Abstract
The purpose of this study was to investigate the drug-resistant phenotypes and genes of Escherichia coli in animal, environmental, and human samples before and after antibiotic use at a large-scale broiler farm to understand the respective effects on E. coli resistance during the broiler feeding cycle. The antibiotic use per broiler house was 143.04 to 183.50 mg/kg, and included tilmicosin, florfenicol, apramycin, and neomycin. All strains isolated on the first day the broilers arrived (T1; day 1) were antibiotic-resistant bacteria. E. coli strains isolated from animal samples were resistant to ampicillin, tetracycline, and sulfamethoxazole (100%), and those isolated from environmental samples were resistant to 5 different drugs (74.07%, 20 of 27). E. coli strains isolated on the last day before the broilers left (T2; day 47) had a higher resistance rate to florfenicol (100%, 36 of 36) than at T1 (P < 0.05). Multidrug resistance increased from T1 (84.21%, 32 of 38) to T2 (97.22%, 35 of 36). Most strains were resistant to 5 classes of antibiotics, and 2 strains were resistant to 6 classes of antibiotics. Among 13 identified drug resistance genes, 11 and 13 were detected at T1 and T2, respectively. NDM-1 was detected in 4 environmental samples and 1 animal sample. In conclusion, the use of antibiotics during breeding increases E. coli resistance to antibacterial drugs. Drug-resistant bacteria in animals and the environment proliferate during the feeding cycle, leading to the widespread distribution of drug resistance genes and an increase in the overall resistance of bacteria.
Key words: Escherichia coli, animal-environment-human, drug resistance, drug resistance gene
Introduction
Bacterial antibiotic resistance has posed significant challenges in the animal breeding industry. The misuse and abuse of veterinary antibiotics have contributed to the development of bacterial resistance (Garcia-Migura et al., 2014). Escherichia coli, one of the most common bacteria in farms, is present in animal intestines, excrement, soil, water, and air (Adachi et al., 2013; Sun et al., 2017; Yang et al., 2018; Falgenhauer et al., 2019; Qiu et al., 2019; Zhang et al., 2019). E. coli can transmit drug resistance genes between drug-resistant and non–drug-resistant bacteria, contributing to the spread of drug resistance genes. In recent years, studies have shown that bacteria have developed resistance to commonly used clinical antibiotics. From 2006 to 2016, the resistance rate to ampicillin, tetracycline, and sulfadiazine of E. coli strains isolated from samples in a veterinary laboratory of Minnesota, United States, was >50% (Hayer et al., 2020). Similarly, from 2008 to 2015, >80% of E. coli strains isolated from chickens and pigs in China showed resistance to ampicillin, tetracycline, and sulfadiazine (Zhang et al., 2017). β-lactams (Klimiene et al., 2018; Ye et al., 2018; Alegría et al., 2020; Wang et al., 2020), tetracycline (Hu et al., 2013; Seifi et al., 2016; Bourély et al., 2019), and plasmid-mediated resistance genes such as PMQR (Hricová et al., 2017; Seo et al., 2019a; Seo et al., 2019a,b) are prevalent among E. coli isolated from poultry.
Several studies have focused on E. coli resistance; however, there is little information on E. coli resistance during breeding (Montoro-Dasi et al., 2020). In this study, we investigated a large-scale broiler farm in Shandong Province, China. The objectives of this study were to 1) assess the effect of E. coli resistance from different aspects (animal, environment, human, and antibiotic use) and 2) to evaluate E. coli resistance in depth to provide a reference for controlling bacterial resistance and promote healthy animal breeding.
Materials and methods
Sample Collection and E. coli Isolation and Identification
Animal samples (poultry cloacal swabs), environmental samples (ground swabs, cage swabs, drinking water, air, and feed), and hand swabs of breeders were collected from 6 houses at a large-scale broiler farm in Shandong Province, China, on July 10, 2019 (T1; the first day after the broilers arrived) and on August 26, 2019 (T2; the last day before the broilers left). The swab samples were stored in Cary-Blair Transport Medium (Beijing Lu Qiao Technology, Beijing, China), and the air samples were collected directly in MacConkey culture medium (Beijing Lu Qiao Technology). All samples were stored at 4°C during transportation.
