Abstract
Background:
Concentric and eccentric cardiac hypertrophy are associated with pressure and volume overload, respectively, in cardiovascular disease both conferring an increased risk of heart failure. These contrasting forms of hypertrophy are characterized by asymmetric growth of the cardiac myocyte in mainly width or length, respectively. The molecular mechanisms determining myocyte preferential growth in width versus length remain poorly understood. Identification of the mechanisms governing asymmetric myocyte growth could provide new therapeutic targets for the prevention or treatment of heart failure.
Methods:
Primary adult rat ventricular myocytes, adeno-associated virus (AAV)-mediated gene delivery in mice, and human tissue samples are used to define a regulatory pathway controlling pathological myocyte hypertrophy. Chromatin Immunoprecipitation Assays with Sequencing (ChIP-seq) and Precision Nuclear Run-On Sequencing (PRO-seq) are used to define a transcriptional mechanism.
Results:
Here we report that asymmetric cardiac myocyte hypertrophy is modulated by serum response factor (SRF) phosphorylation, constituting an epigenomic switch balancing the growth in width versus length of adult ventricular myocytes in vitro and in vivo. SRF Ser103 phosphorylation is bidirectionally regulated by p90 ribosomal S6 kinase type 3 (RSK3) and protein phosphatase 2A (PP2A) at signalosomes organized by the scaffold protein muscle A-kinase anchoring protein β (mAKAPβ), such that increased SRF phosphorylation activates Activator Protein 1 (AP1)-dependent enhancers that direct myocyte growth in width. AAV are used to express in vivo mAKAPβ-derived RSK3 and PP2A anchoring disruptor peptides that block the association of the enzymes with the mAKAPβ scaffold. Inhibition of RSK3 signaling prevents concentric cardiac remodeling due to pressure overload, while inhibition of PP2A signaling prevents eccentric cardiac remodeling induced by myocardial infarction, in each case improving cardiac function. SRF Ser103 phosphorylation is significantly decreased in dilated human hearts, supporting the notion that modulation of the mAKAPβ-SRF signalosome could be a new therapeutic approach for human heart failure.
Conclusions:
We have identified a new molecular switch, namely mAKAPβ signalosome-regulated SRF phosphorylation, that controls a transcriptional program responsible for modulating changes in cardiac myocyte morphology that occur secondary to pathological stressors. Complementary AAV-based gene therapies constitute rationally-designed strategies for a new translational modality for heart failure.
Keywords: heart failure, hypertrophy/remodeling, signal transduction, transcription factors, PP2A, mAKAP, SRF, RSK3
Introduction
In the heart, the contractile myocytes contribute the majority of adult organ mass. As ventricular dimensions match cardiac pumping force and stroke volume to demands for systemic perfusion, both ventricular myocyte volume and length:width ratio that determine ventricular wall thickness and internal dimensions are tightly controlled.1, 2 Myocyte hypertrophy is the principal compensatory response to increased wall stress. Pressure overload induces myocyte growth mainly in width, while volume overload induces growth mainly in length.1, 2 The growth of physiologic hypertrophy is adaptive and reversible. In contrast, pathological hypertrophy is concomitant with abnormal gene expression and metabolism, impaired contractility, increased cell death, and interstitial myocardial fibrosis.1 Consequently, pathological left ventricular hypertrophy is a major risk factor for heart failure, whether concentric hypertrophy that progresses to ventricular dilation and heart failure with reduced ejection fraction (HFrEF), concentric hypertrophy present in heart failure with preserved ejection fraction (HFpEF), or eccentric hypertrophy resulting in HFrEF. Thus, strategies to inhibit pathological myocyte hypertrophy are being sought to prevent heart failure, including interventions that may normalize myocyte length:width ratio and improve cardiac structure and function.1, 2
An extensive intracellular signaling network of calcium, cyclic nucleotide, phosphoinositide, and mitogen-activated protein kinase (MAP-kinase) -dependent pathways has been described that regulates myocyte volume, including pathways that preferentially induce physiological or pathological hypertrophy.1 However, the molecular mechanisms determining myocyte length:width ratio remain unclear. Signaling by extracellular signal-regulated kinases (ERK) have been implicated in the regulation of asymmetric myocyte growth, particularly ERK1/2 that promotes decreased length:width ratio,3 but relevant ERK substrates have not been described. We reported that the ERK effector RSK3 was required in mice for pathological hypertrophy in response to pressure overload and catecholamine infusion, while negligibly contributing to baseline cardiac phenotype and physiological hypertrophy (swim training).4 In addition, RSK3 (Rps6ka2) knock-out inhibited the concentric hypertrophy present in a Raf1L613V Noonan Syndrome mouse model.5 RSK3 comprises a small fraction of the p90RSK enzyme in myocytes, and its action is facilitated by binding the perinuclear mAKAPβ scaffold protein that has a prominent role in the regulation of myocyte gene expression and hypertrophy.4, 6 We now test the hypothesis that the transcription factor SRF is a RSK3 substrate whose post-translational modification directs a gene transcription program promoting pathological myocyte growth associated with a decreased length:width ratio.
RSK family members phosphorylate diverse substrates that regulate transcription and translation and cellular proliferation, growth, survival and motility.7 Of the known RSK substrates, SRF is an attractive candidate for a signal-dependent regulator of myocyte phenotype; SRF serves important roles in myocyte development and maturation, and small differences in SRF activity dramatically affect cardiac phenotype.8, 9 While moderate SRF transgenic expression in mice induced concentric hypertrophy and interstitial fibrosis,10 higher SRF expression resulted in dilated cardiomyopathy.9, 11 Paradoxically, myocyte-specific SRF gene deletion also induced dilated cardiomyopathy.12 SRF contains a conserved MADS (MCM1, agamous, deficiens, SRF) domain that mediates both DNA binding to CArG box [CC(A/T)6GG] serum response elements (SREs) and dimerization with other transcription factors. SRF has low intrinsic transcriptional activity, and interaction with cofactors is important for SRF-dependent gene expression in a temporally regulated and tissue-specific manner.8 Canonical SRF cofactors include the MAP-kinase-regulated Ets domain ternary complex transcription factors (Elk-1, Net, and Sap-1) important for “immediate early” gene expression and myocardin and the Rho and actin-regulated myocardin-related factors (MRTF-A and MRTF-B), that regulate cytoskeletal and muscle-specific genes and contribute to the regulation of hypertrophy.8 As determined by phospho-peptide mapping, RSK phosphorylates SRF solely on Ser103, increasing its binding to the Fos promoter.13 Taken together, these observations make SRF a candidate effector for RSK3-dependent asymmetric myocyte growth.
We now report that SRF Ser103 phosphorylation is bidirectionally regulated in the adult cardiac myocyte by mAKAPβ multimolecular signaling complexes (“signalosomes”) that include RSK3 and PP2A.4, 14 We show that SRF Ser103 phosphorylation induces myocyte growth in width, surprisingly, via cooperative transcriptional activation of a cohort of Activator Protein-1 (AP-1) motif-enriched enhancers. Inhibition or promotion of SRF Ser103 phosphorylation by targeting of mAKAPβ signalosomes using AAV gene therapy vectors improved cardiac structure and function in mouse models of concentric and eccentric disease, respectively. Thus, regulation of SRF phosphorylation by mAKAPβ signalosomes could serve as a molecular switch in the pathological hypertrophic signaling network responsible for determining myocyte growth that is predominantly in length or width, providing a new therapeutic target for the prevention or treatment of heart failure.
Methods
Complete detailed methods are provided in the On-line Data Supplement. The NCBI GEO accession number for PRO-seq and ChIP-seq data reported in this paper is GSE134801. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
Animal and Human Studies:
Animal research was approved by the Institutional Animal Care and Use Committee at the University of Miami. Sprague-Dawley rats were used for primary ventricular myocyte cultures. Wildtype mice were C57BL/6. AAV were injected intraperitoneally (1011 vg i.p.) into neonatal (2-3 day old) mice or intravenously (1012 vg i.v.) via the tail vein into adult mice. All experiments involving human tissue were approved by the Institutional Review Board at Stanford University.
Statistical Analysis:
Except as described in the Data Supplement for ChIP-seq and PRO-seq datasets, statistics were computed using Graphpad Prism 8. n refers to the number of individual mice or myocyte preparations. All data are expressed as mean ± s.e.m. D’Agostino-Pearson omnibus (K2) normality testing was performed for pairwise comparisons and experiments analyzed by 1-way ANOVA. Data failing normality testing were analyzed by non-parametric Mann-Whitney test or Kruskal-Wallis Test followed by Dunn’s Post-hoc testing. ANOVA was performed with matching for experiments involving biological replicates based upon separate myocyte preparations and for in vivo experiments involving serial measurements. 2-way ANOVA was used for experiments with 2-factor design. All datasets involving multiple comparisons for which p-values are provided were significant by ANOVA or Kruskal-Wallis testing, α = 0.05. p-values for experiments involving multiple comparisons were obtained by Tukey, Sidak, or Dunn’s post-hoc testing, albeit p-values for all comparisons are not necessarily shown. Log-rank (Mantel-Cox) test was used to analyze Kaplan-Meier survival curves.
