Abstract
The purpose of this study was to identify the genetic environment of optrA gene in linezolid (LZD)-resistant Enterococcus faecalis from chicken meat and to describe the probable mechanism of dissemination of the optrA gene through plasmid or chromosomal integration. Whole genome sequencing and analysis revealed that all 3 E. faecalis isolates confirmed as LZD- and chloramphenicol-resistant carried fexA adjacent to the optrA gene as well as a variety of resistance genes for macrolides, tetracyclines, and aminoglycosides, simultaneously. But, the other genes conferring LZD resistance, cfr and poxtA, were not detected in those strains. Two isolates harboring the optrA gene in their chromosomal DNA showed >99% similarity in arrangement to the transposon Tn6674 and the transposase genes, tnpA, tnpB, and tnpC and were located in the first open reading frame for transposase. One isolate harboring an optrA-carrying plasmid also showed >99% similarity with the previously reported pE439 plasmid but had 2 amino acid changes (Thr96Lys and Tyr160Asp) and a higher minimum inhibitory concentration against LZD of 16 mg/L than that of pE439 (8 mg/L). Mobile genetic elements such as transposons or plasmids flanking the optrA gene conduct a crucial role in the dissemination of antimicrobial resistance genes. Further investigations are required to identify the way by which optrA is integrated into chromosomal DNA and plasmids.
Key words: optrA, fexA, Enterococcus faecalis, antimicrobial resistance, linezolid-resistance
Introduction
Enterococci are significant pathogens in that they may transfer their antimicrobial resistance genes to other animals or humans via the food chain (Ogier and Serror, 2008). In particular, animal-origin Enterococcus faecalis seems to be a zoonotic threat as it has been reported to express similar phenotypes in animals and humans (Hammerum, 2012; Hasan et al., 2018). The ability to acquire and transfer plasmids and transposons carrying antimicrobial resistance genes and virulence genes of E. faecalis has enabled them to act as multidrug-resistant pathogens, which are of significant concern in many countries (Freitas et al., 2017; Tyson et al., 2018b).
Linezolid (LZD) is a member of a class of oxazolidinones that is used for the treatment of infections in human caused by vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus, and multidrug-resistant gram-positive bacteria including enterococci (Bozdogan and Appelbaum, 2004; O'Driscoll and Crank, 2015). Although LZD is not used in food-producing animals, the appearance of LZD-resistant isolates in animals has been reported in China (Wang et al., 2015), South Korea (Tamang et al., 2017), the United States (Tyson et al., 2018a), Europe (De Jong et al., 2019), and Africa (Elghaieb et al., 2019). The most common mechanisms associated with oxazolidinone resistance among enterococci include mutations in the central loop of domain V of the 23S rRNA (Arias et al., 2010) and the plasmid-mediated antimicrobial genes such as cfr, which contributes resistance to oxazolidinones, phenicols, lincosamides, and streptogramin A (Long et al., 2006). Recently, a transferable oxazolidinone resistance gene, optrA, from E. faecalis of human and animal origins was reported worldwide such as in China (Wang et al., 2015), Italy (Brenciani et al., 2016), Spain (Ca'Mara et al., 2019), and Ireland and Malaysia (Mendes et al., 2014, 2016). The optrA gene encodes for an ATP-binding cassette F protein mediating resistance to both phenicols and oxazolidinones through target protection (Sharkey and O'Neill, 2018), which contributes antimicrobial resistance against LZD, tedizolid, and phenicols (Wang et al., 2015). Moreover, the location of the optrA gene on the plasmids of E. faecalis was revealed to be adjacent to the phenicol resistance gene, fexA, based on sequence analysis of optrA-positive E. faecalis (He et al., 2016; Kang et al., 2019). The optrA-fexA genes have also been reported to be present of chromosomal DNA (He et al., 2016). Although oxazolidinone resistance mediated by plasmids and transposons has been previously described (Bender et al., 2018; Chen et al., 2019), the genetic environment of chromosomal DNA or plasmids harboring optrA in Korea has not been reported.