An aliquot of the water samples (100 mL) was centrifuged, and the resulting precipitate was cultured for 12 h in tryptone soybean broth (Beijing Lu Qiao Technology) and subsequently in MacConkey culture medium. Feed sample (5 g) was mixed with 45 mL of tryptone soybean broth, cultured in 50 mL centrifuge tubes and placed in an incubator at 37°C and 120 rpm for 4 h, and subsequently in MacConkey culture medium. Swab samples were placed directly in MacConkey culture medium. All the samples in MacConkey medium were cultured for 18 to 24 h at 37°C and purified 3 times. The suspected colonies were analyzed using indicator tests (Qingdao Hai Bo Biotechnology, Qingdao, China) of E. coli (e.g., trisaccharide iron test, Kovacs' indigo matrix test, methyl red test, V-P test, and Simon's hydrochloric acid test). All strains identified as E. coli were stored in serum at −40°C.
Medication Survey
A questionnaire was administered to the breeders and managers of the farms to investigate disinfection protocols, drug use, and epidemic prevention of broilers during the breeding period.
Antimicrobial Susceptibility Testing
Representative E. coli strains isolated from T1 and T2 samples were screened for their resistance to antibiotics using the broth dilution method (Clinical and Laboratory Standards Institute, 2017). The antibiotics were ampicillin, amoxicillin-clavulanate, gentamicin, spectinomycin, tetracycline, florfenicol, sulfafurazole, sulfamethoxazole, ceftiofur, ceftazidime, enrofloxacin, ofloxacin, meropenem, and colistin. E. coli strain ATCC 25922 was used for quality control purposes. The strains with simultaneous resistance to 3 classes or more antibiotics were classified as multidrug resistant.
Thirteen antibiotic resistance genes in T1 and T2 samples were analyzed by PCR using the primers listed in Table 1. The antibiotic resistance genes were CMY, OXA, CTX-M, TEM, and NDM-1 (β-lactams); parC, gyrA, qnrS, and aac(6′)-Ib-cr (quinolones); and tetA, tetB, tetM, and tetX (tetracycline). PCR products were sequenced by Shanghai Bioengineering Co. (Shanghai, China), and the resulting sequences were compared against the GenBank database.
Table 1.
Antibiotic resistance genes.
| Gene name | Primer | sequences (5′-3′) | Primer size (bp) | References |
|---|---|---|---|---|
| CMY | F | CAATGTGTGAGAAGCAGTC | 1,432 | Hanson et al., 2002 |
| R | CGCATGGGATTTTCCTTGCTG | |||
| OXA | F | TTCAAGCCAAAGGCACGATAG T | 814 | Briñas et al., 2002 |
| R | TCCGAGTTGACTGCCGGGTTG | |||
| CTX-M | F | AGTGAAAGCGAACCGAATC | 365 | Tian et al., 2011 |
| R | CTGTCACCAATGCTTTACC | |||
| TEM | F | CAGAAACGCTGGTGAAAGTA | 719 | |
| R | ACTCCCCGTCGTGTAGATAA | |||
| NDM-1 | F | GGTTTGGCGATCTGGTTTTC | 621 | Wang et al., 2012 |
| R | CGGAATGGCTCATCACGATC | |||
| parC | F | TGGGCTTAAAACCCACCACT | 319 | Shimada et al., 2010 |
| R | CGGGTTTCTGTGTAACGCAT | |||
| gyrA | F | CGTCGTGTTCTTTATGGTGC | 230 | |
| R | ATAACGTTGTGCAGCAGGTC | |||
| qnrS | F | ACCTTCACCGCTTGCACATT | 571 | Jiang et al., 2008 |
| R | CCAGTGCTTCGAGAATCAGT | |||
| aac (6′)-Ib-cr | F | TGACCTTGCGATGCTCTATG | 616 | |
| R | TTAGGCATCACTGCGTGTTC | |||
| tetA | F | GGCCTCAATTTCCTGACG | 372 | Guillaume et al., 2000 |
| R | AAGCAGGATGTAGCCTGTGC | |||
| tetB | F | GAGACGCAATCGAATTCGG | 228 | |
| R | TTTAGTGGCTATTCTTCCTGCC | |||
| tetM | F | ACAGAAAGCTTATTATATAAC | 171 | Aminov et al., 2001 |
| R | TGGCGTGTCTATGATGTTCAC | |||
| tetX | F | CGCGGATCCATGACAATGCGAATAGATACAG | 1,167 | Wen et al., 2020 |
| R | CCGCATATGTTATACATTTAACAATTGCT |
Statistical Analysis
SPSS 22 (IBM, Qingdao, China) was used for statistical analysis. A 2-sample t test was used to investigate the differences in antibiotic resistance between T1 and T2 samples. Statistical significance was set P < 0.05.