Results
SRF is Phosphorylated by mAKAPβ-RSK3 Signalosomes Inducing Myocyte Growth in Width
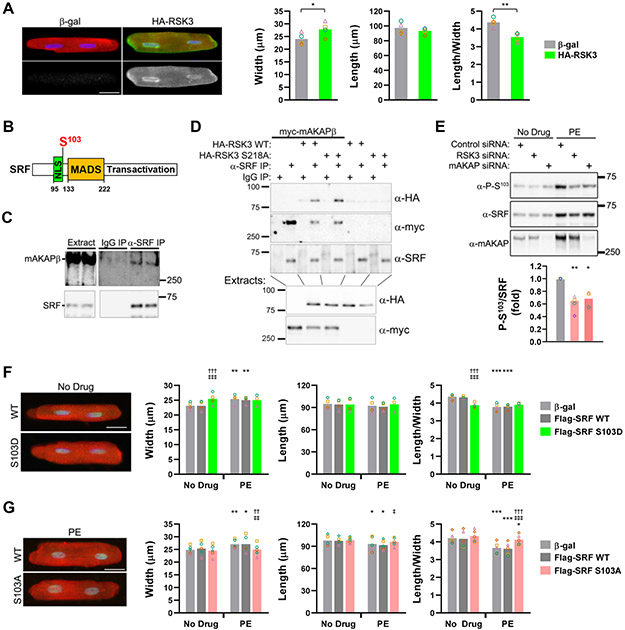
We previously showed that RSK3 knock-out inhibited pathological cardiac hypertrophy in vivo.4, 5 To test whether RSK3 gain-of-function induces adult myocyte hypertrophy, HA-tagged RSK3 was expressed in cultured primary adult rat ventricular myocytes. HA-RSK3 expression increased myocyte width, without affecting myocyte length, resulting in a significantly decreased length:width ratio (Figure 1A).
Figure 1. mAKAPβ-anchored RSK3 regulates SRF S103 phosphorylation, a determinant of myocyte growth in width vs. length.
A, Adult rat ventricular myocytes were infected with adenovirus expressing untagged β-galactosidase (β-gal, control) or HA-tagged RSK3 and maintained in minimal media for 24 hours before immunocytochemistry and measurement of cell width and length (maximum dimension parallel or perpendicular to striations; Bar - 25 μm). Anti-actinin antibody - red; nuclei - blue; anti-HA antibody – green and grayscale panels below. n = 4. Bars and colored symbols indicate average mean and means of independent experiments using different myocyte preparations, respectively. p-values obtained by paired t-test. B, SRF domain structure, showing nuclear localization site (NLS), MADS DNA-binding and heterodimerization domain, and C-terminal transactivation domain.15 C, Co-immunoprecipitation of endogenous complexes from mouse heart extracts. n = 3. D, HA-tagged RSK3 WT or S218A inactive mutant4 and/or myc-mAKAPβ were expressed in COS-7 cells for co-immunoprecipitation assay of ternary complexes. n = 3. E, Neonatal rat ventricular myocytes were transfected with siRNA and treated +/− 10 μmol/L phenylephrine (PE) for 2 days. n = 3-4. * vs. control siRNA + PE. Bar graph below blots shows ratio of phosphorylated to total SRF for PE-treated samples above. F,G, Myocytes were infected with adenovirus expressing Flag-tagged SRF WT, S103D, S103A or control β-galactosidase virus and treated +/− 20 μmol/L phenylephrine (PE) for 1 day. Flag-SRF – green, sarcomeric α-actinin – red, nuclei – blue. * vs. no drug for same protein; † vs. β-gal under the same treatment condition; ‡ vs. SRF WT under the same treatment condition. Bars - 25 μm. F: n = 3-4; G: n = 5. Data in E-G were analyzed by matched, two-way ANOVA with Tukey post-hoc testing. *,‡ p < 0.05; **,††,‡‡ p < 0.01; ***,†††,‡‡‡ p < 0.001.
Given the role of SRF in myocyte gene expression,8, 15 we considered that SRF might be a mediator of RSK3-induced myocyte growth in width. Using recombinant proteins, we confirmed that, like other RSK family members,13 RSK3 can phosphorylate SRF Ser103 (Figure IA in the Data Supplement). Small interfering RNA (siRNA)-mediated RSK3 depletion inhibited serum response element-dependent transient reporter activity induced by the α-adrenergic receptor agonist phenylephrine (PE), suggesting that SRF is a bona fide RSK3 substrate in myocytes (Figure IB,C in the Data Supplement). As RSK3 binds mAKAPβ,4 we tested whether SRF is also associated with mAKAPβ signalosomes. Endogenous mAKAPβ co-immunoprecipitated with SRF from adult mouse heart extracts (Figure 1C). In addition, SRF interacted with RSK3 when expressed with mAKAPβ in COS-7 cells, forming ternary complexes (Figure 1D). Accordingly, RSK3 and mAKAPβ siRNA inhibited phenylephrine-induced SRF Ser103 phosphorylation in myocytes (Figure 1E). These results indicate that mAKAPβ-RSK3 signalosomes induce SRF Ser103 phosphorylation in response to hypertrophic stimuli, consistent with SRF serving as an effector for mAKAPβ-RSK3 signaling.
To test whether SRF Ser103 phosphorylation could induce myocyte growth, Flag-tagged SRF mutants were expressed in adult myocytes. Expression of an SRF S103D phosphomimetic mutant increased myocyte width without affecting cell length (Figure 1F and Figure IIA in Data Supplement), as occurred with RSK3 expression. This result was quantitatively similar to that obtained by treatment of myocytes with phenylephrine (8-10% increase in width and 8-14% decrease in length:width ratio in 24 hours). While SRF S103D induced the growth in width of unstimulated myocytes in minimal media, expression of SRF S103A phosphoablative mutant did not affect basal myocyte size, but, instead opposed the effects of phenylephrine, restoring myocyte length:width ratio (Figure 1G and Figure IIB in Data Supplement).
The morphologic changes observed in vitro were corroborated by expression of the SRF mutants in mice using cardiac myocyte-specific adeno-associated virus vectors (AAV9.SRF, Figure 2A,B). Compared to SRF WT and S103A expression, which had no effect on heart morphology when compared to GFP control, SRF S103D expression resulted in a mild concentric hypertrophy, including increased LV anterior wall thickness, wall thickness to interior diameter ratio [(LVPW+LVAW)/LVID;d] (Figure 2C) and LV mass (Figure 2D), as assessed by echocardiography (Figure III and Table I in the Data Supplement). SRF S103D-induced concentric cardiac hypertrophy correlated with selective myocyte growth in width, as shown by wheat germ agglutinin of heart sections (Figure 2E) and morphologic assessment of ventricular myocytes both after acute isolation (Figure 2F) and by in situ 3-dimensional imaging (Figure IV in the Data Supplement). In addition to hypertrophy, mice injected with AAV9.SRF S103D exhibited mild interstitial fibrosis when compared to AAV9.GFP controls (Figure V in the Data Supplement).
Figure 2. A phosphomimetic SRF mutant induces concentric cardiac hypertrophy in vivo.
A, Flag-tagged SRF WT and mutant proteins were expressed by adeno-associated virus (AAV) serotype 9 vectors that direct expression under the control of the cardiac myocyte-specific chicken cardiac troponin T promoter (AAV9.SRF).24 B, Representative anti-SRF immunoblot of heart extracts from C57BL/6 mice injected i.p. as neonates with 1011 vg AAV9.SRF WT, S103A, or S103D or AAV9.GFP control. Equal loading of samples was confirmed by Ponceau S total protein staining (not shown). C, Left ventricular (LV) anterior wall thickness (LVAW) and wall thickness to interior diameter ratio in diastole [(LVPW+LVAW)/LVID;d] were measured by M-mode echocardiography (Figure III and Table I in Data Supplement). D, left ventricular mass (indexed to body weight) based upon B-mode echocardiographic data. n = 18-27 for C,D. E, Tissue sections stained with wheat germ agglutinin were used to measure myocyte cross-section area. Bar – 25 μm. n = 5-9. F, Cardiac myocytes acutely isolated from AAV-injected mice were measured for length and width using bright-field microscopy. Bar – 25 μm. n = 4-6. p-value symbols: * vs. GFP; † vs. SRF WT; ‡ vs. SRF S103A; *,†,‡ p < 0.05; ††,‡‡ p < 0.01; ***,††† p < 0.001. Bars indicate mean +/− s.e.m. Data passed D’Agostino-Pearson omnibus (K2) testing and were analyzed by one-way ANOVA and Tukey Post-hoc testing.
