Abstract
There have been significant advancements in precision medicine and approaches to medication selection based on pharmacogenetic results. With the availability of direct-to-consumer genetic testing and growing awareness of genetic inter-individual variability, patient demand for more precise, individually tailored drug regimens is increasing. The University of Florida (UF) Health Precision Medicine Program (PMP) was established in 2011 to improve integration of genomic data into clinical practice. In the ensuing years, the UF Health PMP has successfully implemented several single-gene tests to optimize the precision of medication prescribing across a variety of clinical settings. Most recently, the UF Health PMP launched a custom-designed pharmacogenetic panel including pharmacogenes relevant to supportive care medications commonly prescribed to patients undergoing chemotherapy treatment, referred to as “GatorPGx”. This tutorial provides guidance and information to institutions on how to transition from the implementation of single-gene pharmacogenetic testing to a preemptive panel-based testing approach. Here, we demonstrate application of the preemptive panel in the setting of an adult solid tumor oncology clinic. Importantly, the information included herein can be applied to other clinical practice settings.
Keywords: pharmacogenomics, cancer, symptom, supportive care, preemptive, precision, medicine, oncology
Introduction
Pharmacogenomics (PGx) focuses on the role of inherited and acquired genetic variation and how it contributes to differential drug response phenotypes.(1) Additionally, genetic inter-individual variation with respect to drug targets or biological pathways can account for an individual’s unique response to certain drugs.(2) Many clinical practice sites have implemented PGx testing services that primarily focus on single-gene tests in reaction to a newly prescribed medication, a lack of response, or an adverse drug reaction.(3–6) Advances in technology, reduction in genotyping cost, and successful implementation of various single-gene PGx testing services has led to a growing interest in preemptive panel-based testing.
Preemptive panel-based testing is a method used to gather pharmacogenetic data for multiple genetic markers and store them in a patient’s electronic health record (EHR) to serve as a resource for future medication prescribing.(7) Preemptive panel-based results that are promptly and consistently available to clinicians provide the opportunity for personalized prescribing for a wide variety of medications, rather than awaiting for results of a reactive single-gene test for a particular medication. Given the relative stability of PGx results over time, panel testing at an early time point in care can have relevancy for future management. Despite the advantages of preemptive panel-based testing, there has been a paucity of data to assess the clinical utility of such services and guide best practices when implemented into a healthcare setting.
To date, the University of Florida (UF) Health Precision Medicine Program (PMP) has successfully implemented several PGx single-gene tests, which are run in a Clinical Laboratory Improvement Amendments (CLIA)-approved pathology lab, and which may be utilized for multiple drugs, to assist providers in optimizing medication therapy for patients within various diverse practice settings.(5, 8–10) Based on these successes, the UF Health organization made the commitment to implement a preemptive panel-based test. This tutorial is intended to provide guidance and information based on our experience during the transition from single-gene testing to implementation of a preemptive panel-based testing approach in an oncology clinic. For purposes of this article, it is assumed that implementation of preemptive panel-based testing will occur within an institution with an established single-gene PGx service. In the absence of an existing framework for PGx consult services, references exist that provide guidance on the infrastructure needed to develop PGx services at an institutional level.(11–13) Throughout this article, a patient case example is described that highlights the role of preemptive PGx-panel-based testing during treatment with chemotherapy.
Patient Case Paul H. - First Office Visit
Paul H., a 73-year old male, presents to the oncology clinic to establish care for locally advanced urothelial cancer. Paul is concerned with starting a new chemotherapy regimen (single-agent carboplatin) and the adverse events he may experience. He is informed about preemptive PGx testing and is hopeful that the results can assist with supportive care medication selection and dosing while undergoing chemotherapy treatment. In addition to the cancer, Paul is being treated for depression and gastroesophageal reflux disease (GERD) which are both commonly treated with medications that have relevant drug-gene interactions.
RATIONALE FOR PREEMPTIVE PANEL-BASED TESTING
Thoughtful decision making by an organization is necessary to identify clinical setting(s) and patient population(s) in which to offer preemptive PGx services. While ideally, every patient would undergo preemptive panel-based testing, many logistical issues, including reimbursement and capacity for the PGx consult service, remain challenging and can be a deterrent for patients and healthcare organizations. Therefore, utilization of a pragmatic approach to identifying the institutional target population is key. Methods exist for identifying patients that may be ideal candidates for preemptive PGx testing and integrating that data into clinical workflow. For example, development of a clinical decision support (CDS) designed to recommend preemptive PGx testing if a certain risk threshold is exceeded may be a practical way to identify patients.(14) Shi et al. describe a method to develop a preemptive PGx program that accounts for societal and institutional value and may be considered when determining risk estimates in certain patient populations.(15) Additionally, a recent study utilized claims data to identify opportunities for genotype-guided prescribing in certain patient populations.(16) There are many different approaches and the PGx service must be uniquely tailored to the specific patient population and healthcare system.
