Abstract
Rationale and Objectives:
To develop classification and regression models interpreting tumor characteristics obtained from structural (T1w and T2w) MRI data for early detection of dendritic cell (DC) vaccine treatment effects and prediction of long-term outcomes for LSL-KrasG12D;LSLTrp53R172H;Pdx-1-Cre(KPC) transgenic mice model of pancreatic ductal adenocarcinoma(PDAC).
Materials and methods:
Eight mice were treated with DC vaccine for three weeks while eight KPC mice were used as untreated control subjects. The reproducibility of the computed 264 features was evaluated using the intra-class correlation coefficient. Key variables were determined using a three-step feature selection approach. Support vector machines classifiers were generated to differentiate treatment-related changes on tumor tissue following 1st and 3rd weeks of the DC vaccine therapy. The multivariable regression models were generated to predict overall survival (OS) and histological tumor markers of KPC mice using quantitative features.
Results:
The quantitative features computed from T1w MRI data has better reproducibility than T2w MRI features. The KPC mice in treatment and control groups were differentiated with a longitudinally increasing accuracy (1st and 3rd weeks: 87.5% and 93.75%). The linear regression model generated with five features of T1w MRI data predicted OS with an RMSE <6 days. The proposed multivariate regression models predicted histological tumor markers with relative error <2.5% for fibrosis percentage (RMSE: 0.414), CK19+ area (RMSE: 0.027), and Ki67+ cells (RMSE:0.190).
Conclusion:
Our results demonstrated that proposed models generated with quantitative MRI features can be used to detect early treatment-related changes in tumor tissue and predict OS of KPC mice following DC vaccination.
Keywords: Dendritic cell vaccine treatment, machine learning, magnetic resonance imaging, structural MRI radiomics analysis, pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive and lethal types of cancer in the United States (1). Due to frequent diagnosis at advanced stages, only 20% of the patients are surgical candidates, and 5-year overall survival (OS) remains ≤9% with a median OS ≤6 months for advanced-stage patients (2). Therefore, developing novel treatment methods and identifying biomarkers for early detection and dynamic monitoring are among the key topics of cancer research.
Many experimental cancer immunotherapies now involve vaccination of cancer patients with autologous dendritic cells (DCs) loaded with tumor antigens (DC vaccines) (3–5). Therapeutic DC vaccination has a clinically relevant mechanism of action with great potential for systemic treatment of many cancers including PDAC patients (6, 7). Clinical trials have not yet demonstrated positive therapeutic efficacy and clinical response has been limited to a minority of patients (3–5). However, recent preclinical studies demonstrate potential approaches to improve the efficiency of immunotherapy (8–10). With the increasing number of studies focusing on immunotherapy, it has become clear that current clinical response evaluation criteria, including response evaluation criteria in solid tumors (RECIST v1.1), immune-RECIST (iRECIST), immune-modified RECIST (imRECIST), and immune-related response criteria (irRC), do not adequately reflect immune response and OS benefit following DC vaccination therapy (11–13). Therefore, noninvasive cancer immunotherapy response evaluation approaches are needed for monitoring tumor microenvironment variation and assessing the DC vaccine response.
Radiomics, high throughput quantitative feature extraction procedure, describes underlying tissue structures and characterizes malignancies into categories. Several studies have already investigated the potential of MRI radiomics for diagnosis, tumor classification, prediction of gene mutation or OS, detection of disease stage, and response assessment after neoadjuvant chemotherapy (14–18). Despite promising results of the studies integrating T1w, T2w, diffusion-weighted and contrast-enhanced MRI imaging approaches, non-standard acquisition or analysis methods restrict validation and qualification of biomarkers based on diffusion-weighted and contrast-enhanced MRI (19, 20). Therefore, clinically standard MRI (T1w and T2w) data might be preferred for the reproducibility of the developed models.
The purpose of our study was to investigate the potential of multi-parametric radiomic analysis for detection of early immunotherapeutic response and prediction of OS of PDAC subjects following DC vaccine therapy by developing classification and regression models with reproducible radiomics features computed from structural (T1w and T2w) MRI data.
Materials and Methods
The procedure followed in our study was performed according to the regulations determined by the Institutional Animal Care and Use Committee of our institution.
Animal Model and DC Vaccine Treatment
Due to similar pathophysiological aspects and biological characteristics to human PDAC, LSLKrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre (KPC) transgenic mouse model of PDAC was utilized in this study. When pancreatic tumors reach a detectable size with MRI (tumor size ≥ 2 mm), eight KPC mice treated with 3×106 DC vaccines via intraperitoneal injection 1, 8, and 15 days after tumor detection while eight KPC mice were used as untreated control subjects.