In this study, we describe the genetic environments of the optrA-fexA genes in LZD-resistant E. faecalis isolated from chicken meat in South Korea and describe the probable mechanism involved in the dissemination of the optrA gene via plasmid and chromosomal integration.
Materials and methods
Bacterial Strains
Among a total of 345 E. faecalis isolates from 200 retail chicken meat samples previously described (Kim et al., 2018), after the disk diffusion test using broth microdilution method, 7 LZD-resistant E. faecalis isolates were analyzed to confirm the presence of the optrA gene by applying a previously published PCR protocol (Wang et al., 2015). Subsequently, 3 E. faecalis were shown to carry optrA and were analyzed for this study.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) were determined by using the broth microdilution method in the Sensititre custom panel KRVP2F (TREK Diagnostic Systems, West Sussex, England), in accordance with the manufacturer's instructions. The antimicrobial agents tested were ampicillin, chloramphenicol (CHL), ciprofloxacin (CIP), daptomycin, erythromycin (ERY), florfenicol (FFN), gentamicin, kanamycin, LZD, quinupristin/dalfopristin (SYN), streptomycin, tetracycline, tylosin tartrate (TYLT), tigecycline, vancomycin, and salinomycin. E. faecalis ATCC 29212 was used as the quality control strain, and the MIC values were interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2019). When breakpoints were unavailable in the CLSI guidelines, the Danish Integrated Antimicrobial Resistance Monitoring and Research Programme or the National Antimicrobial Resistance Monitoring System was applied (CDC, 2013; DANMAP, 2017).
Conjugation Experiment
The transferability of plasmids carrying the optrA gene was assessed by applying the broth-mating protocol using rifampicin-resistant and fusidic acid–resistant E. faecalis FA2-2 as the recipient strain as described previously (Werner et al., 2008; Tamang et al., 2017). Transconjugants were recovered after incubation at 37°C on BHI agar (Becton Dickinson) plates supplemented with 2 μg/mL LZD, 25 μg/mL rifampicin, and 25 μg/mL fusidic acid followed by antimicrobial susceptibility testing and PCR analysis of the optrA gene.
Whole Genome Sequencing and Analysis
Genomic DNA of optrA-positive E. faecalis was extracted from overnight cultures using the MasterPure Gram Positive DNA Purification Kit (Lucigen, WI) in accordance with the manufacturer's instructions. Genome sequencing, accomplished by using an Illumina HiSeq platform in accordance with standard Illumina protocols, was performed at Macrogen (Seoul, South Korea). The reads were de novo assembled using hierarchical genome assembly process 3. Genome annotation was performed using Rapid Annotation by Subsystem Technology version 2.0. The assembled genomes were initially screened for genes encoding antibiotic resistance, virulence, and multilocus sequence typing (MLST) using the in silico genomic tools ResFinder 3.1 and LRE-finder 1.0, VirulenceFinder 1.5, and MLST 2.0, respectively, that are available through the Center for Genomic Epidemiology (http://www.genomicepidemiology.org). Plasmid content associated with optrA was analyzed using the contigs obtained by plasmidSPAdes that were annotated with Prokka v1.12 (http://vicbioinformatics.com). BLAST analysis with default parameters was performed to compare the contigs with known sequences contained in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Artemis tools and DNAPlotter of the Sanger Institute (http://www.sanger.ac.uk/Software/Artemis) were used for drawing the plasmid schemes and comparisons. All representative sequences of the known optrA genes described were obtained from GenBank in FASTA format.
