Abstract
It is thought that spinal cord injury triggers scar formation with little axon regeneration in mammals1–4. Here we report that in neonatal mice, a crush injury to the spinal cord leads to a scar-free healing that permits the growth of long projecting axons through the lesion. Depletion of microglia in neonates disrupts such healing and stalls axon regrowth, suggesting a critical role for microglia in orchestrating the injury response. Using single cell RNA-sequencing and functional analyses, we discovered that neonatal microglia undergo a transient activation and play at least two critical roles in scar-free healing. First, they transiently secrete fibronectin and its binding proteins, to form extracellular matrix bridges that ligate the severed ends. Second, neonatal, but not adult, microglia express a number of extracellular and intracellular peptidase inhibitors, along with other molecules involved in inflammatory resolution. Strikingly, upon transplantation into adult spinal cord lesions, both adult microglia treated with peptidases inhibitors and neonatal microglia significantly improve healing and axon regrowth. Together, our results reveal the cellular and molecular basis underlying the nearly complete recovery after spinal cord injury in neonatal mice, pointing to potential strategies to facilitate scar-free healing in the adult mammalian nervous system.
Spinal cord injury disrupts axonal connections between the brain and the spinal cord below the lesion. In fish and amphibians, meningeal cells and glia form permissive bridges, allowing injured axons to regenerate across the lesion5,6. However, this is not the case in adult mammals. Instead, a spinal cord injury triggers the formation of a scar, consisting of multiple cell types such as reactive astrocytes, fibroblasts, and microglia/macrophages, resulting in no spontaneous axon regeneration1–4. Despite studies about these different cell types and their associated molecules1–4,7, how the spinal cord responds to an injury and organizes this scar-based wound healing is poorly understood. Furthermore, the role of reactive glia in axon regeneration failure remains an open question because mature neurons also exhibit diminished intrinsic regenerative capacity in the adult8. To address these questions, we sought to determine if spinal cord regeneration could occur in neonatal mice, a developmental stage when CNS neurons still have high growth ability.
Axon passing across neonatal injury
To assess age-dependent differences in axon regrowth, we performed spinal cord crush injuries in wild type mice of different developmental stages (Fig. 1 and Extended Data Fig.1a). At 2 weeks after injury at postnatal day 2 (P2), numerous serotonergic axons9 were seen in the spinal cord distal to the lesion (Fig. 1, a and b), different from what occurred after injury at P7, P20 or adult (Fig. 1a, b, and Extended Data Fig.1a). In addition to serotonergic axons, corticospinal axons (CST) also reached the lumbar spinal cord after P2 injury (Fig. 1c, d, and Extended Data Fig.1c). Similar to previous studies on neonatal opossums10, it is difficult to distinguish if these axons are regenerating or uninjured and late-arriving. Nevertheless, these results suggest that only after an early neonatal (P2) injury, a permissive environment for axon growth could be established.
Fig. 1 |. Scar-free wound healing after neonatal spinal cord crush injury.
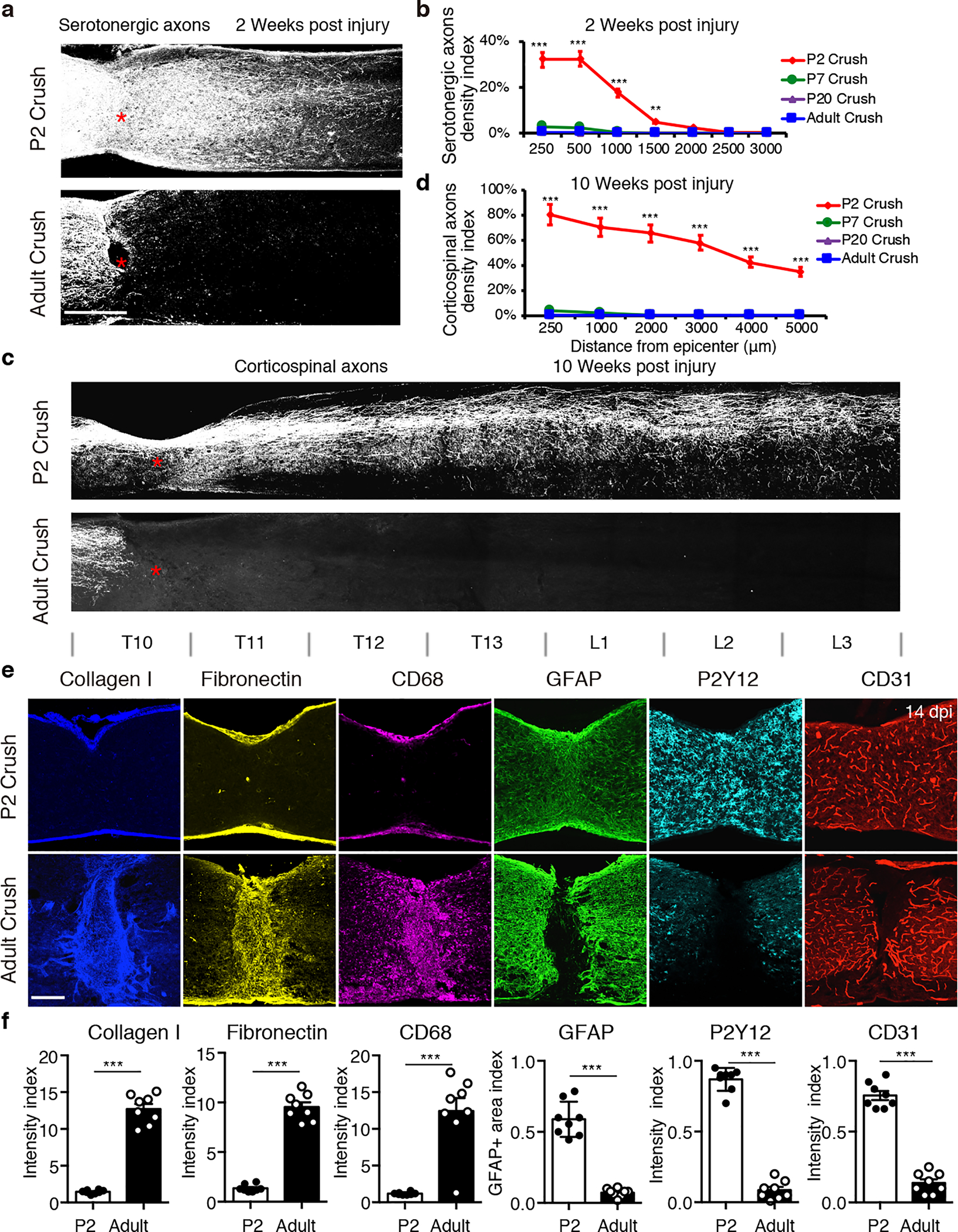
a, Images of spinal sections stained with anti-5HT antibody taken at 2 weeks after P2 (top) or adult (bottom) injury. Red stars indicate the lesion site. b, Quantification of serotonergic axon density (normalized to proximal of lesion) in spinal cord distal to the lesion site. n=8, 5, 5 and 8 for P2, P7, P20 and adult, respectively. **p = 0.0031; ***p < 0.0001. c, Images of spinal sections at 10 weeks post crush showing CST axons labeled by AAV-ChR2-mCherry. d, Quantification of CST axons distal to the lesion site at 10 weeks post crush (n=5). ***p < 0.0001. e, Images of spinal sections at 2 weeks after crush stained with indicated antibodies. f, Quantification of indicated immunoreactive intensity (normalized to the intact region) in the lesion site at 2 weeks after injury (n=8). ***p < 0.0001. b, d, Two-way ANOVA followed by post hoc Bonferroni correction. f, Student’s t test (two-tailed, unpaired). Data shown as mean ± s.e.m. Scale bar: 500μm (a), 1mm (b), 250μm (c, e).
Scar-free healing after neonatal injury
We next compared the characteristics of lesion sites after P2 versus adult injuries. At 2 weeks after adult injury, a typical scar structure formed in the center of lesions with a significant accumulation of CD68+ cells (activated macrophages/microglia11) surrounded by fibroblasts, reactive astrocytes and basal lamina components stained with collagen I, fibronectin, CSPG and laminin (Fig. 1e, f, and Extended Data Fig.1d, e). In contrast, the injury site at 2 weeks after P2 injury had minimal accumulation of CD68+ cells, fibronectin+ and collagen I+, CSPG or laminin+ basal lamina, but with modest accumulation of GFAP+ cells in the lesion center (Fig. 1e, f). Intriguingly, P2Y12, an established marker for homeostatic microglia12, was absent in adult lesions, whereas P2Y12+ cells were evenly distributed throughout the P2 injury site (Fig. 1e, f). Furthermore, immunostaining with anti-CD31 antibodies revealed a continuous vasculature across P2 lesion, in a pattern indistinguishable from the intact spinal cord. The characteristics of the P20 lesions were similar to those in adult injuries, while P7 lesions had reduced scar formation (Extended Data Fig.1b). Thus, the neonatal spinal cord was able to mount a scar-free wound healing response, thereby permitting descending axons to grow across the lesion.
Re-establishing microglia homeostasis
We next examined the lesion site at early time points after P2 injury. At 1–2 days post injury (dpi), a clear gap was present between the two stumps of cut ends and serotonergic axons stopped rostral to the lesion (Fig. 2a). By 3dpi fibronectin+ cells appeared in the gap (Fig. 2a and Extended Data Fig.2b), and by 7dpi the gap was refilled, fibronectin signal disappeared and serotonergic axons grew into and across the lesion. What could be the source of fibronectin+ cells populating the gap at 3dpi? By using Cx3cr1-GFP mice in which microglia are labeled with GFP, we found that microglia accumulated in the stumps at 2 dpi. At this point, they lost the expression of P2Y12, and instead expressed the marker of activated microglia such as SPP1+ and CD68 (Fig. 2b and Extended Data Fig.2). Morphologically, these microglia transformed from highly ramified to amoeboid morphologies (Fig. 2b and Extended Data Fig.2a), consistent with injury-induced microglia activation13. Starting from 3dpi when fibronectin+ bridges form between the two stumps, activated microglia were observed inside the lesion in the absence of GFAP+ astrocytes and collagen I+ fibroblasts (Extended Data Fig.3a). Interestingly, these cells began to regain P2Y12 expression but were still positively stained with SPP1 and CD68 (Fig. 2b and Extended Data Fig.2a). Around 7 dpi, most microglia displayed ramified morphologies and were positive for P2Y12 and negative for SPP1 and CD68 (Fig. 2b, Extended Data Fig.2a). Together with the observations of minimal cell death (Extended Data Fig. 3b) and limited time for cell division (see below), our results suggest that after P2 injury, initially activated microglia switch back to a homeostatic state within the first week.
Fig. 2 |. Microglia are required for bridge formation and rapid healing after neonatal injury.
