Abstract
BACKGROUND:
For more than 16 years, we have selectively bred rats for either high or low levels of exploratory activity within a novel environment. These bred high-responder (bHR) and bred low-responder (bLR) rats model temperamental extremes, exhibiting large differences in internalizing and externalizing behaviors relevant to mood and substance use disorders.
METHODS:
We characterized persistent differences in gene expression related to bHR/bLR phenotype across development and adulthood in the hippocampus, a region critical for emotional regulation, by meta-analyzing 8 transcriptional profiling datasets (microarray and RNA sequencing) spanning 43 generations of selective breeding (postnatal day 7: n = 22; postnatal day 14: n = 49; postnatal day 21: n = 21; adult: n = 46; all male). We cross-referenced expression differences with exome sequencing within our colony to pinpoint candidates likely to mediate the effect of selective breeding on behavioral phenotype. The results were compared with hippocampal profiling from other bred rat models.
RESULTS:
Genetic and transcriptional profiling results converged to implicate multiple candidate genes, including two previously associated with metabolism and mood: Trhr and Ucp2. Results also highlighted bHR/bLR functional differences in the hippocampus, including a network essential for neurodevelopmental programming, proliferation, and differentiation, centering on Bmp4 and Mki67. Finally, we observed differential expression related to microglial activation, which is important for synaptic pruning, including 2 genes within implicated chromosomal regions: C1qa and Mfge8.
CONCLUSIONS:
These candidate genes and functional pathways may direct bHR/bLR rats along divergent developmental trajectories and promote a widely different reactivity to the environment.
Keywords: Anxiety, Bmp4, C1qa, Depression, Etv4, Hyperactivity, Impulsivity, Limbic, Mfge8, Mki67, Ncan, Neonatal, Postnatal, Trhr, Ucp2
The strong pattern of comorbidity among psychiatric disorders is believed to be generated by a spectrum of latent liability (1), arising from a complex interplay of genetic risk and environmental factors such as stress and childhood adversity (1–3). At one end of this spectrum are internalizing disorders, which are associated with neuroticism, anxiety, and depression. At the other end are externalizing disorders, which are associated with risk taking and novelty seeking, as seen in mania, substance abuse, and impulse control disorders (1).
We model the genetic contributions underlying both extremes of this spectrum by selectively breeding rats that react differently to a novel environment. Bred high-responder (bHR) rats are highly exploratory with a disinhibited, novelty-seeking temperament, including hyperactivity, aggression, and drug seeking. Bred low-responder (bLR) rats are highly inhibited, exhibiting reduced locomotor activity and anxious and depressive-like behavior (4–13). These behavioral propensities are robust and stable, beginning early in development (14,15) similar to temperament in humans (16).
This highly differentiated phenotype makes bHR/bLR rats ideal for observing the developmental programming and adult manifestation of neurological factors underlying internalizing/externalizing tendencies (8,11,17). This study focused on the hippocampus, a region important for emotional regulation, behavioral inhibition (18–20), and environmental reactivity (18), including stress-related glucocorticoid release (21,22). We previously observed bHR/bLR differences in hippocampal volume, glucocorticoid receptor and growth factor expression, histone methylation, and cell proliferation and survival (5,9,14,23,24).
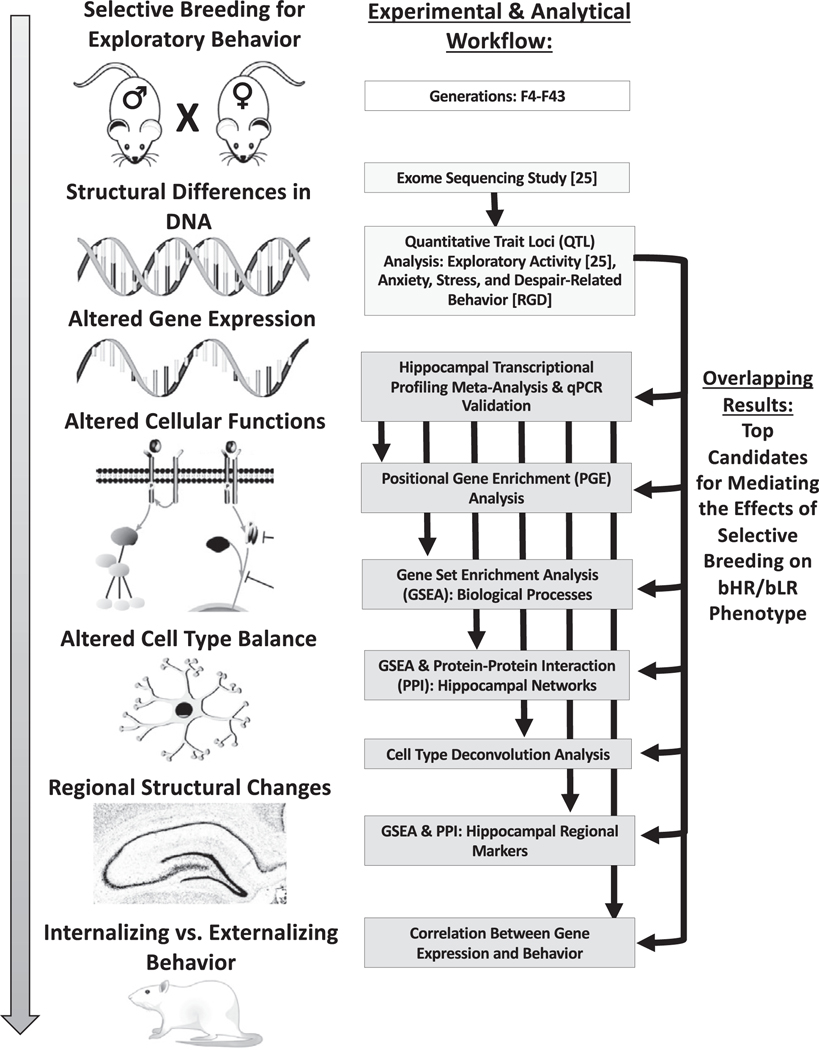
Our current study characterized hippocampal gene expression in bHR/bLR rats across development and adulthood using a meta-analysis of 8 transcriptional profiling datasets spanning 43 generations of selective breeding. Concurrently, we discovered chromosomal regions containing bHR/bLR segregating variants that are likely to contribute to exploratory locomotor phenotype (25). By comparing across these studies, we identified differentially expressed (DE) genes situated within implicated chromosomal regions and hippocampal functional pathways essential for mood and development (Figure 1). These genes are promising candidates for mediating the influence of selective breeding on behavioral phenotype.
Figure 1.