Results
Distribution of Antimicrobial Susceptibility and Genes Conferring Antimicrobial Resistance
Three strains (EFs 116-2, EFs 171-2, and EFs 17-1) were confirmed as LZD- and chloramphenicol-resistant with MICs of 8 to 16 mg/L and >32 mg/L, respectively, and each carried resistance genes for phenicol/oxazolidinone (optrA and fexA) (Table 1). The cat gene, related to phenicol resistance, was detected in only one strain, EFs 171-2. The 3 strains also showed resistance to ciprofloxacin, erythromycin, florfenicol, quinupristin/dalfopristin, tetracycline, and tylosin tartrate, and carried resistance genes for macrolides [erm(B) and Isa(A)] and tetracyclines [tet(M)]. Moreover, 2 strains, EFs 116-2 and EFs 171-2, also carried the erm(A) gene for macrolides and the tet(L) gene for tetracyclines. In particular, one strain, EFs 171-2, showed resistance to gentamicin, kanamycin, and streptomycin with high-level MICs over 2,048 mg/L and carried resistance genes for aminoglycosides such as aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, and aph(3′)-III. Although all 3 strains showed resistance against ciprofloxacin, quinupristin/dalfopristin, and tylosin tartrate, no specific genes related to resistance were detected. Antimicrobial resistance genes, cfr and poxtA, were investigated but not detected in any strain (Table1).
Table 1.
Antimicrobial resistance patterns and genetic features of 3 optrA-positive Enterococcus faecalis isolates from chicken meat.
| Strain | Resistance genes1 | Virulence genes | MLST | Location of optrA | MIC (mg/L)2 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LZD | CHL | AMP | DAP | VAN | ERY | GEN | KAN | STR | SAL | SYN | CIP | TYLT | FFN | TET | TGC | |||||
| EFs 116-2 | optrA, fexA,ant(9)-Ia, aph(3′)-III, erm(A),erm(B), Isa(A),tet(L), tet(M) | ace, agg,cad, camE, cCF10, cOB1, ebpA, ebpB, ebpC, efaAfs,ElrA,hylA,SrtA,tpx | ST476 | Chromosomal DNA | 8 | >32 | <1 | 4 | <2 | >64 | <128 | 1,024 | <128 | 4 | 16 | >16 | >64 | >32 | 128 | <0.12 |
| EFs 171-2 | optrA, fexA,aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, erm(A),erm(B), Isa(A),tet(L), tet(M) | ace, cad, camE, cCF10, cOB1, ebpA, ebpB, ebpC, efaAfs, hylA, tpx | ST476 | Chromosomal DNA | 8 | >32 | <1 | 1 | <2 | >64 | >2,048 | >2,048 | >2,048 | <2 | 16 | >16 | >64 | >32 | >128 | <0.12 |
| EFs 17-1 | optrA, fexA, erm(B), Isa(A), tet(M) | ace, agg,cad,camE,cCF10, cOB1, ebpA, ebpB, ebpC, efaAfs, ElrA, fsrB, gelE, hylA, hylB, SrtA,tpx | ST729 | Plasmid | 16 | >32 | <1 | 1 | <2 | >64 | <128 | <128 | <128 | 4 | 16 | >16 | >64 | >32 | 64 | <0.12 |
| transconjugant of EFs 17-1 | optrA, fexA, erm(B) | -3 | - | - | 16 | >32 | <1 | 1 | <2 | >64 | <128 | <128 | <128 | 4 | 4 | 2 | 4 | >32 | 64 | <0.12 |
Genes detected from all 3 isolates are highlighted in bold.
Resistance genes, cfr and poxtA, were tested but not detected.
LZD, linezolid; CHL, chloramphenicol; AMP, ampicillin; DAP, daptomycin; VAN, vancomycin; ERY, erythromycin; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; SAL, salinomycin; SYN, quinupristin/dalfopristin; CIP, ciprofloxacin; TYLT, tylosin tartrate; FFN, florfenicol; TET, tetracycline; TGC, tigecycline. MIC values indicating resistance to antimicrobials are highlighted in bold.
Not tested.
Molecular Characteristics of optrA-positive E. faecalis and Conjugation Experiments
All 3 strains carried virulence genes related to adhesion (ace), biofilm production (ebpA, ebpB, and ebpC), cell wall adhesion expressed in serum (efaAfs), protection against oxidative stress (tpx), hyaluronidase gene (hylA), and sex pheromone-associated genes involved in the transfer of pheromone-responsive plasmids (cad, camE, cCF10, and cOB1). In addition, 2 strains, EFs 17-1 and EFs 116-2, also harbored supplementary virulence genes for adhesion (agg and ElrA) and protease (SrtA). Other virulence factors such as hyaluronidase (hylB), protease (gelE), and quorum sensing–related (fsrB) genes were observed only in the EFs 17-1 strain. One strain, EFs 17-1, among the 3 optrA-positive E. faecalis isolates, successfully cotransferred the optrA and fexA genes. The transconjugant showed resistance against LZD, CHL, ERY, and FFN, but it was susceptible to CIP, TYLT, and SYN unlike the characteristics of the donor strain.
