearson correlation coefficients using the ‘cor’ function in R. We defined species-specific gene expression as those enriched 20-fold in either direction (mouse > human or human > mouse) with a p-value less than 10−5 (calculated by ‘MAST’ as above) from all cells for the indicated types in the SS2 dataset. Correlations and age-specific genes were obtained the same manner using all cells from 3-month and 24-month in the combined SS2 mouse dataset.
To compare the expression pattern of each gene across species we binarized genes as expressed (1) or not expressed (0) in each cell type’s average expression profile calculated from all mouse and human SS2 cells of the types compared above. A cell type “expressed” a gene if the median of that gene’s non-zero expression values across the constituent cells was greater than the median of every non-zero expression value for all other genes plus/minus two standard deviations (varied in 0.25 increments) and if the percentage of cells within the cell type with non-zero expression values was greater than the median percent of non-zero expression values for all other genes plus/minus two standard decisions (varied in 0.25 increments). These cutoffs were varied independently to ensure genes were robustly categorized. We then ordered these gene vectors to match homologous cell types between species with at least five cells and combined them to a single vector for each gene (V = (a – b) + 2ab, where a is the ordered human vector and b is the ordered mouse vector) that indicated for each cell type whether: Both mouse and human expressed the gene (2), only human (1), only mouse (−1), or neither (0). We then classified genes by the following: Conserved if any element of V equaled 2 and all other elements equaled 0, Type 2 if any element equaled 2 and any other equaled 1 or −1, not expressed if all elements equaled 0, Type 3 if elements were both positive and negative, and Type 1 if elements were either positive or negative and 0.
Statistics and reproducibility
All heatmaps and plots with single cell expression data include every cell from indicated types (numbers available in Supplementary Table 2 for human and Supplementary Table 6 for mouse) for sequencing technology specified (SS2 or 10x), unless otherwise stated. Scatter plots were generated with ggplot2’s ‘geom_point’ function. Dot plots were generated using a modified version of Seurat’s ‘DotPlot’ function (available on GitHub). Violin plots were created with Seurat’s ‘VlnPlot’ function and show proportion of single cells at indicated expression levels. Box-and-whisker plots were generated with ggplot2’s ‘geom_boxplot’ function; lower and upper hinges correspond to first and third quartiles, whiskers extend from hinge to the largest/smallest value no further than 1.5 times the interquartile range. Data beyond whiskers are shown as outlying points. Correlations use Pearson’s coefficient. Differentially expressed genes were identified using the ‘MAST’ statistical framework47 implemented in Seurat’s ‘FindMarkers’ function. Immunostaining and smFISH experiments were performed on at least 2 human or mouse subjects distinct from the donors used for sequencing, and quantifications were based on at least 10 fields of view in each. For smFISH, fields of view were scored manually, calling a cell positive for each gene probed if its nucleus had >3 associated expression puncta.
Data availability
Counts/UMI tables, cellular metadata, Seurat objects, and scanpy objects are available on Synapse (https://www.synapse.org/#!Synapse:syn21041850). The data can be explored in a browser using cellxgene at https://hlca.ds.czbiohub.org/. Human sequencing data is available by data access agreement on the European Genome-phenome Archive (EGA) under accession EGAS00001004344. Use of human sequencing data is restricted to not for profit research only and requires approval or a waiver from requesting investigator’s institutional review board. Mouse sequencing data is available on the National Institute of Health’s Sequence Read Archive (SRA) under BioProject accession PRJNA632939. Source data behind immunostaining or smFISH quantification (Figure 1; Extended Data Figures 3 and 4) are available within the manuscript files.
Code availability
The code for demultiplexing counts/UMI tables, clustering, annotation, downstream analyses, and obtaining source data/generating fiGures that include single cell expression data is available on GitHub (https://github.com/krasnowlab/HLCA).