MRI Acquisition and Processing
Tumor progression was monitored via biweekly MRI scans performed with a 7T Bruker preclinical scanner. After tumors were detected via MRI (tumor size ≥ 2 mm), KPC mice were continued to scan weekly using MRI sequences listed in Table 1 with respiratory triggering.
Table 1.
MRI acquisition parameters on 7T Bruker MRI scanner
| MRI Sequence | TR (ms) | TE (ms) | FA (°) | Thickness/Gap (mm) | Averages | Resolution (μm) |
|---|---|---|---|---|---|---|
| T1w (axial) | 630 | 20 | 90 | 0.7/0.7 | 2 | 156.25 |
| T2w (coronal) | 1600 | 37 | 180 | 1.0/2.0 | 1 | 96.35 |
| T2w (axial) | 2066 | 40 | 180 | 0.8/0.8 | 2 | 117.19 |
T1w and T2w MRI data were co-registered by performing 2D affine transformation with 3DSlicer (v4.10.2). On the basis of T1w and T2w MRI data, pancreatic tumors were delineated on the slice with maximal tumor diameter using ITK-Snap (v3.8.0) by consensus of two experienced radiologists in abdominal MRI. Two weeks after initial segmentation, the first radiologist outlined the tumors again to assess the reproducibility of the radiomic features. A sample MRI slice and tumor masks for selected samples were presented in Figure 1A and agreement of the tumor mask images were visualized with a Bland-Altman plot in Figure 1B.
Figure 1.
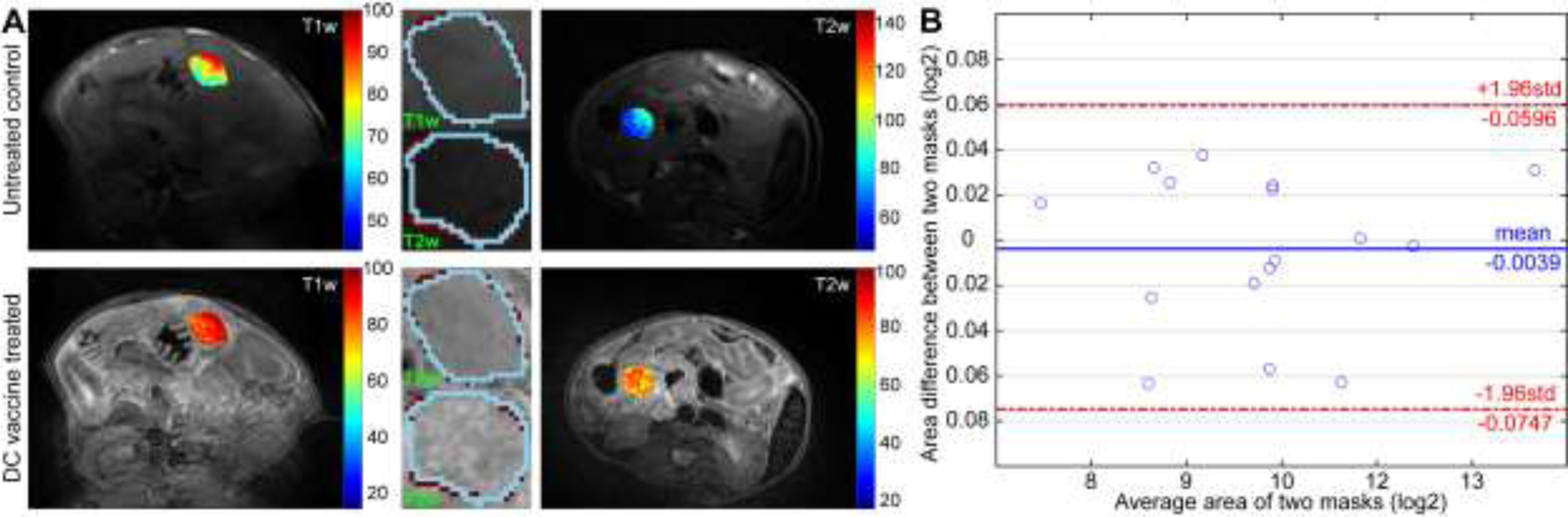
Reproducibility of the mask images. Representation of the T1w and T2w MRI images of samples from untreated control (top row) and DC vaccine (bottom row) group were presented in (A). Bland-Altman plot to show the inter-observer reproducibility of the masks that generated 2-week apart.