Whole Genome Sequencing Analysis
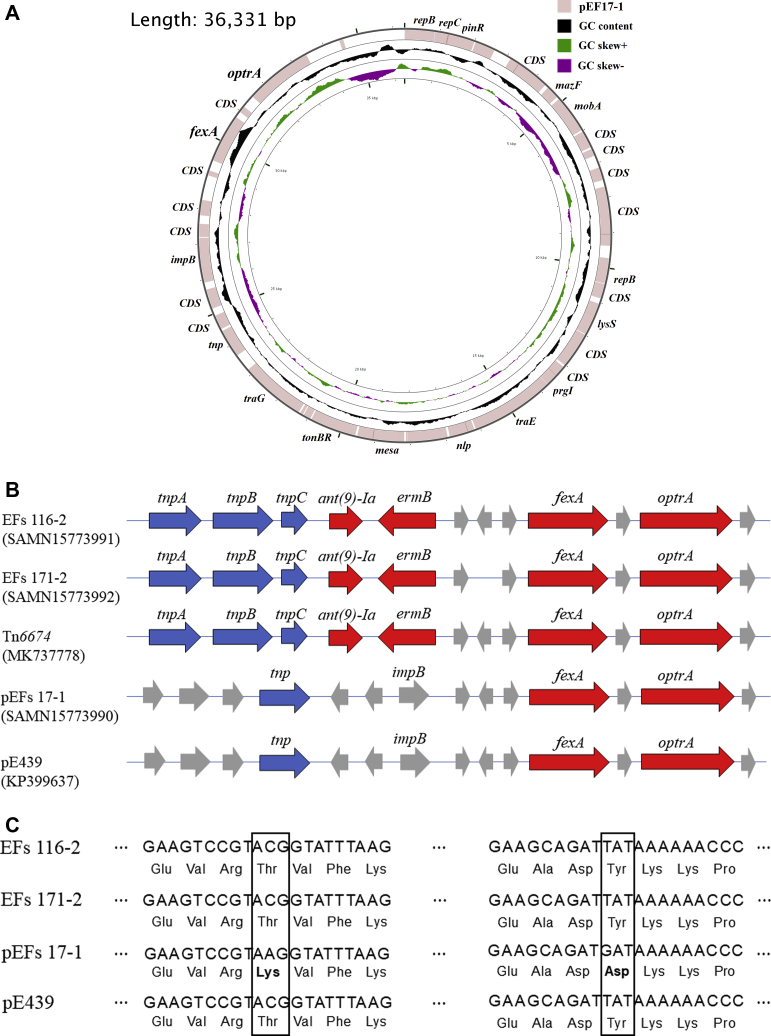
Based on in silico analysis of the assembled genomes, the EFs 116-2 (GenBank accession number SAMN15773991) and EFs 171-2 (GenBank accession number SAMN15773992) strains were identified as ST476, and the optrA genes of these strains were located at their chromosomal DNAs. On the other hand, optrA gene of the EFs 17-1 strain (GenBank accession number SAMN15773990), belonging to the ST729 lineage, was observed in plasmid DNA. The optrA-carrying complete plasmid sequence detected in the EFs 17-1 strain showed >99% identity (query cover, 100%) with the sequence of pE349 (GenBank accession number KP399637) and was deposited under the name of pEFs17-1 (GenBank accession number MT223178). Among the 39 open reading frames (ORFs) identified in the plasmid (36,331 bp), 21 encoded hypothetical proteins with no previously reported functions, whereas 18 products matched with proteins that had identified functions in the database such as antimicrobial resistance (oprA and fexA), replication (repB and repC), plasmid modulator (mazF, lysS), and conjugal transfer (mobA, prgI, traD, traE, traG, and tnp) (Figure 1A).
