Abstract
Membrane protein pores have emerged as powerful nanopore sensors for single-molecule detection. OmpG, a monomeric nanopore, is comprised of fourteen β-strands connected by seven flexible extracellular loops. The OmpG nanopore exhibits pH-dependent gating as revealed by planar lipid bilayer studies. Current evidence strongly suggests that the dynamic movement of loop 6 is responsible for the gating mechanism. In this work, we have shown that enhancing the electrostatic repulsion forces between extracellular loops suppressed the pH-dependent gating. Our mutant containing additional negative charges in loop 6 and loop 1 exhibited minimal spontaneous gating and reduced sensitivity to pH changes compared to the wild type OmpG. These results provide new evidence to support the mechanism of OmpG gating controlled by the complex electrostatic network around the gating loop 6. The pH-independent quiet OmpG pores could potentially be used as a sensing platform that operates at a broad range of pH conditions.
Graphical Abstract
1. Introduction
In nature, protein channels and pores operate as molecular gates, regulating the passage of diverse substances through the cell membrane [1–6]. Thus, they carry out important roles in nutrient uptake[7], maintaining viable salt concentrations[8–10], conferring drug resistance[11] or mediating cell death in cells[12–14]. In biotechnology, protein pores have been used as nanopore sensors for detecting a broad spectrum of molecules[15–17]. Protein nanopores have become powerful bioanalytical tools for studying fundamental questions in chemistry and biology, such as chemical reaction[18], protein-protein interaction[19], protein folding and unfolding[20, 21], and enzymatic activity[22, 23]. Furthermore, nanopores hold tremendous promise in biotechnology with their ability to sequence genomic DNA[24].
Outer membrane protein G (OmpG) is a 33 kDa non-selective monomeric β–barrel pore responsible for the uptake of oligosaccharides in Escherichia. coli (E. coli)[25, 26]. The pore is composed of fourteen β–strands connected by seven flexible extracellular loops and six short turns on the periplasmic side[27–29]. Unlike multimeric biological pores such as α-hemolysin, complex alterations via chemical or genetic modifications are straightforward with OmpG, making it an appealing platform for developing nanopore sensing technology[30–34].
Planar lipid bilayer studies have shown that under an applied potential, OmpG adopts two major conformations denoted as the ‘open’ and ‘closed’ conformations[35]. Previous structural and single-channel studies have greatly aided our understanding of the dynamics of OmpG’s extracellular loops and their role in maintaining open and closed pore conformations[29, 30, 36–38]. Evidence suggests that the extracellular loops on OmpG, particularly the longest, loop 6 (L6) is responsible for the pH-dependent conformational change[29, 30, 38]. At acidic pHs, OmpG mostly adopts a closed conformation where L6 occludes the lumen of the pore; while at neutral pH (pH 7.0), L6 occupancy within the lumen of the pore is significantly reduced causing OmpG shift to more open pore conformations.
Because L6 is the key component for mediating OmpG gating, different engineering approaches to reduce OmpG’s gating activity have been explored to create a quiet pore for sensing purposes[30, 38–40]. For example, a quiet OmpG (qOmpG) was engineered by introducing a disulfide bond between the β-strands 12 and 13 that effectively restricts the flexibility of L6 and is resistant to pH-dependent conformational changes [30]. Likewise, Zhuang and Tamm used lipid anchors to pin L6 to the membrane bilayer as a means to limit the flexibility of L6[38]. Furthermore, Grosse et al. opted to shorten all seven extracellular loops, specifically removing 6–12 amino acids within loop 6, that significantly decreased the OmpG gating[39].
Recently, we combined computational and single-channel studies to further unravel the mechanism of OmpG pH-dependent gating[41]. Our work denoted that the gating loop 6 and charged residues within the barrel of the pore form an electrostatic interaction network that dominantly controls the L6 gating equilibrium. Specifically, the positively charged clusters in the lumen attract L6 toward the lumen while the negatively charged ones repel the loop 6 outward. pH modulates the gating equilibrium primarily through altering the protonation state of the negative patch residues, consequently weakening the repulsion force that stabilizes the open state.
Although we found that the loop-lumen electrostatic interaction dominates the OmpG gating behavior, there was indication that loop-loop interaction may also have an effect on the pH sensitivity of OmpG[37]. In this work, we probed the ability to influence the gating equilibrium of OmpG by adjusting the repulsive effect between one or more extracellular loops. Our work demonstrates that the loop-loop interactions of OmpG also contributes to the electrostatic network controlling the gating equilibrium, albeit to a lesser degree compared to the loop-lumen interaction. The pH-independent quiet OmpG pore created in this study can be easily modified with other recognition groups through thio-chemistry for developing new sensing strategies.
2. Materials and Methods
2.1. Cloning of OmpG mutants
All single or multiple mutations: L1 (V19D, G21E, Y22E, G23D); L2 (S58D, A59E, G60E, K61D); L6 (I226D, R228D); were produced using primers (Eurofins MWG Operon) in Table S1. Briefly, mutagenesis PCRs were carried out with the forward and reverse primers at a 1:1 molar ratio to pT7-OmpGwt template. The PCR mixture was subjected to DpnI digestion for 3 h at 37°C to degrade the template plasmid. Afterwards, the PCR mixtures were transformed into chemically competent E. coli DH5α cells under ampicillin selection (100 ug/mL). Colonies containing the desired mutant were confirmed by DNA sequencing.