While targeted therapies for somatic mutations have primarily been the focus of genetic profiling in the oncology setting, the use of germline genetic information to guide chemotherapy treatment selection has been slower to implement into oncology clinical practice, and as such is the focus of this tutorial.(17) However, during chemotherapy treatment, patients often suffer from disease- and chemotherapy- related symptoms such as pain, nausea, depression, gastrointestinal complications, among others, which affect overall well-being and quality of life.(18) Many of these treatment-related symptoms are managed with medications commonly referred to as “supportive care medications”, such as clopidogrel, omeprazole, ondansetron, voriconazole, warfarin and others. Most supportive care regimens for patients undergoing chemotherapy include multiple drugs with well characterized drug-gene prescribing guidance.(19–25)
The benefits of preemptive panel-based testing and the potential to improve patient outcomes is increasing as more data is made available.(16, 26, 27) While there is increasing evidence to support the feasibility of preemptive PGx testing, a significant amount of debate amongst clinicians exists about the clinical utility of preemptive genotyping.(28, 29) Much of the doubt and skepticism surrounding the value of preemptive genotyping is due to lack of research that accurately demonstrate improvements in patient outcomes. To address this gap in the literature, UF Health is actively conducting a pragmatic clinical trial (PCT) to evaluate multi-symptom patient reported outcomes in patients who undergo preemptive PGx testing while receiving chemotherapy treatment (NCT03924557). The PCT and preemptive panel of pharmacogenes were implemented simultaneously. We hypothesize that patients with PGx test results that are available at the time of supportive care medication prescribing will have decreased symptom distress and improved quality of life compared with those who do not have PGx test results immediately available. Additionally, the PCT will collect pharmacoeconomic data associated with treatment changes related to genotype intervention. We anticipate the trial to conclude by summer of 2022. While conducting a PCT is not a necessary component of a preemptive panel based PGx implementation, the PCT is a way to allow providers to become more comfortable and confident with use of the PGx test results.
Patient Case: Paul H. - Current Medications
Paul is asked to bring a list of all his current medications. Paul’s current medication list includes the following: Atorvastatin 40 mg daily, Fenofibrate 160 mg daily, Lisinopril 20 mg daily, Omeprazole 20 mg daily*, Sertraline 100 mg daily*, Tamsulosin 0.4 mg daily, Ondansetron 8 mg twice daily*. The latter was prescribed by his oncologist for chemotherapy-induced nausea and vomiting prophylaxis.
A medication reconciliation is performed and three of his medications have established drug-gene prescribing guidance. *Indicates evidence-based guidelines available
PREPARATION FOR IMPLEMENTATION
Implementation Team
Assembling a group of clinicians from various practice backgrounds to serve as your implementation team is important for cross-collaboration and to obtain support for preemptive panel-based testing from the providers involved in the clinic workflow. We acknowledge there are various models when establishing a PGx consult service and each is unique and has its own merits.(13, 27) Determining which clinician(s) will lead the implementation is specific to the practice setting, staffing demands, level of training in PGx and various other factors. In our pharmacist-led model, clinical pharmacists with specialized PGx training work with physician champions to establish a framework within the clinic and serve as a resource when selecting genes to consider for inclusion on the panel.(11) The implementation team, similar to a Pharmacy and Therapeutics (P&T) Committee, should consist of a diverse group of clinicians who collectively determine which drugs, genes and variants are selected for testing. The process includes reviewing the applicable literature, translating genotype results into relevant phenotypes, and approving drug therapy recommendations. At a minimum, implementation team members should consist of a PGx knowledgeable leader, physician champion(s), pharmacist(s), and other key stakeholders including clinicians, laboratory personnel, and informatics/information technology (IT) representatives.(11) It is important to note, recommendations for implementation team members are institution-specific. For example, inclusion of laboratory personnel may not be needed if utilizing an outsource laboratory.
Selection of Drug-Gene Pairs
Drug-gene pairs with evidence-based guidelines and demonstrative clinical validity are ideal choices to implement and include on a multi-pharmacogene preemptive panel. The Clinical Pharmacogenetics Implementation Consortium (CPIC) publishes peer-reviewed clinical guidelines that serve as a guide to assist clinicians in prescribing medication therapy with available genetic test results.(30 ) CPIC defines drug-gene pairs as levels A-D, with level A being the highest level of evidence and a strong recommendation that genetic information should be used to change prescribing of the medication when an actionable genotype is detected, and D as the lowest level of evidence with no specific prescribing actions provided. Supportive care medications included in our implementation are all CPIC level A or B drugs.