Results
Isolation of E. coli Isolates and Medication Survey
The farm adopted a cage feeding mode, with 1 full-time keeper in charge of each henhouse. However, there was a situation in which a husband and wife managed 2 henhouses together. After a batch of chickens were raised, the henhouses were kept free for 15 D (known as the vacancy period), during which the henhouses were disinfected twice. Henhouses were disinfected for the last time 2 to 3 D before entering a new batch of chickens. The chickens were caged the following day for prevention and treatment. During the feeding cycle, the main antibiotics used were tilmicosin, florfenicol, apramycin, and neomycin, which belong to the macrolide, amphenicol, and aminoglycoside antibiotic classes.
A total of 468 samples were collected: 234 samples from T1 and 234 samples from T2 (Table 2). The overall E. coli isolation rate was higher in T2 (58.12%, 136 of 243) than in T1 (30.77%, 72 of 234). Except for hand swabs, the E. coli isolation rate was higher in T2 than in T1. The separation rate of cloacal swabs increased from 28.33% (17 of 60) to 90.00% (54 of 60), which was the highest increase among the sample types. One E. coli strain was isolated from a water sample at T1, and at T2, 2 strains were isolated from a water sample and 5 strains were isolated from feed.
Table 2.
Escherichia coli isolated from samples collected at T1 and T2.
| House collection sample type | T1 |
T2 |
||||
|---|---|---|---|---|---|---|
| Sample size | Positive detection | Isolation rate (%) | Sample size | Positive detection | Isolation rate (%) | |
| Animal | ||||||
| Cloacal1 | 60 | 17 | 28.33 | 60 | 54 | 90.00 |
| Environment | ||||||
| Ground | 60 | 22 | 36.67 | 60 | 29 | 48.33 |
| Cage | 60 | 25 | 41.67 | 60 | 32 | 53.33 |
| Air1 | 30 | 3 | 10.00 | 30 | 11 | 36.67 |
| Water | 6 | 1 | 16.67 | 6 | 2 | 33.33 |
| Feed1 | 6 | 0 | 0.00 | 6 | 5 | 83.33 |
| Human | ||||||
| Hand | 12 | 4 | 33.33 | 12 | 3 | 25.00 |
| Total | 234 | 72 | 30.77 | 234 | 136 | 58.12 |
Abbreviations: T1, first day the broilers’ arrival; T2, last day before the broilers left.
P < 0.05.
Antimicrobial Resistance
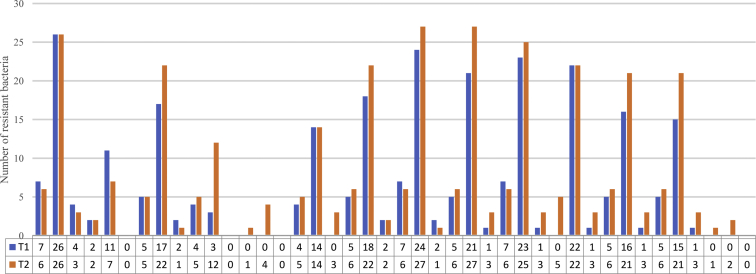
The antibiotic resistance results are shown in Figure 1. All 38 strains of E. coli in T1 were resistant to 12 drugs, but not to meropenem and colistin. E. coli strains were resistant to ampicillin, tetracycline, and sulfafurazole (100%, 7 of 7) in animal samples; ampicillin, tetracycline, florfenicol, sulfafurazole, and sulfamethoxazole (74.07%, 20 of 27) in environmental samples; and ampicillin (100%, 4 of 4), ceftiofur, spectinomycin, and tetracycline (50%) in human samples. Among the environmental samples, the resistance rate to tetracycline and florfenicol was 100% (27 of 27), followed by ampicillin 96.30% (26 of 27). The resistance rate to 6 drugs was >74.07% (20 of 27). E. coli strains from human samples had 100% resistance to 7 drugs (3 of 3) and 100% nonresistance to meropenem and ceftazidime.
Figure 1.
Antibiotic resistance of Escherichia coli isolated from samples collected at T1 and T2. ∗P < 0.05. Abbreviations: A, animal; AMP, ampicillin; A/C, amoxicillin clavulanate; CAZ, cefazolin; CEF, cephalothin; CL, colistin; E, environment; ENR, enrofloxacin; FFC, florfenicol; GEM, gentamicin; H, human; MEM, meropenem; OFL, Ofloxacin; SPT, spectinomycin; SF, sulfafurazole; SXT, sulfamethoxazole; TE, tetracycline; T1, first day the broilers’ arrival; T2, last day before the broilers left.