2.2. Expression and Purification of OmpG Proteins
All OmpG proteins were prepared by following the established protocol[31]. The pT7-OmpGwt and mutant plasmids were transformed into chemically competent BL21(pLysS) E. coli. Cells were grown in Lennox broth (LB) medium at 37°C until the OD600 reached 0.6. IPTG (final concentration 0.5 mM) was added to the culture to induce the protein expression at 37°C for 3 hrs. The harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 200 μg/ml lysozyme, 1 mM EDTA) and lysed by sonication on ice. The lysate was cleared by centrifugation at 20 000g for 30 min. The pellet was washed once with 30 ml wash buffer (50 mM Tris-HCl, pH 8.0, 1.5 M Urea). Thereafter, the inclusion body pellet was solubilized in 50 mM Tris-HCl, pH 8.0 and 8 M Urea and passed through a 0.45 μm filter before FPLC purification. OmpG proteins were purified using a pre-packed 5ml Q-sepharose anion exchange column (GE Healthcare). OmpG proteins were eluted in 50 mM Tris-HCl, pH 9.0, 200 mM NaCl, 8 M Urea. Purified OmpG proteins were diluted in refolding buffer 50 mM Tris-HCl, pH 9.0, 3.25% (w/v) octyl-glucoside (GoldBio) in a 2:3 protein: buffer (v/v) ratio. Samples were then incubated at 37°C for 3 days to allow the OmpG proteins to refold. The refolding efficiency was determined by heat denatured mobility shift by 12% SDS-polyacrylamide gel electrophoresis (Fig. S1).
2.3. Single-Channel Recording of OmpG Proteins
Single-channel recording of OmpG was similar to the previous study[31]. Briefly, experiments were performed in an apparatus containing two chambers separated by a 25 μm thick Teflon film. An aperture of approximately 100 μm diameter was generated near the center of the film with an electric spark. The aperture was pretreated with a hexadecane in pentane (10% v/v) solution before each chamber was filled with buffers as indicated specifically. After allowing the pentane to evaporate, either acidic buffer (20 mM sodium acetate, pH 5.0, 1 M KCl) or neutral buffer (20 mM Tris-HCl, pH 7.0, 1 M KCl) was added to each chamber. An Ag/AgCl electrode was immersed in each chamber with the cis chamber grounded. 1,2-Diphytanoyl-sn-glycerol-3-phosphocholine (Avanti Polar Lipids, USA) dissolved in pentane (10 mg/mL) was deposited on the surface of the buffer in both chambers and monolayers were formed after the pentane evaporated. The lipid bilayer was formed by raising the liquid level up and down across the aperture. Refolded OmpG proteins (~1 nM, final concentration) were added to the cis chamber. Typically, a voltage potential of 200 mV was applied to facilitate OmpG insertion. After a single OmpG pore inserted, the applied voltage was lowered to 50 mV for recording. Current was amplified with an Axopatch 200B integrating patch clamp amplifier (Axon Instruments, Foster City, CA). Signals were filtered with a Bessel filter at 2 kHz (unless otherwise stated) and then acquired by a computer (sampling at 50 μs) after digitization with a Digidata 1320A/D board (Axon Instruments).
2.4. Single-Channel Current Trace Analysis
Current traces were analyzed by Clampfit 10.7 (Molecular Devices) using single channel search of a 30 s section of each pore’s trace. The extracted dwell times of the open and closed state were used to calculate the open probability Po, the inter-event duration, and the gating event duration. To determine the open probability, the sum of the inter-event times was divided by the total time analyzed. To determine the average open state duration τopen, the inter-event duration times were plotted in a histogram that was fitted with a single exponential function (Fig. S2). The average closed state duration τclosed was similarly calculated from the event duration times.
3. Results and Discussions
Introducing single negative charge to loops
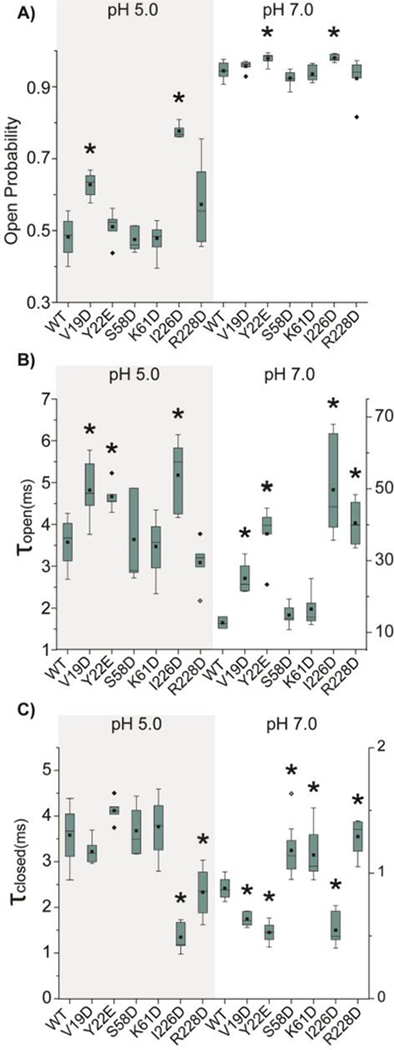
Previous studies indicated that the dynamic movement of loop 1 (L1), loop 2 (L2) and loop 6 (L6) are involved in a concerted motion [37]. Crosslinking experiments revealed that the residue S58 at the center of L2 could directly contact the residue I228 in the middle of L6 when L6 is in the closed conformation [37]. Consequently, we hypothesized that increasing the net negative charge of L6 and opposite loops will cause the loops to repel each other which can drive L6 to adopt more open conformation. We first introduced single negatively charged residues to L1, L2 and L6 by creating the following mutants V19D and Y22E on L1, S58D and K61D on L2, I226D and R228D on L6 (Fig. 1). Single channel recording experiments were performed to investigate how the addition of negative charges at the three loops affected the gating of OmpG. Because the gating properties of OmpG are asymmetric at the positive and negative potentials, we will first focus on testing the gating behaviors of all mutants at the positive potential, which is defined as when the extracellular loops are exposed to the positive side. Data obtained at the negative potential are included in the supporting information document. Under acidic buffer conditions, introducing single negatively charged residues within L1 and L2 results in a negligible change in the open probability (Po), except V19D at L1 and I226D mutation at L6 (Fig. 2A and S3, Table S2). The open probability of V19D construct increases approximately by 25% compared with OmpGwt, from 0.50 ± 0.06 to 0.63 ± 0.04. Among single point mutations within L6, I226D exhibits a substantial increase in the Po from the 0.50 ± 0.06 for OmpGwt to 0.78 ± 0.02 at pH 5.0 while the effect of R228D is negligible (Fig. 2A). We analyzed two kinetic parameters τopen and τclosed, which measure the stability of the open or closed conformation respectively. Both V19D and I226D have larger τopen and smaller τclosed, indicating the mutations stabilize the open pore state and destabilize the closed state. Especially, I226D shows a profoundly destabilized closed state with its τclosed decreased nearly 3-fold compared with OmpGwt. We also noticed that two constructs, Y22E and R228 exhibited a detectable change in either τopen or τclosed, but the open probabilities of the two constructs are not significantly different from OmpGwt (Fig 2A).