Histology
Pancreatic tumor tissues were dissected via surgery, fixed in 10% formalin, and embedded in paraffin. A representative 5μm thick tumor tissue for each sample slide was quantitatively analyzed by an experienced pathologist using ImageJ (v1.52q) to measure fibrosis percentage, CK19+ area, and Ki67+ cells.
Feature Extraction
Since MRI voxel intensity is relative and not absolute, acquired data are not directly comparable across different samples. Therefore, MRI images are standardized with z-score normalization and quantized using fixed bin-size approach suggested by image biomarker standardization initiative (21). To determine optimal bin-size, experiments were repeated using four different bin-sizes (8, 16, 32 and 64).
Two hundred sixty-four features were extracted from each MRI modality (T1w and T2w) using four types of image characteristics (intensity, texture, binary-pattern and shape) and two filters (wavelet and gradients). Briefly, 6 intensity (first-order statistics), 40 texture (co-occurrence, run-length, size-zone, and neighborhood gray-tone difference matrix, 10 binary-pattern, fractal dimension, and 9 shape features were computed from raw MRI images (22–24). Moreover, 6 intensity features were calculated from oriented gradient histogram and 192 features (variance, power, intensity, and texture) were extracted from four wavelet coefficient (approximate, vertical, horizontal, and diagonal) images (17). All radiomic features were extracted from tumor region (ROIs) determined with a consensus of two radiologists and transformed into the range of [0, 1] by performing min-max normalization.
Statistical Analysis
The reliability of the generated mask images was assessed by computing the Dice similarity metric that measures the overlap of two binary mask images. Besides, the reproducibility of the quantitative features extracted from T1w or T2w MRI data was evaluated with an intraclass correlation coefficient (ICC) using features computed over two tumor masks. The variables with an ICC value larger than 0.75 were considered reproducible and other variables were removed from the feature set. Besides, variables correlated (r>0.75) with tumor size were also removed from the feature set. The remaining variables were clustered based on Pearson correlation coefficients of the features. An individual radiomic feature with excellent reproducibility from each cluster was selected as a representative variable. Afterward, the importance of the selected variables was computed using RELIEFF algorithm which calculates the importance of each variable by analyzing interactions with its nearest neighbors (25). The top-ranked twenty features were selected from feature sets of T1w and T2w MRI data. For the combination model (T1w+T2w), the representative features of T1w and T2w MRI were combined, and top-ranked 20 features were determined with RELIEFF algorithm.
The support vector machines (SVM) with a leave-one-out cross-validation approach was employed to construct classifiers to identify treatment-related changes on tumor tissue following the first and third dosages of DC vaccine therapy. A multivariable regression model was generated to predict the OS of KPC mice using MRI features acquired one week after tumor detection. Moreover, histological tumor markers were predicted with regression models generated with features of the last MRI acquisition of the KPC mice. The performance of the classifiers was evaluated with accuracy while regression models were assessed with root-mean-squared error (RMSE), adjusted r2 value, and Pearson’s correlation coefficient. The survival function of the KPC mice was evaluated using Kaplan Meier analysis. A p-value of less than 0.05 was considered statistically significant.
Results
The generated mask images obtained a dice similarity metric of 0.94 which corresponds to 94% similarity and a standard deviation of 0.01 (1%). An average of 163 features was identified as reproducible among 264 variables for T1w MRI data while 154 features were reproducible for T2w MRI data. The number of reproducible features increased with the bin-size for T1w MRI (154, 154, 168 and 173 features) while reproducibility was intended to decrease as increasing the bin-size larger than thirty-two levels for T2w MRI (144, 152, 165, and 155 features). The ICC scores of the variables computed from T1w and T2w MRI data with a bin-size of eight were presented in Figure 2 and ICC scores of variables with remaining quantization level were presented in Supplementary Figures 1–3.
Figure 2.