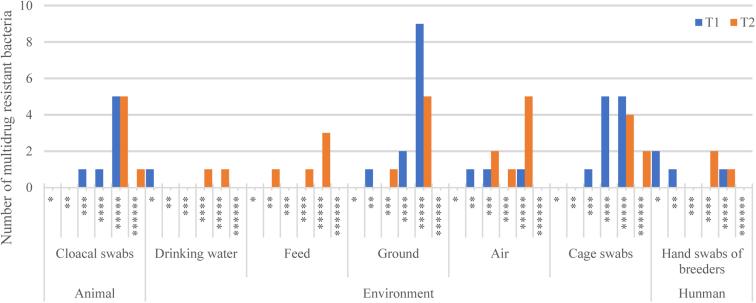
The multidrug resistance of E. coli isolated from T1 and T2 samples is shown in Figure 2. The overall multiple drug resistance rate in T1 samples was 84.21% (32 of 38). There were no multidrug-resistant bacteria in either feed or water (environmental samples) of T1. Multidrug-resistant bacteria were collected from animal and human samples (most had resistance to 4 or 5 drugs). In T2 samples, multidrug resistance increased to 97.22% (35 of 36), whereas multi-drug resistance to 5 classes of antibiotics increased in both water and feed. The other environmental samples showed an increase in the number of resistant bacteria, among which 2 strains with resistance to 6 classes of antibiotics were identified in cage samples. We obtained a wider distribution of drug-resistant bacteria and a higher number of drug-resistant bacteria in T2 samples than in T1 samples.
Figure 2.
Distribution of multidrug resistant bacteria in samples from animals, environment, and humans. The number of ∗ indicates the multiplicity of drug resistance.
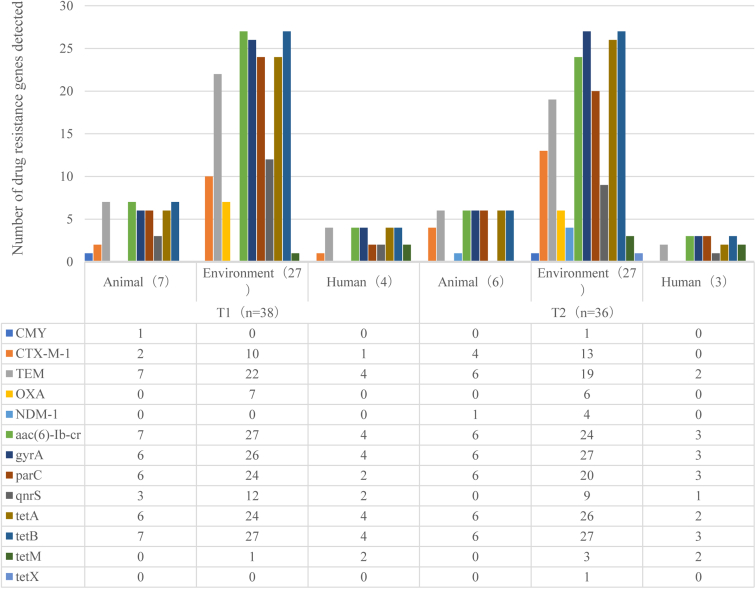
Prevalence of Antibiotic Resistance Genes
Several antibiotic resistance genes were identified among the E. coli isolates (Figure 3). Among the 13 drug resistance genes, 11 were detected to be of different degrees in T1, except NDM-1 and tetX. All 13 drug resistance genes were detected in T2. Furthermore, NDM-1 was detected in 1 animal and 4 environmental samples, and tetX was detected in 1 environmental sample. Among the β-lactam resistance genes in T1 and T2, TEM was the most predominant followed by CTX-M, CMY, and OXA. NDM-1 was not detected in T1 but was detected in T2 animal and environmental samples. In T1, gyrA and parC (quinolone resistance genes) were second in prevalence to aac (6′)-Ib-cr (100% detection rate). Among the tetracycline resistance genes, tetB was the most predominant in T1 and T2 (100%) followed by tetA (100%) in T1 human samples and T2 animal samples. The detection rates of tetM and tetX were relatively low; tetM was identified only in human samples.
Figure 3.
Detection results of antibiotic resistance genes in Escherichia coli isolated from samples collected at T1 and T2. Abbreviations: T1, first day the broilers’ arrival; T2, last day before the broilers left.