Extended Data
Extended Data Figure 1. Strategy for single cell RNA sequencing and annotation of human lung and blood cells.
a, Workflow for capture and mRNA sequencing of single cells from the healthy unaffected regions indicated (D, distal; M, medial; P, proximal lung tissue; see panel d) of fresh, surgically resected lungs with focal tumors from three subjects (1, 2, 3) and their matched peripheral blood. Cell representation was balanced among the major tissue compartments (Endo, endothelial; Immune; Epi, epithelial; Stroma) by magnetic and fluorescence activated cell sorting (MACS and FACS) using antibodies for the indicated surface markers (CD31, CD45, EPCAM; +, marker-positive; -, marker-negative). Cell capture and single cell RNA sequencing (scRNAseq) was done using 10x droplet technology or SmartSeq2 (SS2) analysis of plate-sorted cells. Number of profiled cells from each compartment are shown in parentheses. For blood, immune cells were isolated on a high density Ficoll gradient, and unsorted cells profiled by 10x and sorted cells (using canonical markers for the indicated immune populations) by SS2. Total cell number (all 3 subjects) and median number of expressed genes per cell are indicated for each method. b, Cell clustering and annotation pipeline. Cell expression profiles were computationally clustered by nearest-neighbor relationships and clusters were then separated into tissue compartments based on expression of compartment-specific markers (EPCAM (blue), CLDN5 (red), COL1A2 (green), and PTPRC (purple)), as shown for tSNE plot of lung and blood cell expression profiles obtained by 10x from Patient 3. Cells from each tissue compartment were then iteratively re-clustered until differentially-expressed genes driving clustering were no longer biologically meaningful. Cell cluster annotation was based on expression of canonical marker genes from the literature, markers found through RNA sequencing of purified cell populations (Bulk RNA markers), ascertained tissue location, and inferred molecular function from differentially-expressed genes. c, Heatmap of pairwise Pearson correlations of the average expression profile of each cluster in the combined 10x dataset plus SS2 analysis of neutrophils. n, given in Supplementary Table 2. Tissue compartment and identification number of each of the 58 clusters are indicated. For more details on statistics and reproducibility, please see Methods. d, Representative micrographs of donor lungs from formalin-fixed, paraffin-embedded (FFPE) sections stained with haematoxylin and eosin showing bronchi, bronchioles, submucosal glands, arteries, veins, and alveoli near regions used for single cell RNA sequencing. Staining repeated on at least 5 sections (encompassing different anatomical regions) from each subject used for scRNAseq. Bar, 100 μm.
Extended Data Figure 2. Selectively-expressed RNA markers of human immune cell types from bulk mRNA sequencing of FACS-purified immune cells.
a, Heatmap of RNA expression of the most selectively-expressed genes from bulk mRNA sequencing of the indicated FACS-sorted immune populations (see Supplementary Table 3). This dataset provided RNA markers for human immune cell populations that have been classically defined by their cell surface markers. b, Heatmap of pairwise Pearson correlation scores between the average expression profiles of the immune cell types indicated that were obtained from bulk mRNA sequencing (BulkSeq, panel a) to the average scRNAseq profiles of human blood immune cells in the SS2 dataset annotated by canonical markers and enriched RNA markers from the bulk RNA-seq analysis. The highest correlation in overall gene expression (white dot) of each annotated immune cell cluster in the SS2 dataset (columns) was to the bulk RNA-seq of the same FACS-purified immune population (rows), supporting the scRNAseq immune cluster annotations (red squares). Cell numbers are given in Supplementary Table 2. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 3. Expression differences and localization of lung cell states and canonical epithelial and endothelial subtypes.