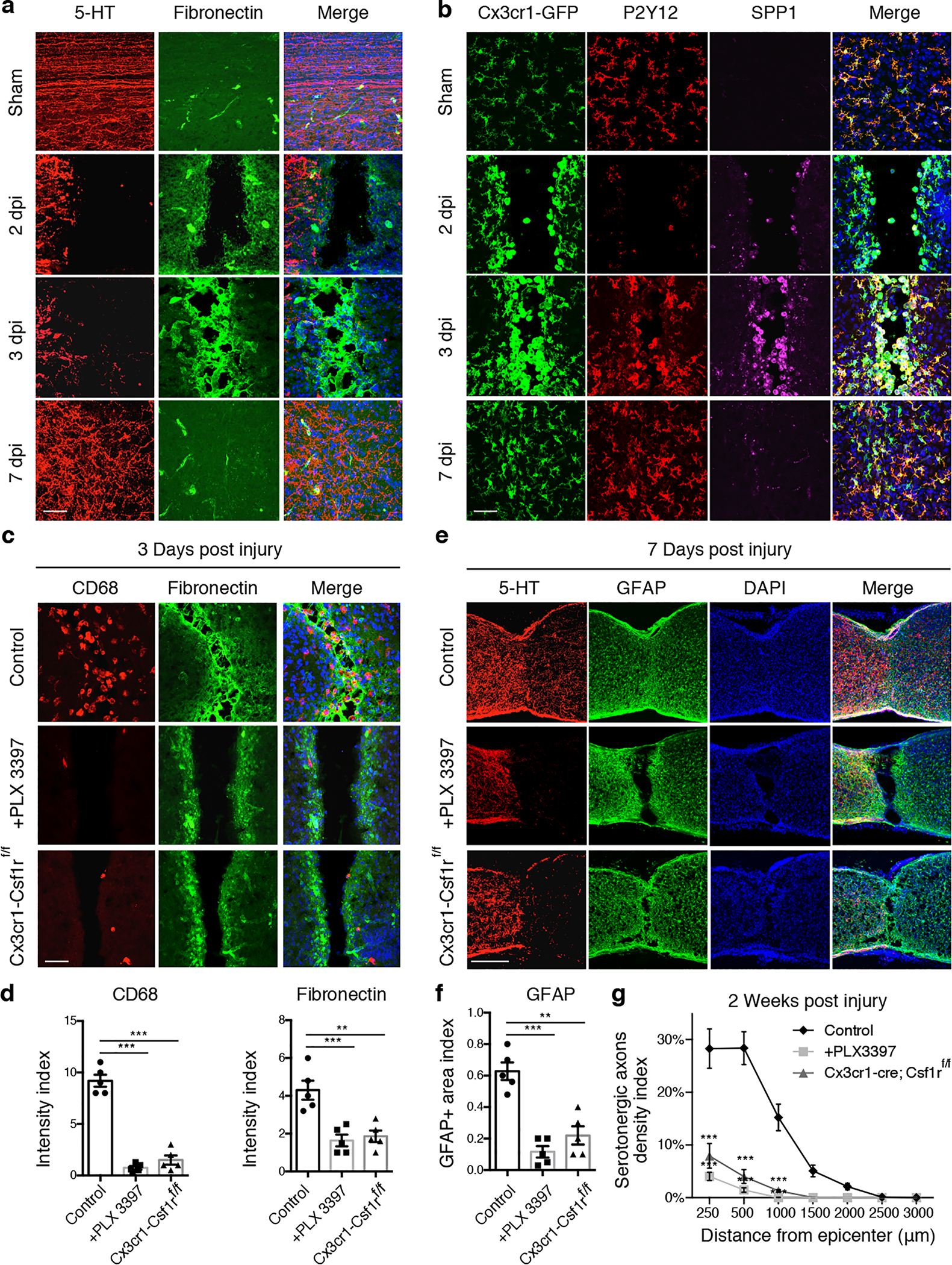
a, Images of spinal lesions after P2 crush stained with indicated antibodies or DAPI (blue). b, Images of spinal cord lesion at different time points after injury stained with antibodies against P2Y12, SPP1 and DAPI (blue) in Cx3cr1-GFP mice. c, Images of spinal sections at 3 dpi from different groups of mice stained with indicated antibodies or DAPI (Blue). d, Quantification of CD68 and Fibronectin immunoreactive intensity (normalized to the intact region) in the lesion site at 3 days after P2 crush (n=5). **p = 0.0014. ***p < 0.0001. e, Images of spinal sections at 7 dpi in different groups of P2 crushed mice stained with antibodies against 5-HT, GFAP and DAPI. f, Quantification of GFAP immunoreactive intensity in the lesion at 7 days after P2 crush (n=5). **p = 0.0003, ***p < 0.0001. g, Quantification of serotonergic axons density in spinal cord distal to the lesion site at 2 weeks after P2 crush (n=5). ***p < 0.0001. d, f, One-way ANOVA followed by post hoc Bonferroni correction. g, Two-way ANOVA followed by post hoc Bonferroni correction. Data shown as mean ± s.e.m. Scale bar: 50μm (a-c), 250μm (e).
A hallmark feature of the injured adult spinal cord is the accumulation and persistence of blood monocyte-derived macrophages in the lesion14. After P2 injury, CCR2-RFP+ monocyte-derived macrophages accumulate in the lesion within 3d, but were absent by 14 days (Extended Data Fig.4a). In contrast, in adult spinal lesions, monocyte-derived macrophages persist in the lesions and activated microglia continuously express CD68 without re-expressing P2Y12 (Extended Data Fig.2c and 4b). Thus, our results suggest a transient accumulation of blood-derived macrophages in neonatal, but not adult, spinal cord injury sites.
Efficient Healing requires microglia
To assess the role of microglia, we first used PLX 3397, a colony-stimulating factor 1 receptor (CSF1R) inhibitor15, to deplete microglia (Extended Data Fig.5a). Strikingly, microglia depletion in neonatal mice impaired bridge formation between the two stumps at 3 or 7 dpi (Fig. 2c–f). At 14 dpi, the gap was closed with GFAP+ astrocytes in the epicenter and blood vessels were absent from the astroglial scar (Fig. 2g, Extended Data Fig.5c). However, limited basal lamina components such as CSPG and laminin were detected (Extended Data Fig.5c), reminiscent of previous findings that swirls of basal lamina structures formed after lesions in the mature, but not the neonatal, cortex16. Most axons stalled at the lesion epicenter abutting the GFAP+ cells. However, a few axons penetrated into and through the lesion, perhaps due to the lack of additional inhibitory basal lamina components17. Importantly, similar results (Fig. 2c–g, Extended Data Fig.5c) were observed after P2 injury in mice with conditional knockout of CSF1R, which removed about 70% of microglia (Extended Data Fig.5b). Thus, microglia are essential for scar-free wound healing after neonatal injury.
Signature of repair-promoting microglia
To gain molecular insights into the neonatal microglia-mediated injury responses, we conducted single cell RNA-seq analysis (scRNA-Seq). At different times after P2 injury (0, 3, and 5 dpi), dissociated cells from the spinal cord tissue around the lesion were FACS sorted with CD11b; CD45low gating (Extended Data Fig.6a) and subjected to 10x scRNA-Seq. Unsupervised clustering revealed 14 cell clusters (Extended Data Fig.6b), including non-dividing microglia cells (C0, C1, C2 and C6), dividing-microglia cells (C3, C7 and C8), macrophages (C4), monocytes (C5), neutrophils (C9), B cells (C10), T cells (C11), astrocytes (C12), and oligodendrocytes (C13) (Extended Data Fig.6b, e).
Among the microglia analyzed, 28.2% at 0 dpi were dividing, and this number reduced to 15.1% at 3 dpi and 9.5% at 5 dpi (Extended Data Fig.6d), suggesting that microglial proliferation gradually declines in the lesion site after injury. As cell cycle genes can overload the major principal components that underlie cell-to-cell variations18,19, we regressed out cell cycle effects and re-clustered all microglia cells into five transcriptionally distinct cell clusters: MG0-MG4 (Fig. 3a–e). They likely represent different states of the same cell type, but some cells could be new ones arising by mitosis or migrating from elsewhere. Most microglia (91.9%) from intact spinal cord are in the cluster MG0, which express homeostatic microglia genes, such as P2ry12, Tmem119 and Siglech. At 3 dpi most microglia fall into two new states MG1 (72.6%) and MG3 (16.1%). Both MG1 and MG3 cells express typical microglial activation markers (e.g., Spp1, Igf1)13,19, but MG3 express additional genes such as Ms4a7 (Extended Data Fig.7), known to be expressed in embryonic microglia13 and border macrophages20,21. On the other hand, at 5dpi microglia are either in the cluster MG2 (64.1%) or in homeostatic MG0 (21.9%). MG2 cells express activation genes at lower levels and are phenotypically between MG1 and MG0 on the UMAP plot (Fig. 3d), supporting the notion that initially activated microglia, both MG1 and MG3, transit back to the homeostatic stage at this time point.
Fig. 3 |. scRNA-seq analysis of microglia isolated from lesion site after P2 injury.
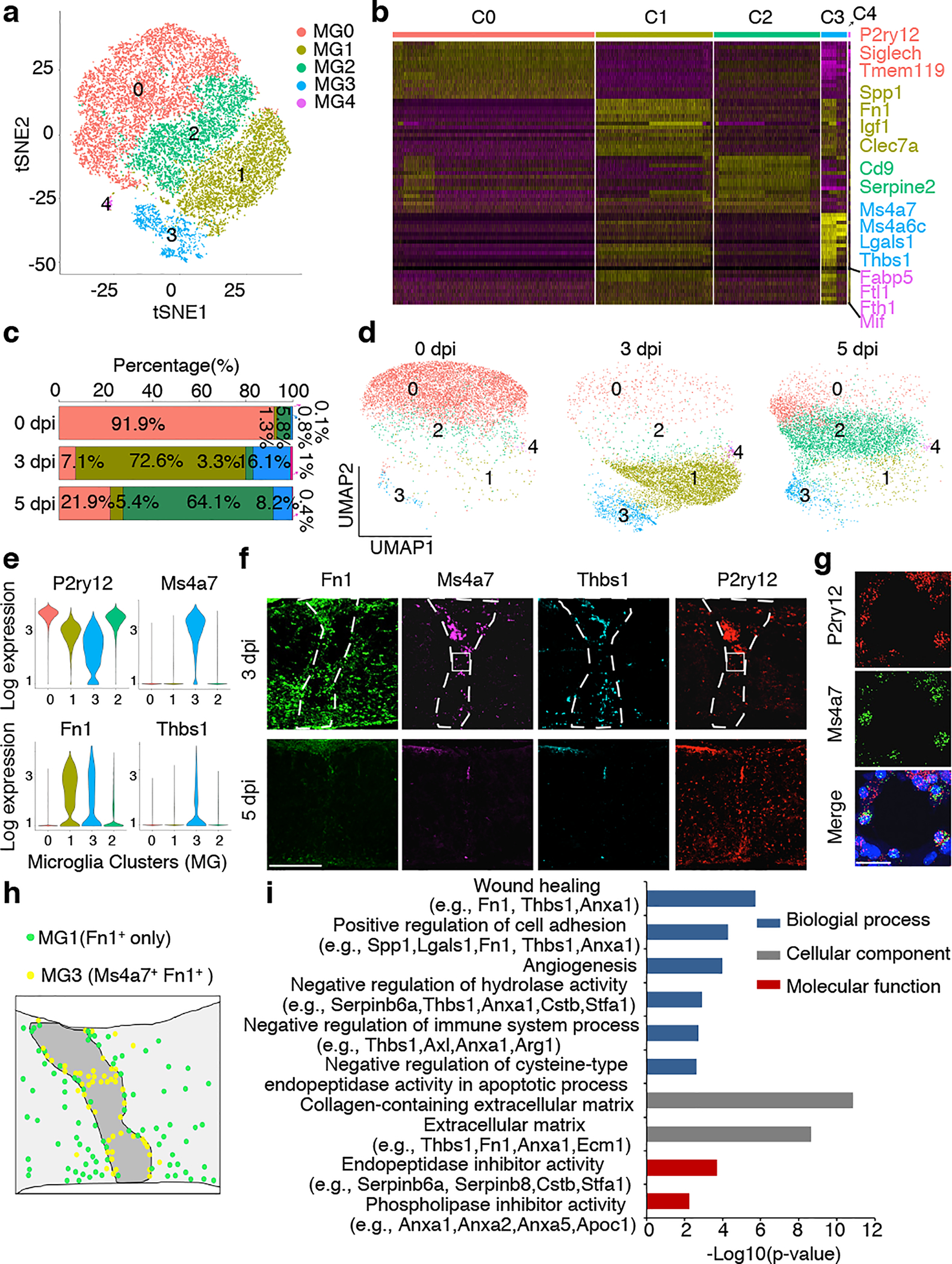
a, tSNE plot showing 5 clusters of microglia isolated at different time points after P2 crush. b, Heatmap showing the top 15 markers for individual clusters. c, Bar plot or d, UMAP of different clusters of microglia at different time points after injury. e, Violin plots showing high-level expression of Ms4a7 and Thbs1 in MG3 and Fn1 in both MG1 and MG3. f, RNA in situ hybridization showing Ms4a7 and Thbs1 enrichment in the lesion epicenter and expression of Fn1 in and around the lesion site. g, Higher magnification images from f, showing co-expression of Ms4a7 and P2ry12 in microglia in the lesion site. All experiments shown in f, and g, were independently repeated 3 times with similar results. h, Schematic showing distribution of MG1 and MG3 microglia. i, Selected GO terms and associated genes enriched in cluster 3 microglia (MG3), two-sided statistical test in function enrichGO was used. Scale bar: 200μm (f), 20μm (g).
By in situ hybridization to examine the expression patterns of several top-ranked markers in each cluster (Fig. 3e), we found that Fn1, encoding fibronectin, is expressed by the cells in and around the lesion at 3dpi, but not 5dpi (Fig. 3f, g). In contrast, Ms4a7+ and Thbs1+, two unique genes expressed in MG3 cells, are concentrated in the microglia immediately in the lesion and in the bridge only at 3dpi (Fig. 3f, g). Thus, MG1 and MG3 cells exhibited a complementary distribution, with MG3 cells immediately in and around the lesion and MG1 cells in the spinal cord around the lesion (Fig. 3h).