An overview of the experimental and analytical workflow used to identify top candidate genes for mediating the effects of selective breeding on bHR/bLR phenotype. Left: Many generations of selective breeding based on exploratory locomotor behavior drove segregation of genetic variants that contribute to internalizing and externalizing behavior within our bHR and bLR rats. The effect of these variants on behavior is mediated by alterations in gene expression and cellular function, which produce local changes in cell type balance and structure within brain regions responsible for affective behavior such as the hippocampus. Right: Our concurrent genetic study used exome sequencing to identify genetic variants that segregated bHR/bLR rats in our colony and then used a sampling of those variants to locate regions of the genome (QTL) that might contribute to exploratory locomotor activity (25). Our current study used meta-analyses of hippocampal transcriptional profiling studies to identify bHR/bLR DE genes, pathways, cell types, and networks in development and adulthood. In our results, we emphasize DE genes that were 1) consistently DE across multiple developmental stages, 2) central to differential expression pathways, cell types, and networks, and 3) located near genetic variants that segregated bHR/bLR rats in our colony and/or within QTL for exploratory locomotion. These genes are the top candidates for mediating the effects of selective breeding on bHR/bLR phenotype. bHR, bred high-responder; bLR, bred low-responder; DE, differentially expressed; GSEA, gene set enrichment analysis; PPI, protein–protein interaction; qPCR quantitative polymerase chain reaction; QTL, quantitative trait loci.
METHODS AND MATERIALS
Full methods for the individual experiments and analyses are provided in Supplement 1. The datasets have been released on GEO (Gene Expression Omnibus) (Table 1) and FigShare (https://doi.org/10.1101/774034). Analysis code (R Studio version 1.0.153; R version 3.2.2) is available at https://github.com/isabellie4/PhenotypeProject and https://github.com/hagenaue/HRLR_MetaAnalysisProject.
Table 1.
An Overview of the 8 Transcriptional Profiling Studies Included in Our Current Meta-analysis of Differential Gene Expression in the bHR and bLR Hippocampus at 4 Developmental Time Points: P7, P14, P21, and Adulthood
| MBNI_AffymetrixRae230_F4 | MBNI_AffymetrixRgU34A_F6 | MBNI_AffymetrixRae230_F15 | MBNI_IlluminaRatRef12v1_F15 | MBNI_RNASeq_F29 | Alabama_Nimblegen_F34 | MBNI_RNASeq_F37 | MBNI_RNASeq_F43 | |
|---|---|---|---|---|---|---|---|---|
| Generation | 4 | 6 | 15 | 15 | 29 | 34 | 37 | 43 |
| Laboratory | MBNI | MBNI | MBNI | MBNI | MBNI | Alabama | MBNI | MBNI |
| Lead Scientist | Dr. John Stead | Dr. Sarah Clinton | Dr. Sarah Clinton | Dr. Sarah Clinton | Dr. Sarah Clinton | Dr. Sarah Clinton | Dr. Peter Blandino | Dr. Cigdem Aydin |
| Age | Adult | P7, P14, P21 | P14 | P14 | P14, adult | P7, P14, P21, adult | Adult | Adult |
| n per Group | 6 | 6 | 6 | 6 | 2 | 5 | 6 | 5 |
| Platform | Affymetrix Rat Expression Set 230 A | Affymetrix Rat Genome U34A GeneChips | Affymetrix Rat Expression Set 230 A | Illumina RatRef-12 v1 BeadChip | RNA-Seq | NimbleGen Rat Gene Expression 12x135 | RNA-Seq | RNA-Seq |
| Tissue Extraction | Whole hippocampus | Whole hippocampus | Whole hippocampus | Whole hippocampus | Whole hippocampus | Dorsal hippocampus tissue punch | Whole hippocampus | Whole hippocampus |
| Treatment | Basal | Basal | Basal | Basal | Basal | Basal | Basal | Vehicle injections |
| Behavioral Testing | Basal | Basal | Basal | Basal | Basal | Basal | Locomotor, anxiety | Social interaction after mild stress |
| GEO Data Release | GSE140596 | GSE29552 | GSE140595 | GSE140594 | GSE140597 | GSE88874 | GSE140598 | GSE140287 |
| Citation | Clinton et al. (14) | Cohen et al. (28) | ||||||
See (14,28). Note that the MBNI_RNASeq_F37 dataset was collected from a subset of bHR and bLR rats included in the exome sequencing study (25).
bHR, bred high-responder; bLR, bred low-responder; GEO, Gene Expression Omnibus; MBNI, Molecular Behavioral Neuroscience Institute; P, postnatal day; RNA-Seq, RNA sequencing.
The bHR/bLR Rat Colony
All experiments were approved by the local university committee on the use and care of animals following the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Selective Breeding.
We began selectively breeding bHR/bLR rats in the Molecular Behavioral Neuroscience Institute (MBNI) at the University of Michigan in 2003 [protocol: (11)]. Later, a second colony was begun at the University of Alabama at Birmingham using generation F30 bHR/bLR rats from MBNI. Our meta-analyses use datasets derived from male bHR/bLR rats spanning generations F4 to F43. We refer to these datasets according to their respective institution, transcriptional profiling platform, and generation (Table 1). MBNI_RNA-Seq_F37 also included a bHRxbLR cross called bred intermediate-responder rats.
Behavioral Testing.
For each generation, locomotor response to a novel environment was assessed between postnatal day 50 (P50) and P75 (11). For MBNI_RNASeq_F37, we measured anxiety-like behavior in adulthood using the percentage time spent in the open arms of an elevated plus maze (5-min test) [protocol: (26)]. For MBNI_RNASeq_F43, we measured social interaction in adulthood [protocol: (4)] after 15 minutes of exposure to the anxiogenic open arms of the elevated plus maze [protocol: (27)].
Hippocampal Gene Expression Analyses
Broad Overview of the Datasets.
Our meta-analyses included 8 datasets from bHR/bLR rats aged P7, P14, P21, and adult (Table 1). The rats were housed in standard conditions with minimal intervention besides behavioral testing or saline injections, and they were sacrificed by rapid decapitation without anesthesia. The whole hippocampus was dissected except in Alabama_Nimblegen_F34, which used dorsal hippocampal tissue punches (28). The extracted RNA was profiled using microarray (earlier generations: F4, F6, F15, and F34) or RNA sequencing (RNA-Seq) (later generations: F29, F37, and F43).
Broad Overview of the Data Preprocessing.
The preprocessing steps for each study varied with platform but involved common steps, including reannotation, normalization to reduce technical variation, and quality control. Microarray data were typically summarized into log(2)-transformed expression sets using robust multiarray average (29). Genelevel RNA-Seq read count summaries were converted to log(2) fragments per million. When applicable, transcript data were averaged by gene symbol to obtain a single expression value per sample per gene.
Meta-analyses.
Within each dataset, we calculated the effect size (Cohen’s d and associated variance) of bHR/bLR phenotype on the expression data for each gene within each age group. This output was aligned across datasets using official gene symbols. The meta-analysis for each age group was performed using rma.mv() (metafor) (30) and was corrected for false discovery rate (FDR) (Benjamini–Hochberg method; multtest) (31). Including generation as a covariate provided little additional insight (generation: all genes FDR > .30).
Overlap With Hippocampal DE Genes in Other Bred Rat Models.
We cross-referenced our top findings with published results (32–39) or reanalyzed data (40) (GEO Accession No. GSE20388) from 9 publications profiling hippocampal expression in other bred rat models targeting behavioral traits that we considered extremes on the internalizing/externalizing spectrum.
Overlap With Previously Identified Genetic Variants.