Discussion
Aminoglycosides and amido alcohols (e.g., florfenicol, apramycin, and neomycin) are used in the prevention and/or treatment of avian colibacillosis (Wang et al., 2003; Becker et al., 2013). In our study, gentamicin resistance was not observed in T1 human samples. However, gentamicin resistance was observed in T1 environmental samples (51.85%, 14 of 27) and T1 animal samples (57.14%, 4 of 7). However, all T2 human samples showed drug resistance. Gentamicin resistance did not increase in environmental samples but increased to 83.33% (5 of 6) in animal samples. In T1 and T2 environment samples, streptomycin resistance was 66.67% (18 of 27) and 81.48% (22 of 27), respectively. Drug resistance increased from 71.43% (5 of 7) at T1 to 100% (6 of 6) at T2 in animal samples and from 50% (2 of 4) at T1 to 66.67% (2 of 3) at T2 in human samples. In general, resistance toward aminoglycosides was more evident at T2 than at T1 probably owing to the use of apramycin and neomycin that resulted in cross-resistance of E. coli during breeding. Streptomycin-resistant E. coli have different degrees of resistance to the other 6 aminoglycoside drugs (Lin et al., 2012; Reeves et al., 2013). The overall resistance toward florfenicol was 71.05% (27 of 38) at T1 and 100% (36 of 36) at T2 (P < 0.05). Our findings revealed the effect of antibiotic use on bacterial drug resistance during breeding process (Mhondoro et al., 2019) and the resistance of E. coli to aminoglycosides and amide alcohols.
At T1, there was a large number of isolates on the ground and cages (environmental samples). Therefore, the disinfection measures were not sufficient to destroy E. coli in the environment, and these isolated strains were drug resistant (Figure 1). At T1, the detection rate of aac (6′)-Ib-cr and tetB was 100% and remained high at T2. Therefore, E. coli had several drug resistance genes before the administration of antibacterial drugs. In addition, most of the drug resistance genes tested in this study can be mediated by plasmids (Park et al., 2019; Silvester et al., 2019; Suzuki et al., 2019; Cui et al., 2020; Koyama et al., 2020; Nair et al., 2020; Nishikawa et al., 2019), which is likely to be the source of bacterial drug resistance transmission during feeding. Newborn chicks had a certain carrier rate (Table. 1), and these strains were drug resistant, which may be related to farm breeding conditions (Zhao et al., 2019). After a breeding cycle, the number of bacteria carried by the environment and chicken population increases, resulting in a higher detection rate of E. coli in water and air samples. The isolation rate of E. coli from animals increased from 28.33 to 90.00% (P < 0.01); therefore, there was considerable E. coli proliferation in chickens.
The E. coli strain with meropenem resistance was isolated at T2. NDM-1 was detected in all the positive strains. The number of bacteria resistant to 5 classes of antibiotics was the largest at T2, followed by the number of bacteria resistant to 4 classes of antibiotics, consistent with past studies. In addition, there were 3 colistin-resistant strains at T2, which were also resistant to 6 classes of antibiotics.
Among the β lactam resistance genes, the most prevalent was TEM, which was owing to the long-term use of this antibiotic (Sun et al., 2019). The main quinolone resistance genes were gyrA, parC, and aac (6′)-Ib-cr. Mutations in gyrA, accompanied by mutations in parC (or parE), can result in quinolone resistance (Bagel et al., 1999). The aac (6′)-Ib-cr gene may be helpful for strains to capture other resistant plasmids and improve their competitiveness (Zhang et al.,2016). Therefore, aac (6′)-Ib-cr was widely present in multidrug-resistant strains. Among the tetracycline resistance genes, tetA and tetB were the most predominant ones. The detection rate was between 80 and 100%, as previously reported (Wu et al., 2017).
In conclusion, to improve the rational use of veterinary antibacterial drugs and achieve the antibacterial target as early as possible, it is important to implement strict disinfection measures to destroy residual bacteria in the environment. In addition, it is necessary to select breeding farms with a standardized use of antibacterial drugs. Drug-resistant bacteria affect the bacterial resistance of chicks. During daily feeding, it is essential to pay considerable attention to the disinfection of cages and ground in the enclosure, increase the disinfection measures of feed and water, and maintain adequate air circulation.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of China (31772707, 31972644, and 31802161] and the National Key Research and Development Program of China (2018YFD0500505). We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Contributor Information
Tianfei Han, Email: qkz@ahau.edu.cn.
Zhina Qu, Email: 641117207@qq.com.