a, Proliferative signature score (based on expression of indicated genes in cells from 10x dataset, cell numbers given in Supplementary Table 2) of each cluster of basal cells, T and NK cells, and macrophages. Three clusters had high scores: basal-proliferative (Bas-p), NK/T-proliferative (NK/T-p), and macrophage-proliferative (MP-p). b, Dot plot of mean level of expression (dot intensity, gray scale) of indicated basal cell markers and percent of cells in population with detected expression (dot size) for 10x dataset. Note partial overlap of markers among different basal populations. c, Immunostaining of adult human pseudostratified airway for differentiation marker HES1 (green) in basal cells (marked by KRT5, red) with DAPI (nuclear) counter stain (blue). Bars, 10 μm. Note apical processes extending from HES1+ basal cells (arrowheads) indicating migration away from basal lamina as they differentiate. Other HES1+ cells have turned off basal marker KRT5. Dashed outlines, basal cell nuclei. Quantification shows fraction of basal cells (Bas, cuboidal KRT5+ cells on basement membrane) and Bas-d cells (KRT5+ cells with apical processes) that were HES1+. n, KRT5+ cells scored in sections of 2 human lungs with staining repeated on 4 subjects. d, Immunostaining of adult human pseudostratified airway for proliferation marker MKI67 (green) in basal cells (marked by KRT5, red) with DAPI counter stain (blue). Bars, 5 μm. Quantification shows abundance of proliferating (MKI67-expressing) basal cells (Bas-p) in pseudostratified (pseudo) and simple epithelial airways; n, KRT5+ cells scored in sections of 2 human lungs with staining repeated on 4 subjects. e, Relative abundance of epithelial and stromal cell types in scRNAseq analysis of human lung samples obtained from proximal (blue; 10x cells from P3) and distal (red; 10x cells from D1a, D1b, D2, D3) lung sites. In addition to the expected proximal enrichment of some airway cell types (goblet, gob; ionocytes, ion, neuroendocrine, NE) and distal enrichment of alveolar cell types (AT1, AT2, AT2-s, myofibroblasts), note three bracketed pairs of related cell types (ciliated (cil) and ciliated-proximal (cil-px); basal (bas) and basal-proximal (bas-px); myofibroblasts (MyoF) and fibromyocyte (FibM)) with one of them proximally-enriched. Relative enrichment values are provisional because they can be influenced by efficiency of harvesting during cell dissociation and isolation. Cell number for proximal cells are 357; 275; 73; 175; 153; 191; 39; 145; 57; 24; 20; 10; 328; 1,505; 235; 25; and 70 and for distal cells are 537; 806; 15; 197; 4; 58; 6; 14; 336; 0; 2; 1; 467; 2,095; 434; 198; and 28. f, RNAscope single molecule fluorescence in situ hybridization (smFISH) and quantification for general basal marker KRT5 (red) and Bas-px marker SERPINB3 (white) with DAPI counter stain (blue) and extracellular matrix autofluorescence (ECM, green) on proximal, pseudostratified bronchi and distal, simple bronchioles. Bars, 20 μm (inset, 10 μm). Note Bas-px cell (KRT5 SERPINB3 double positive, yellow arrowhead and box) enrichment at base of pseudostratified airways. SERPINB3 was not detected in simple airways, indicating Bas (but not Bas-px) cells are present there. Staining repeated on 2 subjects. g, Dot plot of expression in ciliated (Cil) and proximal ciliated (Cil-px) cells of canonical (general) ciliated cell markers and specific Cil-px (proximal) markers (in 10x dataset). h, smFISH and quantification of human pseudostratified epithelial (left panel) and simple epithelial (right panel) airways for general ciliated marker C20orf85 (white) and proximal (Cil-px) marker DHRS9 (red) with DAPI counterstain (blue) and ECM autofluorescence (green). Note Cil-px cell restriction to pseudostratified airways. Bars, 10 μm. Staining repeated on 2 subjects. i, Heatmap of expression of representative general AT2, AT2 selective, and AT2-s selective marker genes in AT2 and AT2-s human lung cells (SS2 data). AT2 selective markers include negative regulators of Hedgehog and Wnt signaling pathways (e.g., HHIP, WIF1, highlighted red) and AT2-s selective markers include Wnt ligands, receptors, and transcription factors (e.g., WNT5A, LRP5, TFC7L2 highlighted green). Values shown are ln(CPM+1) for 50 randomly-selected cells in each cluster (SS2 data). j, Dot plot of expression of endothelial markers (10x dataset). k, Micrograph (low magnification, left) of bronchial vessel (boxed region) showing vessel location near airway (dotted outline). smFISH for general endothelial marker CLDN5 (red, center panel), bronchial vessel-specific markers MYC (green) and Bro1-specific marker ACKR1 (red, right panel) on serial sections of bronchial vessel cells (arrowheads), co-stained for DAPI (blue). Bar, 10 μm. Quantification shows relative abundance of Bro1 and Bro2 cells. Staining repeated on 2 subjects. l-n, smFISH and quantification of vessel types indicated (dotted outlines) showing vein marker ACKR1 (red, panel l), artery marker GJA5 (red, m), lymphatic marker CCL21 (red, n), and general endothelial marker CLDN5 with DAPI counter stain (blue) and ECM autofluorescence (green). Bars, 50 μm (l), 30 μm (m), and 40 μm (n). Staining repeated on 2 subjects. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 4. Markers and lung localization of stromal and dendritic subtypes.