Our exome sequencing study identified bHR/bLR segregating variants (single nucleotide variants) within the F37 generation (n = 12/group) and used a sampling of those variants to pinpoint quantitative trait loci (QTLs) for exploratory locomotion using a bHRxbLR F2 intercross (25). In our current study, we identified all DE genes nearby (±1 MB) segregating variants and QTL peaks (logarithm of the odds score [LOD] > 3) and determined overlap with additional QTLs relevant to bHR/bLR behavioral phenotype from the Rat Genome Database (41) (accessed August 8, 2019, using the following keywords: anxiety, stress, and despair).
Positional Gene Enrichment Analysis.
To explore which genes might be either co-regulated or in linkage disequilibrium with a causal genetic variant, we evaluated the clustering of top bHR/bLR DE genes (nominal p < .01 in P14 and adult meta-analyses; duplicated Entrez IDs removed) within chromosomal regions using positional gene enrichment (http://silico.biotoul.fr/pge) (42).
Gene Set Enrichment Analysis and Protein–Protein Interaction Networks.
To elucidate functional trends within the largest sets of results (P14 and adult), we used gene set enrichment analysis (GSEA) [(43,44); fgsea (45)] and gene set matrix files (.gmt) containing standard gene ontology for rats (http://www.bioinformatics.org/go2msig/) (46) or customized hippocampal-specific gene sets (Table S1 in Supplement 2), including previously identified coexpression modules (47,48) and expression specific to hippocampal neuronal subtypes or subregions (Hipposeq) (49). We further explored top DE genes (adult meta-analysis FDR ˂ .10) and implicated hippocampal gene sets using predicted protein–protein interaction (PPI) networks (https://string-db.org) (50).
Cell Type Data Deconvolution.
To interrogate the relative cell type composition of our samples, we used BrainInABlender methodology (51). Data for genes previously identified as having cell type–specific expression were extracted, normalized, and averaged to produce a cell type index. For this analysis, we excluded the small MBNI_RNASeq_F29 dataset (n = 2/group). We meta-analyzed the effects of bHR/bLR phenotype on these cell type indices using aforementioned methods.
Quantitative Polymerase Chain Reaction Validation.
Hippocampal tissue from bHR and bLR male rats (n = 6/group) was collected at P14 (generation F55) and P90 (generation F51) (Figure S1 in Supplement 1). Following complementary DNA synthesis, Bmp4 (bone morphogenetic protein 4) was quantified with quantitative polymerase chain reaction (qPCR) and custom-designed primers using the Livak method (52) and Gapdh as reference. Group differences in ‒ΔCq at each age were assessed using Welch’s two-sample t test (53).
RESULTS
Selective Breeding Amplifies the Propensity for Internalizing Versus Externalizing Behavior
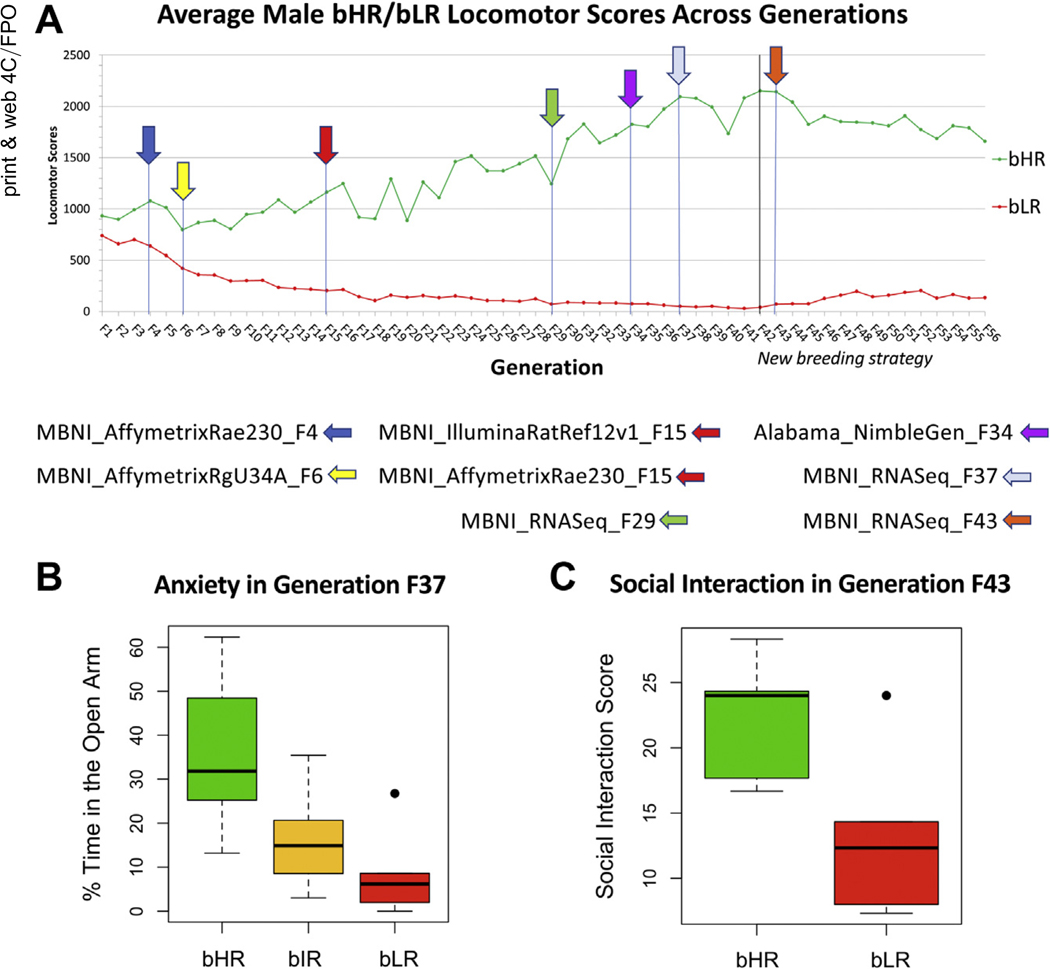
The divergence of bHR/bLR exploratory activity in response to selective breeding happened rapidly (Figure 2A), implying oligogenic inheritance (25). This divergence was accompanied by an amplification of internalizing and externalizing tendencies (11,14,54,55). For example, in behavioral data accompanying our transcriptional profiling datasets, bLRs showed more anxiety-like behavior than bHRs (Figure 2B) and spent less time interacting socially following a stressful challenge (Figure 2C). Therefore, we expected that examining gene expression across bHR/bLR generations would reveal a convergence of effects within implicated chromosomal regions and pathways essential to affective behavior and reactivity to the environment.
Figure 2.