The proximity of MG3 cells to the lesion prompted us to explore their molecular signatures. These cells expressed several genes enriched in proliferative-region-associated microglia (PAM)13,19 and disease-associated microglia (DAM)22, such as Spp1, Igf1, Clec7a19, and embryonic microglia13, such as Ms4a7, Ms4a6c, Lgals1 (Extended Data Fig.7), suggesting that neonatal injury converts homeostatic microglia into a state with both activation and de-differentiation signatures. However, MG3 cells also exhibited unique gene expression (Extended Data Fig.8), with significant enrichment of functions in extracellular matrix (ECM), wound healing, phagocytosis, angiogenesis, and negative regulation of immune responses and endopeptidases (Fig. 3i, Extended Data Fig.9a). Consistently, gene regulatory network analysis indicated up-regulation of ECM genes (e.g., fibronectin 1 or Fn1 and thrombospondin 1 or Thbs1) and anti-inflammatory genes (e.g., serine-type and cysteine-type endopeptidase inhibitor activity, phospholipase A2 inhibitor activity) in MG3 microglia cells (Fig. 3i). For example, Fn1, together with Thbs1, an extracellular matrix molecule that can bind to fibronectin23, are highly expressed in MG3 and might mediate the role of MG3 in bridge formation. MG3 cells also have enriched expression of the inhibitors of peptidases and endopeptidases, such as Cstb, Stfa1 and Serpinb6a, as well as Anxa1, a pro-resolving mediator, which may contribute to their rapid inflammation resolution. Importantly, these genes had strikingly different expression patterns in the microglia from adult spinal cord injury sites: adult microglia have persistent expression of Fn1 and no significant induction of proteinase inhibitors (Extended Data Fig9b, c and Fig. 1e). Together, our results suggested that these injury-induced neonatal MG3 microglia might have unique molecular properties suited for promoting scar-free wound healing.
Bridge forming by microglia-derived Fn1
Based on the molecular signatures of MG3, we first assessed the role of microglia-derived fibronectin as a component of the bridge formed between the two severed spinal ends. In addition to microglia, fibronectin in the lesion site could be from blood (synthesized in liver) and/or endothelial cells24. To selectively delete fibronectin from different cell types, we crossed Fn1flox/flox mice with different Cre drivers (Cx3cr1-Cre for microglia, Albumin-Cre for liver, and Tie2-Cre for endothelial cells) and then performed P2 spinal crush injury in respective mice. As shown in Fig.4, only Cx3cr1-Cre; Fn1flox/flox mice, but not others, exhibited a defect in the bridge formation at 3dpi and reduced axon growth at 14dpi. Thus, microglia-derived fibronectin is crucial for mediating the bridge formation as an initial yet key step in wound healing after spinal cord injury.
Fig. 4 |. Deletion of fibronectin in microglia impairs wound healing and axon regrowth after P2 injury.
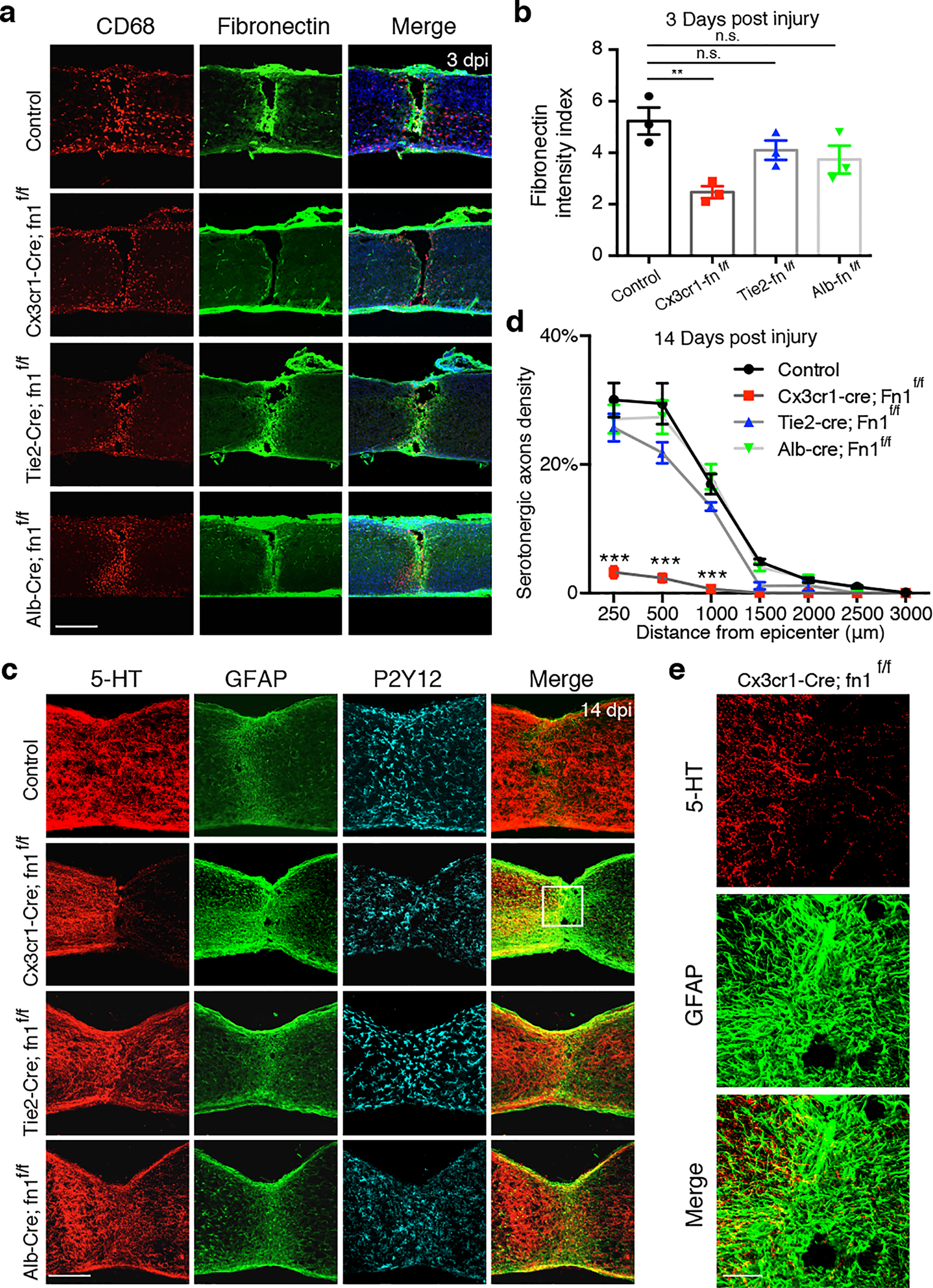
a, Images of spinal sections stained with indicated antibodies taken from 3 dpi of control (Fn1f/f) or different conditional knockout mice. b, Quantification of Fibronectin intensity in the lesion site (3 dpi in P2 injury) in different groups of mice (n=3). One-way ANOVA followed by post hoc Bonferroni correction. **p = 0.0063, n.s., not significant. c, Images of spinal sections stained with antibodies against 5-HT or GFAP, or P2Y12 taken in different groups of mice at 14dpi. d, Quantification of serotonergic axons density (normalized to proximal of lesion) in the spinal cord distal to the lesion (n=5). Two-way ANOVA followed by post hoc Bonferroni correction. ***p < 0.0001. e, Higher magnification images from c, showing 5-HT axon terminals and GFAP+ astrocytes in the lesion site. Data shown as mean ± s.e.m. Scale bar: 250μm (a, c), 50μm (e).
Proteinase inhibitors promote healing
Importantly, we reasoned that if the transiently expressed proteinase inhibitors in neonatal spinal cord play a role in resolving inflammation, preventing scar formation, and thereby facilitating axon regrowth, exogenously provided proteinase inhibitors should alter injury responses in the adult spinal cord injury site. To test this, we utilized two chemical proteinase inhibitors: E64, a membrane-permeable irreversible inhibitor of a wide range of cysteine peptidases and serpina3n, a serine protease inhibitor. We isolated microglia from adult Cx3cr1GFP/+ transgenic mice (Extended Data Fig.10a), treated them with either vehicle or E64/serpina3n combination, and then transplanted them (with or without the inhibitors) into the adult spinal cord lesion site. We expected that such treatments would mimic the transient expression of proteinase inhibitors in MG3. As a control, microglia isolated from neonatal mice were also transplanted into the spinal cord lesion in a separate group of adult mice.
At 2 days after transplantation most microglia were negatively stained with P2Y12 (Extended Data Fig.10b), suggesting their activation in the adult lesion. At 2 weeks after transplantation, both groups with adult microglia transplantation (with or without proteinase inhibitors treatments) exhibited significantly reduced infiltrated immune cells as indicated by Ly6G immunoreactivity and increased GFAP+ region (Fig. 5b and Extended Data Fig.10c). Notably, transplanted adult microglia without proteinase inhibitors remain CD68+ and accumulated in the lesion (Fig. 5a), consistent with the corralling effects of adult microglia and perhaps macrophages in the lesion25. By contrast, many proteinase inhibitor-treated microglia expressed less CD68, re-expressed P2Y12, and some migrated into the host spinal tissues, similar to transplanted neonatal microglia (Fig. 5a, c). Importantly, in these mice, the lesion had significantly less deposition of collagen I and CSPG but more serotonergic axons crossing the lesion (Fig. 5, b-f and Extended Data Fig.10c). However, the number of regenerating axons in the mice with peptidase inhibitors-treated microglia was less than those observed with neonatal microglia transplantation (Fig. 5e, f), implying that other as yet unidentified factors also contribute to the beneficial role of neonatal microglia. Nevertheless, our results indicate that proteinase inhibitors can facilitate the reestablishment of homeostatic microglia and a permissive environment for axon regrowth after adult spinal cord injury.
Fig. 5 |. Transplanted neonatal or proteinase inhibitor-treated adult microglia improve wound healing and axon regeneration in adult mice.

a, b, Images of spinal cord sections taken 2 weeks after adult injury with or without transplantation of adult microglia treated with vehicle or E64/SerpinA (Combination), P1 microglia, and stained with respective antibodies. c, d, Quantification of the results from a, b and Extended data Fig. 10c. n=6. One-way ANOVA followed by post hoc Bonferroni correction. *p = 0.03 (CSPG), 0.0156 (GFAP), **p = 0.0077, ***p < 0.001. n.s., not significant. e, Images of spinal sections from different groups of mice at 4 weeks after adult crush stained with anti-5-HT. Red stars indicate the lesion site. f, Quantification of serotonergic axon density (normalized to proximal of lesion) in spinal cord distal to the lesion site at 4 weeks (n=7). Two-way ANOVA followed by post hoc Bonferroni correction. *p = 0.0192; ***p < 0.0001. Data shown as mean ± s.e.m. Scale bar: 250μm (a, b, e).
Discussion
Together, our results suggest that similar to fish and amphibians, neonatal mice initiate scar-free wound healing with the capacity of spontaneous axon regrowth. Similar to neonatal injured rats26, these mice were able to achieve hindlimb stepping and certain degree of interlimb coordination. However, it is unknown as to the contribution of descending axons and/or intraspinal reorganization. Remarkably, microglia in neonatal mice appear to be the primary coordinator of this reparative injury response. In the acute phase of injury responses, these healing-promoting microglia play multiple roles, not only orchestrating efficient bridging between the cut ends, but also exhibiting remarkable inflammation resolution properties. Different from the permanent activation of microglia in the adult lesions, these initially activated neonatal microglia undergo a rapid and spontaneous state conversion back to a homeostatic state. Our results suggest that such temporal control of microglia activation is crucial for scar-free wound healing process. For example, transient expression of fibronectin and other related molecules is required for forming a bridge across the lesion site, but preventing scar formation. This is reminiscent of CTGF whose transient expression mediates bridge forming and scar-free wound healing in zebrafish5, but its persistent expression in the lesions of adult rat spinal lesions contributes to fibrotic scar formation27. Our results suggested a role of proteinase inhibitors in re-establishing homeostatic microglia in both neonatal and adult lesions. Future studies should identify other molecular players in restoring homeostatic microglia and test their functions in spinal cord injury models. Finally, in light of relationship between chronically-activated and dysfunctional microglia and neurodegeneration28,29, it will be interesting to explore the applications of proteinase inhibitors and other molecules unique to scar free healing-promoting neonatal microglia in these disease conditions.
Methods
Animals.
All experimental procedures were performed in compliance with animal protocols approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital. Wild-type C57BL/6 mice, Cx3cr1-Cre (JAX#025524)30, Tie2-Cre(JAX#008863)31, Alb-Cre(JAX#003574)32, Fn1flox (JAX#029624)33, Csf1rflox(JAX#021212)34, Ccr2RFP/RFP (JAX#017586)35 and Cx3cr1GFP/GFP mice (JAX #005582)36 were obtained from The Jackson Lab. Ccr2RFP/RFP and Cx3cr1GFP/GFP mice were bred to C57BL/6 background, and only heterozygous mice were used.
Surgeries.