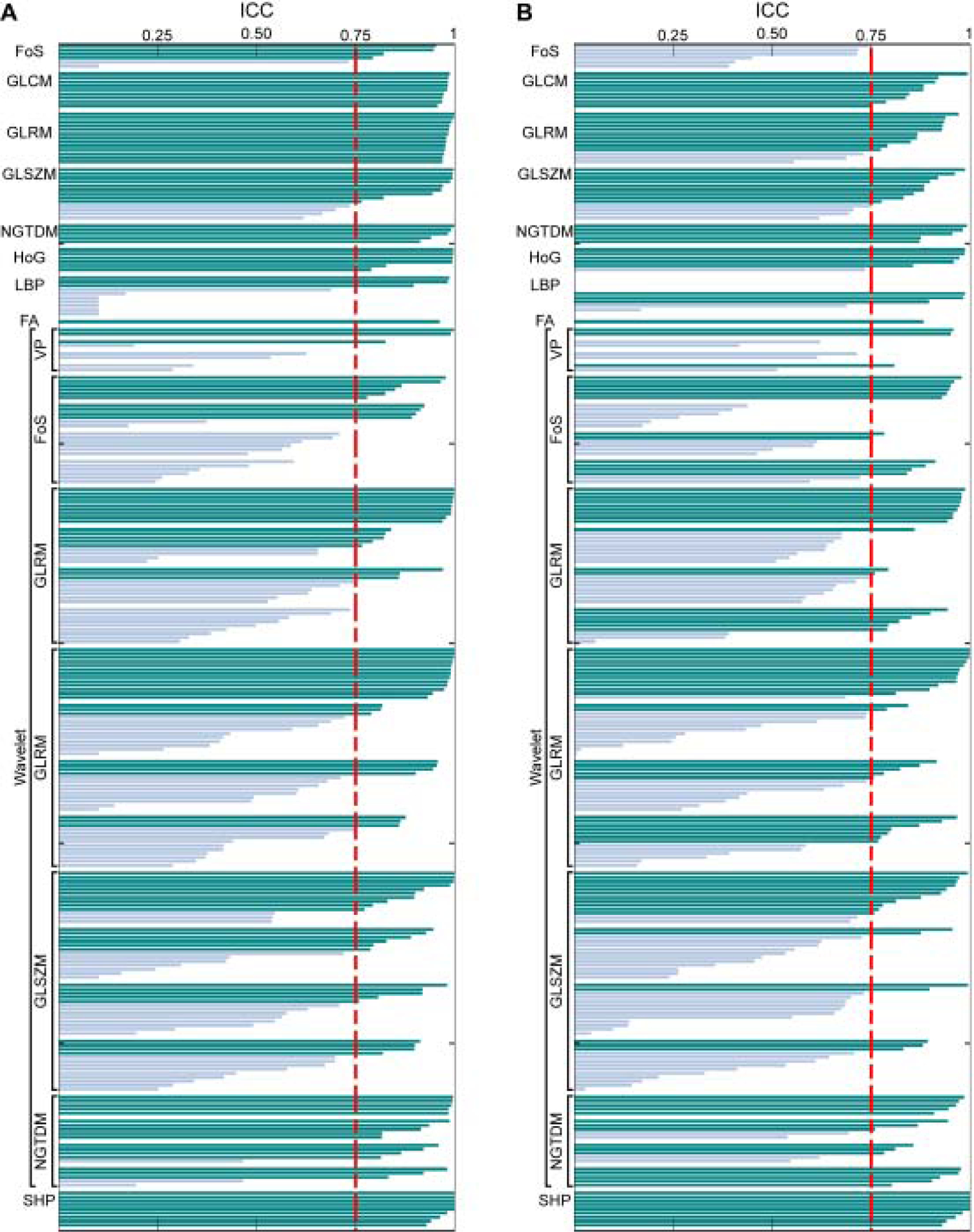
The ICC measurements of the quantitative features extracted from T1w (A) and T2w MRI (B) with a bin size of eight. The ICC values were measured using two masks generated in 2 weeks apart by the same radiologist to determine the reproducibility of the features.
The important radiomics features were identified by performing a multi-step feature reduction process. Initially, an average of 24 features extracted from T1w MRI data and 35 features of T2w MRI data were removed from feature sets due to correlation with tumor size. The remaining variables were clustered according to inter-correlation among the features. An average of 126 clusters was generated for features of T1w MRI data while radiomics features of T2w MRI data were grouped in 61 clusters. The top-ranked twenty features for MRI modalities were further processed in a multivariable analysis.
Four features were identified among twenty variables for MRI modalities via analysis of leave-one-out cross-validation accuracy of the generated classifiers. The classifier generated with four features of T1w MRI data detected vaccine-related changes in the tumor microenvironment with an increasing accuracy during the treatment (1st week: 87.5%, and 3rd week: 93.8%). Besides, classifier generated with T2w MRI features had a diagnostic accuracy of 81.3% and 87.5% for the first and third weeks of the DC vaccine therapy, respectively. Moreover, the combined classification model obtained an improving accuracy from 87.5% to 93.8% for the first and third weeks of treatment, respectively. The best performance was obtained with the features computed from MRI data quantized with a bin size of eight (Figure 3).