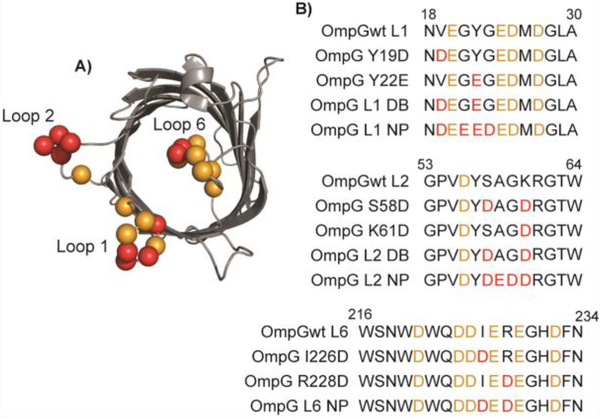
Fig. 1.
Structure and sequences of OmpG mutants. A) Top view of the OmpG structure (pdb: 2IWW) in the closed conformation. B) Sequence alignment of the mutated loop regions of OmpG constructs. The negatively charged residues in the wild type OmpG are highlighted in orange while mutated acidic residues in red.
Fig. 2.
The gating kinetic analysis of single mutant OmpG constructs. Box plots of A) the open probability, B) the inter-event duration (τopen) and C) the event duration (τclosed) of OmpG constructs. Buffers used were 1M KCl in 20 mM sodium acetate pH 5.0 or 20 mM Tris HCl pH 7.0. The applied potential was +50 mV. All data were derived from at least five independent recordings. Two-tailed student’s t-tests were performed to compare each single mutation construct and OmpGwt with stars (*) marking the results with p-values < 0.05. Detailed statistics were shown in Table S3.
Due to the high Po of OmpGwt at neutral pH 7.0, the effect of mutations on the open probability is not obvious (Fig. 2A, Table S2). In general, all mutants show mostly open pores with the Po larger than 0.92. While V19D, S58D, K61D and R228D show a Po almost identical to that of OmpGwt, Y22E and I226D reveal a slightly elevated Po. The τopen of these constructs exhibit a more notable difference from OmpGwt. For example, mutations at L1 and L6 increase τopen by 1.6 to 3.6-fold while S58D and K61D at L2 have almost no effect on (Fig. 2B). The τclosed of three mutations, V19D, Y22E, and I226D, present around 50% reduction at pH 7.0 (Fig. 2C) with other mutants also showing significant variation from OmpGwt. To summarize, the data demonstrate that the gating behavior of OmpG can be altered by adding single negatively charged residues in loop 1 and 6.
Progressive addition of negative charges to loops
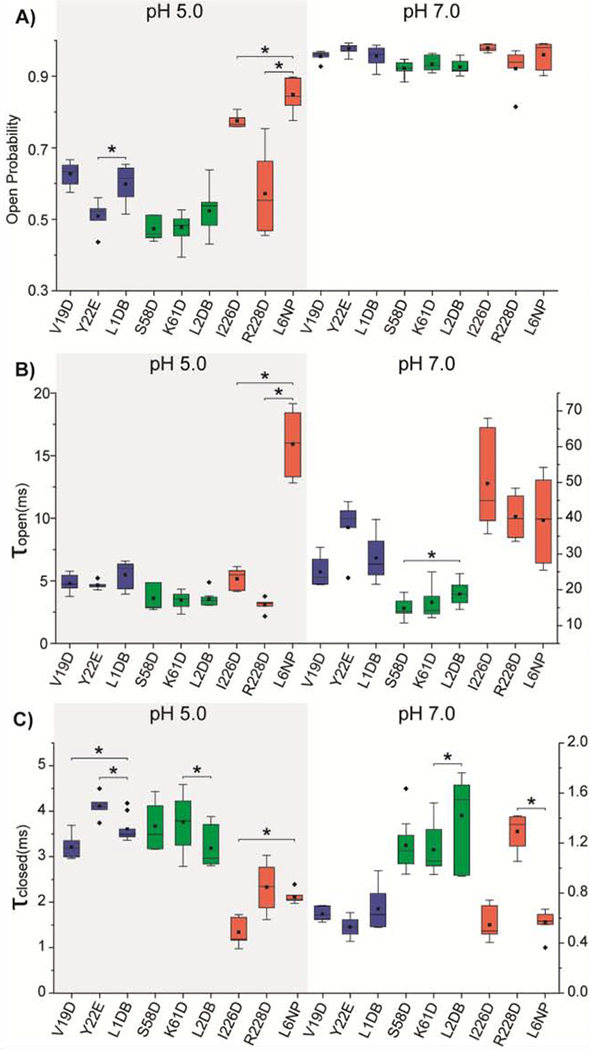
We then tested if gradually adding more charges to the loops would further promote higher open probability. We coupled the aforementioned single charge mutants within their respective loop to generate double negatively charged mutants, L1DB (V19D&Y22E), and L2DB (S58D&K61D) and L6 NP (I226D&R228D). The L2DB pore showed no significant change in the open conformation at both pH conditions compared to the single mutant L2 constructs (Fig. 3A, Fig. S3, Table S2). The open probability of L1DB is similar to that of V19D, higher than Y22E (Fig. 3A, Fig. S3, Table S2). In contrast, the L6 NP mutant shows a considerably enhanced open pore conformation compared with the two single charge mutants I226D and R228D (Fig S3, Table S2), with an open probability of 0.85 ± 0.04 at pH 5.0 and 0.96 ± 0.04 at pH 7.0 (Fig. 3A). Notably, the pH-gating is mostly prohibited in the L6 NP mutant.