Selectively bred bHR and bLR rats model an extreme propensity for internalizing vs. externalizing behavior. (A) Over the course of 56 generations of selective breeding (F1–F56), the bHR rats (green) have developed increasingly elevated exploratory activity in a novel field (y-axis: average total locomotor score), whereas the bLR rats (red) have become less exploratory. These trends plateaued after F42, when our breeding strategy changed to deaccelerate divergence. Arrows indicate the generations during which hippocampal transcriptomic profiling datasets were collected along with a name indicating the respective laboratory, platform, and generation for each dataset. (B) bLR rats have been highly anxious since the initiation of our breeding colony. The example above is from the behavioral data accompanying the MBNI_RNASeq_F37 transcriptomic dataset showing bLRs spending a smaller percentage of time in the anxiogenic open arms of the elevated plus maze than cross-bred (bHRxbLR) bIR rats or bHR rats (effect of phenotype: F2,15 = 6.72, p = 8.25 × 10−3; boxes = first quartile, median, and third quartile; whiskers = range or 1.5× the interquartile range; dot = outlier datapoint falling beyond the whiskers of the boxplot). (C) bLR rats are more reactive to stressors. This example is from the behavioral data accompanying the MBNI_RNASeq_F43 transcriptomic dataset showing bLR rats spending a smaller percentage of time interacting socially following exposure to a single mild stressor in the form of a brief exposure to the anxiogenic open arms of the EPM (F1,8 = 5.86, p = .0418, boxplot follows the conventions of panel B). bHR, bred high-responder; bIR, bred intermediate-responder; bLR, bred low-responder; EPM, elevated plus maze.
Selective Breeding for Exploratory Locomotion Alters Hippocampal Gene Expression
Between generations F4 and F43, we conducted 8 exploratory studies transcriptionally profiling the hippocampus of bHR/bLR rats at 4 ages (P7, P14, P21, and adult). These small studies individually produced few reliable results (Figures S2 and S3 in Supplement 1). Nevertheless, a formal meta-analysis revealed multiple genes with consistent differential expression across generations (Figure 3 and Table S2 in Supplement 2). These results can be explored interactively at https://mbni.org/dashboard/huzefak/hrlrMetaAnalysis/.
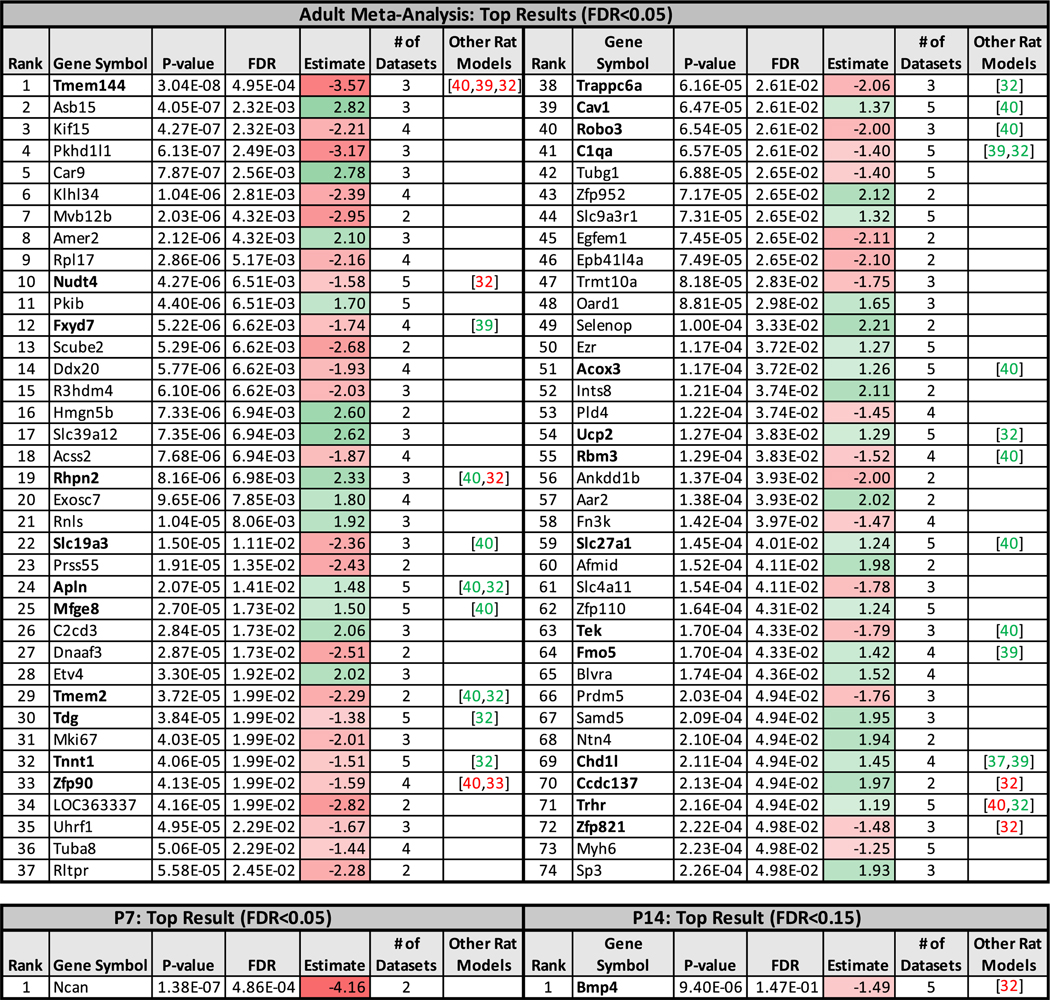
Figure 3.
The top DE genes within the bHR/bLR hippocampal gene expression meta-analyses and their convergence with DE genes identified in other bred models targeting behavioral traits that represent extremes on the internalizing/externalizing spectrum. “Estimate” shows the estimated effect size (i.e., the difference in expression between bHR and bLR rats in units of standard deviation, where green/positive = higher expression in bHRs and red/negative = higher expression in bLRs), and “# of datasets” shows the number of datasets included in the meta-analysis for that gene. Bold gene symbols show genes identified as DE in previous hippocampal transcriptional profiling studies using other bred rat models targeting behavioral traits that could be considered extremes on the internalizing/externalizing spectrum, with the specific citations provided in the “Other Rat Models” column: (32) Wistar Kyoto vs. Fischer 344; (33) Wistar more immobile vs. Wistar less immobile; (37) congenitally helpless vs. helpless resistant; (39) Flinders sensitive vs. Sprague-Dawley; (40) Flinders sensitive vs. Flinders resistant. Not included due to lack of overlapping findings with our model: (34) Roman low avoidance vs. Roman high avoidance; (35) NIH-HS high anxiety vs. NIH-HS low anxiety; (38) Syracuse low avoidance vs. Syracuse high avoidance. bHR, bred high-responder; bLR, bred low-responder; DE, differentially expressed; FDR, false discovery rate; NIH-HS, National Institutes of Health heterogeneous stock; P-value, nominal p value.
Adulthood.
The effect of bHR/bLR phenotype on gene expression was significant for 74 of 16,269 genes (FDR ˂ .05) (Figure 3). About one third of these genes (25/74) had been previously identified as DE in the hippocampus of other bred rat models related to internalizing/externalizing behaviors (32–40) (Figure 3, Table S4 in Supplement 2, and Figure S11 in Supplement 1), and 11% (8/74) had been identified by more than one study, suggesting that our findings represent a rich collection of novel and previously identified candidates that could provide broad insight into internalizing/externalizing behavior.
The estimated effect sizes (βs) were often more extreme for genes exclusively represented in RNA-Seq data from later generations (Figure S4 in Supplement 1), but owing to smaller sample size, these effects were not overrepresented in the top results. To rule out bias due to platform, we ran a meta-analysis using only recent RNA-Seq data (F37/F43) and confirmed that similar DE genes and pathways were identified (Figure S5 in Supplement 1). In the following discussion, we emphasize candidate genes that have evidence that expression diverged during the earliest generations.