Supportive care drug-gene pairs with associated evidence-based guidelines are summarized by indication in TABLE 1. Although there are a variety of supportive care medications commonly prescribed, we elected to preemptively test for drugs that are primarily metabolized by drug-metabolizing enzymes, cytochrome P450 (CYPs). The genes that encode for CYPs are highly polymorphic, resulting in variable pharmacokinetics of relevant substrates.(35) It is expected that certain patients will have genotypes that warrant modification of one or more medication classes to avoid treatment failure or adverse drug effects. Therefore, preemptive PGx testing will likely optimize drug dosing selection and decrease adverse events in this population.
TABLE 1.
List of commonly used supportive care medications with evidence-based drug-gene prescribing guidance
| Drug Classification | Drug(s) | Gene | Guidelines |
|---|---|---|---|
| Antifungal | Voriconazole | CYP2C19 | https://cpicpgx.org/guidelines/guideline-for-voriconazole-and-cyp2c19/ |
| Platelet Inhibitor | Clopidogrel | CYP2C19 | https://cpicpgx.org/guidelines/guideline-for-clopidogrel-and-cyp2c19/ |
| Anticoagulant | Warfarin | CYP2C9, VKORC1, CYP4F2, CYP2C cluster | https://cpicpgx.org/guidelines/guideline-for-warfarin-and-cyp2c9-and-vkorc1/ |
| Proton Pump Inhibitors (PPIs) | Omeprazole, Pantoprazole, Lansoprazole, Rabeprazole, Esomeprazole, and Dexlansoprazole | CYP2C19 | https://www.pharmgkb.org/chemical/PA164713207/guidelineAnnotationa |
| Selective Serotonin Reuptake Inhibitors (SSRIs) | Citalopram, Escitalopram, Fluvoxamine, Paroxetine, Sertraline | CYP2D6, CYP2C19 | https://cpicpgx.org/guidelines/guideline-for-selective-serotonin-reuptake-inhibitors-and-cyp2d6-and-cyp2c19/ |
| Opiates | Codeine, tramadol, hydrocodone, oxycodone | CYP2D6 | https://cpicpgx.org/guidelines/guideline-for-codeine-and-cyp2d6/ |
| Antiemetic | Ondansetron | CYP2D6 | https://cpicpgx.org/guidelines/guideline-for-ondansetron-and-tropisetron-and-cyp2d6-genotype/ |
| Tricyclic Antidepressants (TCAs) | Amitriptyline, Clomipramine, Desipramine, Imipramine, Doxepin, Nortriptyline, Trimipramine | CYP2C19, CYP2D6 | https://cpicpgx.org/guidelines/guideline-for-tricyclic-antidepressants-and-cyp2d6-and-cyp2c19/ |
During the planning phase of our implementation, UF-Health Pathology Laboratories (UFHPL) created a custom clinical laboratory developed test (LDT) to analyze a panel of genes called “GatorPGx” (Table 2). The GatorPGx panel consists of 32 variant alleles across eight pharmacogenes (CYP2C19, CYP2C9, CYP2D6, CYP3A5, CYP4F2, CYP2C Cluster, SLCO1B1 and VKORC1) as well as copy number variation for CYP2D6. GatorPGx includes clinically actionable variant alleles that are relevant to our UF Health population and may be used to optimize over 20 different commonly prescribed supportive care drugs (Table 1). GatorPGx is the first panel-based testing service UF Health has offered in an oncology clinic setting and the decision to select genes relevant to commonly prescribed supportive care medications in the clinic correlates with our active PCT. Outcomes data collection with respect to the PCT is ongoing and will be evaluated at the conclusion of the study and may lead to modifications to the panel. The GatorPGx panel can be used in the clinical practice setting because it provides information for a wide variety of medications that are currently prescribed as well as those that may be prescribed in the future.
Table 2.