Figure 1.
(A) Circular view of the mapped BLAST result of the EFs 17-1 plasmid compared with the originally reported optrA-carrying plasmid pE439 sequence. (B) Linear representation of the optrA genetic environment in the 3 isolates in this study and the optrA-carrying transposon Tn6674 and the plasmid pE439 (GenBank accession number MK737778 and KP399637, respectively). The GenBank accession numbers are written below the strain name or the plasmid names. The transposase genes, antimicrobial resistance genes, and genes that code for other functions are shown in arrow boxes filled in blue, red, and gray, respectively. (C) Comparison of sequences of the optrA gene among the 3 Enterococcus faecalis strains and pE439.
Analysis of regions flanking the optrA gene in the chromosomes of 2 strains, EFs 116-2 and EF171-2, revealed that they displayed >99% nucleotide sequence identity (query cover, 100%) with the sequences of Tn6674 (GenBank accession number MK737778). The transposon genes, tnpA, tnpB, and tnpC, were located in the first ORFs for transposase, similar to that in Tn6674. The antimicrobial resistance genes ant(9)-Ia, ermB, and fexA were also located adjacent to the optrA gene (Figure 1B).
When the OptrA amino acid sequence was compared with that of E. faecalis pE439 (MIC of LZD = 8 mg/L), the 3 strains represented >99% amino acid identity, but pEFs17-1 showed 2 amino acid changes (Thr96Lys and Tyr160Asp) and a higher MIC of LZD (16 mg/L) (Figure 1D).
Discussion
To the best of our knowledge, this study is the first to identify the whole genome sequences of optrA-positive E. faecalis isolated from retail chicken meat in South Korea. Although only 3 optrA-positive E. faecalis strains were tested in this study, the results showed that the optrA gene could be carried by both chromosomal and plasmid DNAs. When DNA sequences of the optrA gene in chromosomal DNA were compared with the available optrA sequences, 2 isolates harboring the optrA gene in their chromosomal DNA showed >99% similarity to previously reported optrA sequences. In particular, the genes adjacent to the optrA gene exhibited an analogous arrangement to the transposon Tn6674 (Li et al., 2019). In addition, the transposase genes, tnpA, tnpB, and tnpC, were conserved in both isolates. The active Tn6674 can excise from the host DNA and produce circular forms that proceed to the integration of the transposon into a new target sequence (Freitas et al., 2020). This mechanism suggests that optrA can be disseminated to a variety of bacteria via transposon mediation (Kehrenberg and Schwarz, 2005; Chen et al., 2019).
In this study, one isolate harbored an optrA-carrying plasmid and also showed >99% similarity to the previously reported pE439 plasmid (Wang et al., 2015). But, when DNA sequences of the optrA gene were compared with that of pE439, the optrA-carrying plasmid tested in this study showed 2 amino acid changes (Thr96Lys and Tyr160Asp) and a higher MIC against LZD (16 mg/L) compared to that of pE439 (8 mg/L). Although Bender et al. (2018) reported amino acid substitution in the optrA gene was associated with both LZD-resistant and -susceptible E. faecalis isolates, Elghaieb et al. (2020) reported that strains could have the same MIC for LZD resistance irrespective of their variable optrA gene mutation profiles (Bender et al., 2018; Elghaieb et al., 2020). Therefore, the contribution of these specific amino acid substitutions to LZD resistance remains unclear.