Neonatal crush injury:
Newborn pups (P2/P7) were anesthetized by isoflurane. A laminectomy was performed at T10 until the spinal cord was exposed completely from side-to-side. The spinal cord was then fully crushed for 2 seconds with forceps with a width of 0.1mm in the last 5 mm of tips. The muscles and skin were sutured in layers with 6–0 sutures. EMLA topical cream was used for post-surgery analgesia. Animals then were warmed up and kept in a box containing the bedding from the original cage for at least 30 minutes. After that, the closed surgical site was rubbed with the feces from the mom using a Q-tip and then returned to the mom. Feeding was closely monitored in the first week after surgery. Nutra-Gel diets (Bio-Serv) or breeder chow diets were provided to avoid cannibalism. Pups were sacrificed when they showed weight loss (more than 10%). In case of bladder dysfunction, bladder emptying was performed once a day until bladder function was restored.
Adult surgeries:
Adult (and young adult) T10 crush injury was performed at thoracic level 10 (T10) similar to that described before37,38. Briefly, a midline incision was made over the thoracic vertebrae, followed by a T9–11 laminectomy. Tips of forceps were carefully inserted on either side of the cord to include the full width of the cord and then to gently scrape them across the bone on the ventral side to not spare any tissue ventrally or laterally. The spinal cord was fully crushed for 2 s with 0.1mm forceps. The muscles were sutured, and the skin was closed with wound clips. Mice were placed on a warming pad after surgery until fully awake and given buprenorphine for pain relief (2 times per day for 3 days after SCI surgery, 2 times per day for 2 days after brain injection). Their bladders were manually expressed 2 times per day for the duration of the experiments. To label CST axons, AAV-ChR2-mCherry was injected to the mouse sensorimotor cortex as described previously39,40. AAVs were generated at the viral core of the Boston Children’s Hospital and their titers were adjusted to 5×1012 copies/ml for injection.
Perfusion and Immunohistochemistry.
Mice were given a lethal dose of anesthesia and transcardially perfused with PBS followed by 4% paraformaldehyde (PFA). PFA fixed tissues were incubated in 30% sucrose in PBS for 3 days at 4°C, embedded in OCT (Tissue-Tek), frozen in a dry ice/ethanol bath, and stored at -80°C. Transverse and sagittal sections were cut on a cryostat at 30μm thick and stored at -20°C until processed. Before staining, sections were warmed to room temperature and dried on a 37°C slide warmer for 2h. Sections then were treated with a blocking solution containing 10% normal donkey serum and 0.5% Triton-100 for 2 hours at room temperature. The primary antibodies used were: rabbit/goat anti-5-HT [Immunostar (20080/20079), 1: 5,000]; rabbit anti-GFAP [DAKO (Z0334), 1:600]; rat anti-GFAP [Thermo Fisher (13–0300), 1:600]; rabbit anti-P2Y12 [AnaSpec (AS-55043A), 1:200]; rat anti-CD68 [Bio-Rad (MCA1957), 1:600]; rabbit anti-Fibronectin [Millipore (AB2033), 1:200]; rabbit anti-Collagen I [Abcam (ab21286), 1:200]; goat anti-CD31 [R&D Systems (AF3628), 1: 200]; chicken anti-GFP [Abcam (ab13970), 1:400]; goat anti-SPP1 [R&D (AF808), 1:400]; mouse anti-Ly6G/Ly6C [R&D (MAB1037), 1:200]; mouse anti-CSPG [Sigma-Aldrich (C8035), 1:200]; rabbit anti-Laminin [Sigma-Aldrich (L9393), 1:500]; rabbit anti-RFP [Abcam (ab34771), 1:400]. Secondary antibodies (all from Invitrogen) included: Alexa Fluor 488-conjugated donkey anti chicken/rabbit (SA1–72000/A-21206); Alexa Fluor 555-conjugated donkey anti goat/rabbit/mouse (A-21432/A-31572/A-31570) and Dylight 650-conjugated donkey anti rat (SA5–10029). Spinal cord transverse and sagittal sections were imaged with a confocal laser-scanning microscope (Zeiss 700 or Zeiss 710). Figures showing large longitudinal spinal cord section were acquired using Zeiss 710 with image stitching and/or stack software.
Lesion analysis and quantification.
The lesion site was defined and traced using Collagen I, Fibronectin, GFAP, P2Y12, CD68, CD31, CSPG, Laminin or Ly6G/Ly6C staining. Mice were perfused separately at 3, 7 or 14 days after spinal cord crush as described above to visualize lesion site. Cell nuclei were stained by incubation with DAPI for 30 s. Immunostaining intensity in the lesion site (100μm width region in the epicenter) was measured using ImageJ software and normalized to the intact (proximal) region of the spinal cord.
Axon Counting and Quantification.
To quantify the regenerating/late-arriving 5-HT/CST axons after spinal cord crush, sagittal sections through the lesion were stained with antibodies against 5-HT or RFP. Images of 5–6 sections for each animal were taken under a 10X objective and used for quantification. A series of rectangular segments 100μm wide and 400μm high covering the dorsal-ventral aspect of the cord were superimposed onto the sagittal sections, starting from lesion center up to a defined distance caudal. After subtracting the background, the pixel value of each segment was normalized by dividing with the rostral segment (1 mm rostral). The results were presented as a ratio at different distances (axons density index).
Microglia depletion.
To deplete microglia in vivo, adult mice were given the CSF1R inhibitor PLX3397 mixed into AIN-76A standard chow (Research Diets). The dose of PLX3397 was 290 mg/kg and respective controls received AIN-76A standard chow. For neonatal microglia depletion, pregnant mice were given the PLX3397 chow from E14 and newborn pups continued to receive subcutaneous injection of PLX3397 (50mg/kg/day) on daily basis. To accomplish microglia depletion by genetic means, Cx3cr1-Cre mice were crossed with Csf1rflox/flox mice to generate Cx3cr1-Cre; Csf1rflox/flox mice.
RNA Isolation, Bulk RNA-Seq Analysis.
After a quick PBS perfusion, spinal cords of adult mice were rapidly dissected out before (intact), 3 days after or 5 days after the crush. The central 1 mm of the lower thoracic lesion including the lesion core and 0.5 mm rostral and caudal were then rapidly removed after PBS perfusion. Tissues were dissociated using Neural Tissue Dissociation Kit (P) (Miltenyi Biotec) and stained with CD45 and CD11b for 10 min at 4°C. Microglia were then FACS purified using the markers CD45 and CD11b. RNAs of CD45low CD11bhi cells were extracted using RNeasy Micro Kit following the manufacturer’s instructions. RNA quality was verified with an Agilent BioAnalyzer 2100. All experimental steps through RNA extraction were performed in triplicate for the control and experimental groups, with each replicate performed on a different day. The samples within a replicate were prepared on the same day, in a different order each replicate, to avoid any systemic errors from differences in timing.
RNA-sequencing was carried out by the UCLA Neuroscience Genomics Core. Briefly, RNA samples were sequenced by the UNGC. Samples were pooled and barcoded. Library was prepared using Nugen Ovation RNA Ultra Low Input (500 pg) +Kapa Hyper. Preparation included 75bp pair-end reads and sequencing run was carried out over 5 lanes. An average of 55M reads were obtained. Reads were aligned to mouse GRCm38 reference genome using STAR (ver 2.4.0) and an average of 80.9±1.1(SE)% uniquely aligned rate were obtained. Read counts for refSeq genes (mm10) were generated by HT-Seq 0.6.1. Low count genes were filtered and FPKM values were generated. Differentially expressed genes were identified using Limma package.
Single Cell RNAseq Analysis.
Single cell suspension from FACS-sorted microglia cells was prepared using 10x Genomics Chromium Single Cell 3’ Reagent Kits v3 (10X Genomics, Pleasanton, CA) according to manufacturer’s protocol. Quantity and quality of cDNA were assessed by Agilent 2100 expert High Sensitivity DNA Assay. cDNA samples were sequenced on 1 lane of NovaSeq 6000 S2 flowcell at UCLA Technology Center for Genomics and Bioinformatics. Reads were mapped to Mouse GrCm38 genome using Cell Ranger v.3.0.2. More than 380 million reads were obtained for each sample. Average number of genes detected was 3571 (SE±255). Confident read mapping rates were 91.7–93.4% with over 86.8% of reads in cells. Filtering genes and cells: Seurat package v3.1.1 was used to do analysis. For each condition (0dpi, 3dpi, 5dpi), genes expressing in less than 5 cells were removed. Cells with mitochondria express level less than 5%, ribosome express level less than 30%, and number of features larger than 2000 were used.
Removal of doublets:
Package DoubletFinder was used to remove doublet. SCTransform was applied in each condition. PCs 1 to 30 were used for peak identification. A doublet formation rate of 7.5% was considered as threshold. Cells identified as doublet were removed.
Clustering & cell type classification: After removing doublet, all Seurat objects were merged together. Cells with number of features larger than 500, number of RNA count smaller than 80000, mitochondria express percent less than 15% were used. LogNormalize method with scale factor of 10000 was used for normalization. FindVariableFeatures function was used to extract top 5000 variable features. Data was scaled according to mitochondria percent by ScaleData function. Clustering results were visualized using tSNE/UMAP plot. Function FindAllMarkers was used to find significant genes in each cluster. Cell types were identified by using several marker genes from literature.
Subset and Cell cycle regression:
Clusters identified as microglia-like cells were used for further analysis. Cell cycle related genes were regressed out to remove cell cycle effects according to cell cycle phase scores, as instructed in Seurat vignette (https://satijalab.org/seurat/v3.1/cell_cycle_vignette.html). Same method was used to cluster and identify subtypes of microglia cells.
GO enrichment analysis:
Package DOSE 3.8.2 and database org.Mm.eg.db 3.7.0 were used for GO enrichment analysis. Genes with adjusted p-value less than 0.05 and log fold change larger than 0.25 were selected. Top 300 genes ranked by log fold change in cluster 3 (compare with cluster 0) were used as input. All three ontologies (BP, CC, MF) were exported.
In situ hybridization in sample on a slide.
To assess the expression pattern of Fn1, Ms4a7, P2ry12 and Thbs1, we performed in situ hybridization by hybridization chain reaction (HCR). Commercial in situ kit was purchased from Molecular Instruments (Molecularinstruments.org). Each kit containing a DNA probe set, a DNA HCR amplifier (comprising a pair of fluorophore-labeled DNA hairpins), and hybridization, wash and amplification buffers. The designed probes were synthesized by Integrated DNA Technologies. Each probe was designed for 20 or 40 probe sets, and the size of each probe set was based on the expression level of the target. Mice at multiple stages after crush were anesthetized with ketamine/xylazine and perfused with DEPC-PBS followed by 4% paraformaldehyde (PFA). Spinal cord was dissected and fixed in 4% PFA overnight. Dehydrated in 30% sucrose/DEPC-PBS at 4°C, embedded in OCT and cryosectioned at 30 μm, and slices were kept in −20°C. Tissues were permeabilized in 5% SDS for 30 min at room temperature (RT) and pre-hybridized in hybridization buffer for 3 hour at 37°C. Then the slides were incubated in pre-warmed hybridization buffer including probes (2.5 nM for each) at 37°C overnight. After hybridization, slices were washed for 1 hour at 37°C with wash buffer followed by 2xSSC for 15 min at RT. The amplification step was performed with HCR amplifiers (B1, B2, B3 or B4) for overnight at RT.
Neonatal/adult microglia isolation and transplantation.
Neonatal/adult microglia were isolated from the brain of CX3CR1GFP/+ mice. Mice were anesthetized with ketamine/xylazine (100/10 mg/kg) and perfused with cold PBS. Cerebral cortices from postnatal day 1 (P1) or 2 months old mice brain were dissected without Dura. Enzymatic digestion with Neural Tissue Dissociation Kit (P) (Miltenyi Biotec) was performed, dissociated cells were re-suspended in PBS buffer with 0.5% BSA and then were passed through a 70μm cell strainer (Falcon). Myelin was removed using myelin removal beads II and the MACS system (Miltenyi Biotec). After myelin depletion, microglial cells were positively selected from cell suspensions using CD11b MicroBeads and MACS system (Miltenyi Biotec). Neonatal microglial were purified directly without the myelin removal step.
To assess the role of proteinase inhibitors, we utilized two chemical compounds: E64, a membrane-permeable irreversible inhibitor of a wide range of cysteine peptidases and serpina3n, a serine protease inhibitor41–43. For the combination treatment group, adult microglial cells (1*106/ml) were incubated with 10μM E64 and 100ng/ml Serpina3n for 30min, centrifuge at 300g for 10min and re-suspended to ~2*105 /μl cells. 30μM E64 and 500ng/ml Serpina3n were added to the cells again during transplantation. Microglia transplantations were performed one hour after spinal cord injury. One microliter containing ~2*105 /ul CD11b+ purified mouse microglia in PBS were slowly injected into the crushed sites by a Nanoliter injector (WPI) with a pulled glass microcapillary tube (WPI).