GatorPGx Panel of Pharmacogenes
| Gene Symbol | SNP rs# | Star Allele | Allele Functional Status |
|---|---|---|---|
| CYP2C19 | rs4244285 | *2 | No function |
| rs4986893 | *3 | No function | |
| rs28399504 | *4 | No function | |
| rs72552267 | *6 | No function | |
| rs41291556 | *8 | No function | |
| rs6413438 | *10 | Decreased function | |
| rs12248560 | *17 | Increased function | |
| CYP2C9 | rs1799853 | *2 | Decreased function |
| rs1057910 | *3 | No function | |
| rs28371686 | *5 | Decreased function | |
| rs9332131 | *6 | No function | |
| rs7900194 | *8 | Decreased function | |
| rs28371685 | *11 | Decreased function | |
| CYP2C Clustera | rs12777823 | NA | Decreased Function |
| CYP2D6b,c | |||
| rs16947 | *2 | Normal function (AV=1) | |
| rs35742686 | *3 | No function (AV =0) | |
| rs3892097 | *4 | No function (AV =0) | |
| rs5030655 | *6 | No function (AV =0) | |
| rs5030867 | *7 | No function (AV =0) | |
| rs5030865 | *8 | No function (AV =0) | |
| rs5030656 | *9 | Decreased function (AV =0.5) | |
| rs1065852 | *10 | Decreased function (AV =0.25) | |
| rs28371706 | *17 | Decreased function (AV =0.5) | |
| rs59421388 | *29 | Decreased function (AV =0.5) | |
| rs28371725 | *41 | Decreased function (AV =0.5) | |
| CYP3A5 | rs776746 | *3 | No function |
| rs10264272 | *6 | No function | |
| rs41303343 | *7 | No function | |
| CYP4F2 | rs2108622 | NA | Decreased function |
| SLCO1B1 | rs4149056 | *5 | Decreased function |
| VKORC1 | rs9923231 | NA | Decreased activity |
SNP, Single Nucleotide Polymorphism; AV, Activity Value; NA, Not Applicable.
rs12777823 is a SNP located within the CYP2C gene cluster near the CYP2C18 gene on chromosome 10.
Each allele is assigned an “activity value” ranging from 0−1 (e.g., 0 for no function, 0.25 or 0.5 for decreased function, and 1.0 for normal function).
The assay can identify gene rearrangements associated with the deletion (*5) and copy number variations, which are defined as two or more gene copies per allele.
Laboratory
Selecting a laboratory to process and analyze patient samples is a critical decision point prior to implementation of a PGx service. If your institution does not have an American College of Pathologists accredited/ CLIA-licensed facility able to process and analyze genetic samples, then a rigorous review and selection of a commercial reference laboratory to outsource testing must be made.(36, 37) Outsource laboratories offer a myriad of services such as creation of a custom-panel, interpretation of results based on evidence, and prediction of drugs that may lead to poor patient outcomes. However, when an outside laboratory is considered, it is important to determine how the genotype data will be returned to the provider. If the data cannot be integrated as a discrete field in the EHR, it may not be possible to utilize automated CDS alerts based on genotype results. Ultimately, the work conducted by the outsource facility will depend on the needs of the institution, staff knowledge of PGx testing, and budgetary constraints. Information on CLIA-certified laboratory locations and questions about the CLIA program can be accessed on the Centers for Medicare and Medicaid Services webpage – (www.CMS.gov/Regulations-andGuidance/Legislation/CLIA/index.html).
When developing or selecting a commercially available and CLIA-approved PGx panel, determining drug-gene pairs relevant to your patient population and formulary is imperative to the overall utility and clinical adoption of the preemptive panel. Similar to implementing a single gene-drug PGx consult service, allele frequency variation within genes on your panel are factors to consider when selecting single nucleotide polymorphisms (SNPs) to test. It is important to note that some assays incorporate variants that are common in certain populations (most often Caucasians); however, variants that are specific to other minority populations may be missing. CPIC guidelines include information, if available for the gene, on allele frequency and presence of variants in ethnically diverse groups, and recently published recommendations on standardizing CYP2D6 genotype to phenotype translation.(30, 38) Additionally, the Pharmacogenomics Knowledgebase (PharmGKB) website contains links to gene-specific tables that support CPIC guidelines and provide information on allele star (*) mapping, allele functionality and frequency, and diplotype to phenotype mapping.(31) Furthermore, the Association for Molecular Pathology and College of American Pathologists issued a joint recommendation for designing assays for CYP2C9 and CYP2C19 that may be utilized as a reference guide for laboratories developing assays.(37–39) Appropriate resources pertaining to allele frequencies among racial groups should be consulted when determining which variants to include on your panel. SNP selection should be based on population race/ethnic makeup as well as common and rare alleles that account for variants that are associated with drug response extremes or adverse effects.