Fig. 3.
The gating kinetic analysis of double mutant OmpG constructs compared to single mutant. Box plots of A) the open probability, B) the inter-event duration (τopen) and C) the event duration (τclosed) of OmpG constructs. Buffers used were 1M KCl in 20 mM sodium acetate pH 5.0 or 20 mM Tris HCl pH 7.0. The applied potential was +50 mV. All data were derived from at least five independent recordings. Two-tailed student’s t-tests were performed to compare each single mutation construct with the double mutation construct at the same loop with stars (*) marking the results with p-values < 0.05. L1 mutations are presented in blue, L2 mutations in green, and L6 mutations in red. Detailed statistics were shown in Table S3.
All OmpG variants adopt a largely open conformation (Po>0.94) at neutral pH with kinetic analysis revealing minor differences between constructs; thus, we focus on the gating behavior of OmpG mutants at pH 5.0. The kinetic analysis shows that L6 NP exhibits a profound increase in τopen, approximately 4-fold higher than OmpGwt (Table S2), 3-fold higher than single mutants I226D and R228D at L6 (Fig. 3B), suggesting that the extra negative charge in L6 significantly stabilizes the open state. None of the τclosed of L1DB, L2DB and L6NP shows a significant decrease compared with their respective single mutation constructs at each loop (Fig 3C, Table S2), indicating that the addition of a negative charge to L1, L2 or L6 does not destabilize the closed state further.
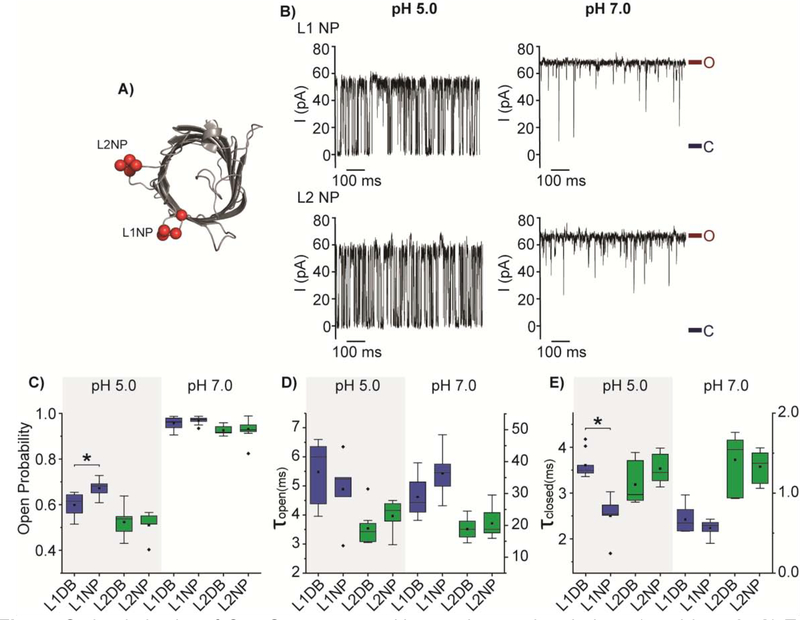
We further introduced two more negative charges to L1DB and L2DB generating a negative patch at the two loops. These new constructs are denoted L1 negative patch (NP) (V19D, G21E, Y22E, G23D) and L2 negative patch (NP) (S58D, A59E, G60E, K61D) (Fig. 4A). The L1 NP shows a higher open probability than L1 DB while L2 NP mutant does not exhibit any substantial change in the gating equilibrium compared with L2DB (Fig. 4B). The Po of the L1 NP slightly rises to 0.67 ± 0.04 compared with the 0.60 ± 0.05 of the L1 DB mutant at the acidic pH (Fig. 4C). Kinetic analysis the increased open probility of L1NP is primarily due to the destabilized closed state as (L1 NP and L1 DB) have similar τopen (Fig. 4D). Not surprisingly, the introduction of two additional negative charges into L2 (A59E and G60E) has not any noticeable effect on the dwell times of the open and closed states compared to L2 DB (Fig. 4D&E). In conclusion, these results have supported our hypothesis that increasing the negative charge at L1, L2, and L6 indeed shifts the gating equilibrium toward the open state. By comparison, the addition of the acidic residues to L6 induces the largest shift in the equilibrium, while L1 exhibits less influence and L2 the least.
Fig. 4.
Gating behavior of OmpG constructs with negative patches in loop 1 and loop 2. A) Top view of the cartoon representation of OmpG (PDB:2IWW). Red balls represent the mutated residues at the loops. B) Representative single channel recording traces of L1 NP and L2 NP. The open and closed states were marked with maroon and navy lines, respectively. Box plots of C) the open probability, D) the inter-event duration (τopen) and E) the event duration (τclosed) of OmpG constructs. Buffers used were 1M KCl in 20 mM sodium acetate pH 5.0 or 20 mM Tris HCl pH 7.0. The applied potential was +50 mV. All data were derived from at least five independent recordings. Two-tailed student’s t-tests were performed to compare DB constructs and NP constructs of each loop with stars (*) marking the results with p-values < 0.05. L1 mutations are represented in blue and L2 mutations in green. Detailed statistics were shown in Table S3.