Effects of Ser103 phosphorylation on SRF-Dependent Transcription Activation
To induce myocyte growth in width, SRF Ser103 phosphorylation was hypothesized to fine-tune SRF-dependent gene expression. To test whether Ser103 phosphorylation alters the binding of SRF to chromatin, the phospho-SRF cistrome in rat ventricular adult myocytes was defined by chromatin immunoprecipitation assays with sequencing (ChIP-seq) using a phospho-Ser103 antibody. ChIP-seq revealed that 12.1% of total detected sites were significantly increased in Ser103-phosphorylated SRF binding following acute α-adrenergic stimulation (false discovery rate < 0.05; defined hereafter as Group 1 sites), while 78.2% and 9.7% of total sites bound similar or decreased levels of Ser103-phosphorylated SRF, respectively (defined hereafter as Groups 2 and 3, respectively) (Figure 3A,B). ChIP-seq was also performed using SRF antibodies to determine SRF occupancy regardless of post-translational modification, finding that ~95% of the 15,872 SRF antibody ChIP-seq peaks corresponded to phospho-Ser103 antibody sites. Comparison of the SRF occupancy for the three phospho-Ser103 groups revealed that increased Ser103 phosphorylation correlated with increased SRF occupancy (Figure 3C). The apparent increase in binding affinity of phosphorylated SRF was consistent with early reports of phosphorylation-induced serum response element binding in electromobility shift assays13, 16 and was further supported by ChIP-seq using a Flag-tag antibody and myocytes expressing Flag-tagged SRF WT, S103A, and S103D proteins (Figure VIA in the Data Supplement). The relatively weaker signals by ChIP-seq using the SRF antibody for Group 1 sites in unstimulated cells (Figure 3C) also suggested that binding of SRF at those sites was more sensitive to Ser103 phosphorylation. Together, these observations indicate that α-adrenergic stimulation differentially alters SRF phosphorylation and occupancy at specific genomic loci.
Figure 3. SRF S103 phosphorylation induces transcriptional activity at select enhancers in adult rat ventricular myocytes.
A, Volcano plot showing the binding sites for Ser103 phosphorylated-SRF increased (Group 1), unchanged (Group 2), or decreased (Group 3) by false discovery rate < 0.05 in ChIP-seq using phospho-Ser103-specific antibody for myocytes treated for 1 hour +/− phenylephrine (PE). B, Heatmaps showing phospho-Ser103-specific antibody ChIP-seq signals over individual sites in Groups 1-3 as defined in A. C, Violin and box-and-whisker plots showing normalized SRF antibody ChIP-seq tags for each Group. D, Average enrichment profile (sites with significant counts only) for H3K27Ac ChIP-seq signals relative to the center of H3K27Ac-marked enhancers within the indicated Groups. E, Plots showing normalized Pol II antibody ChIP-seq tags for H3K27Ac-marked enhancers within the indicated Groups. F, Same as E for myocytes expressing Flag-tagged SRF WT, S103A, and S103D protein. For all violin blots, center lines show the medians (values in parentheses below), box limits indicate the 25th and 75th percentiles, and whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles.
Genomic location analysis of the SRF myocyte cistrome revealed that most of the phospho-SRF-bound chromatin sites were within potential enhancers located in introns or intergenic regions (Figure VIB in the Data Supplement). We employed histone H3 lysine 27 acetylation (H3K27Ac), a mark of active enhancers often highly sensitive to regulatory signaling,17 to identify phospho-SRF enhancers that were most likely to be functional. ~70% (2,351/3,358) of Group 1, 51% (11049/21612) of Group 2 and 58% (1,551/2,669) of Group 3 sites were marked with H3K37Ac ChIP-seq signals. Following phenylephrine treatment, H3K27Ac signal was increased at Group 1 enhancers, unaltered at Group 2 enhancers, and actually decreased at Group 3 enhancers, revealing a positive correlation between H3K27 acetylation (enhancer activation) and SRF phosphorylation (Figure 3D). The implied association of increased enhancer activity with increased SRF phosphorylation was corroborated by profiling the enrichment of RNA polymerase II (Pol II), which is associated with transcriptional activity, including transcription of enhancer RNAs. Pol II ChIP-seq signal was found to be elevated at Group 1 and depressed at Group 3 enhancers in response to phenylephrine (Figure 3E). In addition, in comparison to SRF WT and S103A controls, SRF S103D expression resulted in increased Pol II loading at Group 1 SRF-bound enhancers (Figure 3F). Together, these data suggest that increased Ser103 phosphorylation increases the binding intensity of SRF at specific enhancers and potentiates the activities of those enhancers.
Direct measurement of gene transcription levels, independent of any post-transcriptional effects on mRNA stability, constitutes a definitive assay for direct transcriptional effects and is best performed by the quantitative detection of nascent pre-mRNA transcripts by precision nuclear run-on sequencing (PRO-seq).18 PRO-seq (and GRO-seq) using isolated primary cardiac myocytes has not previously been reported, reflecting the limited number of myocytes typically available for such studies. By modifying PRO-seq to permit the assay of ~5-10-fold less nuclei than previously reported (Figure VII in the Data Supplement), we detected that 1,101 and 962 genes were upregulated and downregulated (FDR<0.05), respectively, in myocytes following phenylephrine treatment for 1 hour (Figure 4A). Phenylephrine-induction of known immediate early genes19 and markers of hypertrophy20 were detected by PRO-seq, including Egr1, Jun, Junb, Fos, Nr4a1, Acta1, Myh7, and Nppa (p < 10−7). As expected, fewer genes were affected by SRF S103D expression (Figure 4B). As was the case for morphologic phenotype (Figure 1F), there was a significant positive correlation between the effects of phenylephrine and S103D expression (Figure 4C and Figure VIII in the Data Supplement), including the induction of known SRF targets Actc1, Des, Myh6, Myh7 and Tpm1 (p < 10−3 for both phenylephrine and S103D).21 Conversely, the effects on gene expression of the S103A mutant were inversely correlated to those for phenylephrine and the S103D mutant, further confirming that a subset of the phenylephrine-induced gene program is dependent upon SRF phosphorylation. Gene ontology and network analysis using the gene list upregulated by both SRF S103D and phenylephrine revealed that these genes included those highly associated with the myocyte sarcomere and cytoskeleton and muscle metabolism (Figure 4D and Figure IX in the Data Supplement), as might be expected in pathological cardiac hypertrophy.22
Figure 4. Phosphorylated SRF activates a gene program that in concert with AP-1 at Group 1 enhancers promotes the preferential growth in width of adult rat ventricular myocytes.
A, Volcano plot showing genes significantly upregulated (green, n = 1,101) or downregulated (red, n = 962) with a false discovery rate < 0.05 in PRO-seq for myocytes treated for 1 hour +/− 20 μmol/L phenylephrine (PE). B, Volcano plot showing myocyte genes significantly upregulated (green, n = 287) or downregulated (red, n = 289) with a false discovery rate < 0.05 in PRO-seq by expression of Flag-tagged SRF S103D in comparison to control Flag-SRF WT protein. C, Heatmap showing fold-change (Log2) expression due to PE treatment or Flag-SRF S103D or S103A expression as assayed by PRO-seq for the PE-regulated genes in A. D, Network of genes upregulated by both PE and S103D expression analyzed by Cytoscape.40 Each term is represented by a circle node, where its size is proportional to the number of input genes falling into that term, and its color represents its cluster identity. Terms with a similarity score > 0.3 are linked by an edge, and the thickness of the edge represents the similarity score. E, HOMER motif analysis of the enhancers in Groups 1-3 showing the most enriched motifs for the different Groups as present in each Group. F, Myocytes transfected with control (Cont) siRNA or siRNA for individual AP-1 family members were cultured for 24 hours +/− 20 μmol/L PE before morphometric assay (Cf. Figure X in Data Supplement). Representative polarized light microscopy images are shown. Bar - 25 μm. Bars and colored symbols indicate average mean and means of independent experiments using different myocyte preparations, respectively. Data analyzed by matched two-way ANOVA and Tukey post-hoc testing. † vs. no drug for same siRNA; * vs. control siRNA under the same treatment condition; n = 4. *,† p < 0.05; ††† p < 0.001.
We next performed Hypergeometric Optimization of Motif EnRichment (HOMER) analysis23 to identify potential emergent properties of the regulatory program. DNA binding site motifs for SRF and myocyte enhancer factor 2 (MEF2) family members were relatively evenly distributed among all three enhancer groups and comprised the most common motifs for Groups 2 and 3 enhancers (Figure 4E). Surprisingly, motifs for Activator Protein-1 (AP-1) family members ATF3, Fra1/Fosl1, BATF, JunB, and Jun/c-Jun were dramatically enriched in Group 1 enhancers. In addition, further analysis revealed that AP-1 motifs were enriched in Group 1 enhancers close to phenylephrine- and SRF S103D-upregulated transcriptional start sites (Table II in the Data Supplement). To investigate whether AP-1 family members actually regulate myocyte morphology, adult myocytes were transfected with AP-1 siRNAs. Jun and JunB siRNAs inhibited phenylephrine-induced growth in width and the corresponding decrease in length:width ratio, without affecting baseline cell dimensions, similar to SRF S103A expression (Figure 4F, Figure X in the Data Supplement). BATF siRNA significantly increased the length:width ratio for phenylephrine-treated myocytes. Interestingly, ATF3 siRNA increased myocyte width and decreased myocyte length:width ratio at baseline, with no significant effect on phenylephrine-treated myocytes, while Fra1 siRNA exerted no morphologic effect. Taken together, these data reveal that Jun, JunB and potentially BATF contribute to the phenylephrine-dependent induction of myocyte growth in width, while ATF3 serves as a baseline repressor of myocyte growth in width. Thus, Group 1 enhancers appear to represent a set of transcriptional regulatory elements in which phosphorylated SRF and select AP-1 family members synergistically control gene expression programs responsible for the stimulus-dependent growth in width of the cardiac myocyte.