Prior to starting patient testing, any clinical assays assay must be validated to ensure accuracy of results before implementation in a practice setting. The National Institutes of Health (NIH), National Human Genome Research Institute provides information as to how the Food and Drug Administration (FDA) and the Centers for Medicare and Medicaid Services (CMS) have the authority to regulate genetic testing.(40) Laws relating to quality assurance and other pertinent federal regulations are located at (CLIA 42 CFR 493.1253 and College of American Pathologists GEN 42020–42163). Discussion with the contracted laboratory or your institution’s respective department regarding validation of results and compliance with applicable regulations is recommended well in advance before genetic testing implementation. The GatorPGx assay uses QuantStudio 12K Flex Real Time PCR System (Applied Biosystems by Life Technologies) and Lifetechnology TaqMan® SNP Genotyping Assays with a 384-well microtiter plate robotic pipetting qPCR setup. Genotype information is translated into enzyme activity category calls (i.e., phenotype) and the phenotype calls are based on CPIC Guidelines.(41)
As part of the selection of a laboratory, institution or clinic preferred method(s) of sample collection should be determined. Population specific parameters will also dictate sample collection methods. For example, will testing require a separate encounter for a blood draw from the patient or will a saliva or buccal cell collection kit need to be mailed to patients? Discussions with your laboratory and clinical providers on sample type (i.e. blood, saliva, or buccal cell) will reveal which is preferred. If your laboratory has the capability to analyze a variety of sample types with high accuracy, then selection should be based on ease of integration. Several buccal and saliva sample collection kits are commercially available; however, recent studies show variability among the yield and quality of DNA isolated from buccal cell and saliva kits and should be considered when determining a preferred collection method.(42–44) Because patients in our clinic are beginning chemotherapy regimens and require routine pre-treatment blood sampling for organ function assessments, we utilized blood specimens to extract DNA for the purposes of the GatorPGx panel. However, the GatorPGx panel is also validated for buccal cell and saliva samples for use in other populations as appropriate.
Laboratory Report and Consult Note
The format and contents of the laboratory report were developed by the UF Health Pathology Laboratory in collaboration with UF Health PMP PGx clinical pharmacists, with feedback from health care providers. The final laboratory report is organized by gene and includes the patient’s genotype and phenotype (based on genotype), and non-normal results are clearly identified (FIGURE 1). Within the laboratory report, each gene has a clinical implication section, which is helpful for clinicians to understand the meaning of the phenotype. The clinical implication section, named as “comment” on the report, defines the level of activity of the particular phenotype, lists CPIC level A drugs (but does not indicate the drug-gene relationship nor expected clinical response) and drugs that may cause phenoconverison (if applicable for specific gene and phenotype).
Figure 1. GatorPGx Laboratory Report.
– Abbreviated laboratory report corresponding to the patient case: Paul H.
For preemptive PGx testing to be successful, the results should be made available to the clinician quickly and accurately, and formatted for ease of understanding. A common challenge facing clinicians is locating PGx results within the EHR and being reminded of the potential for PGx results to inform drug therapy over time. As a result, the UF Health PMP PGx pharmacists add to the patient’s problem list the International Classification of Diseases, Tenth Revision (ICD-10) code “Z13.79: Encounter for pharmacogenetic testing”, which serves as a reminder to the clinician that PGx results are available and language is added to direct them to the location of the consult note.
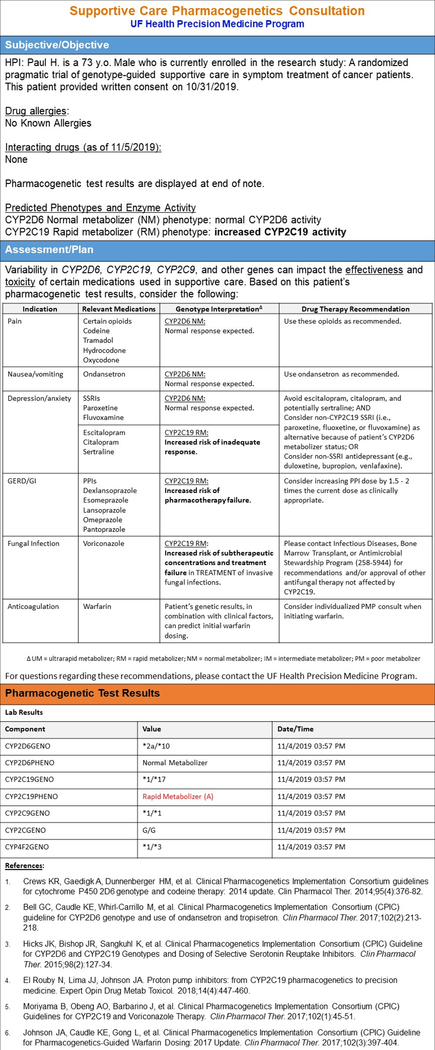
Results are generally released into the EHR within 3 – 5 days after sample collection. Given that there is typically a delay from a cancer diagnosis to initiation of therapy, this delay does not negatively impact the clinical use of the PGx data. At UF Health, the provider who ordered the test and the PGx pharmacists are notified that the results have been returned via an in-basket message in the EHR. The pharmacist then reviews the results and a consultation note is prepared and subsequently routed to the provider. The UF Health PMP consult note utilizes an abbreviated Subjective-Objective-Assessment-Plan (SOAP) note format with a nested table to display the gene-drug results, which provides key recommendations on the clinical phenotype (i.e., consideration of drug interactions that may change the phenotype reported by the laboratory) for all supportive care medications impacted by the genes tested, independent of whether the patient is on the medication at the time of consult (FIGURE 2.). Specifically, the consult note includes information on history of present illness (HPI), drug allergies, and interacting drugs. For clinically actionable drug-gene pairs, recommendations to optimize drug therapy for genotype/phenotype are included based on CPIC guidelines. A tabular format for the consultation note to improve readability and organization is utilized. For patients being genotyped on the GatorPGx panel outside of the supportive care area, a similarly formatted consult note is provided with inclusion of additional gene-drug pairs as applicable.