Negative charged loops in tandem
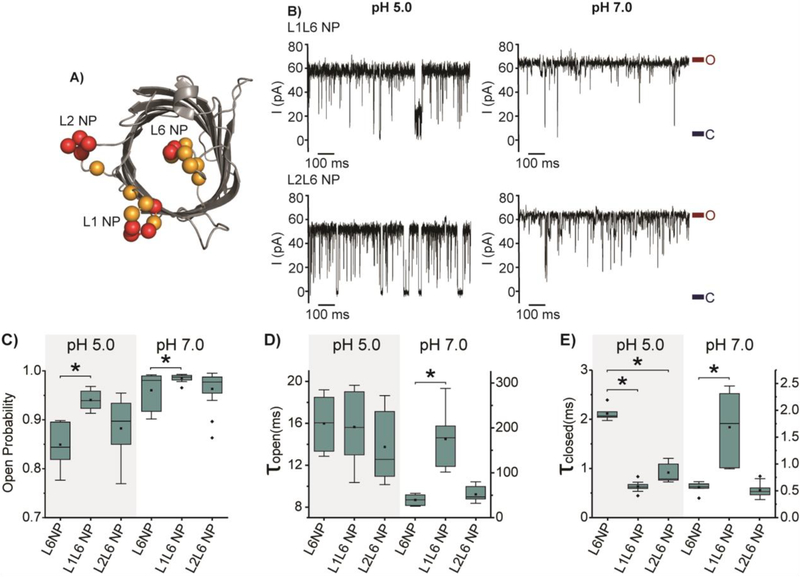
Although the pH-dependent gating of OmpG L6 NP is substantially inhibited, the protein still has a considerable level of gating activity at the acidic pH. We decided to couple L6 NP with either the L1 NP or L2 NP mutations to see if the mutants L1L6 NP and L2L6 NP would further reduce the gating to create a quiet pore at pH 5.0 (Fig. 5A). In general, L1L6 NP and L2L6 NP generate mostly open pores at both acidic and neutral pH conditions (Fig. 5B and C). The Po of L1L6 NP significantly increases by around 10% compared with L6 NP, while L2L6 NP shows no significant change in the open probability (Fig. 5C, Table S2). Notably, L1L6 NP has a Po of 0.94 ± 0.02 at pH 5.0, comparable to that of OmpGwt at pH 7.0. The kinetic analysis shows that the τclosed of L1L6 NP and L2L6NP decreases approximately by 4 and 2-fold compared with L6 NP, while the τopen of L6 NP, L1L6 NP, and L2L6 NP are similar at pH 5.0 (Fig. 5D and E). This result suggests that the negative charges at L1 and L2 mainly play the role of destabilizing the closed state of L6NP at the acidic condition.
Fig. 5.
Gating behavior of OmpG constructs with coupled negative loop patches. A) Cartoon representation of OmpG showing the native negatively charged residues (orange balls) and introduced negatively charged residues (red balls) at L1, L2 and L6 (PDB:2IWW). B) Representative single channel current traces of OmpGwt, L1L6 NP, and L2L6 NP. The open and closed states were marked with maroon and navy lines, respectively. Box plots of C) the open probability, D) the inter-event duration (τopen) and E) the event duration (τclosed) of OmpG constructs. Buffers used were 1M KCl in 20 mM sodium acetate pH 5.0 or 20 mM Tris HCl pH 7.0. The applied potential was +50 mV. All data were derived from at least five independent recordings. Stars (*) mark the results with p-values < 0.05 for two-tailed student’s t-tests comparing the double loop constructs and L6NP. Detailed statistics were shown in Table S3.
At pH 7.0, the change in open probability shows a similar trend, albeit to a lesser level, due to the already large τopen and small τclosed of L6 NP. Interestingly, L1L6NP shows a different effect on the kinetic parameters at the two pHs. For example, introducing charges to L1 substantially increase τopen from 39.42 ± 11.86 ms to 175.19 ± 55.8 ms at pH 7.0 (Fig. 5D). One possible explanation for the difference is that the negative charge states of OmpG loops at the two pH conditions. At low the pH, some acidic residues at the OmpG loops may be protonated. Therefore adding two charges at L1 could create greater repulsion among the loops at pH 7.0 than pH 5.0, explaining the drastic increase of τopen at the neutral pH.
Interestingly, L1L6NP also causes a slight increase in τclosed at pH 7.0 (Fig. 5E). We surmise these gating events may originate from the dynamic behavior of loops other than L6. For example, the higher net negative charges of L1 and L2 may strongly attract positive residues at the OmpG barrel[41], forming L1 or L2- mediated closed conformations distinct from the L6 occluded state.
To summarize, we have shown that adding negative charges to the gating loop 6 and the opposite loops induce a more open conformation at both acidic and neutral buffering conditions. This observation can be explained by the fact that increasing the net negative charge within OmpG’s extracellular loops creates enhanced repulsive force to trigger the loop 6 to swing away from the lumen. Our data also shows that the addition of four extra negative charges into L1 causes a more profound change in gating behavior compared to L2. We believe that the shorter distance between L1 and L6 than that of L2 and L6, as observed in the solid-state NMR structure of OmpG (5MWV, Fig. S4) may account for such an observation[42]. Note that L1 and L6 of the wild type OmpG are considerably negatively charged. Our data also suggests that the L1-L6 interaction/repulsion contributes to keeping the native OmpG porin in the open state.