(a-d) smFISH for RNA of indicated marker genes of alveolar fibroblasts (AlvF, a, b) and adventitial fibroblasts (AdvF, c, d) in adult human (a, c) and mouse (b, e) alveolar (a, b) and pulmonary artery (c, d) sections. ECM autofluorescence (green, panels a, c) to show blood vessels; Elastin (green, panels b, d); DAPI counterstain (blue, all panels). Staining repeated on 2 human subjects or 3 mice. a, smFISH probes: general fibroblast marker COL1A2 (white) and AlvF-selective marker GPC3 (red). Arrowheads, AlvF cells. Inset, close-up of boxed region showing merged (top) and split channels of an AlvF. Bars, 20 μm (inset 60 μm). b, smFISH probes: AlvF-selective markers Slc7a10 (white) and Frfr4 (red). Elastin (green) shows alveolar entrance ring. Arrowheads, AlvF cells. Bar, 5 μm. c, smFISH probes: general fibroblast marker COL1A2 (white) and AdvF-selective marker SERPINF1 (red). AdvF (some indicated with arrowheads) localize around blood vessels (ECM, green). Inset, close-up of boxed region showing merged (top) and split channels of an AdvF. Dashed line, artery boundary. Bars, 30 μm (inset 90 μm). d, smFISH probes: AdvF-selective markers Pi16 (white) and Serpinf1 (red). AdvF (arrowheads) surround artery (marked by Elastin, green). Bar, 10 μm. e, Heatmap of expression of representative general, adventitial-selective, and alveolar-selective fibroblast markers in 50 randomly-selected cells from AdvF (left) and AlvF (right) clusters (SS2 dataset). Note specialization (highlighted red) in growth factors (AdvF: PDGFRL, IGFBP4; AlvF: FGFR4, VEGFD) and morphogen (AdvF: SFRP2; AlvF: NKD1, DKK3) signaling/regulation. f, g, smFISH and quantification of cell abundance in human alveolar (Alveoli, f) and pseudostratified epithelial airway (Pseudo, g) sections probed for myofibroblast (MyoF) and fibromyocyte marker ASPN (red), and for fibromyocyte (FibM) and airway smooth muscle (ASM) markers COX4I2 (white, f) and ACTG2 (white, g). ECM autofluorescence, green; DAPI counter stain, blue. Inset (f), boxed region showing close-up of merged (top) and split channels of ASPN+ COX4I2− myofibroblast. Myofibroblasts and fibromyocytes (see below) likely make up remaining cells in Figure 1f quantification. Inset (g), boxed regions showing close-up of merged (top) and split channels of FibM (white box) and ASM (yellow box) cells. FibM (white arrowheads) and ASM (yellow arrowheads) are intermingled in wall of pseudostratified airway (dotted outline). Staining repeated on 2 subjects. h, i, smFISH of human alveolar sections probed for general stromal marker COL1A2 (white), pericyte (Peri) marker COX4I2 (red, panel h), lipofibroblast (LipF) marker APOE (red, panel i). ECM autofluorescence, green; DAPI counter stain, blue. Inset (h), boxed region showing close-up of pericyte. Inset (i), boxed region showing close-up of COL1A2 APOE double-positive LipF. LipF cells are intermingled among other stromal cells (single-positive COL1A2) and macrophages (single-positive APOE). Quantification in Fig. 1f. Bars, 20 μm. Staining repeated on 2 subjects. j, Dot plot of COX4I2 expression in alveolar stromal cell types (10x dataset). k, Heatmap of expression of dendritic cell marker genes in 50 randomly-selected cells from indicated dendritic cell clusters (human blood and lung 10x datasets). Cells in all clusters express general dendritic markers including antigen presenting genes but each cluster also has its own selective markers. Red-highlighted markers distinguishing the newly-identified dendritic cell clusters (IGSF21+, EREG+, TREM2+) suggest different roles in asthma (IGSF21+), growth factor regulation (EREG+), and lipid handling (TREM2+). l-n, smFISH of adult human lung proximal and alveolar (Alv) sections as indicated probed for IGSF21+ DC markers IGSF21 (red) and GPR34 (white) (panel l), EREG+ DC marker EREG (red) and general DC marker GPR183 (white) (panel m), and TREM2+ DC markers TREM2 (red) and CHI3L1 (white) (panel n). DAPI counter stain, blue. (Non-punctate signal in red channel (panels l, n) is erythrocyte autofluorescence. Insets, boxed regions showing merged and split channels of close-up of single dendritic cell of indicated type. Bars, 20 μm. Arrowheads, double-positive cells. Quantification shows distribution of each dendritic type; note IGSF21+ and EREG+ dendritic cells show strong proximal enrichment. Staining repeated on 2 subjects. o, tSNE of expression profile clusters of monocytes and B, T, and NK cells (10x dataset, subject 1, 2,622 cells). Note separate cell clusters of each immune cell type isolated from lung (no outline) and blood (dashed outline). Asterisk, small number of B cells isolated from the lung that cluster next to blood B cells. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 5. Markers and transcription factors that distinguish human lung cell types.