Statistical analysis.
The normality of distributions and the equality of variances were tested by SAS before we applied any parametric analyses. Two-tailed Student’s t test was used for the pairwise comparison between two groups. The rest of the data were analyzed using one-way or two-way ANOVA depending on the appropriated design. Multiple comparison procedures were carried out to identify specific between-group differences using Bonferroni’s post hoc. Error bars in all figures represent mean ± SEM. Differences were considered statistically significant at a p value below 0.05. All data were analyzed using GraphPad Prism and SAS.
Extended Data
Extended Data Fig.1 |. Age-dependent decline in serotonergic axon regrowth and wound healing.
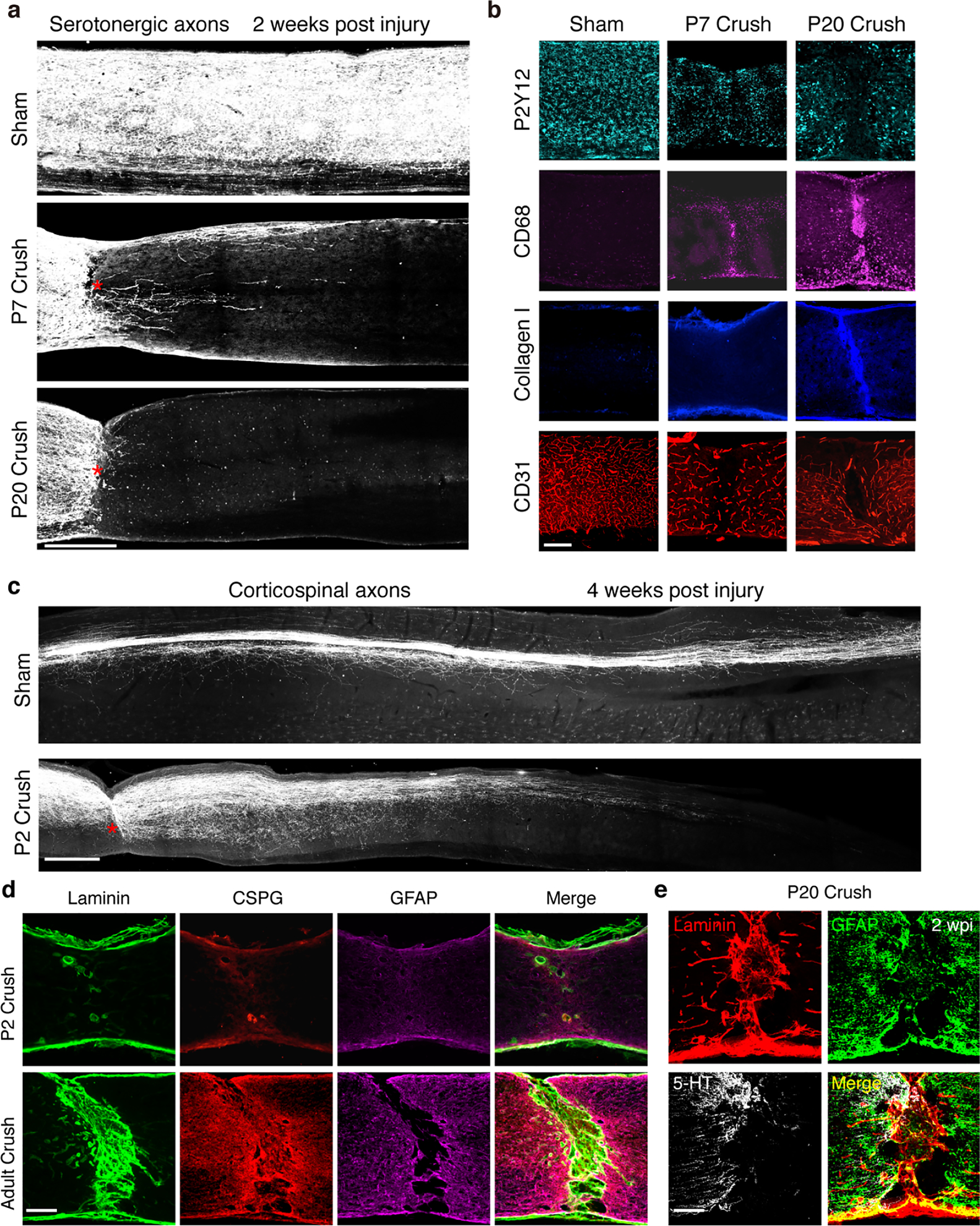
a, Representative images of the spinal cord sagittal sections showing 5HT-labeled axons from sham, P7 or P20 mice at 2 weeks after crush. Scale bar: 500μm. b, Representative images of the spinal cord sections from sham, P7 or P20 mice at 2 weeks after crush stained with antibodies against Collagen I, CD68, P2Y12 or CD31. Scale bar: 250μm. c, Representative images of spinal sagittal sections at 4 weeks after Sham control or P2 crush stained with corticospinal axon tracing by AAV-ChR2-mCherry. Red stars indicate the lesion site. Scale bar: 500μm. d, Representative images of sagittal spinal cord sections at 2 weeks after crush stained with antibodies against Laminin, CSPG and GFAP, respectively. Scale bar: 250μm. e, Representative images of sagittal spinal cord sections at 2 weeks after P20 crush stained with antibodies against Laminin, CSPG and 5-HT, respectively. Scale bar: 250μm. All experiments shown in this figure were independently repeated 3 times with similar results.
Extended Data Fig.2 |. Distinct microglia/macrophage responses after neonatal or adult spinal cord crush.
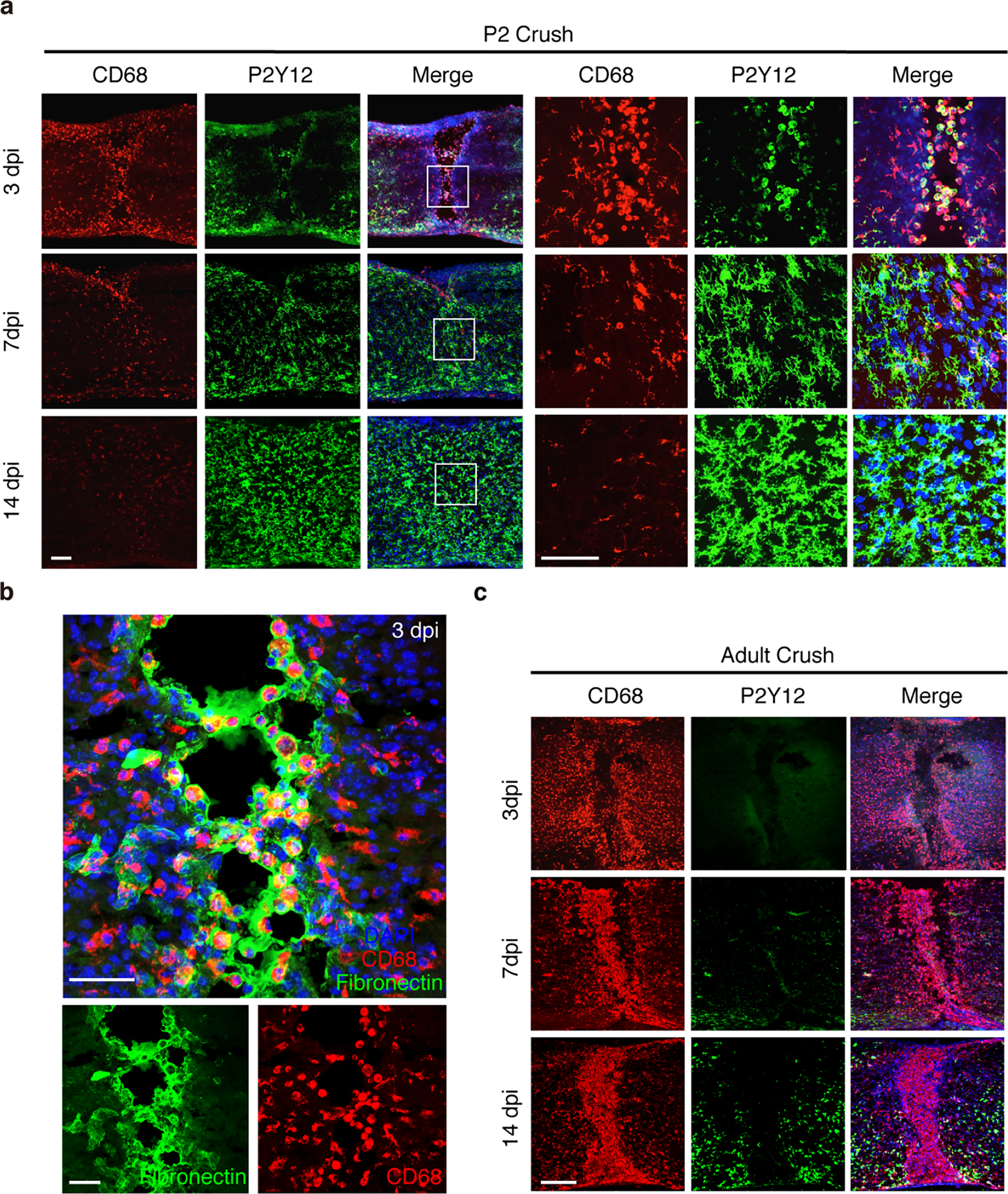
a, Images of spinal cord sections stained with antibodies against CD68 and P2Y12 mice at 3 dpi, 7 dpi or 14dpi. Higher magnification images showing P2Y12+ cells were co-labeled with CD68 at 3 dpi. Cells with highly ramified morphology at 7 dpi, and 14dpi, around lesion sites. Scale bar: 100μm. b, Higher magnification images from Figure 2a showing CD68+ cells and Fibronectin matrix form bridges between gap at 3 dpi. Scale bar: 50μm. c, Immunolabeling for CD68 and P2Y12 in adult mice at 3 dpi, 7 dpi or 14dpi showing CD68-positive cells lacking P2Y12 expression. Scale bar: 200μm. All experiments shown in this figure were independently repeated 3 times with similar results.
Extended Data Fig.3 |. Histological assessments of bridges formed after neonatal spinal cord crush.
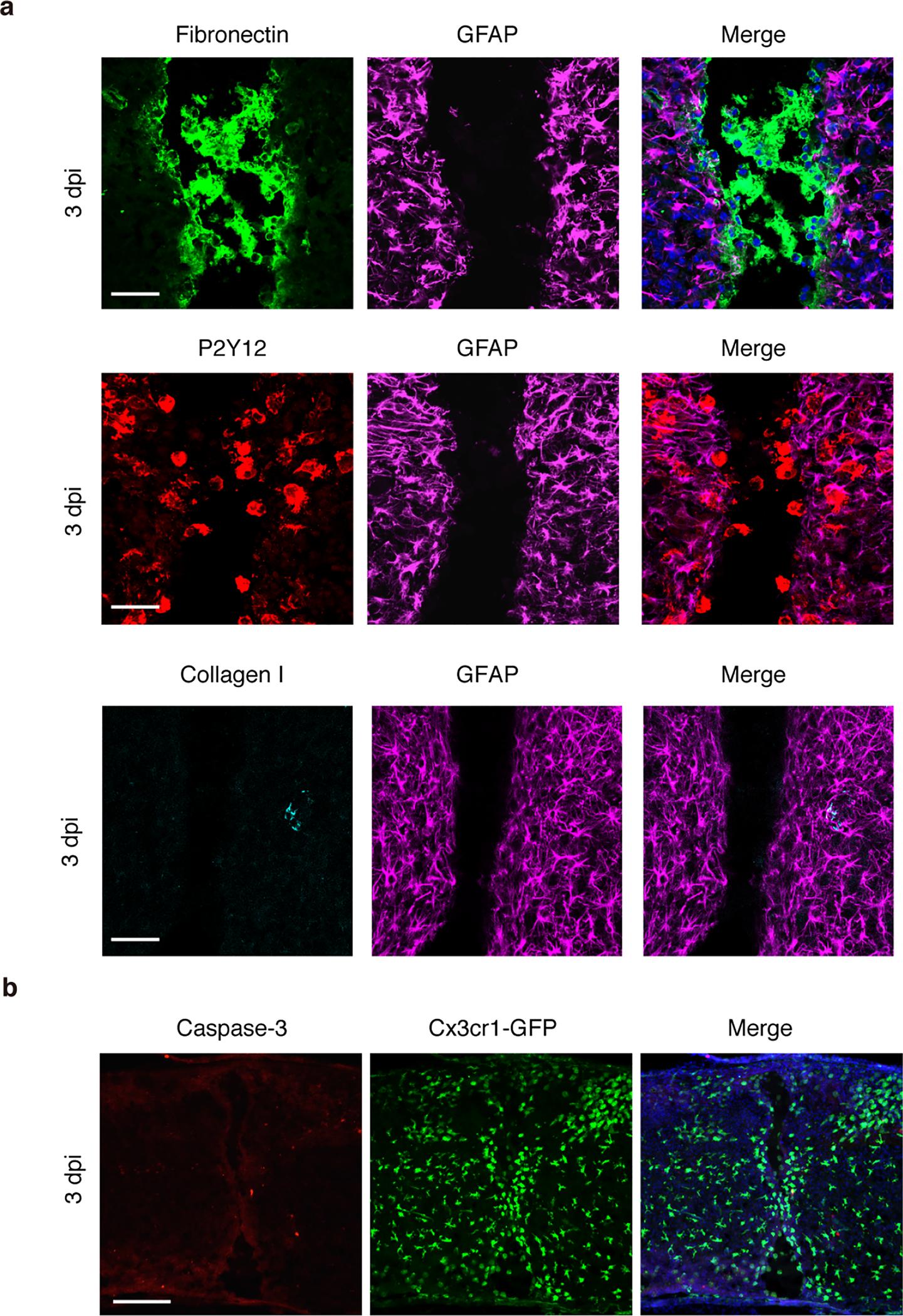
a, Higher magnification images of spinal sections of spinal cord bridges area stained with antibodies against Fibronectin, GFAP, P2Y12, Collagen I and DAPI (Blue) at 3 dpi in P2 injury. Scale bar: 50μm. b, Representative images of spinal sections of Cx3cr1-GFP mice immunolabeled with Caspase-3 showing cells around the lesion sites at 3 dpi in P2 injury. Scale bar: 200μm. All experiments shown in this figure were independently repeated 3 times with similar results.