Development.
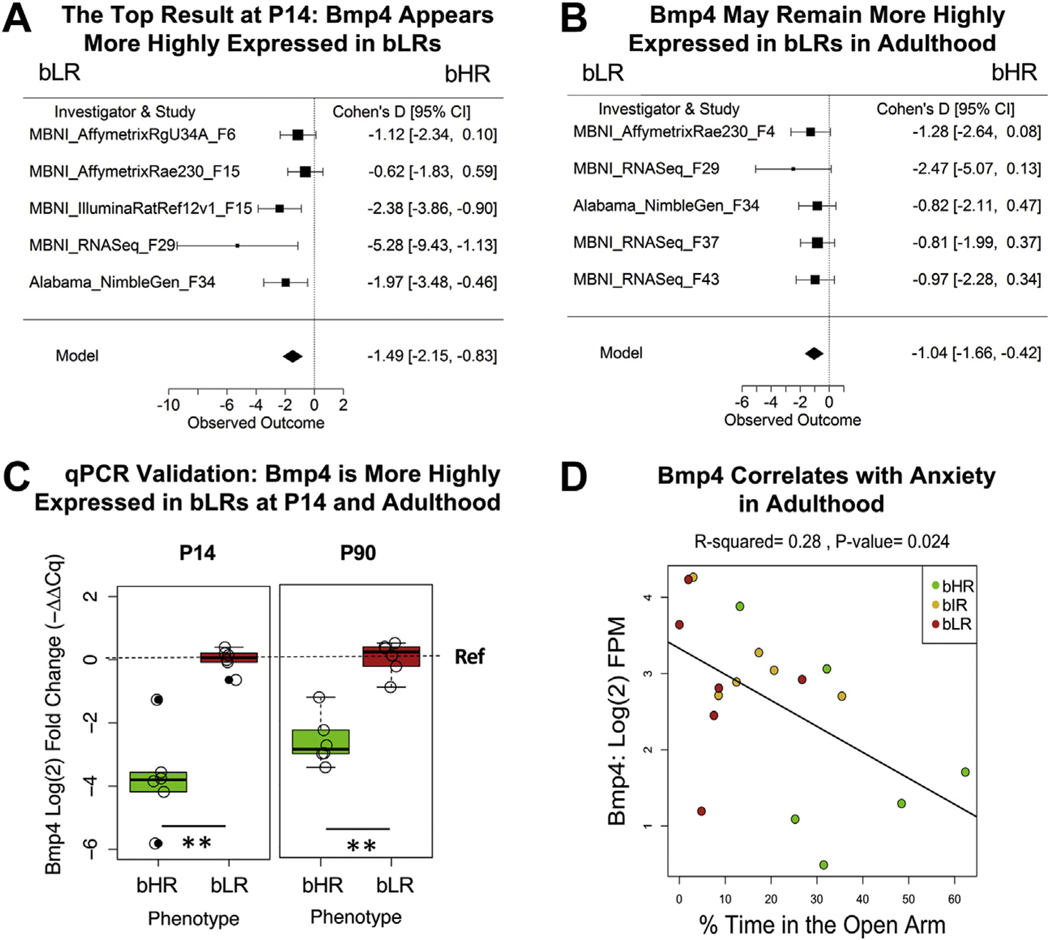
The developmental meta-analyses depended on data from earlier generations and produced less robust results. However, the top genes had consistent effects across age groups. Of 3257 genes included in the P7 meta-analysis, only Ncan (neurocan) was DE, with higher expression in bLRs than in bHRs since an early generation (Figure 4B, C) in a manner that nominally persisted at P14 (Figure 4D). Within the P14 and P21 meta-analyses, none of the effects survived multiple comparison correction (for 15,682 and 3257 genes, respectively). However, the top gene at P14, Bmp4, was consistently expressed at higher levels in bLRs than in bHRs within both the P14 (Figure 5A) and adult datasets since the F4 generation (Figure 5B). We confirmed differential expression of Bmp4 at these ages using qPCR (Figure 5C and Figure S1 in Supplement 1).
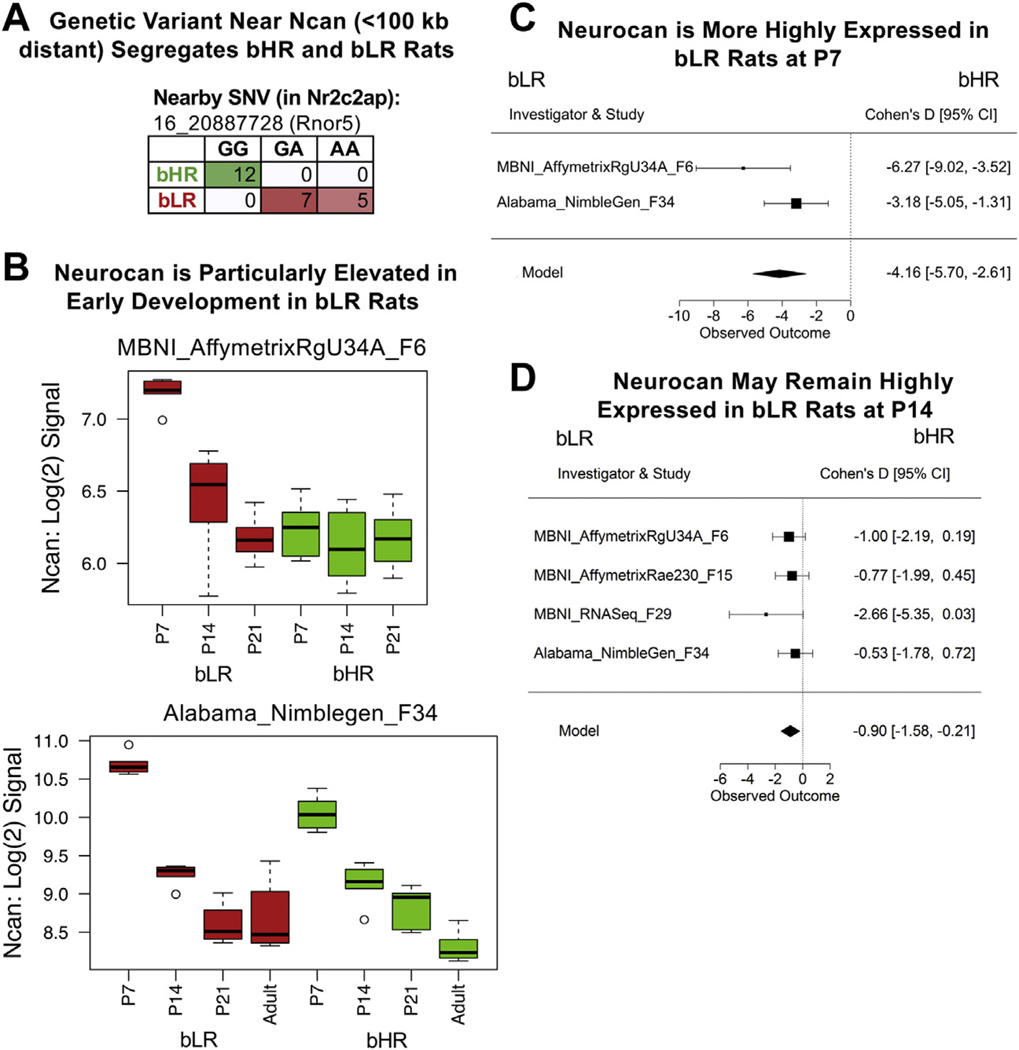
Figure 4.