a, Violin plots of expression levels (ln(UP10K + 1)) of the most sensitive and specific markers (gene symbols) for each human lung cell type in its tissue compartment (10x dataset). Cell numbers given in Supplementary Table 2. b, Scheme for selecting the most sensitive and specific marker genes for each cell type using Matthews Correlation Coefficient (MCC). Box-and-whisker plots below show MCCs, True Positive Rates (TPR), and False Discovery Rates (FDR) for each cell type (n=58) using indicated number (nGene) of the most sensitive and specific markers (10x dataset). Note all measures saturate at approximately 2–4 genes, hence simultaneous in situ probing of a human lung for the ~100–200 optimal markers would assign identity to nearly every cell. c, Alveolar section of human lung probed by smFISH for AT1 marker AGER and transcription factor MYRF. MYRF is selectively expressed in AT1 cells (arrowheads; 97% of MYRF+ cells were AGER+, n=250 scored cells). Inset, boxed region showing merged and split channels of AT1 cell. Bar, 10 μm. Staining repeated on 2 subjects. d, Alveolar section of human lung probed by smFISH for pericyte marker COX4I2 and transcription factor TBX5. TBX5 is enriched in pericytes (arrowheads, 92% of TBX5+ cells were COX4I2+, n=250). Inset, boxed region showing merged and split channels of pericyte. Bar, 5 μm. Staining repeated on 2 subjects. e, Dot plot of expression of enriched transcription factors in each lung cell type (SS2 dataset). Red text, genes not previously associated with the cell type. Red shading, transcription factors including MYRF that are highly enriched in AT1 cells, and TBX5 and others highly enriched in pericytes. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 6. Lung cell targets of circulating hormones and local signals.
a, Dot plot of hormone receptor gene expression in lung cells (SS2 dataset). Type and name of cognate hormones for each receptor are shown at top. Teal, broadly-expressed receptors in lung; other colors, selectively-expressed receptors (<3 lung cell types). Small colored dots next to cell type names show selectively targeted cell types. AA, amino acid; CGRP, Calcitonin gene-related peptide; AM, adrenomedullin; SST, somatostatin; EPO, erythropoietin; GIP, gastric inhibitory peptide; GH, growth hormone; IGF, insulin-like growth factor; MCCT, mineralocorticoid; GCCT, glucocorticoid; RA, retinoic acid. b, Schematic of inferred pericyte cell contractility pathway and its regulation by circulating hormones (AGT, PTH) and capillary-expressed signals (EDN, NO). Dots show expression of indicated pathway genes: values at left (outlined red) in each pair of dots in capillary diagram (top) show expression in Cap-a cells (aerocytes) and at right (outlined blue) show expression in general Cap cells (SS2 dataset). Note most signal genes are preferentially expressed in Cap relative to Cap-a cells. c, Heatmaps showing number of interactions predicted by CellPhoneDB software between human lung cell types located in proximal lung regions (left panel in each pair) and distal regions (right panel) based on expression patterns of ligand genes (“Sending cell”) and their cognate receptor genes (“Receiving cell”) (SS2 dataset). The pair of heatmaps at upper left show values for all predicted signaling interactions (“All interactions”), and other pairs show values for the indicated types of signals (growth factors, cytokines, integrins, WNT, Notch, Bmp, FGF, and TFGB). Predicted interactions between cell types range from 0 (lymphocyte signaling to neutrophils) to 136 (AdvF signaling to Cap-i1). Note expected relationships, such as immune cells expressing integrins to interact with endothelial cells and having higher levels of cytokine signaling relative to their global signaling, and unexpected relationships, such as fibroblasts expressing majority of growth factors and lack of Notch signaling originating from immune cells. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 7. Lung cell expression patterns of genes implicated in lung disease.