Extended Data Fig.4 |. Infiltrated CCR2+ monocytes/macrophages were eliminated after neonatal but not adult spinal cord injury.
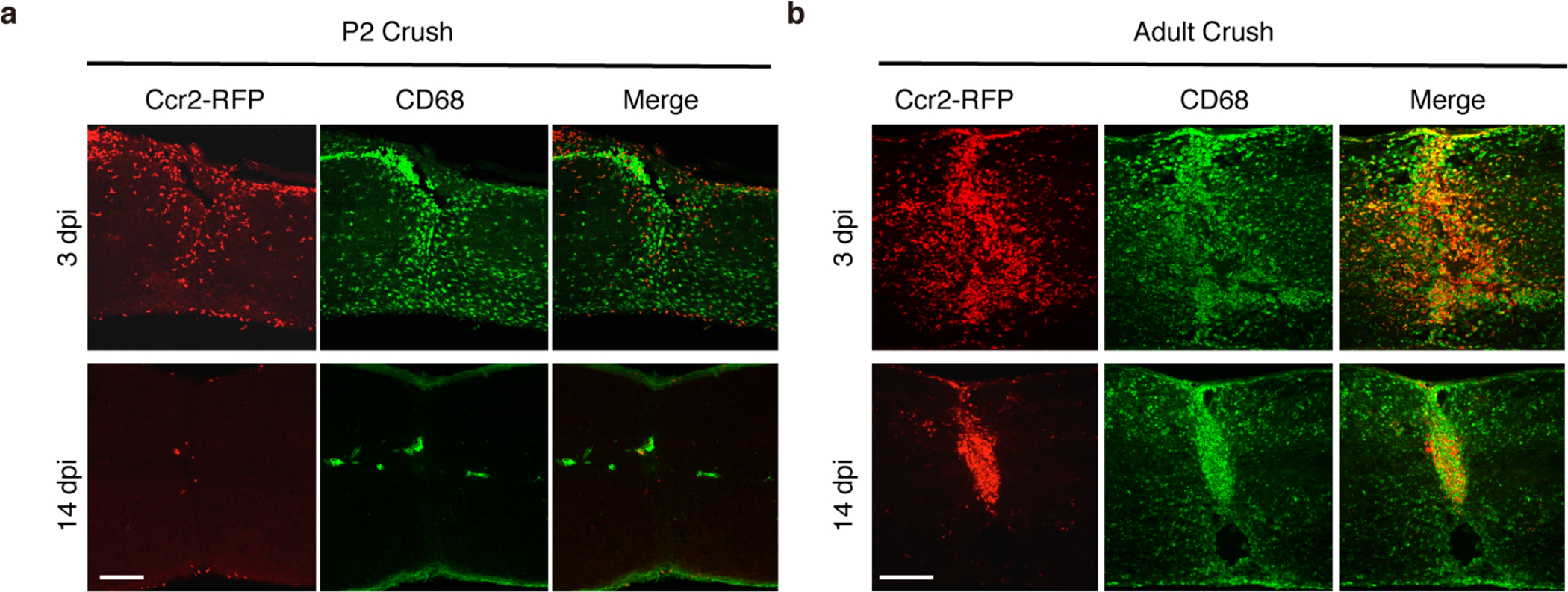
a, b Representative images of sagittal sections of injured spinal cord of Ccr2-RFP mice at 3 or 14 dpi in a, P2 crush or b, adult crush. Sections were immunostained for CD68 and RFP (for Ccr2-RFP). Scale bar: 250μm. All experiments shown in this figure were independently repeated 3 times with similar results.
Extended Data Fig.5 |. Microglia depletion impaired wound healing and axon regrowth after neonatal spinal cord injury.
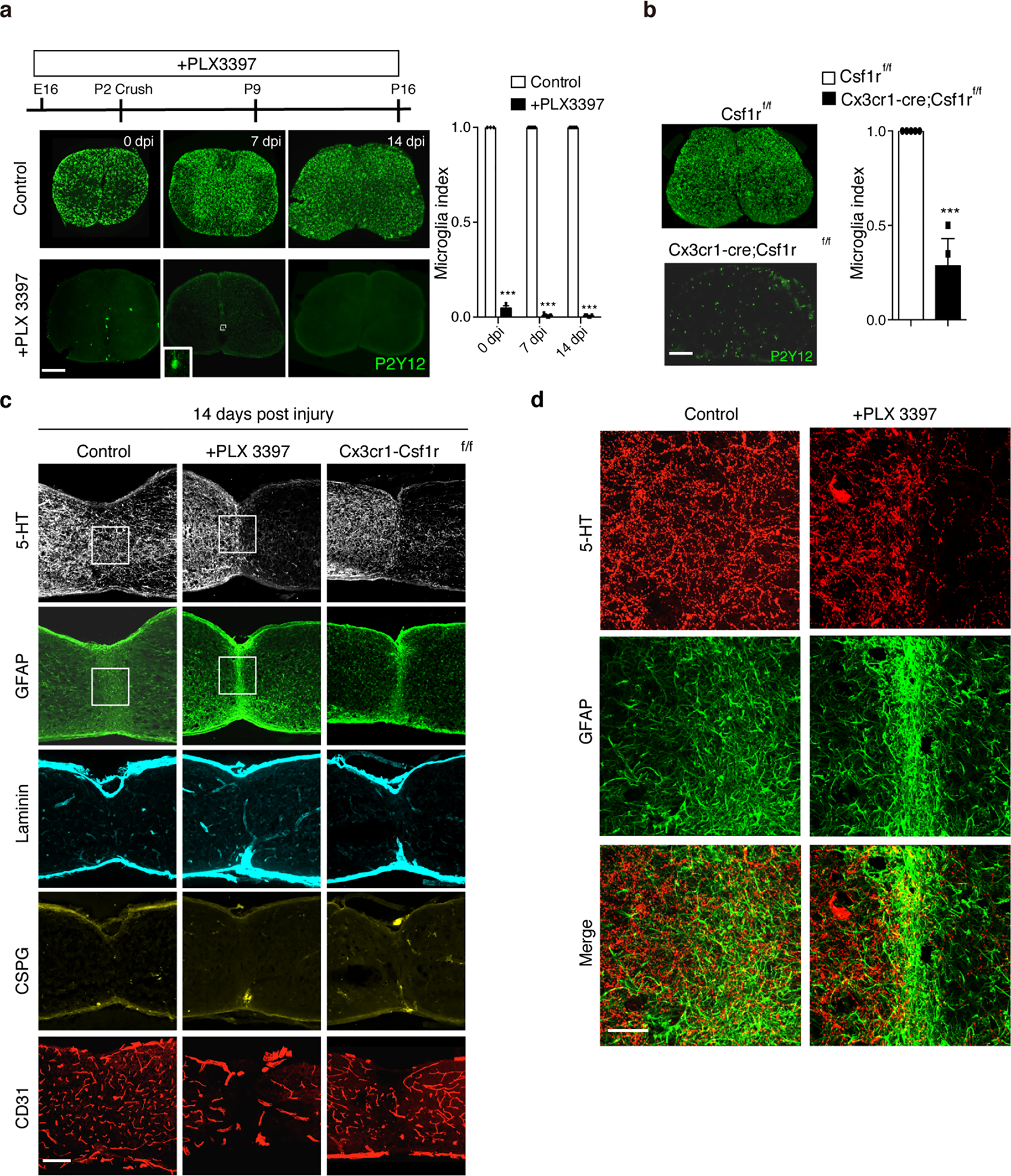
a, Representative P2Y12-stained spinal cord images showing PLX3397-mediated depletion of microglia cells (left). Quantification of microglia depletion in the spinal cord treated with PLX3397 or vehicle at 0, 7 or 14 dpi (right). Student’s t test (two-tailed, unpaired). ***p < 0.0001. Scale bar: 250μm. n=3, 5 and 5 for 0dpi, 7dpi and 14dpi respectively, Data shown as mean ± s.e.m.. b, Representative images of P2Y12-stained spinal cord sections from control (Csf1rf/f) and Csf1r KO (Cx3cr1-Cre; Csf1rf/f) mice showing ~70% reduction of microglia throughout the spinal cord in the mutant mice. Student’s t test (two-tailed, unpaired). ***p = 0.0004. Scale bar: 250μm. n=5 per group. Data shown as mean ± s.e.m. c, Representative images of sagittal spinal sections taken at 14 days after P2 crush and immunostained with antibodies against 5-HT, GFAP, laminin, CSPG or CD31. Scale bar: 200μm. d, Higher magnification images from c, showing 5-HT axon and GFAP+ astrocytes in the lesion site. Scale bar: 50μm.
Extended Data Fig.6 |. Isolation and scRNA-seq results of immune cells after neonatal spinal cord injury.
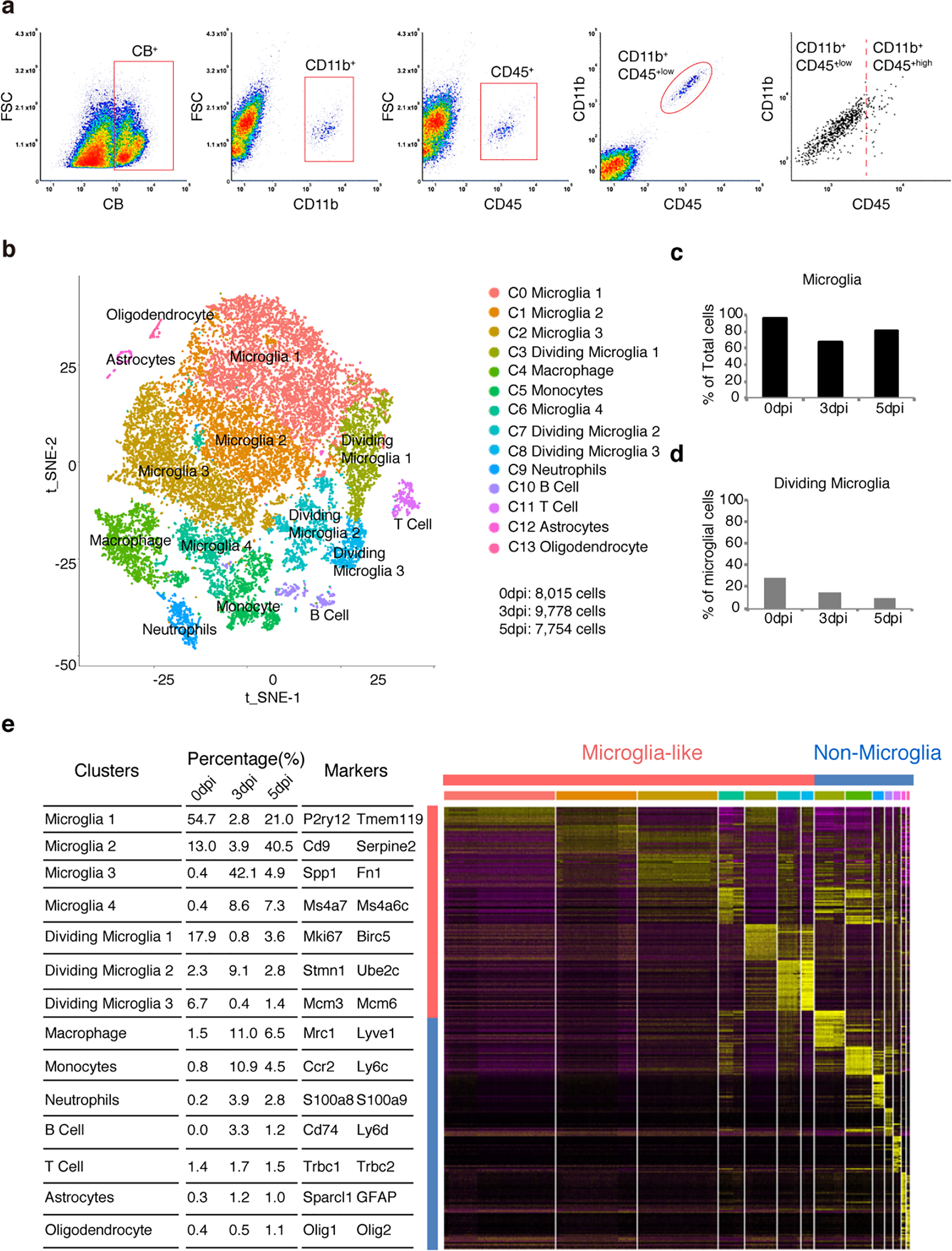
a, Plots of FACS showing selection of CD11b+, CD45+ cells from neonatal spinal cord dissociated cells. b, tSNE plot showing 14 clusters and population annotations. c, d, Relative proportions of microglia among total cells and dividing microglia among microglia cells. e, Table showing the percentage of each cluster and their signature genes (left). Heatmap depicting top 30 DE (differential expression) genes for each of the 14 clusters (right).