Figure 2. Patient Consult Note.
– Pharmacist written consult note corresponding to the patient case: Paul H.
Patient Case: Paul H. - Pharmacogenomic Consult Note
Please refer to Paul’s consult note to see specific drug-gene results that are now available to guide his supportive care medications. (FIGURE 2). Based on Paul’s PGx results, the following prescribed medications have actionable recommendations: sertraline and omeprazole. Currently, Paul’s depression is controlled by 100 mg of sertraline; therefore, no recommendations were made at this time. However, a note was added that if there ever was the need to switch antidepressant medications that non-CYP2C19 SSRIs should be considered as an alternative or non-SSRI antidepressants. For his GERD pharmacotherapy, it was recommended to consider increasing the proton pump inhibitor (PPI) dose as clinically appropriate.
Clinical Decision Support
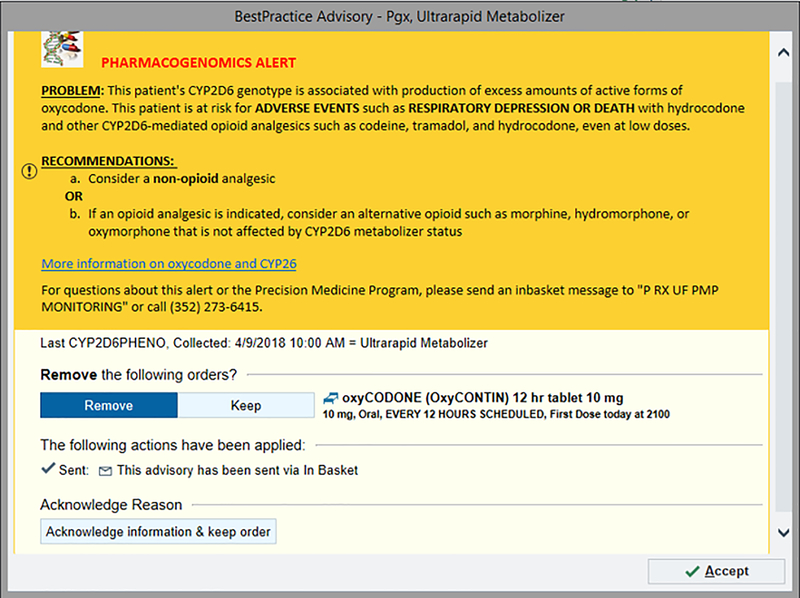
Coordinating the technology needs and software prior to implementation is vital to the success of a clinical service. CDS integrated into the EHR, whether in the form of an alert or consult note, provides meaningful support to clinicians when appropriately nested into the workflow and is a helpful instrument for success of PGx in clinical practice.(45) There are many strategies to use when developing CDS tools in an institution. In our oncology clinic, specific PGx CDS rules were written and implemented to address supportive care drug-gene interactions. When a provider prescribes a supportive care or another medication for a patient that has previously undergone GatorPGx panel testing, CDS rules designed to review the genotype and prescription in real-time, may trigger a best practice alert (BPA) to notify the prescriber if there is an actionable genotype-driven medication change recommended. As appropriate, the BPA notifies the prescriber that an alternative medication, or dosage change is recommended. Alerts can incorporate a single drug-gene interaction or account for multiple genes and/or multiple drugs. For example, a specific alert will fire if a provider orders oxycodone for a patient with an CYP2D6 ultrarapid metabolizer (UM) phenotype (FIGURE 3.). From discussions with our implementation team and to address provider concerns pertaining to workflow, we created concise PGx BPAs that state the problem and recommendation, and use bold terms to bring attention to the most important information in the alert. A hyperlink with “additional information” reference the evidence-based guidelines. The architecture of the alerts should be in place within an existing single-gene infrastructure and can easily be adapted to meet the needs of multi-gene panel testing. For example, we updated existing alerts to fire off two genes for SSRIs and created additional alerts to support the results. Prior to launching our implementation, alerts were in place for all relevant supportive care oncology medications.
Figure 3. Best Practice Alert (BPA).
– Image of alert that appears when patient is a CYP2D6 ultrarapid metabolizer and the provider orders oxycodone.