RSK3 Anchoring Disruptor Therapy for Pressure Overload Disease
As the phosphomimetic SRF S103D mutant induced concentric hypertrophy (Fig. 2), increased SRF phosphorylation was predicted to be associated with early pressure overload disease. Phosphorylation of SRF Ser103 was increased 43% in total left ventricular extracts by acute transverse aortic constriction (Figure XIA in the Data Supplement). In contrast, both phosphorylated and total ventricular SRF were decreased in mice subjected to swim training, a model for physiologic eccentric hypertrophy (Figure XIB and Table III in the Data Supplement). The decrease in total and phosphorylated SRF was not affected by RSK3 gene deletion, that we previously showed did not prevent the hypertrophy in these exercised mice.4
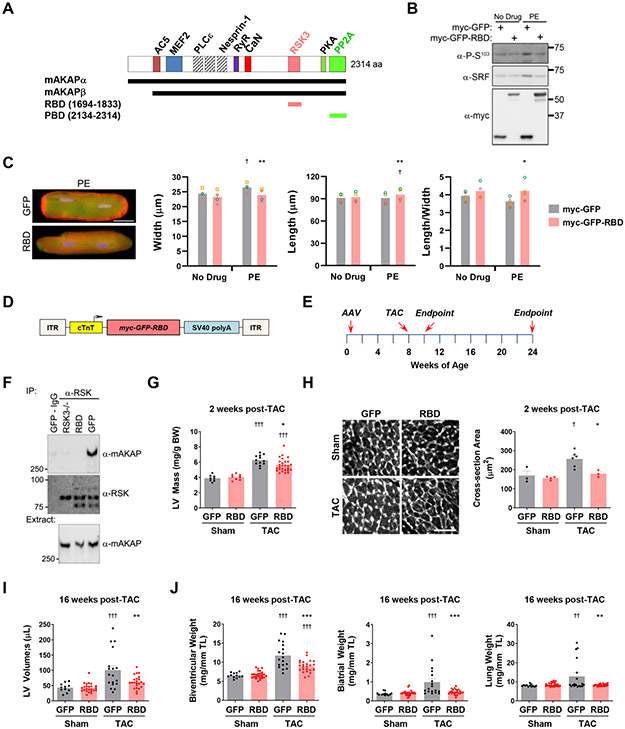
Given the induction of SRF S103 phosphorylation by pressure overload, we next considered that RSK3-mAKAPβ signalosomes might constitute a specific target for inhibition of pathological remodeling. The isoform-specific RSK3 N-terminal domain binds a discrete RSK3-Binding Domain (RBD) located at mAKAPβ residues 1694-1833 (Figure 5A).4 Expression of a myc-tagged, green fluorescent protein (GFP) RBD-fusion protein, that competes mAKAPβ-RSK3 binding,4 inhibited phenylephrine-induced SRF Ser103 phosphorylation in both neonatal and adult rat ventricular myocytes (Figure 5B and Figure XII in the Data Supplement). Accordingly, expression of the RSK3-Binding Domain anchoring disruptor peptide in adult myocytes prevented the increase in width and decrease in length:width ratio induced by phenylephrine in vitro (Figure 5C).
Figure 5. RSK3 anchoring disruption attenuates pressure overload-induced cardiac hypertrophy and heart failure.
A, mAKAP domain structure showing sites for binding partners that have been finely mapped and RBD and PBD peptides used in this project.4, 14, 27, 41-45 Muscle mAKAPβ is identical to neuronal mAKAPα but starting at residue 245. AC5 - adenylyl cyclase 5; MEF2 - myocyte enhancer factor-2; PLCε – phospholipase Cε; RyR – ryanodine receptor; CaN – calcineurin. B, Adult rat ventricular myocytes infected with adenovirus expressing myc-GFP or myc-GFP-RBD were cultured for 24 hours +/− 20 μmol/L phenylephrine (PE). n = 3. Cf. Figure XII in Data Supplement. C, Myocytes as in B were assayed by immunocytochemistry and morphometry. Anti-actinin – red, GFP and GFP-RBD in green, nuclei in blue. Bar - 25 μm. Bars and colored symbols indicate average mean and means of independent experiments using different myocyte preparations, respectively. n = 3. * vs. myc-GFP + PE; † vs. myc-GFP + no drug. D, myc-GFP-RBD (AAV9.RBD) and myc-GFP (AAV9.GFP) were expressed in mice using AAV9 and the cardiac myocyte-specific chicken troponin T promoter.24 E, Timeline for AAV9.RBD prevention studies shown in F-J. F, Co-immunoprecipitation of mAKAPβ-RSK3 complexes using heart extracts obtained from adult RSK3−/− and AAV9.RBD and AAV9.GFP injected mice and using a pan-RSK antibody (C-20). n: RSK3−/− - 5, AAV mice - 8. G, left ventricular mass (normalized by body weight) based upon M-mode echocardiography 2 weeks post-TAC. n = 8-27. H, Wheat-germ agglutinin staining 2 weeks post-TAC. Bar - 50 μm. n = 3-6. I, left ventricular volume in systole 16-weeks post-TAC. J, gravimetric analysis 16-weeks post-TAC. I,J: n = 11-24. G-J: Bars indicate mean +/− s.e.m. * vs. GFP-TAC cohort; † vs. Sham treatment for same AAV. Data for C and G-J analyzed by matched two-way ANOVA and Tukey post-hoc testing. *,† p < 0.05; **,†† p < 0.01; ***,††† p < 0.001.
These in vitro results suggested that RSK3 anchoring disruption might be beneficial in pressure overload disease in vivo. AAV9 virus were generated to express myc-GFP-RBD (AAV9.RBD) and control myc-GFP (AAV9.GFP) under the control of the cardiac myocyte-specific cardiac troponin T promoter (Figure 5D).24 Neonatal mice were injected with AAV and at 8 weeks of age subjected to pressure overload by transverse aortic constriction for 2 weeks to induce concentric hypertrophy or for 16 weeks to allow the development of heart failure with reduced ejection fraction (Figure 5E). Highly penetrant delivery of the RSK3-Binding Domain peptide to the left ventricle was confirmed by inhibition of RSK3-mAKAPβ co-immunoprecipitation (Figure 5F). 2 weeks post-TAC, AAV9.RBD-injected mice had decreased left ventricular hypertrophy (Figure 5G). Inhibited myocyte growth in width was confirmed by histological analysis of myocardium using wheat germ agglutinin staining (Figure 5H), and improvement of pathological gene expression was demonstrated by mRNA profiling (Table IV in the Data Supplement). After 16 weeks of pressure overload, control AAV9.GFP injected mice exhibited left ventricular dilation (both systolic and diastolic) and hypertrophy that were attenuated in AAV9.RBD mice (Figure 5I,J and Figure XIII and Table V in the Data Supplement). In addition, AAV9.RBD inhibited the development of atrial hypertrophy, a morphologic change that is often associated with diastolic dysfunction.25 Importantly, AAV9.RBD prevented the development of heart failure as indicated by measurement of wet lung weight, an assay for pulmonary edema. These results show that, in mice, inhibition of anchored RSK3 signaling will attenuate pathological cardiac remodeling and prevent heart failure in the context of persistent pressure overload.
We also performed a study in which mice were treated with AAV after induction of pressure overload (Figure XIVA and Table VI in the Data Supplement). AAV9.RBD treated mice had significantly decreased left ventricular posterior wall thickness 4, 8 and 12 weeks post-transverse aortic constriction surgery (Figure XIVB,C in the Data Supplement). This decreased concentric hypertrophy was accompanied by a persistently lower left ventricular mass compared to controls (Figure XIVD,E in the Data Supplement). In addition, AAV9.RBD-treated mice lacked significant development of late systolic dysfunction (Figure XIVF in the Data Supplement). Gravimetric analyses were limited by the high mortality in this study (Table VI in the Data Supplement), such that ventricular and atrial hypertrophy and wet lung weights exhibited improvement, but failed to reach significance at endpoint (e.g. wet lung weight indexed to tibial length - p = 0.09 between TAC cohorts). Histologic analysis confirmed AAV9.RBD-mediated inhibition of myocyte growth in width (Figure XIVH in the Data Supplement). Taken together, these data suggest that RSK3 anchoring disruption is capable of inhibiting the development of pathological cardiac remodeling when given as a treatment in the context of persistent pressure overload.
PP2A Anchoring Disruptor Therapy for Ischemic Heart Disease
Prolonged pressure overload in mice is associated with a transition from concentric hypertrophy to ventricular dilation, resulting in heart failure (Figure XVA in the Data Supplement).2, 26 Examination of control mouse hearts 16 weeks after transverse aortic constriction surgery demonstrated that phosphorylated SRF was suppressed 30% below that in sham-operated controls (Figure XVB in the Data Supplement). These results raised the possibility that a phosphatase opposes SRF S103 phosphorylation when stressed myocytes are not preferentially growing in width, such as during ventricular dilation and the onset of heart failure.