The extracellular matrix constituent Ncan (neurocan) has elevated expression in bLR rats at P7. (A) A genetic variant on chromosome 16 nearby Ncan (˂100 kb) segregated bHR and bLR rats (Fisher’s exact test: p = 1.63 × 10−7). (B) Box-plots illustrating the effect of age and bHR/bLR phenotype on Ncan expression [log(2) signal] in 2 microarray studies (boxes = median and interquartile ranges; whiskers = range; red = bLR; green = bHR). The effect of phenotype is most obvious at an age when Ncan is elevated in development (P7). (C, D) Forest plots showing that Ncan was more expressed in bLRs than in bHRs (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis) in the P7 meta-analysis (β = 24.16, p = 1.38 × 10−7, FDR = 4.86 × 10−4) (C) and nominally in the P14 meta-analysis (β = 0.90, p = 1.01 × 10−2, FDR = 7.24 × 10−1) (D). bHR, bred high-responder; bLR, bred low-responder; CI, confidence interval; FDR, false discovery rate; P, postnatal day; SNV, single nucleotide variant.
Figure 5.
A regulator of proliferation and differentiation, Bmp4, is more highly expressed in bLR rats than in bHR rats at P14 and adulthood. (A, B) Two forest plots showing that Bmp4 appeared to be consistently elevated in bLR rats (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis) at P14 (β = –1.49, p = 9.40 × 10−6, FDR = 1.47 × 10−1) (A) and adulthood (β = –1.04, p = 1.01 × 10−3, FDR = 9.38 × 10−2) (B). This direction of effect mirrors findings in the literature showing that blocking the expression of Bmp4 in mice reduces anxiety and depressive-like behavior (108). (C) Using qPCR, we confirmed that bLRs showed greater Bmp4 expression than bHRs at P14 and adulthood (P90) using hippocampal tissue from later generations (F51 and F55). Log(2) FC in Bmp4 expression was calculated with the Livak method [–ΔΔCq (52)] using Gapdh as the reference housekeeping gene and bLRs as the reference group (therefore, the bLR mean is set to 0 in all panels). **p ˂ .005. (Data release: https://doi.org/10.6084/m9.figshare.10321658; P14: log(2) FC = –3.74, t5.60 = –6.10, p = .00115; P90: t8.74 = –6.87, p = 8.44 3 10-5.) Note that for both ages, the magnitude of effect in the qPCR data appears to be greater than what was observed in the meta-analysis, which may reflect differences in technology or generation. Similarly, in the qPCR data, the log(2) FC at P14 appears to be larger than the log(2) FC in adults in a manner that seems to replicate the larger effects observed in the P14 vs. adult meta-analysis, but these differences should be interpreted with caution owing to the technical factors that differed between batches. (D) Within the behavioral data accompanying the MBNI_RNASeq_F37 dataset, we found that Bmp4 showed a negative relationship with percentage time in the open arms (β = –0.034, R2 = .28, p = .024) and a positive relationship with the number of fecal boli produced on the EPM (β = 0.32, R2 = .29, p = .020; data not shown). bHR, bred high-responder; bIR, bred intermediate-responder; bLR, bred low-responder; CI, confidence interval; EPM, elevated plus maze; FC, fold change; FDR, false discovery rate; FPM, fragments per million; P, postnatal day; P-value, nominal p value; qPCR, quantitative polymerase chain reaction.
Many bHR/bLR DE Genes Are Located Within Implicated Chromosomal Regions
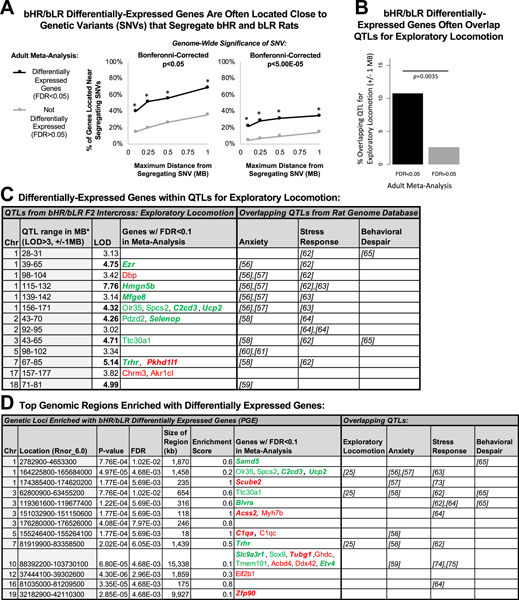
Our exome sequencing study identified bHR/bLR segregating genetic variants and then used a sampling of those variants to identify chromosomal regions that are likely to contribute to exploratory locomotor phenotype (QTL peaks). These implicated regions overlapped extensively with QTLs relevant to internalizing/externalizing behavior (41), including anxiety (56–61), stress response (62–64), and behavioral despair (65). In our current study, 68% of the DE genes in adulthood (FDR ˂ .05) were within ±1 MB of a bHR/bLR segregating variant, and 21% were within ±100 kB of a highly segregating variant (Bonferroni-corrected α = 5.00 × 10−5), a 4.7-times enrichment compared with non-DE genes (Figure 6A) (Fisher’s exact test: p = 3.50 × 10−6). DE genes were also 4.5 times more likely to be located within a QTL for exploratory locomotion (LOD > 4) (Figure 6B, C). These results fit our expectation that the influence of bHR/bLR segregating genetic variants on exploratory locomotion is at least partially mediated by effects on gene expression within the hippocampus.
Figure 6.