In this study, all 3 strains harboring the optrA gene in their chromosomal or plasmid DNAs also carried the fexA gene adjacent to optrA. Ca'Mara et al. reported that the presence of both optrA and fexA in the same isolates occurred at a high frequency (Ca'Mara et al., 2019); moreover, cotransfer of the fexA gene with optrA in E. faecalis has been already shown in many countries such as China (Wang et al., 2015), Spain (Ca'Mara et al., 2019), and the United States (Wardenburg et al., 2019). In this study, the strains with the embedded fexA-optrA complex, which is highly similar to those of Tn6674 and pE439, also induced simultaneous resistance to phenicols and oxazolidinones as has been previously described (Wang et al., 2015; Cai et al., 2018; Li et al., 2019). Moreover, mobile genetic elements such as transposons or plasmids flanking the optrA gene can be mediated by the horizontal transfer of a variety of resistance genes in the absence of antimicrobial drugs (Morroni et al., 2018; Wardenburg et al., 2019). Mobile genetic elements have frequently carried not only optrA but also other resistance determinants such as fexA, ermA, and Isa(A) genes. Freitas et al. (2017) and Tamang et al. (2017) have reported that oxazolidinones-resistant isolates linked to animals in which phenicols, marcolides, and streptogramins may be used might coselect resistance to other antibiotic families (Freitas et al., 2017; Tamang et al., 2017). In this study, 3 strains showed chromosomal- or plasmid-mediated oxazolidinone resistance as indicated by the coresistances to CHL, ERY, SYN, CIP, TYLT, and FFN. Although LZD has not been used in the poultry industry in South Korea, the emergence of an LZD-resistant E. faecalis in chickens could be the result of horizontal transfer of resistance genes and coselection of phenicol-resistant strains as has been previously described (Wang et al., 2015; Ca'Mara et al., 2019).
The analysis of 3 optrA-positive E. faecalis in this study showed the presence of a number of virulence genes in common, which is consistent with the results obtained from other E. faecalis strains of food origin (Vidana et al., 2016; Cavaco et al., 2017). Virulence factors contribute to adaptation to different environments and can enable the sharing of various genes such as virulence determinant and antimicrobial resistance genes (Coburn et al., 2007; Vidana et al., 2016). Especially, the expressions of different families of efflux pumps could contribute to the antimicrobial resistance and their pathogenesis (Martinez et al., 2009). As the association of these virulence factors with optrA is unclear, further monitoring of food animals is required to ascertain the role of virulence factors affecting the resistances determined by optrA.
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agriculture, Food and Rural Affairs Convergence Technologies Program for Educating Creative Global Leader, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA; 716002-7).
Conflict of Interest Statement: There are no conflicts of interest for this paper.
References
- Arias C.A., Contreras G.A., Murray B.E. Management of Multi-Drug resistant enterococcal infections. Clin. Microbiol. Infect. 2010;16:1–13. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J.K., Fleige C., Lange D., Klare I., Werner G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents. 2018;52:819–827. doi: 10.1016/j.ijantimicag.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Bozdogan B., Appelbaum P.C. Oxazolidinones : activity, mode of action , and mechanism of resistance. Antimicrob. Agents. 2004;23:113–119. doi: 10.1016/j.ijantimicag.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Brenciani A., Morroni G., Vincenzi C., Manso E., Mingoia M., Giovanetti E., Varaldo P.E. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 2016;71:1118–1119. doi: 10.1093/jac/dkv438. [DOI] [PubMed] [Google Scholar]
- Ca'Mara J., Camoez M., Pujol M., Ayats J., Ardanuy C., ngeles Domıńguez M.A. Detection of the novel optrA gene among linezolid-resistant enterococci in Barcelona, Spain. Microb. Drug Resist. 2019;25:87–93. doi: 10.1089/mdr.2018.0028. [DOI] [PubMed] [Google Scholar]