Figure 3.
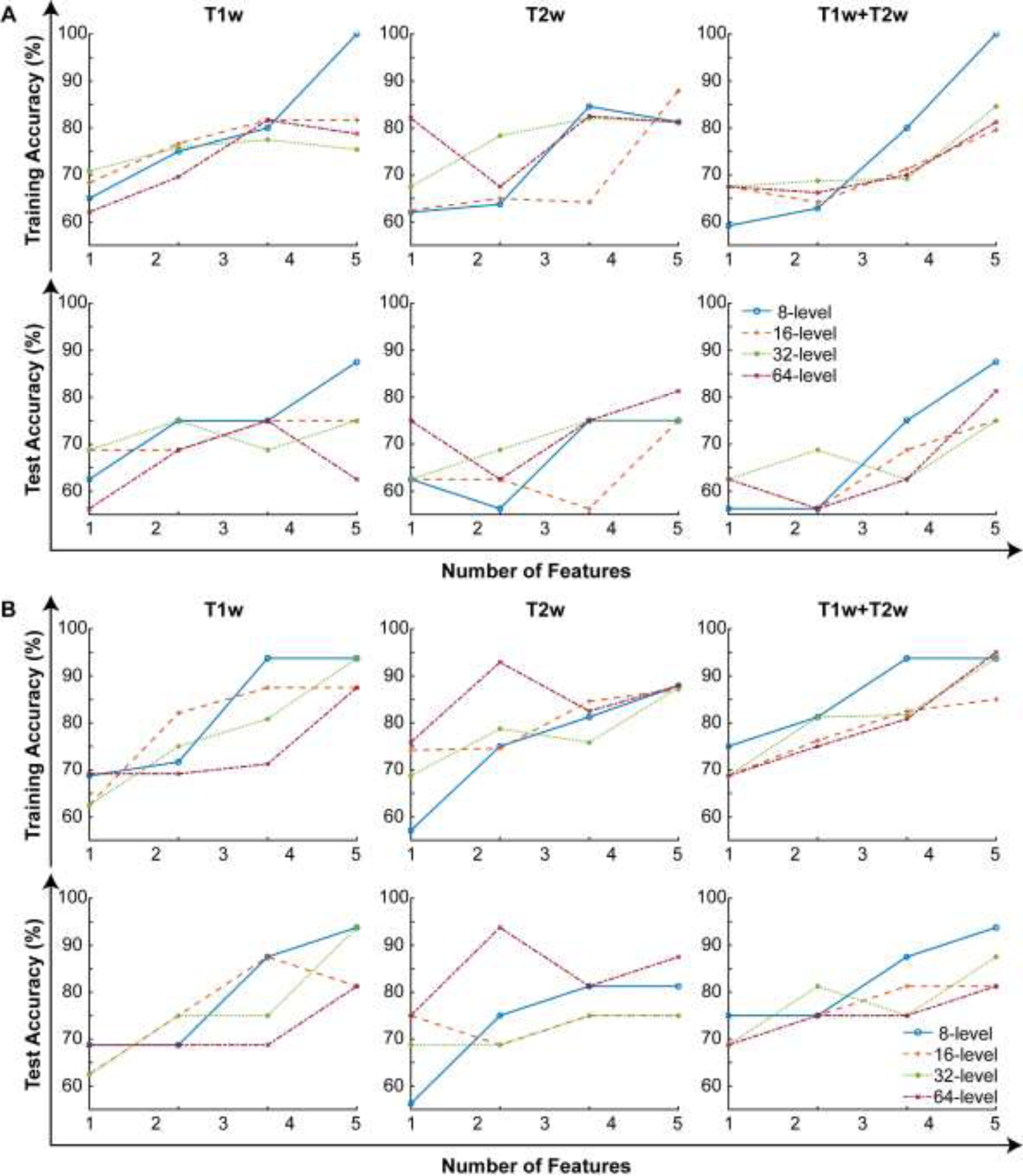
The behavior of the generated three SVM classifiers (T1w, T2w, T1w+T2w) while an increasing number of features. (A) represents the training and test accuracies after the first dosage of the DC vaccine treatment and the results for the data acquired after the 3rd dosage of the DC vaccine are presented in (B).