Gating behavior of OmpG variants at the negative potential
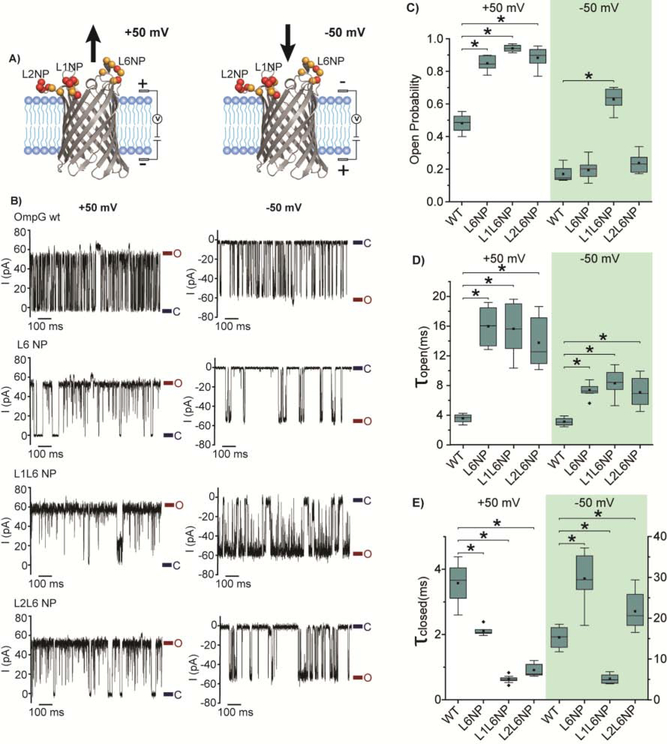
In nature, the gating behavior of OmpG is controlled by the electrostatic network formed between OmpG loops and charged residues in the lumen[41]. However, the external electric potential could also impact the movement of charged loops of OmpG in single channel recording experiments. We anticipate that the observed gating equilibrium is controlled by a combination of the electrophoretic effect exerted by external potentials and the internal electrostatic network. Here we investigated the electrophoretic effect on gating behavior of all mutants by reversing the applied potential to negative (Fig. S3, 5–9), which is defined as when the extracellular loops are exposed to the negative voltage chamber (Fig. 6A). Under a positive potential, the electrophoretic force drives the negatively charged loops out of the lumen and consequently stabilizes the open state (Fig. 6A). Conversely, the negative potential pushes the loops towards the lumen and stabilizes the closed state (Fig. 6A). Thus, it is expected that the OmpG proteins exhibit higher open probability at +50 mV than at −50 mV. At pH 7.0, OmpG variants predominantly occupy the open state (Po>0.94) at ± 50 mV (Fig. S3, S5–9, Table S2), and the polarity of the potential has a small effect on the gating kinetics indicating the interaction within the electrostatic network of OmpG greatly exceeds the electrophoretic effect.
Fig. 6.
Voltage-effect on the gating behavior of three OmpG constructs. A) Cartoon representation of OmpG pore in the lipid bilayer with the different voltage applied. The native negatively charged residues are represented as orange balls and the introduced negatively charged residues as red balls at L1, L2 and L6 (PDB:2IWV). The arrow indicates the electric force applied to OmpG Loop 6 by the external potential. B) Representative single channel current traces of OmpG constructs. Open state and closed state were marked with maroon and navy lines, respectively. Box plots of C) the open probability, D) the inter-event duration (τopen) and E) the event duration (τclosed) of OmpG constructs. Buffers used were 1 M KCl 20 mM sodium acetate pH 5.0. All data were derived from at least five independent recordings. Stars (*) mark the results with p-values < 0.05 for two-tailed student’s t-tests comparing each construct and OmpG WT. Detailed statistics were shown in Table S3.
Under the acidic pH 5.0, all 14 OmpG variants show significantly reduced open probability at −50mV compared at +50mV, albeit to a different extent (Table S2). For example, the open probability of OmpGwt reduces from 0.50 ± 0.06 to 0.17 ± 0.04, nearly 3-fold when switching the potential from + 50 mV to −50 mV (Fig. 6). Similarly, the L6 NP and L2L6NP also show strongly reduced Po at −50 mV compared with +50 mV (Fig. 6A&B). Notably, these two constructs only show slightly larger Po than that of OmpGwt at −50 mV (Fig. 6C, table S2). Nevertheless, the kinetic analysis reveals an increased τopen for both L6 NP and L2L6 NP compared with OmpGwt at −50 mV, indicating the mutations stabilize the open state, presumably by enhancing loop-loop repulsion (Fig. 6D). Notably, both mutants also have a higher τclosed than OmpGwt, which can be explained by the elevated electrophoretic force induced by the additional negative charges at these loops (Fig. 6E). Thus, we conclude that the mutations of L6 NP and L2L6 NP intensify the electrophoresis and the repulsion simultaneously, which results in the cancellation of the two opposing effects. Interestingly, the L1L6 NP still mostly occupies the open state with a Po of 0.63 ± 0.06 at −50mV, suggesting that the strong L1-L6 repulsion overweighs the electrophoretic force. In summary, an enhanced loop-loop repulsion, against the electrophoretic effect, can keep the OmpG at more open states at the acid pH under an applied positive potential.