- Cai J., Schwarz S., Chi D., Wang Z., Zhang R., Wang Y. Faecal carriage of optrA -positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. 2018;25:2–7. doi: 10.1016/j.cmi.2018.07.025. [DOI] [PubMed] [Google Scholar]
- Cavaco L.M., Bernal J.F., Zankari E., Le M., Hendriksen R.S., Aarestrup F.M. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia) J. Antimicrob. Chemother. 2017;72:678–683. doi: 10.1093/jac/dkw490. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). CDC; Atlanta, GA: 2013. The National Antimicrobial Resistance Monitoring System (NARMS) integrated report. [Google Scholar]
- Chen H., Wang X., Yin Y., Li S., Zhang Y., Wang Q., Wang H. Molecular characteristics of oxazolidinone resistance in enterococci from a multicenter study in China. BMC Microbiol. 2019;19:1–9. doi: 10.1186/s12866-019-1537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). 2019. M100 Performance Standards for Antimicrobial Susceptibility Testing, Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- Coburn P.S., Baghdayan A.S., Dolan G.T., Shankar N. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol. Microbiol. 2007;63:530–544. doi: 10.1111/j.1365-2958.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP). 2017. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark, National Food Institute, Kongens Lyngby, Denmark. [Google Scholar]
- Elghaieb H., Freitas A.R., Abbassi M.S., Novais C., Zouari M., Hassen A., Peixe L. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J. Antimicrob. Chemother. 2019;74:2865–2869. doi: 10.1093/jac/dkz263. [DOI] [PubMed] [Google Scholar]
- Elghaieb H., Tedim A.P., Abbassi M.S., Novais C., Duarte B., Hassen A., Peixe L., Freitas A.R. From farm to fork: identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals. J. Antimicrob. Chemother. 2020;75:30–35. doi: 10.1093/jac/dkz419. [DOI] [PubMed] [Google Scholar]
- Freitas A.R., Elghaieb H., Leo R., Abbassi M.S., Novais C., Coque T.M., Hassen A., Peixe L. Detection of optrA in the African continent ( Tunisia ) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J. Antimicrob. Chemother. 2017;72:3245–3251. doi: 10.1093/jac/dkx321. [DOI] [PubMed] [Google Scholar]
- Freitas A.R., Tedim A.P., Novais C., Lanza V.F., Peixe L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genomics. 2020;6:1–17. doi: 10.1099/mgen.0.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012;18:619–625. doi: 10.1111/j.1469-0691.2012.03829.x. [DOI] [PubMed] [Google Scholar]
- Hasan K.A., Ali S.A., Rehman M., Bin-Asif H., Zahid S. The unravelled Enterococcus faecalis zoonotic superbugs : Emerging multiple resistant and virulent lineages isolated from poultry environment. Zoonoses Public Heal. 2018;65:921–935. doi: 10.1111/zph.12512. [DOI] [PubMed] [Google Scholar]
- He T., Shen Y., Schwarz S., Wu C., Shen J., Wang Y. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 2016;71:1466–1473. doi: 10.1093/jac/dkw016. [DOI] [PubMed] [Google Scholar]
- De Jong A., Simjee S., Rose M., Moyaert H., El Garch F., Youala M., Butty P., Haag-Diergarten S., Klein U., Pellet T., Schiffer G., Serreyn P.J., Vila T. Antimicrobial resistance monitoring in commensal enterococci from healthy cattle, pigs and chickens across Europe during 2004-14 (EASSA Study) J. Antimicrob. Chemother. 2019;74:921–930. doi: 10.1093/jac/dky537. [DOI] [PubMed] [Google Scholar]
- Kang Z., Lei C., Kong L., Wang Y., Ye X., Ma B., Wang X., Li C., Zhang Y., Wang H. Detection of transferable oxazolidinone resistance determinants in Enterococcus faecalis and Enterococcus faecium of swine origin in Sichuan Province, China. J. Glob. Antimicrob. Resist. 2019;19:333–337. doi: 10.1016/j.jgar.2019.05.021. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S. Florfenicol-chloramphenicol Exporter gene fexA is Part of the novel transposon Tn 558. Antimicrob. Agents Chemother. 2005;49:813–815. doi: 10.1128/AAC.49.2.813-815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. Bin, Seo H.J., Seo K.W., Jeon H.Y., Kim D.K., Kim S.W., Lim S., Lee Y.J. Characteristics of high-level ciprofloxacin-resistant Enterococcus faecalis and Enterococcus faecium from retail chicken meat in Korea. J. Food Prot. 2018;81:1357–1363. doi: 10.4315/0362-028X.JFP-18-046. [DOI] [PubMed] [Google Scholar]