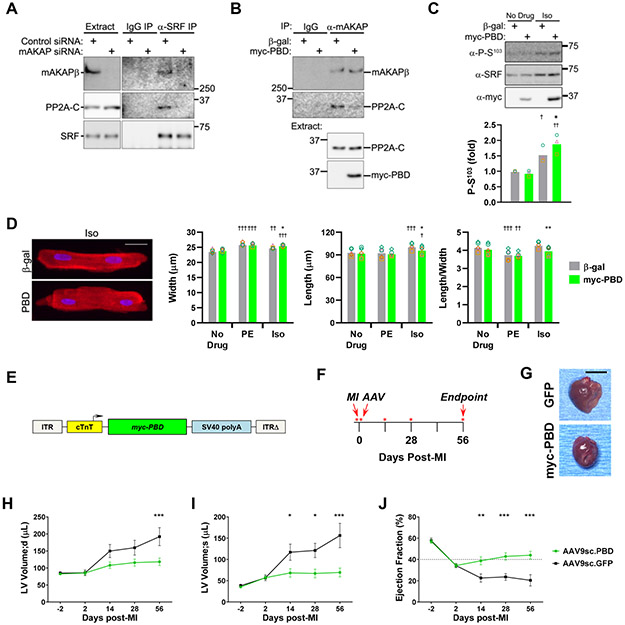
mAKAPβ binds both the Ca2+/calmodulin-dependent phosphatase calcineurin and a protein kinase A (PKA)-activated isoenzyme of PP2A that contains B56δ-subunit (PPP2R5D).14, 27 Treatment of neonatal myocytes with the PP1/PP2A inhibitor okadaic acid, but not the calcineurin inhibitor cyclosporin A, promoted phosphorylation of SRF Ser103 (Figure XVIA in the Data Supplement). Accordingly, purified PP2A dephosphorylated SRF Ser103 (Figure XVIB in the Data Supplement). Analogous to findings with RSK3, we observed that PP2A forms ternary complexes with SRF and mAKAPβ, such that SRF and PP2A co-immunoprecipitated from myocytes only in the presence of mAKAPβ (Figure 6A). PP2A binds a C-terminal domain of mAKAPβ,14 and expression of the PP2A-Binding Domain (PBD, Figure 5A) inhibited endogenous mAKAPβ-PP2A association in myocytes (Figure 6B). Consistent with our previously published finding that cAMP activates mAKAPβ-bound PP2A,14 PP2A-Binding Domain expression potentiated SRF Ser103 phosphorylation in adult myocytes stimulated with the β-adrenergic agonist isoproterenol (Figure 6C). In aggregate, these results indicate that by including both RSK3 and PP2A, mAKAPβ signalosomes can regulate SRF Ser103 phosphorylation in a bidirectional manner in response to different upstream stimuli.
Figure 6. SRF dephosphorylation by mAKAPβ-bound PP2A promotes myocyte hypertrophy with increased length:width ratio.
A, Neonatal myocytes transfected with control or mAKAP siRNA were used for co-immunoprecipitation assay of endogenous SRF-mAKAPβ-PP2A complexes. PP2A holoenzyme contains an A- and catalytic C-subunit homodimer core and a scaffolding B-subunit.14 PP2A C-subunit (PP2A-C) was detected by immunoblot. n = 3. B, Neonatal myocytes infected with adenovirus expressing myc-tagged PP2A binding domain (myc-PBD) or β-galactosidase (β-gal) before co-immunoprecipitation of endogenous mAKAPβ-PP2A complexes . n = 3. C, Adult myocytes infected with myc-PBD or β-gal adenoviruses were cultured for 24 hours +/− 10 μmol/L isoproterenol (Iso) before detection of Ser103 phosphorylated SRF. n = 3. D, Adult rat ventricular myocytes were infected with adenovirus expressing myc-PBD or β-gal control and cultured for 24 hours +/− 20 μmol/L phenylephrine (PE) or 10 μmol/L Iso before immunocytochemistry and morphometry. C,D: Bars and colored symbols indicate average mean and means of independent experiments using different myocyte preparations, respectively. n = 5. † vs. no drug condition for same protein; * vs. Iso-treated β-gal control. E, myc-PBD (AAV9sc.PBD) and myc-GFP (AAV9sc.GFP) were expressed in mice using a self-complementary AAV9 and the cardiac myocyte-specific chicken troponin T promoter.24 F, Timeline for PBD treatment study shown in G-J. Mice were randomized by echocardiography two days post-MI for i.v. treatment with AAV the following day. Echocardiography was performed on days marked by red asterisks. G, Representative whole heart pictures at endpoint. Bar - 5 mm. H-J, Serial M-mode echocardiography. n: AAV9sc.PBD – 8 (green); AAV9sc.GFP – 5 (black). Mean +/− s.e.m. indicated. * p-value for difference in cohorts at given time point. Data were analyzed by two-way ANOVA followed by Tukey (C,D) and Sidak (H-J) post-hoc testing. *,† p < 0.05; **,†† p < 0.01; ***,††† p < 0.001.
In contrast to phenylephrine, chronic stimulation of cultured adult myocytes with isoproterenol increases both myocyte length and width (Figure 6D), resulting in a symmetric hypertrophy (no change in length:width ratio), similar to the effects of chronic isoproterenol infusion in vivo that promotes cardiac dilation in addition to wall thickening.4 Displacement of PP2A phosphatase from mAKAPβ signalosomes had no effect on basal myocyte morphology, nor on the increase in myocyte width induced by phenylephrine. However, in the presence of isoproterenol, PP2A-Binding Domain expression resulted in a decreased length:width ratio, with the isoproterenol-induced increase in myocyte width and length greater and lesser, respectively, in the presence of PP2A displacement.
Given the effects of the PP2A-Binding Domain peptide on isoproterenol-induced hypertrophy in vitro, we considered that the peptide might oppose eccentric remodeling in vivo. Coronary heart disease is a leading cause of eccentric hypertrophy and heart failure with reduced ejection fraction.28 To test whether PP2A-Binding Domain expression would improve outcome after myocardial infarction, adult mice were subjected to permanent left coronary artery ligation. Two cohorts of mice to be treated with either a PP2A-Binding Domain self-complementary AAV vector (AAV9sc.PBD) or AAV9sc.GFP control were defined that had mean ejection fraction of 34% (Figure 6E,F). While control GFP mice exhibited progressively increased intraventricular volumes (both diastolic and systolic), AAV9sc.PBD treatment significantly inhibited ventricular dilation (Figure 6G-I). In addition, AAV9sc.GFP mice exhibited progressively decreased ejection fraction, while AAV9sc.PBD mice exhibited persistent restoration of ejection fraction (Figure 6J and Figure XVIIA in the Data Supplement). Moreover, AAV9sc.PBD mice had preserved anterior wall thickness and LV remodeling index (Figure XVIIB,C in the Data Supplement). These results suggest that PP2A anchoring disruptor therapy, that displaces PP2A from mAKAPβ where it would otherwise dephosphorylate SRF, will inhibit eccentric cardiac hypertrophy in ischemic heart disease.
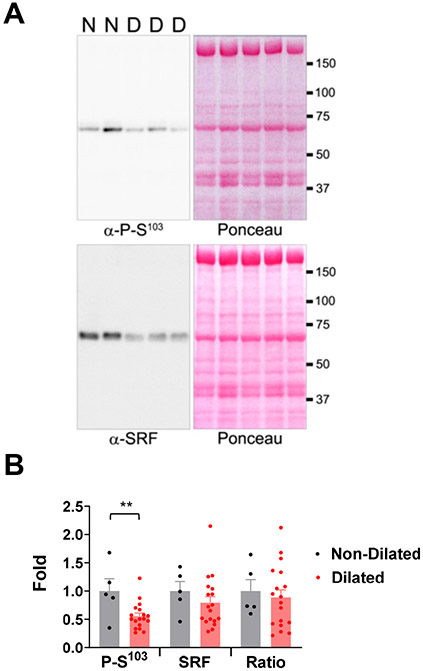
Finally, the potential relevance of these findings to human disease was assessed using patient tissue samples. When compared to left ventricular tissue from patients with normal left ventricular interior diameter, Ser103 phosphorylated SRF in patients with dilated hearts was reduced 44% (p = 0.008, Figure 7, Table VII in Data Supplement).
Figure 7. Dilated human hearts have reduced S103 phosphorylated SRF.
A, Left ventricular tissue from human patients with dilated cardiomyopathy (including non-ischemic and ischemic cardiomyopathies, n = 18, “D”) was compared with tissue from those without LV dilation (both non-dilated heart disease and normal controls, n = 5, “N”). Patient cohorts are described in Table VII in the Data Supplement. Immunoblot signals for phospho-S103 and total SRF antibodies (blots on left) were normalized by total protein content detected by Ponceau S stain (stained identical blots on right). B, Data are expressed as fold to mean for control cohort, with bars indicating mean +/− s.e.m. SRF and P- S103/SRF ratio datasets passed D’Agostino-Pearson omnibus (K2) normality testing and were compared using unpaired t-tests, while the SRF datasets were compared by Mann-Whitney test. ** p = 0.008.