Dot plots of expression (in SS2 dataset) of 233 lung disease genes curated from Genomewide Association Studies (GWAS, genome-wide association genes ≥10−20 significance) and Online Mendelian Inheritance in Man (OMIM). For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 8. Mapping cellular origins of lung disease by cell-selective expression of disease genes.
a, Dot plots of expression of lung disease genes (numbered, associated disease shown above) enriched in specific lung cell types (SS2 datasets). Red, novel cell type association of gene/disease; gray, diseases with developmental phenotype. BBS, Bardet-Biedl syndrome; Dys, dysplasia; IPF, idiopathic pulmonary fibrosis; SMD, surfactant metabolism dysfunction; PH, pulmonary hypertension; SM, smooth muscle; SGB, Simpson-Golabi-Behmel; TB, tuberculosis; AWS, Alagille-Watson syndrome; VDES, Van den Ende-Gupta syndrome; EDS, Ehlers-Danlos syndrome; CF, Cystic fibrosis; Fam Med, Familial Mediterranean; COPD, Chronic Obstructive Pulmonary disease. b, Dot plot of expression (SS2 dataset) of all genes implicated in PH, TB, and COPD/emphysema (OMIM, Mendelian disease genes from OMIM database; GWAS, genome-wide association genes ≥10−20 significance). Note canonical AT2 cells (red shading) express all and AT2-s cells (blue shading) express most. c, smFISH of alveolar section of adult human lung probed for PH disease gene KCNK3 (red) and pericyte marker COX4I2 (white) with DAPI counterstain (blue) and ECM autoflourescence (green). Note pericyte-specific expression (arrowheads, 91% of COX4I2+ pericytes were KCNK3+, n=77). Bar, 5 μm. Cell numbers for each type given in Supplementary Table 2. d, smFISH of alveolar section of adult human lung probed for atrioventricular (AV) dysplasia gene ACVRL1 (red), endothelial marker CLDN5 (white) with DAPI counterstain. Note ACVRL1 CLDN5 double-positive capillaries (white arrowheads, 70% of CLDN5+ capillaries were ACVRL1+, n=102) and some CLDN5 single positive capillaries (yellow arrowheads). Bar, 5 μm e, smFISH of alveolar section of adult human lung probed for COPD/emphysema gene SERPINA1 and AT2 marker SFTPC, and DAPI. Note AT2-specific expression (arrowheads; 93% of AT2 cells were SERPINA1+, n=176). Bar, 5 μm. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 9. Lung cell expression patterns of respiratory virus receptors.
a, Dot plot showing expression in human lung cell types of entry receptors (indicated at left) for respiratory viruses (indicated at right, numbers indicate viral families) (SS2 dataset). Red shading, cell types inhaled viruses could directly access (epithelial cells and macrophages); darker red shading shows expression values for measles receptor NECTIN4 and rhinovirus C receptor CDHR3. b, Violin plots (left) and dot plots (immediately above violin plots) showing expression of coronavirus receptors ACE2, DPP4, and ANPEP in lung cell types (10x dataset, cell numbers given in Supplementary Table 2). Grey shading, cell types inhaled viruses can directly access. Donut plots (right) showing relative number of receptor-expressing cells of cell types viruses can directly access (shaded grey in panel a), normalized by their abundance values from Supplementary Table 1 (and refined by the relative abundance values in Figures 2 and S4). Note prevalence of AT2 alveolar cells for ACE2, receptor for SARS-CoV and SARS-CoV-2, and for DPP4, receptor for MERS-CoV, in contrast to prevalence of macrophages for ANPEP, receptor for common cold causing coronavirus 229E. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 10. Lung cell expression patterns of non-respiratory virus receptors.
a, Dot plot of expression of entry receptors for non-respiratory viruses in human lung cell types (compare with Extended Data Figure Fig. 10a showing expression of receptors for respiratory viruses). For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 11. Comparison of mouse and human gene expression profiles in homologous lung cell types and across age.