- Li D., Li X., Schwarz S., Yang M., Zhang S., Hao W., Du X. Tn6674 is a novel enterococcal optrA-carrying multiresistance transposon of the Tn554 family Dexi. Antimicrob. Agents Chemother. 2019;63:1–5. doi: 10.1128/AAC.00809-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K.S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. The cfr rRNA Methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, Pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J.L., Sánchez M.B., Martínez-Solano L., Hernandez A., Garmendia L., Fajardo A., Alvarez-ortega C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Mendes R.E., Hogan P.A., Jones R.N., Sader H.S., Flamm R.K. Surveillance for linezolid resistance via the Zyvoxw Annual Appraisal of Potency and Spectrum (ZAAPS) programme (2014): Evolving resistance mechanisms with stable susceptibility rates. J. Antimicrob. Chemother. 2016;71:1860–1865. doi: 10.1093/jac/dkw052. [DOI] [PubMed] [Google Scholar]
- Mendes R.E., Hogan P.A., Streit J.M., Jones R.N., Flamm R.K. Zyvox® Annual Appraisal of Potency and Spectrum (ZAAPS) Program: report of linezolid activity over 9 years (2004-12) J. Antimicrob. Chemother. 2014;69:1582–1588. doi: 10.1093/jac/dkt541. [DOI] [PubMed] [Google Scholar]
- Morroni G., Brenciani A., Antonelli A., Andrea M.M.D., Di Pilato V., Fioriti S., Mingoia M., Vignaroli C., Cirioni O., Biavasco F., Varaldo P.E., Rossolini G.M., Giovanetti E. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front. Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll T., Crank C.W. Vancomycin-resistant enterococcal infections : epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015;8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier J.C., Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 2008;126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Sharkey L.K.R., O’Neill A.J. Antibiotic resistance ABC-F proteins: Bringing target protection into the Limelight. ACS Infect. Dis. 2018;4:239–246. doi: 10.1021/acsinfecdis.7b00251. [DOI] [PubMed] [Google Scholar]
- Tamang M.D., Moon D.C., Kim S.R., Kang H.Y., Lee K., Nam H.M., Jang G.C., Lee H.S., Jung S.C., Lim S.K. Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet. Microbiol. 2017;201:252–256. doi: 10.1016/j.vetmic.2017.01.035. [DOI] [PubMed] [Google Scholar]
- Tyson G.H., Nyirabahizi E., Crarey E., Kabera C., Lam C., Rice-Trujillo C., McDermott P.F., Tate H. Prevalence and antimicrobial resistance of enterococci isolated from retail meats in the United States, 2002 to 2014. Appl. Environ. Microbiol. 2018;84:1–9. doi: 10.1128/AEM.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.H., Sabo J.L., Hoffmann M., Hsu C.H., Mukherjee S., Hernandez J., Tillman G., Wasilenko J.L., Haro J., Simmons M., Wilson Egbe W., White P.L., Dessai U., McDermott P.F. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J. Antimicrob. Chemother. 2018;73:3254–3258. doi: 10.1093/jac/dky369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidana R., Rashid M.U., Ozenci V., Weintraub A., Lund B. The origin of endodontic Enterococcus faecalis explored by comparison of virulence factor patterns and antibiotic resistance to that of isolates from stool samples, blood cultures and food. Int. Endod. J. 2016;49:343–351. doi: 10.1111/iej.12464. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lv Y., Cai J., Schwarz S., Cui L., Hu Z., Zhang R., Li J., Zhao Q., He T., Wang D., Wang Z., Shen Y., Li Y., Feßler A.T., Wu C., Yu H., Deng X., Xia X., Shen J. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015;70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- Wardenburg K.E., Potter R.F., Souza A.W.D., Hussain T., Wallace M.A. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J. Antimicrob. Chemother. 2019;74:3445–3452. doi: 10.1093/jac/dkz367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G., Coque T.M., Hammerum A.M., Hope R., Hryniewicz W., Johnson A., Klare I., Kristinsson K.G. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance. 2008;13:1–11. [PubMed] [Google Scholar]