Kaplan-Meier analysis was performed to evaluate the survival function of the KPC mice (Figure 4A). The treated KPC mice (median: 57 days) had a longer OS compared to untreated mice in the control group (median: 37 days). The regression models were generated to predict OS with five radiomics features of MRI modalities, separately. The regression model constructed with T1w MRI data had an RMSE of 5.96 days that corresponds to a relative error of 13.1%. Besides, the T2w MRI model obtained an RMSE of 7.20 days and a relative error of 15.9% (Figure 4B). Moreover, the model generated with features of combined MRI had an RMSE of 5.96 days corresponding to a relative error of 13.1%. The optimal bin-size for computing the descriptive features was thirty-two levels. Furthermore, all three regression models resulted in a strong Pearson correlation (r = [0.96, 0.94, 0.96]) and adjusted r-squared values (r2 = [0.89, 0.83, 0.89]).
Figure 4.

The survival function of the KPC model was presented using Kaplan-Meier analysis (A). The prediction performance of the generated multivariable model while increasing the number of features (B).
There were significant differences between the control and treatment groups in terms of histological tumor markers (p < 0.03). KPC mice in the control group had a higher percentage of fibrosis (21.10% ± 3.87% vs. 11.86% ± 2.66%) and the number of Ki67+ cells than KPC mice in the treatment group (218.15 ± 28.41 vs. 85.19 ± 40.05). Besides, untreated KPC mice had a lower CK19+ area than treated KPC mice (16.27 ± 3.35 vs. 30.54 ± 3.16). The regression models were generated to predict the behavior of these markers of KPC mice in the control and treatment groups. A regression model generated with T2w MRI features had the best prediction performance for fibrosis percentage with an RMSE of 0.35 that corresponds to a relative error of 2.1% while the models generated with T1w or combined MRI had lower accuracy (RMSE: [0.41, 0.39] and relative error: [2.5%, 2.4%]). The bin-size of eight was optimum for computing features to predict fibrosis percentage. For CK19 marker, regression models integrating five features extracted from T1w, T2w, and combined MRI data (T1w+T2w) predicted CK19+ area with an RMSE of 0.03, 0.04 and 0.08 that corresponded to a relative error of 1%, 2%, and 4%, respectively. The model generated with T1w MRI features computed from MRI data quantized with eight levels has the lowest RMSE as predicting CK19+ area. Besides, three regression models generated to express Ki67+ cells with features of T1w, T2w, and combined MRI data. These models predicted measured values of the Ki67 marker with an RMSE ≤ 0.21 and a relative error of 0.1%. The best performance was obtained with the model integrating five features of T1w MRI data quantized with thirty-two levels. The performance of the generated regression models was visualized using Pearson correlation and RMSE in Figure 5. The features used to generate classification and regression models were presented in Table 2.
Figure 5.

The fitting performance of the multivariable models concerning observed histology data. Pearson correlation coefficients between the generated model and observed data are presented in (A) and the percentage of the root mean squared error was given in (B) for evaluation of the prediction performance.
Table 2.
MRI radiomic features that used to build classification and regression models
| Features | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Treatment | T1w | SR LGLE | LGL RE (a) | LGL RE | ZS variance | |
| T2w | ZS variance | Dissimilarity (a) | Busyness | Coarseness (d) | ||
| T1w+T2w | LGL RE (a, t1) | LZ HGLE (d, t1) | ZS variance (t1) | ZR LGLE (t1) | ||
| Overall Survival | T1w | LR HGLE (v) | Solidity | HGL ZE | SZ HGLE | HGL RE |
| T2w | RP (v) | LZ LGLE (a) | LBP-6 | RP (a) | Entropy (a) | |
| T1w+T2w | LR HGLE (v, t1) | HGL ZE (t1) | HGL RE (t1) | SZ HGLE (t1) | Solidity (t1) | |
| Trichrome | T1w | GL variance (v) | SZ HGLE (h) | CM energy (v) | FD | ZP (h) |
| T2w | Busyness | FD | LBP-9 | GL NU (d) | CM contrast (a) | |
| T1w+T2w | Dissimilarity (v, t2) | CM variance (a, t1) | SZ LGLE (v, t2) | Busyness (a, t1) | GL NU (a, t1) | |
| CK19 marker | T1w | SZ HGLE (a) | RL variance (h) | SZ LGLE (d) | LBP-4 | SZ HGLE (h) |
| T2w | CM energy (v) | GL variance (v) | GL NU (d) | SRE (a) | LBP-9 | |
| T1w+T2w | ZS variance (a, t1) | RL NU (v, t2) | HoG_mean (t2) | AU (d, t1) | CM energy (a, t1) | |
| Ki67 marker | T1w | CM energy (d) | AU (d) | Kurtosis (h) | Entropy (h) | AU (h) |
| T2w | Kurtosis (v) | Coarseness (d) | Coarseness (h) | LZ LGLE (a) | SR LGLE (a) | |
| T1w+T2w | CM energy (d, t1) | AU (d, t1) | Kurtosis (h, t1) | Entropy (h, t1) | AU (h, t1) | |
Abbreviations
AU: Autocorrelation; CM: co-occurrence matrix; FD: fractal dimension; GL: gray-level; HGLE: high gray-level; HoG: histogram of oriented gradients; LGLE: low gray-level emphasis; LR: long-run; LZ: large-zone; NU: nonuniformity; RE: run emphasis; RL: run-length; RP: run percentage; SR: short-run emphasis, SZ: small-zone; ZE: zone emphasis; ZP: zone percentage; ZS: zone-size.