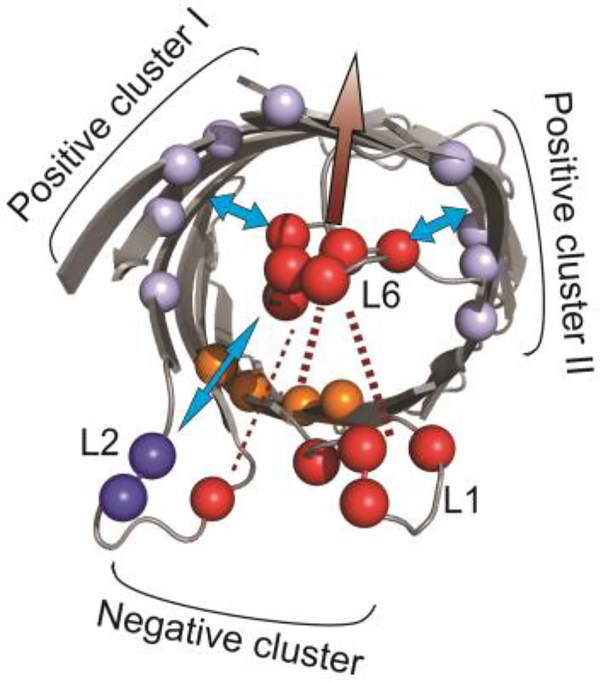
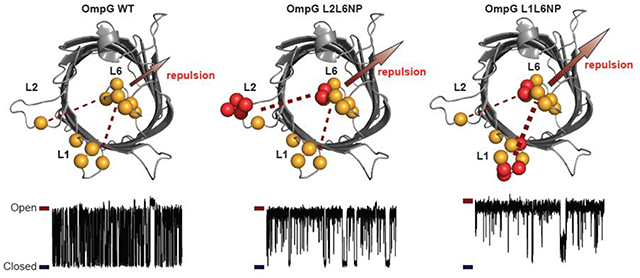
We previously proposed that OmpG conformational changes of loop 6 are controlled by a complex electrostatic interaction network formed around the loop 6 [41]. The central components of the network include three charged clusters in the lumen (Fig.7): the negatively charged cluster within the barrel repels L6 from the lumen. This repulsion constitutes an energy barrier that prohibits L6 from invading the pore. In contrast, the two positively charged clusters in the wall stabilize the closed conformation after L6 enters the lumen (Fig.7). Results obtained in this study provide additional information to support this model. First, this model can explain the greater ability of L6 NP to keep the OmpG pore open compared with L1 NP and L2 NP. It is because adding negative charges to L6 enhances the L6-lumen repulsion and the loop-loop repulsion, whereas more negative charges to L1 and L2 only enhance the loop-loop repulsion. Second, the kinetic analysis reveals that additional charges at L1 and L2 only increase the dwell time of the open state of OmpG but had no effect on the closed state. This result indicates that the loop-loop repulsion mostly contributes to the energy barrier that prevents L6 from invading the lumen. Thus, our study suggests that the loop-loop repulsion belongs to the electrostatic network that controls the L6 open conformation (Fig. 7). In contrast, more negative charges in L6 not only prolong the open state but also shorten the closed state. The enhanced L6-lumen repulsion that pushes L6 outwards accounts for the stabilizing effect of the open state, while the destabilizing effect on the closed state can be explained based on the structure of the closed conformation. In the closed state, L6 collapses into the lumen which brings it near to the negative clusters. This would drastically enhance the L6-lumen repulsion that repels L6 outside of the lumen thus strongly destabilizing the closed state. Based on this study, we propose an updated model for the OmpG pH-gating mechanism: the conformational changes of L6 is controlled primarily by the L6-lumen interactions with L6-loop repulsions/attractions playing the secondary role (Fig. 7). The L6-lumen interactions strongly regulate both the open and closed state while the L6-loop repulsions mostly stabilize the open state.
Fig. 7.
Schematic illustration of the electrostatic network model controlling OmpG gating. The native negatively and positively charged residues in the wild type OmpG loop 1, 2 & 6 are represented as red and blue balls. The negative cluster and the positive cluster I and II at the barrel is colored in orange and light blue respectively (PDB:2IWW). The arrow indicates the attractive (blue arrow) and repulsive (marron arrow) forces applied to L6 by the charged residues within OmpG barrels, L1 and L2.
4. Conclusions
In summary, we demonstrated that enhancing the loop-loop repulsion forces within OmpG can shift the gating equilibrium towards a more open conformation. Our finding adds the loop-loop interaction as an auxiliary factor to the previous pH-gating model. The new model will help guide the design of novel OmpG proteins with enhanced pH-resistance or quietness for sensor development.
Supplementary Material
Acknowledgements
This research was supported by the US National Institutes of Health grant R01-GM115442.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Delcour AH, Outer membrane permeability and antibiotic resistance, Biochim Biophys Acta 1794(5) (2009) 808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Koebnik R, Locher KP, Van Gelder P, Structure and function of bacterial outer membrane proteins: barrels in a nutshell, Mol Microbiol 37(2) (2000) 239–53. [DOI] [PubMed] [Google Scholar]
- [3].Nikaido H, Molecular basis of bacterial outer membrane permeability revisited, Microbiol Mol Biol Rev 67(4) (2003) 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vergalli J, Bodrenko IV, Masi M, Moynie L, Acosta-Gutierrez S, Naismith JH, Davin-Regli A, Ceccarelli M, van den Berg B, Winterhalter M, Pages JM, Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria, Nat Rev Microbiol 18(3) (2020) 164–176. [DOI] [PubMed] [Google Scholar]
- [5].Zeth K, Thein M, Porins in prokaryotes and eukaryotes: common themes and variations, Biochem J 431(1) (2010) 13–22. [DOI] [PubMed] [Google Scholar]
- [6].Pages JM, James CE, Winterhalter M, The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria, Nat Rev Microbiol 6(12) (2008) 893–903. [DOI] [PubMed] [Google Scholar]
- [7].Klebba PE, Newton SM, Mechanisms of solute transport through outer membrane porins: burning down the house, Curr Opin Microbiol 1(2) (1998) 238–47. [DOI] [PubMed] [Google Scholar]
- [8].Alcaraz A, Nestorovich EM, Aguilella-Arzo M, Aguilella VM, Bezrukov SM, Salting out the ionic selectivity of a wide channel: the asymmetry of OmpF, Biophys J 87(2) (2004) 943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alcaraz A, Nestorovich EM, Lopez ML, Garcia-Gimenez E, Bezrukov SM, Aguilella VM, Diffusion, exclusion, and specific binding in a large channel: a study of OmpF selectivity inversion, Biophys J 96(1) (2009) 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Delcour AH, Solute uptake through general porins, Front Biosci 8 (2003) d1055–71. [DOI] [PubMed] [Google Scholar]
- [11].Fernandez L, Hancock RE, Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance, Clin Microbiol Rev 25(4) (2012) 661–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Madesh M, Hajnoczky G, VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release, J Cell Biol 155(6) (2001) 1003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N, VDAC, a multi-functional mitochondrial protein regulating cell life and death, Mol Aspects Med 31(3) (2010) 227–85. [DOI] [PubMed] [Google Scholar]