Patient Communication and Education
Effective communication with the patient regarding the goals of preemptive panel-based genotyping is important prior to ordering a test. Similarly, education is also important for single-gene reactive genotyping, but emphasis on the life-time applicability of results in the preemptive setting is key. The concepts of genetic testing and its benefit can be difficult for patients to comprehend, especially during stressful times such as beginning or restarting chemotherapy treatment. As a result, we developed a short video that can be accessed via our institution’s Precision Medicine Program website that the patient can watch while waiting for their appointment.(46) The video is approximately seven minutes in length and is intended to review basic concepts of pharmacogenomics and expectations of its usefulness. After genetic test results have been returned to the ordering provider, they are released into the patient’s EHR portal where they can be viewed by the patient. Through their online patient portal, accessible via internet browser or mobile application, the patient can download their results as well as send and receive messages about their results and may use them to inform other providers outside of the health system.
Provider Education and Buy-In
Educating providers and nursing staff on the components of the preemptive panel-based PGx testing is essential to create an atmosphere of cooperation and inclusiveness. A barrier that clinicians continue to face is the difficulty in translating the results of PGx testing into useful tools in clinical practice.(45) Health care providers and staff at our clinic were offered personal genotype testing and counseling to increase their knowledge of preemptive panel-based PGx testing. An “unofficial” consult note was created in the same format as patients in the clinic in order for the prescriber to become familiar with it. PGx-trained pharmacists individually reviewed the results of the genetic test with each health care provider and staff member that participated and addressed any questions they had at that time. Additionally, information was given on how to locate evidence-based PGx guidelines. Overall, the providers and staff found the educational component helpful in understanding the benefits of implementing the testing service in clinic. Moreover, we found that providing this educational opportunity increased awareness of the preemptive panel-based PGx testing service.
Patient Case: Paul H. – Follow-up Office Visit
The PGx consult note, which summarizes genotype results and implications for drug response, and provides genotype-guided prescribing recommendations, is electronically routed via an “in-basket” within the EHR software to Paul’s oncologist for review. Paul’s physician reviews the consult note and associated lab report. The physician agreed with the recommendation by the pharmacist to increase the PPI dose based on Paul’s CYP2C19 rapid metabolizer status and persistent symptoms of GERD. At Paul’s next office visit, documentation was noted in his EHR that his symptoms have resolved after the dose increase of his PPI medication.
CONCLUSION
Implementation of panel-based PGx testing requires increased collaboration amongst the various clinicians involved in the workflow compared to single-gene testing; however, with a well-structured clinical team and pharmacogenomic champion, transition to a preemptive panel-based service is feasible. Developing an implementation plan with the proper personnel, preferably those familiar with your institution’s single gene infrastructure, will help ensure the service is appropriate and beneficial to the target patient population. Successful integration of PGx data into a patient’s EHR will serve as an additional tool for clinicians to inform supportive care medication selection and dose optimization. As demonstrated by our patient case, pharmacogenomic data is important to consider when selecting a medication or determining an optimal dose for supportive care medications. Lastly, providing educational opportunities to clinicians across the healthcare system is paramount, since patients often receive care from multiple healthcare providers within a healthcare system. Equipping clinicians with the knowledge and resources to interpret the scientific results and appropriately translate them to meaningful decision making may lead to decreased medication related adverse events and improved patient outcomes in oncology patients.
Acknowledgements
The authors would like to thank Steven Fischer, MS, and the rest of the clinical research coordinator team at UF Health - Medical Oncology for their dedication and commitment to the success of our pragmatic clinical trial.
Funding Information: This project is part of the University of Florida’s “Creating the Healthiest Generation” Moonshot initiative, which is supported by the UF Office of the Provost, UF Office of Research, UF Health, UF College of Medicine, and UF Clinical and Translational Science Institute and is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1 TR001427. Additional support comes from NIH NHGRI 5U01HG007269-06.
Footnotes
Conflicts of Interest: The authors declared no competing interests for this work. As an Editor-in-Training for Clinical Pharmacology & Therapeutics, Emily J. Cicali was not involved in the review or decision process for this paper.