a, Scatter plots showing median expression levels (ln(CPM+1)) in indicated cell types of each expressed human gene and mouse ortholog (mouse and human SS2 datasets, human and mouse cell numbers given in Supplementary Tables 2 and 6, respectively). Note tens to hundreds of genes that show a 20-fold or greater expression difference (and p-value < 0.05, MAST) between species (red dots, gene names indicated for some and total number given above). Bas/Ma 1 cells have the most differentially-expressed genes (343), and CD4+ M/E T cells have the least (79). Pearson correlation scores (R values) between the average mouse and human gene expression profiles for each cell type are indicated. “Mm()” and “Hs()”, genes where duplications between mouse and human were collapsed to HomologyID. b, Heatmap showing global transcriptome Pearson correlation between indicated human and mouse epithelial cells (SS2 dataset, human and mouse cell numbers given in Supplementary Tables 2 and 6, respectively). Red outline, homologous cell types based on classical markers described in Supplementary Table 6. White dot, human to mouse correlation. c, Dot plot of expression of canonical goblet cell markers MUC5B and MUC5AC and transcription factor SPDEF in mouse (left) and human (right) goblet cells. d, Scatter plot showing average expression levels (dots) across all cells (“pseudo-bulk” lung expression) of each expressed human gene and mouse ortholog (mouse and human SS2 datasets). Scale, ln(CPM+1). Pearson correlation (R values) between the average mouse and human gene expression profiles are indicated. e, Scatter plots comparing median expression levels (ln(CPM+1)) in indicated mouse lung cell types of each expressed gene at age 3 months (x-axis) and 24 months (y-axis) in SS2 datasets from Tabula Muris Senis56 (cell numbers given in Supplementary Table 6). Pearson correlation scores between average gene expression profile for each cell type at each age are indicated (R values), along with number of genes (red dots) showing 20-fold or greater expression difference (and p-value < 0.05, MAST) between ages. Names of some genes are given next to the corresponding red dot. For more details on statistics and reproducibility, please see Methods.
Extended Data Figure 12. Patterns of conserved and divergent gene expression across human and mouse lung cell types.
a, Dot plots of PTPRC and MYL6 expression in mouse and human lung cell types (SS2 datasets) showing two examples of conserved (Type 0) expression pattern. Blue shading, homologous cell types with conserved expression. b, Dot plots showing gain of expression (Type 1 change) in multiple human cell types of RNASE1 (left panel) and all human cell types of TRIM38 (right panel). Red shading, cell types with divergent (gained) expression. c, Alveolar section of adult mouse lung probed by smFISH for general alveolar epithelial marker Nkx2–1, AT2 marker Sftpc, and transcription factor Myrf. Note Myrf is selectively expressed in mouse AT1 cells (Nkx2–1+ Sftpc- cells), as it is in humans (Fig. ED6c). Bar, 5 μm. Staining repeated on 3 mice. d, Dot plots of expression of CGRP and ADM hormone receptor genes showing expansion of expression (Type 2 change) in human endothelial cells (10x datasets). e, Dot plots of expression of emphysema-associated gene SERPINA1 showing switched expression (Type 3 change) from mouse pericytes (top) to human AT2 cells (bottom) (SS2 datasets). f, Dot plots comparing expression and conservation of HHIP with those of other Hedgehog pathway genes including ligands (SHH, DHH, IHH), receptors (PTCH1, PTCH2, SMO,), and transducers (GLI1, GLI2, GLI3) (SS2 datasets). g, Dot plots of expression of serous cell markers LTF, LYZ, BPIFBP1, and HP showing switched expression (Type 3 change) from mouse airway epithelial cells to human serous cells, which mice lack (*). Dot plots of expression of lipid handling genes APOE, PLIN2, and FST show switched expression (Type 3 change) from mouse alveolar stromal cells to human lipofibroblasts (LipF), which mice lack (*). “Mm()” or “Hs()”, genes where duplications between mouse and human were collapsed to HomologyIDs (10x and SS2 datasets). h, Pie chart of fraction of expressed genes in lung showing each of the four types of evolutionary changes in cellular expression patterns from mouse to human. Histogram below shows number of lung cell types that the 602 genes with perfectly conserved cellular expression patterns (Type 0) are expressed in; note that almost all are expressed in either a single cell type (67%) or nearly all cell types (33%). For more details on statistics and reproducibility, please see Methods.