Many of the top DE genes are located near genetic variants that segregate bHR/bLR rats within QTLs for exploratory locomotor activity. (A) Our concurrent genetic study used exome sequencing to identify SNVs that segregated bHR/bLR rats in our colony (25). A high percentage of bHR/bLR DE genes (adult meta-analysis: FDR ˂ .05) were found within ±100, 250, or 500 kb or 1 MB of these segregating variants, using either traditional (Bonferroni-corrected p ˂ .05) or more stringent (Bonferroni-corrected p ˂ 5.00 × 10−5) criteria to define segregation. An asterisk (*) designates enrichment (Fisher’s exact test: p ˂ .0001) in comparison with the non-DE genes in our meta-analysis (FDR > .05). (B) Our concurrent genetic study used a sampling of bHR/bLR segregating variants to identify QTLs for exploratory locomotor activity in a novel field using a bHRxbLR F2 intercross (25). bHR/bLR DE genes (adult meta-analysis: FDR ˂ .05) were 4.5 times more likely to overlap (±1 MB) QTLs for exploratory locomotion than other genes included in our meta-analysis (Fisher’s exact test: p = .0035). (C) A table illustrating the top genes from our meta-analyses (FDR ˂ .10; bold + italic = FDR ˂ .05) that overlap (±1 MB) significant (LOD > 4) and putative (LOD > 3) QTLs for exploratory locomotion identified by our concurrent genetic study (25). Also depicted is overlap with QTLs identified in the Rat Genome Database (41) for the following behaviors relevant to the bHR/bLR phenotype: anxiety (56–61), stress-related responses (62–64), and behavioral despair (65). (D) The top chromosomal loci enriched for bHR/bLR DE genes overlap previously identified QTLs relevant to externalizing and internalizing behaviors. The top chromosomal loci enriched for bHR/bLR DE genes were identified using PGE analysis: location (full coordinates, defined to include the start coordinates for all genes with p ˂ .01 in the P14 or adult meta-analysis in the region), p value, FDR, and enrichment score. Also depicted is the overlap (±1 MB) of these enriched chromosomal loci with QTLs for exploratory locomotor activity (25) as well as with QTLs identified in the Rat Genome Database (41) for anxiety (56–59), stress-related responses (62–64,73–75), and behavioral despair (65). Enrichment Score: ratio of genes with p ˂ .01 out of all genes in the region. bHR, bred high-responder; bIR, bred intermediate-responder; bLR, bred low-responder; Chr, chromosome; DE, differentially expressed; FDR, false discovery rate; LOD, logarithm of the odds score; MB, megabase; PGE, positional gene enrichment; P-value, nominal p value; QTLs, quantitative trait loci; SNV, single nucleotide variant.
Positional Gene Enrichment Analysis Specifies Narrower Chromosomal Regions Contributing to bHR/bLR Phenotype
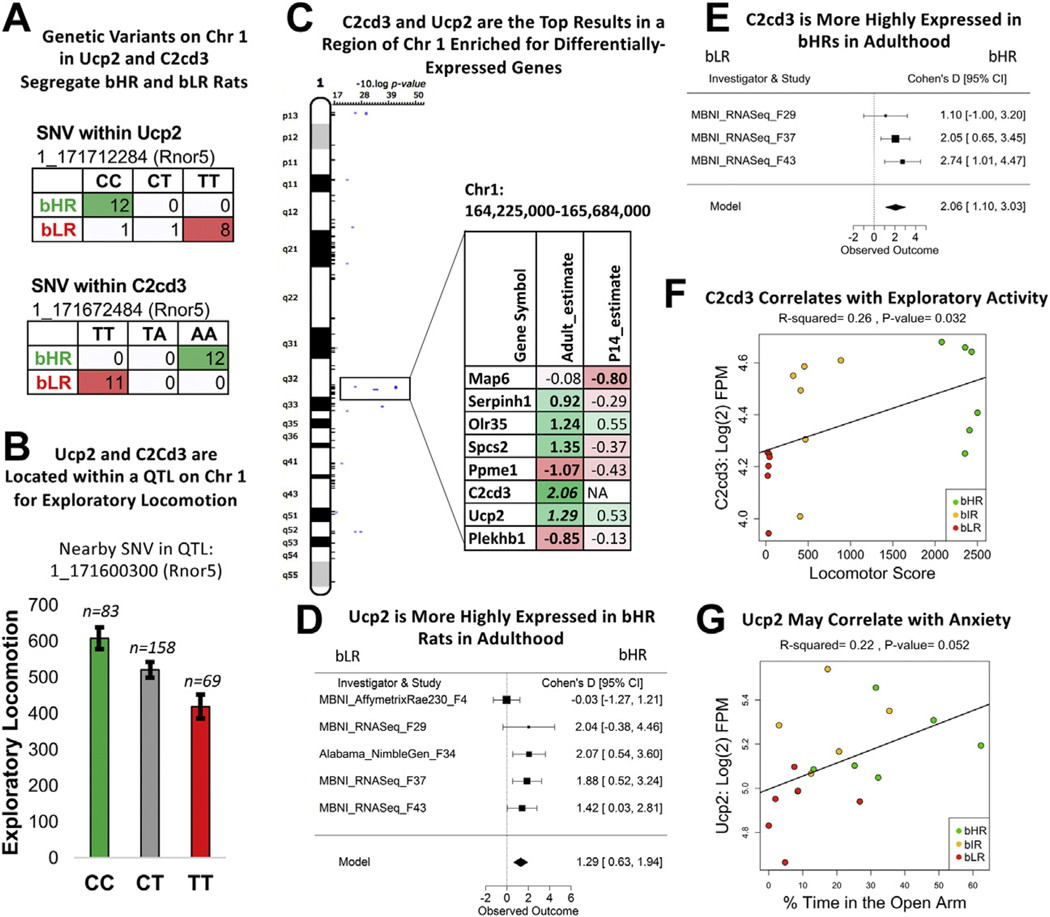
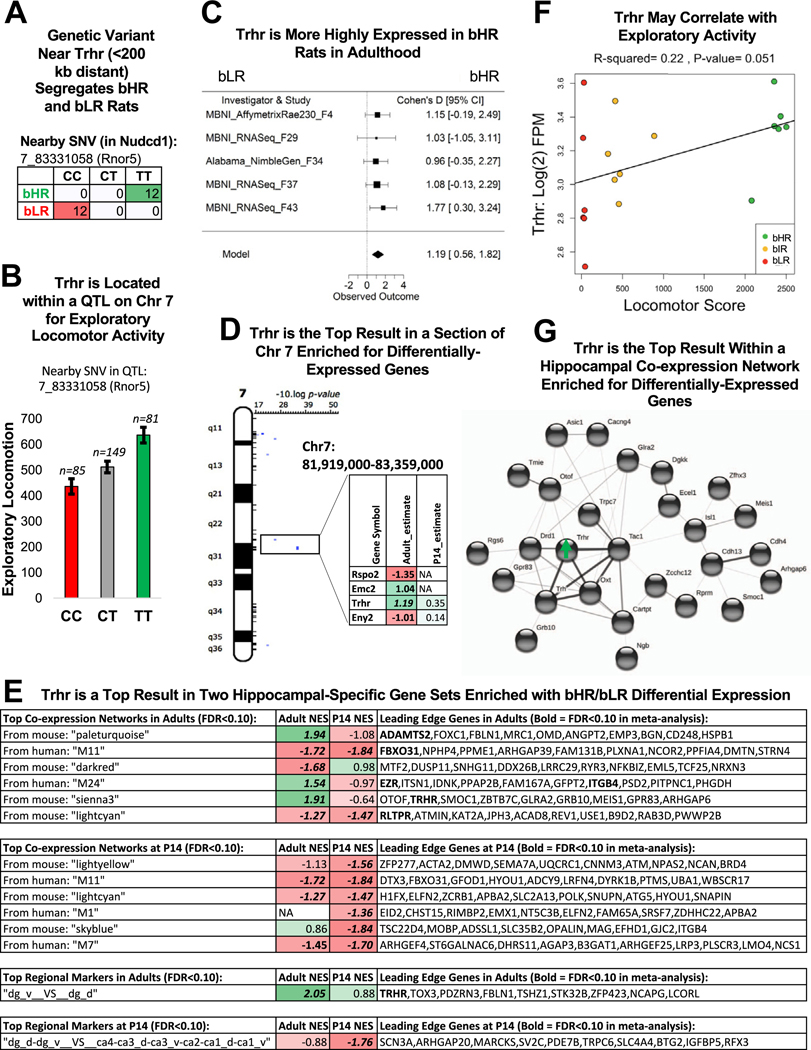
Positional gene enrichment identified 132 chromosomal regions with a significant enrichment (FDR ˂ .05) of DE genes within the P14 and adult meta-analyses. We focused on the top regions (FDR ˂ .001) (Figure 6D), many of which (10/13) could be identified using recent RNA-Seq data (F37/F43), ruling out bias toward regions overrepresented on older microarray platforms. These top loci were narrow chromosomal regions (measured in kilobases) but overlapped a high percentage of QTLs for exploratory activity [37.5%: 3/8 QTLs with LOD > 4 in (25)], encompassing three DE genes previously associated with brain function, development, and internalizing/externalizing behavior: Ucp2 (uncoupling protein 2), Trhr (thyrotropin-releasing hormone receptor), and C2cd3 (C2 calcium-dependent domain containing 3) (Figures 7 and 8) (66–72). These top loci also overlapped a surprising percentage of QTLs in the Rat Genome Database (41) for anxiety [13%: 6/45 QTLs (56–59)], stress-related responses [21%: 8/38 QTLs (62–64,73–75)], and behavioral despair [23%: 3/13 QTLs (65)]. Therefore, these enriched loci could contain genetic variants contributing to internalizing/externalizing aspects of the bHR/bLR behavioral phenotype beyond exploratory locomotion.