Extended Data Fig.7 |.
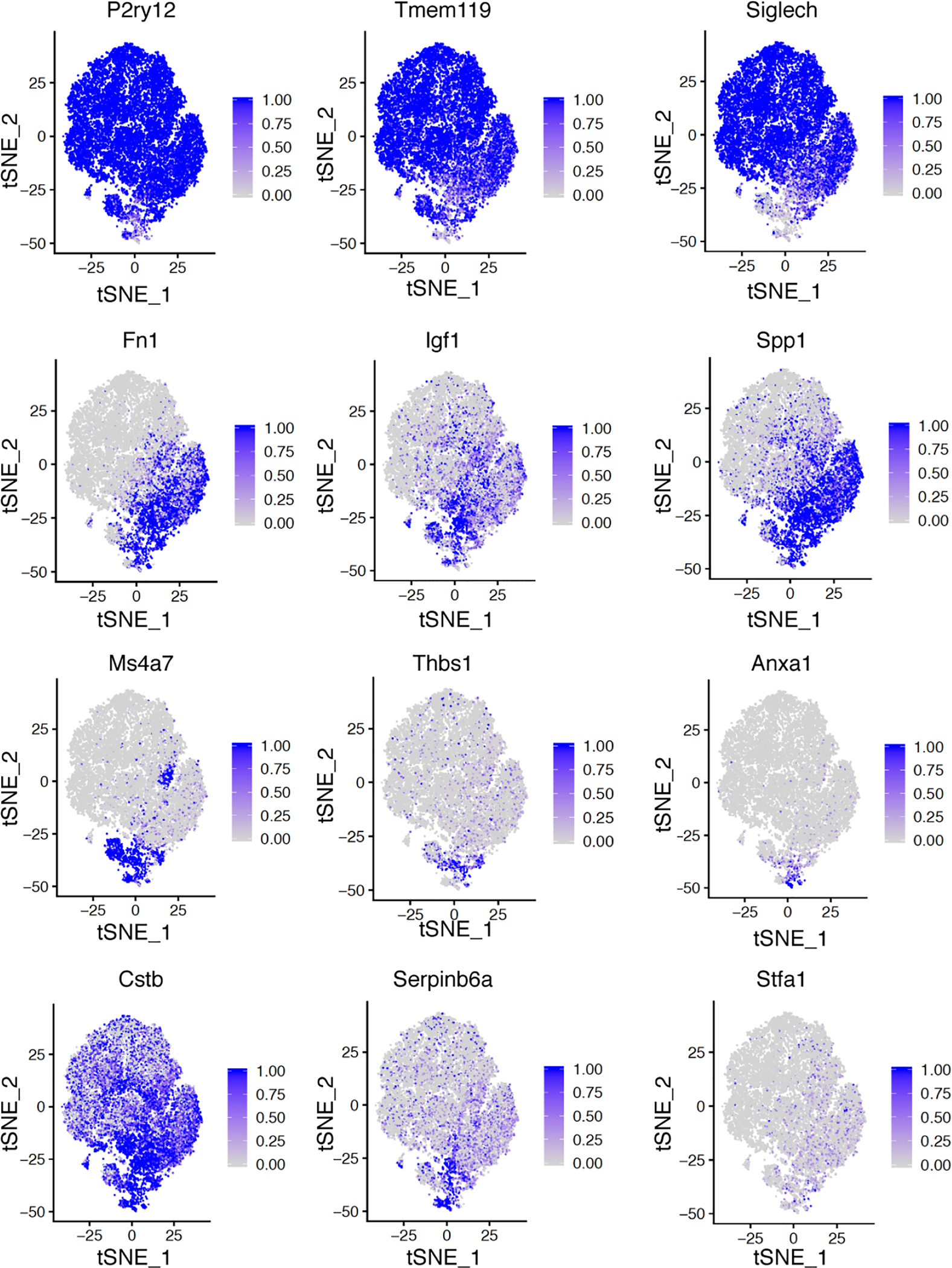
Feature plots showing examples of differentially expressed genes in different clusters.
Extended Data Fig.8 |. Comparison of gene expression changes in PAM, DAM and MG3.
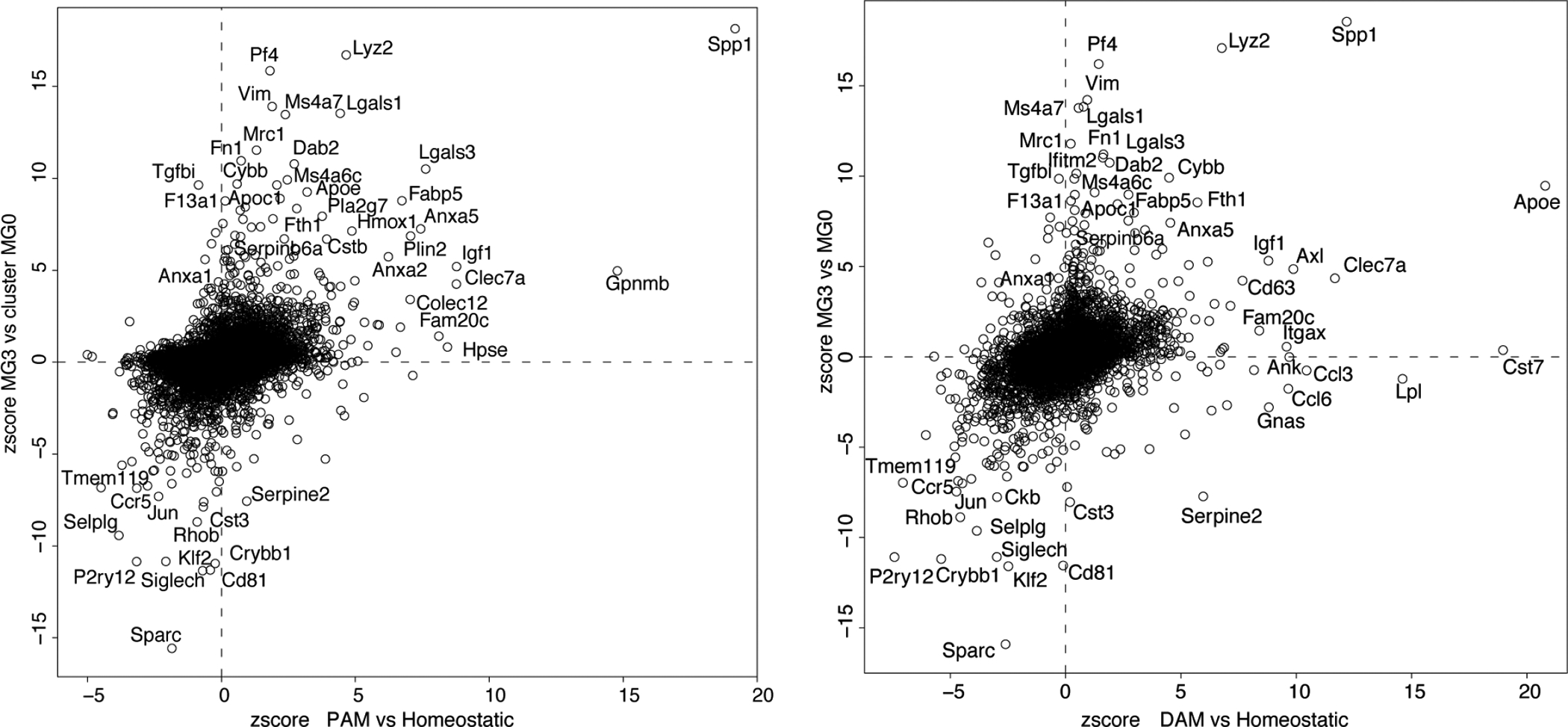
Dot plots showing gene expression correlation between proliferative-region associated microglia (PAM) or disease associated microglia (DAM) (normalized to homeostatic microglia) with MG3 (normalized to MG0) showing different sets of up- and down-regulated genes. n=13755 and 14423 genes, for PAM and DAM, respectively.
Extended Data Fig.9 |. Network analysis and further characterization of MG3 DE genes.
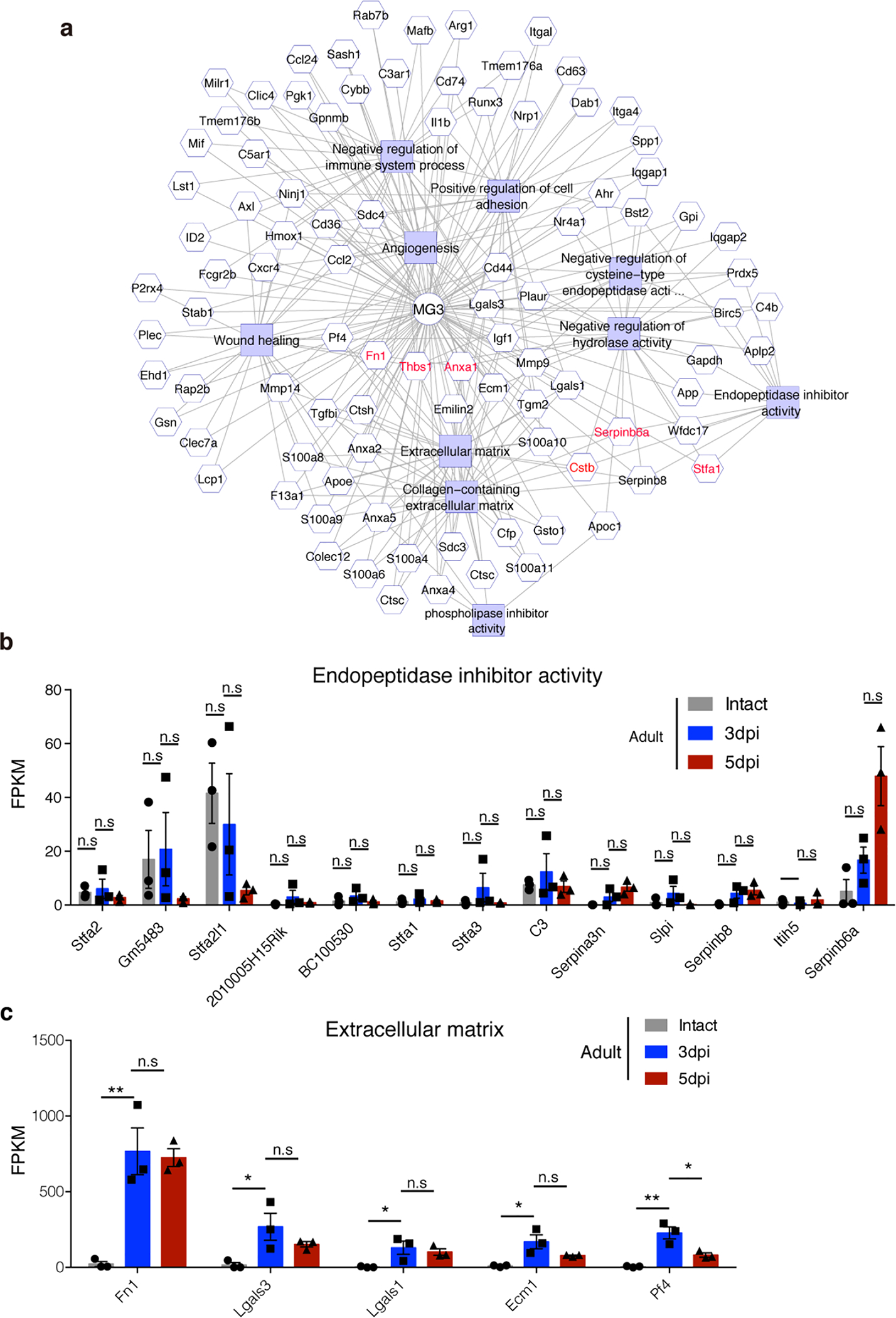
a, Diagram depicting correlated gene modules that underlie cluster identities of MG3 microglia. b-c, The expression of selected genes in microglia isolated at different time points after adult injury using bulk RNAseq. Expression of genes associated with endopeptidase inhibitor activity b, and extracellular matrix c, in adult microglia at 0, 3 and 5dpi. One-way ANOVA followed by post hoc Bonferroni correction. *p = 0.03 (lgals3), 0.04 (lgals1), 0.01 (ecm), 0.01(pf4); **p = 0.003 (fn1), 0.0013 (pf4). n.s., not significant. n=3 per group. Data shown as mean ± s.e.m.