Discussion
Evidence is presented here that SRF Ser103 phosphorylation, regulated by mAKAPβ signalosomes and inducing cooperative binding to a specific subset of enhancers, serves as a determinant of asymmetric myocyte hypertrophy, providing an epigenomic mechanism determining the balance between myocyte growth in width and length in pathological cardiac hypertrophy. We pursued the hypothesis that posttranscriptional modification of a critical DNA-binding transcription factor required for myocyte-specific gene expression serves to fine tune the morphology of that cell type. In contrast to the tight control of SRF expression required to maintain myocyte-specific gene expression responsible for the terminally differentiated state of the cell,11, 12 our study demonstrates that SRF Ser103 phosphorylation determines the relative growth of the myocyte along its short and long axes, with clear pathophysiological consequences (Figure 8).
Figure 8. Model for the role of SRF S103 phosphorylation in the regulation of pathological cardiac hypertrophy.
Increased width and length of the cardiac myocyte results in increased ventricular (LV – left, RV – right) wall thickness and internal diameter in pathological concentric and eccentric hypertrophy, respectively, ultimately leading to heart failure.2 Depending upon the pathophysiologic condition, pressure overloaded hearts can undergo ventricular dilation, resulting in a ventricle like in eccentric hypertrophy. The RSK3 anchoring disruptor peptide RBD, that displaces RSK3 from mAKAPβ signalosomes, will inhibit SRF phosphorylation and concentric hypertrophy. The PP2A anchoring disruptor PBD, that displaces PP2A from mAKAPβ signalosomes, will promote SRF phosphorylation and inhibit ventricular dilation and eccentric hypertrophy.
Accumulating evidence suggests that gene enhancers activated during cell type determination and differentiation participate in the mature cell in the signal-dependent fine tuning of gene expression regulating phenotype in homeostasis and disease.29 The cohort of enhancers linked to SRF Ser103 phosphorylation-dependent myocyte growth in width, referred to here as Group 1 enhancers, were enriched for binding sites for AP-1 family members. The association of AP-1 with Group 1 enhancers was surprising because other transcription factors have classically been associated with SRF-dependent gene expression.8 Ets domain ternary complex transcription factors are closely associated with stress-activated SRF-dependent gene expression.30 Ets family motifs were not, however, highly represented in any of the myocyte enhancer groups defined by phospho-Ser103 antibody ChIP-seq. In addition, a lack of enrichment of MEF2 and GATA sites in Group 1 enhancers suggests that SRF regulation of myocyte growth in width is distinct from SRF function in cardiac myocyte maturation.9
The identification of AP-1 as a co-determinant of asymmetric myocyte hypertrophy was an emergent property of SRF-focused ChIP-seq and Pro-Seq analyses, demonstrating the value of genome-wide studies. Like SRF, AP-1 family members can be activated by stress stimuli. Nuclear activity of Jun and JunB, but not Fra1, is induced rapidly in adult myocytes by α-adrenergic stimulation,31 consistent with our observations that Jun and JunB siRNA, but not Fra1 siRNA, attenuated PE-induced myocyte growth in width in vitro. Although AP1 has not previously been considered a regulator of asymmetric myocyte growth, our results are consistent with previous reports that Jun genetically-deleted mice exhibited prematurely dilated hearts when subjected to pressure overload,32 and that junb morpholinos resulted in a dilated zebrafish cardiac phenotype.33 In contrast, Atf3 apparently serves to oppose growth in width, consistent with previous observations that Atf3 knock-out exacerbated concentric hypertrophy in high-fat diet fed mice.34
SRF Ser103 phosphorylation is regulated by RSK3 and PP2A at perinuclear mAKAPβ signalosomes present in the heart only in myocytes.20 We propose that mAKAPβ-bound RSK3 is activated by pathological stress, such that RSK3 anchoring disruption inhibited phenylephrine-induced SRF phosphorylation and myocyte growth in width in vitro, as well as pressure overload-induced concentric hypertrophy in vivo. These results are consistent with our prior characterization of the RSK3 knock-out mouse.4, 5 In contrast to ERK1/2,3 that can activate RSK3,4 RSK3 and RSK3-dependent SRF phosphorylation do not seem to play a significant role in physiologic growth. Like RSK3 knock-out,4 expression of the RBD anchoring disruptor conferred no significant phenotype on unstressed mice in vivo. Expression of the dominant negative Flag-SRF S103A mutant also did not induce an in vivo phenotype in unstressed mice, only opposing phenylephrine-induced growth in width in vitro. We suggest that mAKAPβ-bound RSK3 is poised to respond to signals induced by pathologic stress that induce myocyte growth in width, such that an increase in RSK3 activity in response to Gq-coupled receptor signaling will induce phosphorylated SRF-dependent morphologic change. Accordingly, we note here that in contrast to the attenuation of TAC-induced hypertrophy by RSK3 gene targeting,4 RSK3 knock-out had no effect on the eccentric phenotype induced by myocardial infarction (Table VIII in the Data Supplement).
SRF Ser103 dephosphorylation also appears to be regulated by a signal-dependent enzyme at mAKAPβ signalosomes, and expression of the PP2A anchoring disruptor similarly had no effect on baseline myocyte morphology. mAKAPβ-bound PP2A includes the PKA-phosphorylated B56δ subunit, conferring β-adrenergic and cAMP-dependent PP2A activity in adult cardiac myocytes and explaining the selective effect of PP2A anchoring disruption on isoproterenol-treated myocytes.14, 35 Thus, by binding both a kinase and a phosphatase that can differentially control SRF S103 phosphorylation, mAKAPβ signalosomes are poised to affect the quality of remodeling that occurs in response to distinct pathological signals and stressors.
The modulation of myocyte morphology via isoform-specific targeting of downstream RSK3-PP2A-mAKAPβ-SRF signalosomes suggested new rationally-designed targeted strategies for the prevention of heart failure. Inhibition of RSK3 signaling in mice using AAV-mediated expression of the RSK3-Binding Domain anchoring disruptor inhibited pressure-overload induced concentric hypertrophy and prevented subsequent systolic dysfunction, ventricular dilation, and heart failure. Although myocyte growth in width may compensate for increased wall stress in pressure overload disease (Law of LaPlace),2 wall stress reduction may not be required for improved long term outcome.36 Blocking pathological remodeling, including concentric hypertrophy, by RSK3 inhibition early in the progression of pressure overload disease could represent an approach to prevent subsequent heart failure.2 Further justifying the consideration of RSK3 as a therapeutic target, RSK3 levels were increased in swine with concentric cardiac hypertrophy due to aortic banding (Figure XVIII in Data Supplement).
The dephosphorylation of SRF phospho-S103 by mAKAPβ-bound PP2A suggests that anchored phosphatase inhibition might be useful in pathological eccentric hypertrophy. In contrast to the dilated cardiomyopathy induced by targeting of PP2A catalytic subunit and loss of all myocyte PP2A,37 isoform-selective inhibition of PP2A by the mAKAPβ-derived anchoring disruptor was beneficial after myocardial infarction. While not tested here, PP2A anchoring disruption may also be useful late in pressure overload disease to prevent systolic dysfunction once concentric hypertrophy has ceased and SRF S103 phosphorylation is decreased. Notably, it has been reported that PKA-phosphorylated PP2A B56δ is elevated in mice subjected to chronic pressure overload35 and that PP2A subunits including B56δ are increased in expression in a canine model for heart failure.38 In addition, SRF Ser103 phosphorylation was observed to be decreased in a mouse model for familial dilated cardiomyopathy, suggesting that inhibition of mAKAPβ-bound PP2A may be broadly applicable.39
In this study, mAKAPβ-derived peptides were used to inhibit select RSK and PP2A isoenzymes. The use of AAV-expressed anchoring disruptor peptides has limitations similar to other pharmacologic agents, including potential for off-target effects. The anchoring disruptors will inhibit the binding of RSK3 and PP2A to any binding partners sharing the mAKAPβ binding interface, potentially affecting enzyme elsewhere in the myocyte and limiting conclusions concerning pathophysiologic mechanism. This includes whether targeting anchored RSK3 and PP2A might have effects under pathophysiologic conditions other than concentric and eccentric hypertrophy, respectively. However, the use of anchoring disruptor peptides allowed a first step in the translation of this mechanism. Future additional animal studies will determine whether AAV9-mediated expression of the RBD and PBD peptides should be pursued as part of a new translational pipeline for the treatment of pathological cardiac remodeling.
Supplementary Material
Clinical Perspective.
What is New?
Phosphorylation of serum response factor Ser103 promotes cardiac myocyte growth in width and concentric cardiac hypertrophy through induction of a gene transcription program in concert with AP1 family transcription factors.
Phosphorylation of serum response factor Ser103 is regulated by RSK3 and PP2A at mAKAPβ signalosomes in cardiac myocytes.
Anchoring disruptor peptides for RSK3 and PP2A modulate myocyte growth in width versus length in vitro and attenuate pathological remodeling in vivo following pressure overload and myocardial infarction in mice, respectively, both improving cardiac function.
What Are the Clinical Implications?
Inhibition of pathological cardiac remodeling can slow or block the development of heart failure, and the development of therapeutic strategies to attenuate pathological hypertrophy is compelling given the morbidity and mortality associated with heart failure despite current therapies.
The identification of a molecular mechanism regulating asymmetric cardiac myocyte growth provides a new target for the inhibition of pathological cardiac hypertrophy.