References
- Adachi F., Yamamoto A., Takakura K., Kawahara R. Occurrence of fluoroquinolones and fluoroquinolone-resistance genes in the aquatic environment. Sci. Total Environ. 2013;444:508–514. doi: 10.1016/j.scitotenv.2012.11.077. [DOI] [PubMed] [Google Scholar]
- Alegría Á., Arias-Temprano M., Fernández-Natal I., Rodríguez-Calleja J.M., García-López M.L., Santos J.A. Molecular diversity of ESBL-producing Escherichia coli from foods of animal origin and human patients. Int. J. Environ. Res. Public Health. 2020;17:E1312. doi: 10.3390/ijerph17041312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R.I., Garrigues-Jeanjean N., Mackie R.I. Molecular ecology of tetracycline resistance: development and validationof primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 2001;67:22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagel S., Hüllen V., Wiedemann B., Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 1999;43:868–875. doi: 10.1128/aac.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B., Cooper M.A. Aminoglycoside antibiotics in the 21st century. ACS Chem. Biol. 2013;8:105–115. doi: 10.1021/cb3005116. [DOI] [PubMed] [Google Scholar]
- Bourély C., Cazeau G., Jarrige N., Jouy E., Haenni M., Lupo A., Madec J.Y., Leblond A., Gay E. Co-resistance to amoxicillin and tetracycline as an indicator of multidrug resistance in Escherichia coli isolates from animals. Front Microbiol. 2019;10:2288. doi: 10.3389/fmicb.2019.02288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briñas L., Zarazaga M., Sáenz Y., Ruiz-Larrea F., Torres C. Escherichia coli isolates from foods, humans,and healthy animals beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Antimicrob. Agents Chemother. 2002;46:3156–3163. doi: 10.1128/AAC.46.10.3156-3163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Clinical and Laboratory Standards Institute; Wayne, PA: 2017. Perfor-mance Standards for Antimicrobial Susceptibility Testing, M100S27. [Google Scholar]
- Cui Z.H., Ni W.N., Tang T., He B., Zhong Z.X., Fang L.X., Chen L., Chen C., Cui C.Y., Liu Y.H., Liao X.P., Sun J. Rapid detection of plasmid-mediated high-level tigecycline resistance in Escherichia coli and Acinetobacter spp. J. Antimicrob. Chemother. 2020;75:1479–1483. doi: 10.1093/jac/dkaa029. [DOI] [PubMed] [Google Scholar]
- Falgenhauer L., Schwengers O., Schmiedel J., Baars C., Lambrecht O., Heß S., Berendonk T.U., Falgenhauer J., Chakraborty T., Imirzalioglu C. Multidrug-resistant and clinically relevant gram-negative bacteria are present in German surface waters. Front Microbiol. 2019;10:2779. doi: 10.3389/fmicb.2019.02779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Migura L., Hendriksen R.S., Fraile L., Aarestrup F.M. Antimicrobial resistance of zoonotic and commensal bacteria in europe: the missing link between consumption and resistance in veterinary medicine. Vet. Microbiol. 2014;170:1–9. doi: 10.1016/j.vetmic.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Guillaume G., Verbrugge D., Chasseur-Libotte M., Moens W., Collard J. PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS. Microbiol. Ecol. 2000;32:77–85. doi: 10.1111/j.1574-6941.2000.tb00701.x. [DOI] [PubMed] [Google Scholar]
- Hanson N.D., Moland E.S., Hossain A., Neville S.A., Gosbell I.B., Thomson K.S. Unusual Salmonella enterica serotype Typhimurium isolate producing CMY-7, SHV-9 and OXA-30 beta-lactamases. J. Antimicrob. Chemother. 2002;49:1011–1014. doi: 10.1093/jac/dkf052. [DOI] [PubMed] [Google Scholar]
- Hayer S.S., Rovira A., Olsen K., Johnson T.J., Vannucci F., Rendahl A., Perez A., Alvarez J. Prevalence and trend analysis of antimicrobial resistance in clinical Escherichia coli isolates collected from diseased pigs in the USA between 2006 and 2016. Transbound Emerg. Dis. 2020;67:1930–1941. doi: 10.1111/tbed.13528. [DOI] [PubMed] [Google Scholar]
- Hricová K., Röderová M., Pudová V., Hanulík V., Halová D., Julínková P., Dolejská M., Papoušek I., Bardoň J. Quinolone-resistant Escherichia coli in poultry farming. Cent. Eur. J. Public Health. 2017;25:163–167. doi: 10.21101/cejph.a4328. [DOI] [PubMed] [Google Scholar]