Extended Data Fig. 10 |. Microglia isolation and transplantation.
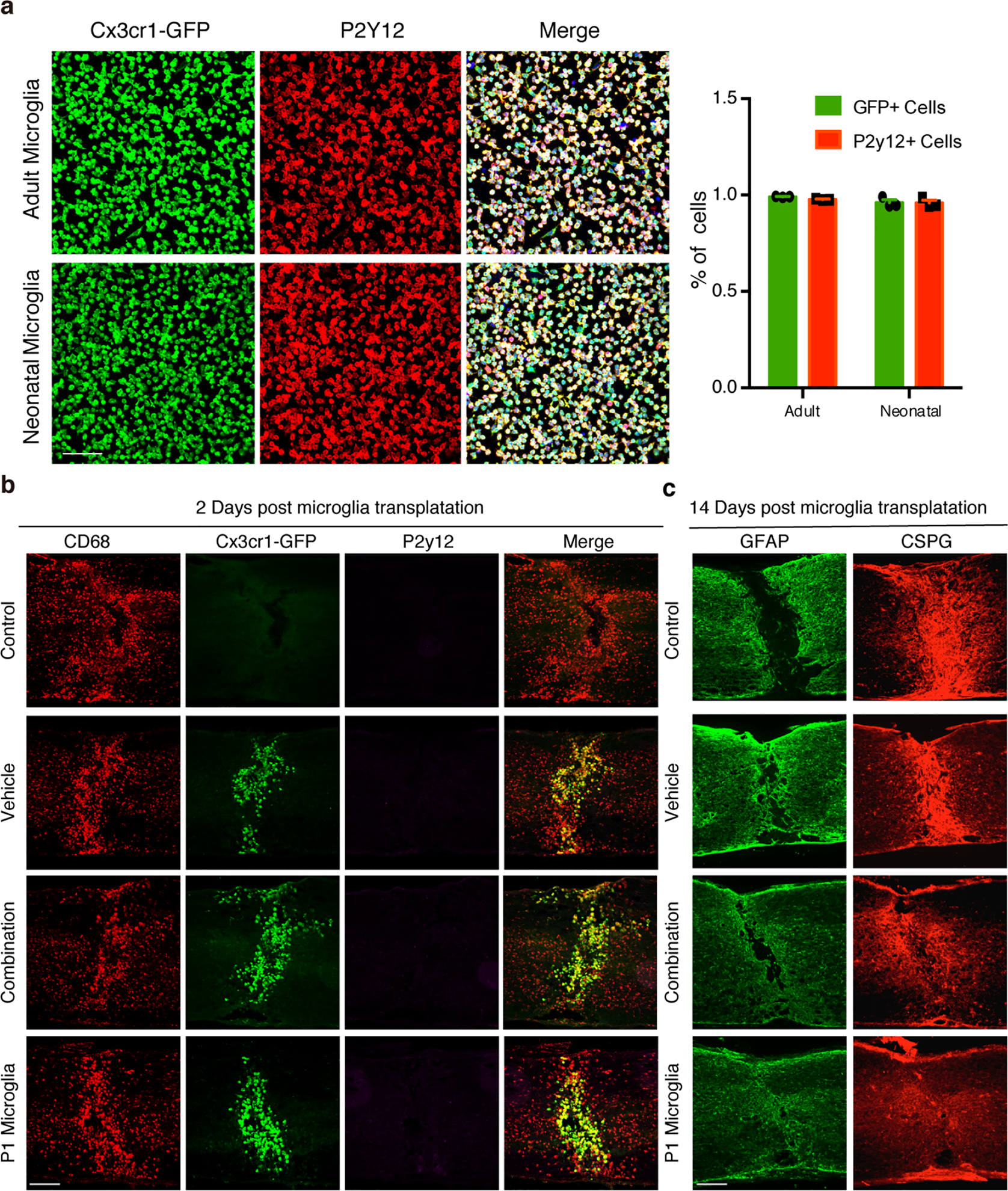
a, Representative images (left) and quantification (right) of isolated microglia (P2Y12+) from neonatal or adult Cx3cr1-GFP mice, n=400 cells examined over 3 independent experiments. Data shown as mean ± s.e.m. Scale bar: 50μm. b, Representative images of spinal cord sections showing the activation of transplanted microglia in the adult lesion at 2 days after grafting. Scale bar: 250μm. All experiments shown in b, were independently repeated 3 times with similar results. c, Representative images of spinal cord sections showing the GFAP and CSPG in the adult lesion at 14 days after grafting, quantification results were shown in Figure 5d. Scale bar: 250μm.
Supplementary Material
Acknowledgments:
We thank J. Sanes and I. Benhar for discussion and S. Hegarty, J. Sanes, B. Stevens, F. Wang, K. H. Wang, C. Winter, and C. Woolf for critically reading the manuscript. This study was supported by NIH R01 grants NS096294 and NS110850 (to ZH), NIH-NINDS R35NS111582 and Ray W. Poppleton endowment (to PGP), grants from Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to ZH, RK, VS, DHG), Craig H. Neilsen Foundation (to YL), and NIH training grant T32NS007473 (to AM). IDDRC and viral cores supported by the grants NIH HD018655 and P30EY012196 were used for this study.
Footnotes
Competing Interests: Z.H. is an advisor of SpineX.
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo), accession number GSE150871.
References
- 1.O’Shea TM, Burda JE & Sofroniew MV Cell biology of spinal cord injury and repair. The Journal of clinical investigation 127, 3259–3270, doi: 10.1172/JCI90608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran AP, Warren PM & Silver J The Biology of Regeneration Failure and Success After Spinal Cord Injury. Physiological reviews 98, 881–917, doi: 10.1152/physrev.00017.2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilton BJ & Bradke F Can injured adult CNS axons regenerate by recapitulating development? Development 144, 3417–3429, doi: 10.1242/dev.148312 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Stenudd M, Sabelstrom H & Frisen J Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol 72, 235–237, doi: 10.1001/jamaneurol.2014.2927 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Mokalled MH et al. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634, doi: 10.1126/science.aaf2679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zukor KA, Kent DT & Odelberg SJ Meningeal cells and glia establish a permissive environment for axon regeneration after spinal cord injury in newts. Neural development 6, 1, doi: 10.1186/1749-8104-6-1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Y & Zheng B Multitasking: Dual Leucine Zipper-Bearing Kinases in Neuronal Development and Stress Management. Annual review of cell and developmental biology 35, 501–521, doi: 10.1146/annurev-cellbio-100617-062644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z & Jin Y Intrinsic Control of Axon Regeneration. Neuron 90, 437–451, doi: 10.1016/j.neuron.2016.04.022 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Bregman BS Spinal cord transplants permit the growth of serotonergic axons across the site of neonatal spinal cord transection. Brain research 431, 265–279, doi: 10.1016/0165-3806(87)90214-8 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Fry EJ, Stolp HB, Lane MA, Dziegielewska KM & Saunders NR Regeneration of supraspinal axons after complete transection of the thoracic spinal cord in neonatal opossums (Monodelphis domestica). The Journal of comparative neurology 466, 422–444, doi: 10.1002/cne.10904 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Schafer DP et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705, doi: 10.1016/j.neuron.2012.03.026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butovsky O et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nature neuroscience 17, 131–143, doi: 10.1038/nn.3599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond TR et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271 e256, doi: 10.1016/j.immuni.2018.11.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popovich PG & Hickey WF Bone marrow chimeric rats reveal the unique distribution of resident and recruited macrophages in the contused rat spinal cord. Journal of neuropathology and experimental neurology 60, 676–685, doi: 10.1093/jnen/60.7.676 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Elmore MR et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397, doi: 10.1016/j.neuron.2014.02.040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudge JS & Silver J Inhibition of neurite outgrowth on astroglial scars in vitro. The Journal of neuroscience : the official journal of the Society for Neuroscience 10, 3594–3603 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKeon RJ, Schreiber RC, Rudge JS & Silver J Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. The Journal of neuroscience : the official journal of the Society for Neuroscience 11, 3398–3411 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buettner F et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nature biotechnology 33, 155–160, doi: 10.1038/nbt.3102 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Li Q et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207–223 e210, doi: 10.1016/j.neuron.2018.12.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mrdjen D et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48, 380–395 e386, doi: 10.1016/j.immuni.2018.01.011 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Jordao MJC et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, doi: 10.1126/science.aat7554 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217, doi: 10.1016/j.cell.2017.05.018 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Tan K & Lawler J The interaction of Thrombospondins with extracellular matrix proteins. J Cell Commun Signal 3, 177–187, doi: 10.1007/s12079-009-0074-2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe EA & Mosher DF Synthesis of fibronectin by cultured human endothelial cells. The Journal of experimental medicine 147, 1779–1791, doi: 10.1084/jem.147.6.1779 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X et al. Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2. Nature neuroscience 23, 337–350, doi: 10.1038/s41593-020-0597-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miya D et al. Fetal transplants alter the development of function after spinal cord transection in newborn rats. The Journal of neuroscience : the official journal of the Society for Neuroscience 17, 4856–4872 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad S, Schluesener HJ, Adibzahdeh M & Schwab JM Spinal cord injury induction of lesional expression of profibrotic and angiogenic connective tissue growth factor confined to reactive astrocytes, invading fibroblasts and endothelial cells. J Neurosurg Spine 2, 319–326, doi: 10.3171/spi.2005.2.3.0319 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Stevens B & Schafer DP Roles of microglia in nervous system development, plasticity, and disease. Developmental neurobiology 78, 559–560, doi: 10.1002/dneu.22594 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Pluvinage JV et al. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192, doi: 10.1038/s41586-019-1088-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yona S et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91, doi: 10.1016/j.immuni.2012.12.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kisanuki YY et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Developmental biology 230, 230–242, doi: 10.1006/dbio.2000.0106 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Postic C et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. The Journal of biological chemistry 274, 305–315, doi: 10.1074/jbc.274.1.305 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Sakai T et al. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nature medicine 7, 324–330, doi: 10.1038/85471 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Li J, Chen K, Zhu L & Pollard JW Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis 44, 328–335, doi: 10.1002/dvg.20219 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Saederup N et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PloS one 5, e13693, doi: 10.1371/journal.pone.0013693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung S et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Molecular and cellular biology 20, 4106–4114, doi: 10.1128/mcb.20.11.4106-4114.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nature neuroscience 13, 1075–1081, doi: 10.1038/nn.2603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zukor K et al. Short hairpin RNA against PTEN enhances regenerative growth of corticospinal tract axons after spinal cord injury. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 15350–15361, doi: 10.1523/JNEUROSCI.2510-13.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B et al. Reactivation of Dormant Relay Pathways in Injured Spinal Cord by KCC2 Manipulations. Cell 174, 521–535 e513, doi: 10.1016/j.cell.2018.06.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y et al. A Sensitized IGF1 Treatment Restores Corticospinal Axon-Dependent Functions. Neuron 95, 817–833 e814, doi: 10.1016/j.neuron.2017.07.037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada K et al. Isolation and Characterization of E–64, a New Thiol Protease Inhibitor. Agricultural and Biological Chemistry 42, 523–528, doi: 10.1080/00021369.1978.10863014 (1978). [DOI] [Google Scholar]
- 42.Vicuna L et al. The serine protease inhibitor SerpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nature medicine 21, 518–523, doi: 10.1038/nm.3852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ou J et al. iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell 173, 851–863 e816, doi: 10.1016/j.cell.2018.03.010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