References
- (1).Weinshilboum RM & Wang L Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin Proc 92, 1711–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Luzum JA et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin Pharmacol Ther 102, 502–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Shuldiner AR et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet 166C, 76–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cavallari LH et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med 16, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Empey PE et al. Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther 104, 664–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hamburg MA & Collins FS The path to personalized medicine. N Engl J Med 363, 301–4 (2010). [DOI] [PubMed] [Google Scholar]
- (8).Weitzel KW et al. Implementation of Standardized Clinical Processes for TPMT Testing in a Diverse Multidisciplinary Population: Challenges and Lessons Learned. Clin Transl Sci 11, 175–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mosley SA et al. Design and rational for the precision medicine guided treatment for cancer pain pragmatic clinical trial. Contemp Clin Trials 68, 7–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cicali EJ et al. Novel Implementation of Genotype-Guided Proton Pump Inhibitor Medication Therapy in Children: A Pilot, Randomized, Multisite Pragmatic Trial. Clin Transl Sci 12, 172–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA & Duarte JD Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin Transl Sci 9, 233–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Weitzel KW et al. A stepwise approach to implementing pharmacogenetic testing in the primary care setting. Pharmacogenomics 20, 1103–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Caraballo PJ et al. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet Med 19, 421–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Schildcrout JS et al. A prognostic model based on readily available clinical data enriched a preemptive pharmacogenetic testing program. J Clin Epidemiol 72, 107–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shi Y et al. A Decision-Theoretic Approach to Panel-Based, Preemptive Genotyping. MDM Policy Pract 4, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).El Rouby N et al. Clinical Utility of Pharmacogene Panel-Based Testing in Patients Undergoing Percutaneous Coronary Intervention. Clin Transl Sci, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gillis NK, Patel JN & Innocenti F Clinical implementation of germ line cancer pharmacogenetic variants during the next-generation sequencing era. Clin Pharmacol Ther 95, 269–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nayak MG et al. Quality of Life among Cancer Patients. Indian J Palliat Care 23, 445–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hicks JK et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 98, 127–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bell GC et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 102, 213–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Birdwell KA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin Pharmacol Ther 98, 19–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hicks JK et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Moriyama B et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy. Clin Pharmacol Ther 102, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Roden DM et al. Benefit of Preemptive Pharmacogenetic Information on Clinical Outcome. Clin Pharmacol Ther 103, 787–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dunnenberger HM et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Janssens AC & Deverka PA Useless until proven effective: the clinical utility of preemptive pharmacogenetic testing. Clin Pharmacol Ther 96, 652–4 (2014). [DOI] [PubMed] [Google Scholar]
- (29).Kasi PM et al. Feasibility of Integrating Panel-Based Pharmacogenomics Testing for Chemotherapy and Supportive Care in Patients With Colorectal Cancer. Technol Cancer Res Treat 18, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).The Clinical Pharmacogenetics Implementation Consortium (CPIC). CPIC Guidelines. <https://cpicpgx.org/guidelines/>. Accessed February 14 2020.
- (31).PGx Gene-specific Information Tables PGx <https://www.pharmgkb.org/page/pgxGeneRef> (2020). Accessed April 18 2020.
- (32).Swen JJ et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther 89, 662–73 (2011). [DOI] [PubMed] [Google Scholar]
- (33).Lima JJ & Franciosi JP Pharmacogenomic testing: the case for CYP2C19 proton pump inhibitor gene-drug pairs. Pharmacogenomics 15, 1405–16 (2014). [DOI] [PubMed] [Google Scholar]
- (34).El Rouby N, Lima JJ & Johnson JA Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin Drug Metab Toxicol 14, 447–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Maitland-van der Zee AH, de Boer A & Leufkens, H.G. The interface between pharmacoepidemiology and pharmacogenetics. Eur J Pharmacol 410, 121–30 (2000). [DOI] [PubMed] [Google Scholar]
- (36).Vo TT, Bell GC, Owusu Obeng A, Hicks JK & Dunnenberger HM Pharmacogenomics Implementation: Considerations for Selecting a Reference Laboratory. Pharmacotherapy 37, 1014–22 (2017). [DOI] [PubMed] [Google Scholar]
- (37).Pratt VM et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 21, 746–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Caudle KE et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 13, 116–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pratt VM et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn 20, 269–76 (2018). [DOI] [PubMed] [Google Scholar]
- (40).National Human Genome Research Institute. Regulation of Genetic Tests. <https://www.genome.gov/about-genomics/policy-issues/Regulation-of-Genetic-Tests> (November 20, 2019). Accessed February 11 2020.
- (41).Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89, 464–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).King IB et al. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev 11, 1130–3 (2002). [PubMed] [Google Scholar]
- (43).Bruinsma FJ, Joo JE, Wong EM, Giles GG & Southey MC The utility of DNA extracted from saliva for genome-wide molecular research platforms. BMC Res Notes 11, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ang JS et al. Evaluation of buccal swabs for pharmacogenetics. BMC Res Notes 11, 382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Johnson JA & Weitzel KW Advancing Pharmacogenomics as a Component of Precision Medicine: How, Where, and Who? Clin Pharmacol Ther 99, 154–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).UF Health Precision Medicine Program. What is pharmacogenetics? <https://precisionmedicine.ufhealth.org/what-is-pharmacogenetics/>. Accessed February 14 2020. [Google Scholar]