- Hu G.Z., Pan Y.S., Wu H., Hu H., Xu R., Yuan L., Liu J.H., Feng J.K. Prevalence of tetracycline resistance genes and identification of tet(M) in clinical isolates of Escherichia coli from sick ducks in China. J. Med. Microbiol. 2013;62(Pt 6):851–858. doi: 10.1099/jmm.0.051896-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Zhou Z., Qian Y., Wei Z., Yu Y., Hu S., Li L. Plasmid-mediated quinolone resistance determinants qnr and aac(6')-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 2008;61:1003–1006. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
- Klimiene I. Evaluation of genotypical antimicrobial resistance in ESBL producing Escherichia coli phylogenetic groups isolated from retail poultry meat. J. Food Saf. 2018;38:e12370. [Google Scholar]
- Koyama S., Murase T., Ozaki H. Research note: longitudinal monitoring of chicken houses in a commercial layer farm for antimicrobial resistance in Escherichia coli with special reference to plasmid-mediated quinolone resistance. Poult. Sci. 2020;99:1150–1155. doi: 10.1016/j.psj.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.C., Yue F.L., Li J., Wei F., Jin S., Cao X.G. Detection of aminoglycoside antibiotic resistance of Escherichia coli from chicken. Guangdong Agric. Sci. 2012;39:90–91. (in Chinese) [Google Scholar]
- Mhondoro M., Ndlovu N., Bangure D., Juru T., Gombe N.T., Shambira G., Nsubuga P., Tshimanga M. Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012-2017: a secondary dataset analysis. BMC Infect Dis. 2019;19:746. doi: 10.1186/s12879-019-4295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro-Dasi L., Villagra A., Sevilla-Navarro S., Pérez-Gracia M.T., Vega S., Marin C. The dynamic of antibiotic resistance in commensal Escherichia coli throughout the growing period in broiler chickens: fast-growing vs. slow-growing breeds. Poult. Sci. 2020;99:1591–1597. doi: 10.1016/j.psj.2019.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Day M., Godbole G., Saluja T., Langridge G.C., Dallman T.J., Chattaway M. Genomic surveillance detects Salmonella enterica serovar Paratyphi A harbouring blaCTX-M-15 from a traveller returning from Bangladesh. PLoS One. 2020;15:e0228250. doi: 10.1371/journal.pone.0228250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R., Murase T., Ozaki H. Plasmid-mediated quinolone resistance in Escherichia coli isolates from commercial broiler chickens and selection of fluoroquinolone-resistant mutants. Poult. Sci. 2019;98:5900–5907. doi: 10.3382/ps/pez337. [DOI] [PubMed] [Google Scholar]
- Park M., Rafii F. The prevalence of plasmid-coded cpe enterotoxin, β2 toxin, tpeL toxin, and tetracycline resistance in Clostridium perfringens strains isolated from different sources. Anaerobe. 2019;56:124–129. doi: 10.1016/j.anaerobe.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Qiu W., Sun J., Fang M., Luo S., Tian Y., Dong P., Xu B., Zheng C. Occurrence of antibiotics in the main rivers of Shenzhen, China: Association with antibiotic resistance genes and microbial community. Sci. Total Environ. 2019;653:334–341. doi: 10.1016/j.scitotenv.2018.10.398. [DOI] [PubMed] [Google Scholar]
- Reeves A.Z., Campbell P.J., Sultana R., Malik S., Murray M., Plikaytis B.B., Shinnick T.M., Posey J.E. Aminoglycoside cross-resistance in mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob. Agents Chemother. 2013;57:1857–1865. doi: 10.1128/AAC.02191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifi S., Khoshbakht R. Prevalence of tetracycline resistance determinants in broiler isolated Escherichia coli in Iran. Poult. Sci. 2016;57:729–733. doi: 10.1080/00071668.2016.1232478. [DOI] [PubMed] [Google Scholar]
- Seo K.W., Lee Y.J. Detection of plasmid-mediated quinolone resistance genes in β-lactamase-producing Escherichia coli isolates from layer hens. Poult. Sci. 2019;98:1480–1487. doi: 10.3382/ps/pey545. [DOI] [PubMed] [Google Scholar]
- Seo K.W., Lee Y.J. Characterization of plasmid mediated quinolone resistance determinants in ciprofloxacin resistant-Escherichia coli from chicken meat produced by integrated broiler operations in Korea. Int. J. Food Microbiol. 2019;307:108274. doi: 10.1016/j.ijfoodmicro.2019.108274. [DOI] [PubMed] [Google Scholar]