Supplementary Material
Acknowledgements
We are grateful to the tissue donors and the clinical staff at Stanford Medical Center who made tissue collection possible, especially Jalen Benson and Emily Chen. We are especially grateful to Jim Spudich who spurred this study. We also thank the Stanford Shared FACS Facility for their expertise and sorting services, especially Dr. Lisa Nichols and Meredith Weglarz; members of Chan Zuckerberg Biohub and Quake Lab who supported this work, particularly Aaron McGeever, Dr. Brian Yu, Bob Jones, and Saroja Kolluru; Dr. Maya Kumar for discussions on annotation of stromal cells; and Maria Petersen for illustrating the lung schematic (Fig. 1b) and Dr. Camilla Kao for help with figure formatting. Some computing for this project was performed on the Sherlock cluster; we thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to the results. We thank Jim Spudich and members of the Krasnow lab for valuable discussions and comments on the manuscript, and Alexander Lozano for discussions on bioinformatic analyses. This work was supported by funding from the Chan Zuckerberg Biohub (S.R.Q.) and the Howard Hughes Medical Institute, National Institutes of Health, and the Vera Moulton Wall Center for Pulmonary Vascular Disease (M.A.K.). K.J.T was supported by a Paul and Mildred Berg Stanford Graduate Fellowship. M.A.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Enge M et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X et al. Mapping the mouse cell atlas by microwell-seq. Cell 173, 1307 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Zeisel A et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders A et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vento-Tormo R et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young MD et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aizarani N et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Young J Malpighi’s “De pulmonibus.”. (1929). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehr P et al. The normal human lung: ultrastructure and morphometric estimation of diffusion capacity. Respir Physiol 32, 121–140 (1978). [DOI] [PubMed] [Google Scholar]
- 12.Balis JU et al. Distribution and subcellular localization of surfactant-associated glycoproteins in human lung. Lab Invest 52, 657–669 (1985). [PubMed] [Google Scholar]
- 13.Hermans C and Bernard A Lung epithelium-specific proteins: characteristics and potential applications as markers. Am J Respir Crit Care Med 159, 646–678 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Franks TJ et al. Resident cellular components of the human lung. Proc Am Thor Soc 5, 763–766 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tang F et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Gawad C et al. Single-cell genome sequencing: current state of the science. Nat Rev Genet 17, 175–188 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Treutlein B et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyfman PA et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med 199, 1517–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braga FAV et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nature Medicine 2018 24:8 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014 9:1 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Blondel VD et al. Fast unfolding of communities in large networks. J Stat Mech 2008, P10008 (2008). [Google Scholar]
- 22.Howitt MR et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rock JR et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639–648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia SR et al. Single-cell RNA sequencing reveals novel cell differentiation dynamics during human airway epithelium regeneration. bioRxiv 140, 451807 (2018). [Google Scholar]
- 25.Nabhan AN et al. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zacharias WJ et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stan RV et al. The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev Cell 23, 1203–1218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan SYS & Krasnow MA Developmental origin of lung macrophage diversity. Development 143, 1318–1327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Brink SC et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017 8 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiow LR et al. CD69 acts downstream of interferon-α/β to inhibit S1P 1 and lymphocyte egress from lymphoid organs. Nature 440, 540–544 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Mackay LK et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Moffitt JR & Zhuang X RNA Imaging with Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH). Methods Enzymol 572, 1–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limjunyawong N et al. Measurement of the pressure-volume curve in mouse lungs. J Vis Exp e52376 (2015). doi: 10.3791/52376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeley RR et al. Essentials of anatomy and physiology. (2005). [Google Scholar]
- 39.Tabula Muris Consortium et al. A single cell transcriptomic atlas characterizes aging tissues in the mouse. bioRxiv 661728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Amerongen R et al. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Greif DM et al. Radial construction of an arterial wall. Dev Cell 23, 482–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muzumdar MD et al. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2009 13:1 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moraga I et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 160, 1196–1208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai TJ, Brownfield DG & Krasnow MA Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler A et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018 36:5 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finak G et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. http://bioinfo.life.hust.edu.cn/AnimalTFDB/ [DOI] [PMC free article] [PubMed]
- 49.Amberger JS et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buniello A et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Counts/UMI tables, cellular metadata, Seurat objects, and scanpy objects are available on Synapse (https://www.synapse.org/#!Synapse:syn21041850). The data can be explored in a browser using cellxgene at https://hlca.ds.czbiohub.org/. Human sequencing data is available by data access agreement on the European Genome-phenome Archive (EGA) under accession EGAS00001004344. Use of human sequencing data is restricted to not for profit research only and requires approval or a waiver from requesting investigator’s institutional review board. Mouse sequencing data is available on the National Institute of Health’s Sequence Read Archive (SRA) under BioProject accession PRJNA632939. Source data behind immunostaining or smFISH quantification (Figure 1; Extended Data Figures 3 and 4) are available within the manuscript files.