Studies in mice using adeno-associated virus mediated expression of RSK3 and PP2A anchoring disruptor peptides provide proof-of-concept that inhibition of these specific enzyme isoforms might be useful in the treatment of pathological cardiac remodeling and the prevention of heart failure.
Acknowledgments
The authors thank Dawn Bruffett, Zoharit Cozacov, and Dominica Passariello for their technical assistance in this project. We thank Bill Hulme of the Center for Genome Technology at the John P. Hussman Institute for Human Genomics, Center for Genome Technology at the University of Miami Miller School of Medicine, and the IGM Genomics Center at the University of California, San Diego for assistance with Nanostring, RNA-seq, and ChIP-seq technologies. Dr. Jinliang Li contributed to the conception of the project and performed in vitro experimentation, with additional contributions of in vitro data by H. Thakur and Dr. Dodge-Kafka; Dr. Tan performed PRO-seq and bioinformatics analyses with the assistance of K. Ohgi; Dr. Passariello and Dr. Martinez performed in vivo experimentation with additional contributions of in vivo data by Dr. Kritzer, Dr. Xueyi Li, Dr. Xiaofeng Li, Dr. Yang Li, and Dr. Yu; Dr. McArthur and Dr. Woo acquired human tissue samples and data; Dr. Emter acquired swine tissue and data with the assistance of J. Ivey; Dr. Kapiloff and Dr. Rosenfeld provided overall supervision for this project and wrote the manuscript with the assistance of the co-authors.
Sources of Funding
This work was supported by NIH Grants R01HL126825 (Dr. Kapiloff and Dr. Dodge-Kafka), R01HL146111 (Dr. Kapiloff and Dr. Dodge-Kafka), R41HL129524 (Dr. Kapiloff), R01HL075398 (Dr. Kapiloff), R01DK018477 (Dr. Rosenfeld), RO1DK039949 (Dr. Rosenfeld), R01NS034934 (Dr. Rosenfeld), R01HL089315 (Dr. Woo), R01HL112998 (Dr. Emter), T32HL094274 (Dr. Yu), and F32HL117537 (Dr. Passariello), the NHLBI Gene Therapy Resource Program, Department of Defense grant W81XWH1810178 (Dr. Kapiloff and Dr. Emter), the California Tobacco-Related Disease Research Grants Program Office of the University of California Grant 27IR-0045 (Dr. Kapiloff); and the Florida Biomedical Research Program 4KB08 (Dr. Kapiloff). Dr. Rosenfeld is an Investigator with HHMI.
Non-standard Abbreviations and Acronyms:
- AAV
adeno-associated virus
- AP1
activator protein 1
- ChIP-seq
Chromatin Immunoprecipitation Assays with Sequencing
- ERK
extracellular signal-regulated kinase
- GFP
green fluorescent protein
- Iso
isoproterenol
- H3K27Ac
histone H3 lysine 27 acetylation
- HFpEF
heart failure with preserved ejection fraction
- HFrEF
heart failure with reduced ejection fraction
- mAKAPβ
muscle A-kinase anchoring protein β
- MAP-kinase
mitogen-activated protein kinase
- MI
myocardial infarction
- PBD
PP2A-Binding Domain
- PE
phenylephrine
- Pol II
RNA polymerase II
- PRO-seq
Precision Nuclear Run-On Sequencing
- PP2A
protein phosphatase 2A
- RBD
RSK3 binding domain
- RSK3
p90 ribosomal S6 kinase type 3
- SRF
serum response factor
- TAC
transverse aortic constriction
Footnotes
Disclosures
Dr. Kapiloff, Dr. Jinliang Li, Dr. Kritzer and Dr. Passariello are inventors of patent-protected intellectual property concerning the targeting of mAKAPβ signalosomes for the treatment of heart failure, by which they, the University of Miami, and Stanford University may gain royalties from future commercialization. Dr. Kapiloff holds equity in Anchored RSK3 Inhibitors, LLC, and Cardiac RSK3 Inhibitors, LLC, companies interested in developing mAKAP signalosome-targeted therapies.
References
- 1.Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nature reviews Cardiology. 2018;15:387–407. doi: 10.1038/s41569-018-0007-y [DOI] [PubMed] [Google Scholar]
- 2.Schiattarella GG, Hill JA. Inhibition of hypertrophy is a good therapeutic strategy in ventricular pressure overload. Circulation. 2015;131:1435–1447. doi: 10.1161/CIRCULATIONAHA.115.013894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehat I, Davis J, Tiburcy M, Accornero F, Saba-El-Leil MK, Maillet M, York AJ, Lorenz JN, Zimmermann WH, Meloche S, Molkentin JD. Extracellular signal-regulated kinases 1 and 2 regulate the balance between eccentric and concentric cardiac growth. Circ Res. 2011;108:176–183. doi: 10.1161/CIRCRESAHA.110.231514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Kritzer MD, Michel JJ, Le A, Thakur H, Gayanilo M, Passariello CL, Negro A, Danial JB, Oskouei B, Sanders M, Hare JM, Hanauer A, Dodge-Kafka K, Kapiloff MS. Anchored p90 ribosomal S6 kinase 3 is required for cardiac myocyte hypertrophy. Circ Res. 2013;112:128–139. doi: 10.1161/CIRCRESAHA.112.276162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passariello CL, Martinez EC, Thakur H, Cesareo M, Li J, Kapiloff MS. RSK3 is required for concentric myocyte hypertrophy in an activated Raf1 model for Noonan syndrome. J Mol Cell Cardiol. 2016;93:98–105. doi: 10.1016/j.yjmcc.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodge-Kafka K, Gildart M, Tokarski K, Kapiloff MS. mAKAPbeta signalosomes - A nodal regulator of gene transcription associated with pathological cardiac remodeling. Cell Signal. 2019;63:109357. doi: 10.1016/j.cellsig.2019.109357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509 [DOI] [PubMed] [Google Scholar]
- 8.Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90:1274–1284. doi: 10.1038/labinvest.2010.104 [DOI] [PubMed] [Google Scholar]
- 9.Guo Y, Jardin BD, Zhou P, Sethi I, Akerberg BN, Toepfer CN, Ai Y, Li Y, Ma Q, Guatimosim S, Hu Y, Varuzhanyan G, VanDusen NJ, Zhang D, Chan DC, Yuan GC, Seidman CE, Seidman JG, Pu WT. Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nature communications. 2018;9:3837. doi: 10.1038/s41467-018-06347-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Azhar G, Furr MC, Zhong Y, Wei JY. Model of functional cardiac aging: young adult mice with mild overexpression of serum response factor. Am J Physiol Regul Integr Comp Physiol. 2003;285:R552–560. doi: 10.1152/ajpregu.00631.2002 [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Azhar G, Chai J, Sheridan P, Nagano K, Brown T, Yang J, Khrapko K, Borras AM, Lawitts J, Misra RP, Wei JY. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280:H1782–1792. doi: 10.1152/ajpheart.2001.280.4.H1782 [DOI] [PubMed] [Google Scholar]
- 12.Parlakian A, Charvet C, Escoubet B, Mericskay M, Molkentin JD, Gary-Bobo G, De Windt LJ, Ludosky MA, Paulin D, Daegelen D, Tuil D, Li Z. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112:2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778 [DOI] [PubMed] [Google Scholar]
- 13.Rivera VM, Miranti CK, Misra RP, Ginty DD, Chen RH, Blenis J, Greenberg ME. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodge-Kafka KL, Bauman A, Mayer N, Henson E, Heredia L, Ahn J, McAvoy T, Nairn AC, Kapiloff MS. cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3. J Biol Chem. 2010;285:11078–11086. doi: 10.1074/jbc.M109.034868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack CP. Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol. 2011;31:1495–1505. doi: 10.1161/ATVBAHA.110.221135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, Hipskind RA, Houthaeve T, Nordheim A, Stunnenberg HG. Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. EMBO J. 1992;11:1045–1054. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J Jr., Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kritzer MD, Li J, Passariello CL, Gayanilo M, Thakur H, Dayan J, Dodge-Kafka K, Kapiloff MS. The scaffold protein muscle A-kinase anchoring protein beta orchestrates cardiac myocyte hypertrophic signaling required for the development of heart failure. Circulation Heart failure. 2014;7:663–672. doi: 10.1161/CIRCHEARTFAILURE.114.001266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balza RO Jr., Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281:6498–6510. doi: 10.1074/jbc.M509487200 [DOI] [PubMed] [Google Scholar]