- Shimada Y., Deguchi T., Nakane K., Masue T., Yasuda M., Yokoi S., Ito S., Nakano M., Ito S., Ishiko H. Emergence of clinical strains of Mycoplasma genitalium harbouring alterations in parC associated with fluoroquinolone resistance. Int. J. Antimicrob. Agents. 2010;36:255–258. doi: 10.1016/j.ijantimicag.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Silvester R., Pires J., Van Boeckel T.P., Madhavan A., Balakrishnan Meenakshikutti A., Hatha M. Occurrence of β-lactam resistance genes and plasmid-mediated resistance among vibrios isolated from southwest coast of India. Microb. Drug Resist. 2019;25:1306–1315. doi: 10.1089/mdr.2019.0031. [DOI] [PubMed] [Google Scholar]
- Sun J., Huang T., Chen C., Cao T.T., Cheng K., Liao X.P., Liu X.H. Comparison of fecal microbial composition and antibiotic resistance genes from swine, farm workers and the surrounding villagers. Sci. Rep. 2017;7:4965. doi: 10.1038/s41598-017-04672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.Y., Jiang Z.N., Shen X., Xu S.X. Report on the use of veterinary antibiotics in China in 2018. China Anim. Health. 2019;21:8–9. (in Chinese) [Google Scholar]
- Suzuki Y., Ida M., Kubota H., Ariyoshi T., Murakami K., Kobayashi M., Kato R., Hirai A., Suzuki J., Sadamasu K. Multiple β-Lactam resistance gene-carrying plasmid harbored by Klebsiella quasipneumoniae isolated from urban sewage in Japan. mSphere. 2019;4 doi: 10.1128/mSphere.00391-19. e00391-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.B., Wang H.N., Zhang A.Y., Zhang Y., Yang X., Xu C.W. Detection of β-lactam drug resistance of Escherichia coli in large-scale pig farms and its extended-spectrum b-lactamase investigation. Chin. J. Prev. Vet. Med. 2011;10:776–780. (in Chinese) [Google Scholar]
- Wang L.P., Chen S.F., Shi X.L., Jiang S.X., Yu G.Z. Therapeutic effect of 5% compound florfenicol oral liquid in chickens infected E. coli by artificial revulsive method. Prog. Vet. Med. 2003:110–112. (in Chinese) [Google Scholar]
- Wang Y.N., Liu F., Zhu B.L., George F.G. Discovery of tigecycline resistance genes tet (X3) and tet (X4) in live poultry market worker gut microbiomes and the surrounded environment. Sci. Bull. 2020;65:340–342. doi: 10.1016/j.scib.2019.12.027. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wu C., Zhang Q., Qi J., Liu H., Wang Y., He T., Ma L., Lai J., Shen Z., Liu Y., Shen J. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One. 2012;7:e37152. doi: 10.1371/journal.pone.0037152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Huang J., Cao J., Xu J., Mi J., Wang Y., Ma B., Zou Y., Liao X., Liang J.B., Wu Y. Heterologous expression of the tetracycline resistance gene tetX to enhance degradability and safety in doxycycline degradation. Ecotoxicol. Environ. Saf. 2020;191:110214. doi: 10.1016/j.ecoenv.2020.110214. [DOI] [PubMed] [Google Scholar]
- Wu H.B., Yang J., Zhou J.L., Wu P.F. Isolation, identification and drug resistance analysis of Escherichia coli from chicken. Heilongjiang Ani. Sci. Vet. Med. 2017;10:97–99. (in Chinese) [Google Scholar]
- Yang Y., Zhou R., Chen B., Zhang T., Hu L., Zou S. Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach. Chemosphere Dec. 2018;213:463–471. doi: 10.1016/j.chemosphere.2018.09.066. [DOI] [PubMed] [Google Scholar]
- Ye Q., Wu Q., Zhang S., Zhang J., Yang G., Wang J., Xue L., Chen M. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front Microbiol. 2018;9:1709. doi: 10.3389/fmicb.2018.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., He L.Y., Liu Y.S., Zhao J.L., Liu W.R., Zhang J.N., Chen J., He L.K., Zhang Q.Q., Ying G.G. Fate of veterinary antibiotics during animal manure composting. Sci. Total Environ. 2019;650(Pt 1):1363–1370. doi: 10.1016/j.scitotenv.2018.09.147. [DOI] [PubMed] [Google Scholar]
- Zhang P., Shen Z., Zhang C., Song L., Wang B., Shang J., Yue X., Qu Z., Li X., Wu L., Zheng Y., Aditya A., Wang Y., Xu S., Wu C. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008-2015. Vet. Microbiol. 2017;203:49–55. doi: 10.1016/j.vetmic.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Zhao S., Wang C.L., Chang S.K., Tsai Y.L., Chou C.H. Characterization of Escherichia coli isolated from day-old chicken fluff in taiwanese hatcheries. Avian Dis. 2019;63(1):9–16. doi: 10.1637/11935-072318-Reg.1. [DOI] [PubMed] [Google Scholar]
- Zhang W.H. South China Agricultural University; China: 2016. (2016) Characteristic of the Aac(6')-Ib-Cr Gene on Fluoroquinolone Resistance. Ph.D. Thesis. [Google Scholar]