- [14].Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS, The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death, Curr Pharm Des 12(18) (2006) 2249–70. [DOI] [PubMed] [Google Scholar]
- [15].Howorka S, Siwy Z, Nanopore analytics: sensing of single molecules, Chem Soc Rev 38(8) (2009) 2360–84. [DOI] [PubMed] [Google Scholar]
- [16].Mirsaidov UM, Wang D, Timp W, Timp G, Molecular diagnostics for personal medicine using a nanopore, Wiley Interdiscip Rev Nanomed Nanobiotechnol 2(4) (2010) 367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Movileanu L, Watching single proteins using engineered nanopores, Protein Pept Lett 21(3) (2014) 235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee J, Bayley H, Semisynthetic protein nanoreactor for single-molecule chemistry, Proc Natl Acad Sci U S A 112(45) (2015) 13768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thakur AK, Movileanu L, Real-time measurement of protein-protein interactions at single-molecule resolution using a biological nanopore, Nat Biotechnol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oukhaled A, Pastoriza-Gallego M, Bacri L, Mathe J, Auvray L, Pelta J, Protein unfolding through nanopores, Protein Pept Lett 21(3) (2014) 266–74. [DOI] [PubMed] [Google Scholar]
- [21].Stefureac RI, Lee JS, Nanopore analysis of the folding of zinc fingers, Small 4(10) (2008) 1646–50. [DOI] [PubMed] [Google Scholar]
- [22].Craig JM, Laszlo AH, Brinkerhoff H, Derrington IM, Noakes MT, Nova IC, Tickman BI, Doering K, de Leeuw NF, Gundlach JH, Revealing dynamics of helicase translocation on single-stranded DNA using high-resolution nanopore tweezers, Proc Natl Acad Sci U S A 114(45) (2017) 11932–11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhao Q, de Zoysa RS, Wang D, Jayawardhana DA, Guan X, Real-time monitoring of peptide cleavage using a nanopore probe, J Am Chem Soc 131(18) (2009) 6324–5. [DOI] [PubMed] [Google Scholar]
- [24].Wanunu M, Nanopores: A journey towards DNA sequencing, Phys Life Rev 9(2) (2012) 125–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fajardo DA, Cheung J, Ito C, Sugawara E, Nikaido H, Misra R, Biochemistry and regulation of a novel Escherichia coli K-12 porin protein, OmpG, which produces unusually large channels, J Bacteriol 180(17) (1998) 4452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Misra R, Benson SA, A novel mutation, cog, which results in production of a new porin protein (OmpG) of Escherichia coli K-12, J Bacteriol 171(8) (1989) 4105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liang B, Tamm LK, Structure of outer membrane protein G by solution NMR spectroscopy, Proc Natl Acad Sci U S A 104(41) (2007) 16140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Subbarao GV, van den Berg B, Crystal structure of the monomeric porin OmpG, J Mol Biol 360(4) (2006) 750–9. [DOI] [PubMed] [Google Scholar]
- [29].Yildiz O, Vinothkumar KR, Goswami P, Kuhlbrandt W, Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation, EMBO J 25(15) (2006) 3702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen M, Khalid S, Sansom MS, Bayley H, Outer membrane protein G: Engineering a quiet pore for biosensing, Proc Natl Acad Sci U S A 105(17) (2008) 6272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fahie M, Chisholm C, Chen M, Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore, ACS Nano 9(2) (2015) 1089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fahie MA, Chen M, Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms, J Phys Chem B 119(32) (2015) 10198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fahie MA, Yang B, Mullis M, Holden MA, Chen M, Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore, Anal Chem 87(21) (2015) 11143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fahie MA, Yang B, Pham B, Chen M, Tuning the selectivity and sensitivity of an OmpG nanopore sensor by adjusting ligand tether length, ACS Sens 1(5) (2016) 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Conlan S, Bayley H, Folding of a monomeric porin, OmpG, in detergent solution, Biochemistry 42(31) (2003) 9453–65. [DOI] [PubMed] [Google Scholar]
- [36].Mari SA, Koster S, Bippes CA, Yildiz O, Kuhlbrandt W, Muller DJ, pH-induced conformational change of the beta-barrel-forming protein OmpG reconstituted into native E. coli lipids, J Mol Biol 396(3) (2010) 610–6. [DOI] [PubMed] [Google Scholar]
- [37].Zhuang T, Chisholm C, Chen M, Tamm LK, NMR-based conformational ensembles explain pH-gated opening and closing of OmpG channel, J Am Chem Soc 135(40) (2013) 15101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhuang T, Tamm LK, Control of the conductance of engineered protein nanopores through concerted loop motions, Angew Chem Int Ed Engl 53(23) (2014) 5897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grosse W, Psakis G, Mertins B, Reiss P, Windisch D, Brademann F, Burck J, Ulrich A, Koert U, Essen LO, Structure-based engineering of a minimal porin reveals loop-independent channel closure, Biochemistry 53(29) (2014) 4826–38. [DOI] [PubMed] [Google Scholar]
- [40].Sanganna Gari RR, Seelheim P, Liang B, Tamm LK, Quiet Outer Membrane Protein G (OmpG) Nanopore for Biosensing, ACS Sens 4(5) (2019) 1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Perez-Rathke A, Fahie MA, Chisholm C, Liang J, Chen M, Mechanism of OmpG pH-Dependent Gating from Loop Ensemble and Single Channel Studies, J Am Chem Soc 140(3) (2018) 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Retel JS, Nieuwkoop AJ, Hiller M, Higman VA, Barbet-Massin E, Stanek J, Andreas LB, Franks WT, van Rossum BJ, Vinothkumar KR, Handel L, de Palma GG, Bardiaux B, Pintacuda G, Emsley L, Kuhlbrandt W, Oschkinat H, Structure of outer membrane protein G in lipid bilayers, Nat Commun 8(1) (2017) 2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.