- 22.Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, Fujita K, Harada M, Toko H, Kobayashi Y, Ito K, Takimoto E, Akazawa H, Morita H, Aburatani H, Komuro I. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nature communications. 2018;9:4435. doi: 10.1038/s41467-018-06639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad KM, Xu Y, Yang Z, Acton ST, French BA. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 2011;18:43–52. doi: 10.1038/gt.2010.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Huertas-Vazquez A, Wang Y, Lusis AJ. Isoproterenol-Induced Cardiac Diastolic Dysfunction in Mice: A Systems Genetics Analysis. Front Cardiovasc Med. 2019;6:100. doi: 10.3389/fcvm.2019.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deAlmeida AC, van Oort RJ, Wehrens XH. Transverse aortic constriction in mice. J Vis Exp. 2010. doi: 10.3791/1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Negro A, Lopez J, Bauman AL, Henson E, Dodge-Kafka K, Kapiloff MS. The mAKAPbeta scaffold regulates cardiac myocyte hypertrophy via recruitment of activated calcineurin. J Mol Cell Cardiol. 2010;48:387–394. doi: 10.1016/j.yjmcc.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Writing Group M, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, American Heart Association Statistics C, Stroke Statistics S. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–360. doi: 10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- 29.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- 30.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028 [DOI] [PubMed] [Google Scholar]
- 31.Taimor G, Schluter KD, Best P, Helmig S, Piper HM. Transcription activator protein 1 mediates alpha- but not beta-adrenergic hypertrophic growth responses in adult cardiomyocytes. Am J Physiol Heart Circ Physiol. 2004;286:H2369–2375. doi: 10.1152/ajpheart.00741.2003 [DOI] [PubMed] [Google Scholar]
- 32.Windak R, Muller J, Felley A, Akhmedov A, Wagner EF, Pedrazzini T, Sumara G, Ricci R. The AP-1 transcription factor c-Jun prevents stress-imposed maladaptive remodeling of the heart. PLoS One. 2013;8:e73294. doi: 10.1371/journal.pone.0073294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meder B, Just S, Vogel B, Rudloff J, Gartner L, Dahme T, Huttner I, Zankl A, Katus HA, Rottbauer W. JunB-CBFbeta signaling is essential to maintain sarcomeric Z-disc structure and when defective leads to heart failure. J Cell Sci. 2010;123:2613–2620. doi: 10.1242/jcs.067967 [DOI] [PubMed] [Google Scholar]
- 34.Kalfon R, Koren L, Aviram S, Schwartz O, Hai T, Aronheim A. ATF3 expression in cardiomyocytes preserves homeostasis in the heart and controls peripheral glucose tolerance. Cardiovasc Res. 2017;113:134–146. doi: 10.1093/cvr/cvw228 [DOI] [PubMed] [Google Scholar]
- 35.Ranieri A, Kemp E, Burgoyne JR, Avkiran M. beta-Adrenergic regulation of cardiac type 2A protein phosphatase through phosphorylation of regulatory subunit B56delta at S573. J Mol Cell Cardiol. 2018;115:20–31. doi: 10.1016/j.yjmcc.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiattarella GG, Hill TM, Hill JA. Is Load-Induced Ventricular Hypertrophy Ever Compensatory? Circulation. 2017;136:1273–1275. doi: 10.1161/CIRCULATIONAHA.117.030730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Fang C, Xu D, Xu Y, Fu H, Li J. Cardiomyocyte specific deletion of PP2A causes cardiac hypertrophy. American journal of translational research. 2016;8:1769–1779. doi: [PMC free article] [PubMed] [Google Scholar]
- 38.DeGrande ST, Little SC, Nixon DJ, Wright P, Snyder J, Dun W, Murphy N, Kilic A, Higgins R, Binkley PF, Boyden PA, Carnes CA, Anderson ME, Hund TJ, Mohler PJ. Molecular mechanisms underlying cardiac protein phosphatase 2A regulation in heart. J Biol Chem. 2013;288:1032–1046. doi: 10.1074/jbc.M112.426957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryba DM, Li J, Cowan CL, Russell B, Wolska BM, Solaro RJ. Long-Term Biased beta-Arrestin Signaling Improves Cardiac Structure and Function in Dilated Cardiomyopathy. Circulation. 2017;135:1056–1070. doi: 10.1161/CIRCULATIONAHA.116.024482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Malik S, Kelley GG, Kapiloff MS, Smrcka AV. Phospholipase C epsilon scaffolds to muscle-specific A kinase anchoring protein (mAKAPbeta) and integrates multiple hypertrophic stimuli in cardiac myocytes. J Biol Chem. 2011;286:23012–23021. doi: 10.1074/jbc.M111.231993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapiloff MS, Piggott LA, Sadana R, Li J, Heredia LA, Henson E, Efendiev R, Dessauer CW. An adenylyl cyclase-mAKAPbeta signaling complex regulates cAMP levels in cardiac myocytes. J Biol Chem. 2009;284:23540–23546. doi: 10.1074/jbc.M109.030072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pare GC, Easlick JL, Mislow JM, McNally EM, Kapiloff MS. Nesprin-1alpha contributes to the targeting of mAKAP to the cardiac myocyte nuclear envelope. Exp Cell Res. 2005;303:388–399. doi: 10.1016/j.yexcr.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 44.Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J Cell Sci. 1999;112 ( Pt 16):2725–2736. doi: [DOI] [PubMed] [Google Scholar]
- 45.Marx SO, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang YM, Rosemblit N, Marks AR. Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treisman R Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5 [DOI] [PubMed] [Google Scholar]
- 47.Kapiloff MS, Mathis JM, Nelson CA, Lin CR, Rosenfeld MG. Calcium/calmodulin-dependent protein kinase mediates a pathway for transcriptional regulation. Proc Natl Acad Sci U S A. 1991;88:3710–3714. doi: 10.1073/pnas.88.9.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pare GC, Bauman AL, McHenry M, Michel JJ, Dodge-Kafka KL, Kapiloff MS. The mAKAP complex participates in the induction of cardiac myocyte hypertrophy by adrenergic receptor signaling. J Cell Sci. 2005;118:5637–5646. doi: 10.1242/jcs.02675 [DOI] [PubMed] [Google Scholar]
- 49.Kapiloff MS, Jackson N, Airhart N. mAKAP and the ryanodine receptor are part of a multi-component signaling complex on the cardiomyocyte nuclear envelope. J Cell Sci. 2001;114:3167–3176. doi: [DOI] [PubMed] [Google Scholar]
- 50.Johansen FE, Prywes R. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol. 1993;13:4640–4647. doi: 10.1128/mcb.13.8.4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho SN, Thomas DJ, Timmerman LA, Li X, Francke U, Crabtree GR. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J Biol Chem. 1995;270:19898–19907. doi: 10.1074/jbc.270.34.19898 [DOI] [PubMed] [Google Scholar]
- 52.Collins KA, Korcarz CE, Shroff SG, Bednarz JE, Fentzke RC, Lin H, Leiden JM, Lang RM. Accuracy of echocardiographic estimates of left ventricular mass in mice. Am J Physiol Heart Circ Physiol. 2001;280:H1954–1962. doi: 10.1152/ajpheart.2001.280.5.H1954 [DOI] [PubMed] [Google Scholar]
- 53.Ghanem A, Roll W, Hashemi T, Dewald O, Djoufack PC, Fink KB, Schrickel J, Lewalter T, Luderitz B, Tiemann K. Echocardiographic assessment of left ventricular mass in neonatal and adult mice: accuracy of different echocardiographic methods. Echocardiography. 2006;23:900–907. doi: 10.1111/j.1540-8175.2006.00323.x [DOI] [PubMed] [Google Scholar]
- 54.Nakamura A, Rokosh DG, Paccanaro M, Yee RR, Simpson PC, Grossman W, Foster E. LV systolic performance improves with development of hypertrophy after transverse aortic constriction in mice. Am J Physiol Heart Circ Physiol. 2001;281:H1104–1112. doi: 10.1152/ajpheart.2001.281.3.H1104 [DOI] [PubMed] [Google Scholar]
- 55.Tan Y, Jin C, Ma W, Hu Y, Tanasa B, Oh S, Gamliel A, Ma Q, Yao L, Zhang J, Ohgi K, Liu W, Aggarwal AK, Rosenfeld MG. Dismissal of RNA Polymerase II Underlies a Large Ligand-Induced Enhancer Decommissioning Program. Mol Cell. 2018;71:526–539 e528. doi: 10.1016/j.molcel.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 58.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 60.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basnet H, Su XB, Tan Y, Meisenhelder J, Merkurjev D, Ohgi KA, Hunter T, Pillus L, Rosenfeld MG. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516:267–271. doi: 10.1038/nature13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis J, Davis LC, Correll RN, Makarewich CA, Schwanekamp JA, Moussavi-Harami F, Wang D, York AJ, Wu H, Houser SR, Seidman CE, Seidman JG, Regnier M, Metzger JM, Wu JC, Molkentin JD. A Tension-Based Model Distinguishes Hypertrophic versus Dilated Cardiomyopathy. Cell. 2016;165:1147–1159. doi: 10.1016/j.cell.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hiemstra JA, Liu S, Ahlman MA, Schuleri KH, Lardo AC, Baines CP, Dellsperger KC, Bluemke DA, Emter CA. A new twist on an old idea: a two-dimensional speckle tracking assessment of cyclosporine as a therapeutic alternative for heart failure with preserved ejection fraction. Physiological reports. 2013;1:e00174. doi: 10.1002/phy2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hiemstra JA, Gutierrez-Aguilar M, Marshall KD, McCommis KS, Zgoda PJ, Cruz-Rivera N, Jenkins NT, Krenz M, Domeier TL, Baines CP, Emter CA. A new twist on an old idea part 2: cyclosporine preserves normal mitochondrial but not cardiomyocyte function in mini-swine with compensated heart failure. Physiological reports. 2014;2doi: 10.14814/phy2.12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.