(a, -): approximate wavelet coefficients; (v,-): vertical wavelet coefficients; (h,-): horizontal wavelet coefficients; (diagonal,-): diagonal wavelet coefficients;
(-, t1): T1w image; (-, t2): T2w image;
Discussion
In this study, we investigated potential learning models generated with reproducible radiomics features computed from structural MRI data for early detection of immune-related treatment effects following DC vaccine and prediction of OS in the KPC mouse model. The results demonstrated that proposed models constructed with structural MRI radiomics features can be used as a noninvasive tool for dynamic monitoring of therapeutic responses and detection of OS following DC vaccination.
In recent years, several studies have investigated the potential of multi-parametric MRI radiomics features for stratifying tumor types and prediction of prognosis, treatment outcomes, and recurrence (26–28). Although the studies demonstrated the potential of multi-parametric MRI radiomics nomograms, histological analysis was not utilized to evaluate the potential of the multi-parametric radiomics models. Therefore, preclinical studies are still required to validate the potential of multi-parametric MRI models before translation to clinical studies.
The purpose of this study was to develop learning (classification and regression) models for early detection of immune-response and prediction OS for dynamic monitoring of tumor microenvironment using reproducible features extracted from clinically standard MRI data. The proposed learning models generated with identified features demonstrated the potential of T1w MRI data for early detection of immune-related treatment effects and prediction of OS, and histopathological tumor markers (fibrosis, CK19+, and Ki67+). Besides, the regression model constructed with T2w features accurately predicted the fibrosis percentage.
There were several limitations to this study. First, a limited number of PDAC subjects were included in our study due to the nature of the preclinical studies. Further studies will be performed increasing the number of samples before translating to clinical studies. Moreover, the tumors were outlined using a manual approach despite that it was widely performed in preclinical studies. An automated segmentation process will be beneficial to minimize the pre-processing time for larger studies. Finally, the reproducibility of the computed features was evaluated by analyzing the data acquired from a single scanner due to the monocentric setting of the study. Multi-institutional studies will further advance the analysis of reproducibility.
In conclusion, our study demonstrated that multivariable learning (classification and regression) models generated with quantitative imaging features extracted from structural MRI data can serve as a noninvasive tool to detect early immunotherapeutic response following the DC vaccine therapy and predict OS of PDAC subjects.
Supplementary Material
Acknowledgment
This study was supported by the National Cancer Institute (grants R01CA209886, R01CA196967), by 2019 Harold E. Eisenberg Foundation Scholar Award and by the Fishel Fellowship Award at the Robert H. Lurie Comprehensive Cancer Center.
Abbreviations
- AUC
Area under the receiver operating curve
- DC
Dendritic cell
- ICC
Intraclass correlation coefficient
- OS
Overall survival
- PDAC
Pancreatic ductal adenocarcinoma
- SVM
Support vector machines
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Conflict of Interest
The authors declare no potential conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. The Lancet. 2016; 388(10039):73–85. [DOI] [PubMed] [Google Scholar]
- 3.Oshita C, Takikawa M, Kume A, et al. Dendritic cell-based vaccination in metastatic melanoma patients: phase II clinical trial. Oncol Rep. 2012; 28(4):1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012; 12(4):265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahradova L, Mollova K, Ocadlikova D, et al. Efficacy and safety of Id-protein-loaded dendritic cell vaccine in patients with multiple myeloma--phase II study results. Neoplasma. 2012; 59(4):440–9. [DOI] [PubMed] [Google Scholar]
- 6.Aarntzen EH, De Vries IJ, Lesterhuis WJ, et al. Targeting CD4(+) T-helper cells improves the induction of antitumor responses in dendritic cell-based vaccination. Cancer Res. 2013; 73(1):19–29. [DOI] [PubMed] [Google Scholar]
- 7.Tada F, Abe M, Hirooka M, et al. Phase I/II study of immunotherapy using tumor antigen-pulsed dendritic cells in patients with hepatocellular carcinoma. Int J Oncol. 2012; 41(5):1601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shangguan A, Shang N, Figini M, et al. Prophylactic dendritic cell vaccination controls pancreatic cancer growth in a mouse model. Cytotherapy. 2020; 22(1):6–15. [DOI] [PubMed] [Google Scholar]
- 9.El-Ashmawy NE, Salem ML, Khedr EG, El-Zamarany EA, Ibrahim AO. Dual-targeted therapeutic strategy combining CSC–DC-based vaccine and cisplatin overcomes chemo-resistance in experimental mice model. Clinical and Translational Oncology. 2019. [DOI] [PubMed] [Google Scholar]
- 10.Orr S, Capelletti M, Ghiasuddin H, et al. Development of Novel Second Generation DC/Tumor Fusion Vaccine in Lymphoma. Blood. 2019; 134(Supplement_1):392-. [Google Scholar]
- 11.Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. The Lancet Oncology. 2017; 18(3):e143–e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodi FS, Ballinger M, Lyons B, et al. Immune-Modified Response Evaluation Criteria In Solid Tumors (imRECIST): Refining Guidelines to Assess the Clinical Benefit of Cancer Immunotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2018; 36(9):850–8. [DOI] [PubMed] [Google Scholar]
- 13.Gerwing M, Herrmann K, Helfen A, et al. The beginning of the end for conventional RECIST - novel therapies require novel imaging approaches. Nature reviews Clinical oncology. 2019; 16(7):442–58. [DOI] [PubMed] [Google Scholar]
- 14.Qihua L, Hongmin B, Yinsheng C, et al. A Fully-Automatic Multiparametric Radiomics Model: Towards Reproducible and Prognostic Imaging Signature for Prediction of Overall Survival in Glioblastoma Multiforme. Sci Rep. 2017; 7(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalvati F, Zhang Y, Baig S, et al. Prognostic value of CT radiomic features in resectable pancreatic ductal adenocarcinoma. Sci Rep. 2019; 9(1):5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min X, Li M, Dong D, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. Eur J Radiol Open. 2019; 115:16–21. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Eresen A, Lu Y, et al. Radiomics signature for the preoperative assessment of stage in advanced colon cancer. Am J Cancer Res. 2019; 9(7):1429–38. [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Li Z, Qu J, et al. Radiomics of multi-parametric MRI for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019; 25(12). [DOI] [PubMed] [Google Scholar]
- 19.Padhani AR, Liu G, Koh DM, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia (New York, NY). 2009; 11(2):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma S, Turkbey B, Muradyan N, et al. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR Am J Roentgenol. 2012; 198(6):1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv e-prints 2016.
- 22.Eresen A, Yang J, Shangguan J, et al. MRI radiomics for early prediction of response to vaccine therapy in a transgenic mouse model of pancreatic ductal adenocarcinoma. Journal of Translational Medicine. 2020; 18(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thibault G, Fertil B, Navarro C,L, et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. 10th International Conference on Pattern Recognition and Information Processing, PRIP 2009. Minsk, Belarus: 2009; 140–5. [Google Scholar]
- 24.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics. 1989; 19(5):1264–74. [Google Scholar]
- 25.Kononenko I, Šimec E, Robnik-Šikonja M. Overcoming the Myopia of Inductive Learning Algorithms with RELIEFF. The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 1997; 7(1):39–55. [Google Scholar]
- 26.Osman AFI. A Multi-parametric MRI-Based Radiomics Signature and a Practical ML Model for Stratifying Glioblastoma Patients Based on Survival Toward Precision Oncology. Frontiers in Computational Neuroscience. 2019; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Tian J, Dong D, et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res. 2017; 23(15):4259. [DOI] [PubMed] [Google Scholar]
- 28.Tang T-Y, Li X, Zhang Q, et al. Development of a Novel Multiparametric MRI Radiomic Nomogram for Preoperative Evaluation of Early Recurrence in Resectable Pancreatic Cancer. J Magn Reson Imaging. 2019; DOI: 10.1002/jmri.27024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