Figure 7.
A region on Chr 1 implicated in the bHR/bLR phenotype contains two genes important for brain function and development: Ucp2 and C2cd3. (A) Genetic variants on Chr 1 within Ucp2 and C2cd3 segregate bHR and bLR rats in our colony (Rnor5 coordinates, Fisher’s exact test: SNV 1_171712284: p = 1.66 × 10−9; SNV 1_171672484: p = 1.27 × 10−13). (B) Ucp2 and C2cd3 are located on Chr 1 within a QTL for exploratory locomotor activity. An example of the correlation between genetic variation in this region and behavior is illustrated using the sequencing results from a nearby SNV and exploratory locomotor activity measured in a bHR×bLR F2 intercross (n = 310; adjusted R2 = .049, p = 2.00 × 10−4, FDR = .0048). (C) C2cd3 and Ucp2 were the top DE genes (FDR < .05) within a segment of Chr 1 enriched for DE genes and containing a QTL for exploratory locomotor activity (25). The table illustrates the DE genes within this region. Estimate shows the estimated effect size (green/positive = higher expression in bHRs). Bold = p < .05, and bold + italic = FDR < .05. (D) A forest plot showing that Ucp2 had higher expression in bHRs in 4 of the 5 adult datasets included in the adult meta-analysis (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis; effect of phenotype: β = 1.29, p = 1.27 × 10−4, FDR = 3.83 × 10−2). This direction of effect mirrors findings in the literature showing that Ucp2 knockout mice have higher anxiety-like behavior and lower locomotor activity as well as greater sensitivity to stress (66,67,69,70), much like our bLR rats. (E) A forest plot showing that C2cd3 had higher expression in bHRs in 3 adult datasets included in the adult meta-analysis (effect of phenotype: β = 2.06, p = 2.84 × 10−5, FDR = 1.73 × 10−2). (F) In the behavioral data accompanying the MBNI_RNASeq_F37 dataset, C2cd3 [units: log(2) FPM] showed a positive relationship with exploratory locomotor activity (β = 0.000109, R2 = .26, p = 3.20 × 10−2). (G) In the behavioral data accompanying the MBNI_RNASeq_F37 dataset, Ucp2 showed a trend toward a positive relationship with percentage of time spent in the anxiogenic open arms of the EPM (β = 0.03, R2 = .22, p = 5.16 × 10−2). bHR, bred high-responder; bIR, bred intermediate-responder; bLR, bred low-responder; Chr, chromosome; CI, confidence interval; DE, differentially expressed; EPM, elevated plus maze; FDR, false discovery rate; FPM, fragments per million; QTL, quantitative trait locus; SNV, single nucleotide variant.
Figure 8.
Trhr was the top gene within 2 hippocampal-specific gene sets and within a region of Chr 7 implicated in the bHR/bLR phenotype. (A) A genetic variant on Chr 7 nearby Trhr (<200 kb distant) segregates bHR and bLR rats in our colony (Fisher’s exact test: p = 6.20 × 10−14). (B) Trhr is located on Chr 7 within a QTL for exploratory locomotor activity. An example of the correlation between genetic variation in this region and behavior is illustrated using the sequencing results from an SNV nearby Trhr (discussed above) and exploratory locomotor activity measured in a bHR×bLR F2 intercross (n = 315, adjusted R2 = .061, p = 5.79 × 10−5, FDR = .00178). (C) A forest plot showing that Trhr expression was consistently elevated in bHR rats since generation F4 (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis; β = 1.19, p = 2.16 × 10−4, FDR = 4.94 × 10−2). This direction of effect mirrors findings in the literature showing that Trhr knockout mice exhibit greater anxiety and depressive-like behavior (71). (D) Trhr was the strongest result within a segment of Chr 7 enriched for DE genes and overlapping a QTL for exploratory locomotor activity. The table illustrates the DE genes within this region (estimate = estimated effect size [green/positive = greater expression in bHRs]; bold = p < .05; bold + italic = FDR < .05). (E) Trhr was a leading gene in 2 of the top hippocampal-specific gene sets identified as enriched for bHR/bLR DE genes by GSEA (bold = p < .05 in GSEA results; bold + italic = FDR < .05 in GSEA results; NES: positive scores [green] indicate higher expression in bHRs and negative scores [red] indicate higher expression in bLRs). The top 10 leading edge genes for each gene set are shown (bold + italic = FDR < .05 in meta-analysis). These genes have large estimated effect sizes within the meta-analysis and help to drive the enrichment of effects within these gene sets. Regional marker gene sets use the following abbreviations: dg, dentate gyrus; v, ventral; d, dorsal; ca, cornu ammonis subregion. (F) In the behavioral data accompanying the MBNI_RNA-Seq_F37 dataset, Trhr [units: log(2) FPM] showed a trend toward a positive relationship with exploratory locomotor activity (β = 4.48 × 10−4, R2 = .22, p = 5.08 × 10−2). (G) Trhr and its ligand, Trh, are hub genes within a hippocampal-specific coexpression network that is enriched for bHR-upregulated genes. Genes within this network with known protein–protein interactions are illustrated above (STRINGdb: confidence setting = .15 due to hippocampal coexpression already suggesting potential interaction). Many of these genes have documented associations with reward behavior. bHR, bred high-responder; bIR, bred intermediate-responder; bLR, bred low-responder; Chr, chromosome; CI, confidence interval; DE, differentially expressed; FDR, false discovery rate; FPM, fragments per million; GSEA, gene set enrichment analysis; NA, not applicable; NES, normalized enrichment score; QTL, quantitative trait locus; SNV, single nucleotide variant.
bHR/bLR Differential Expression Is Enriched Within Hippocampal Functional Pathways
bHR/bLR Phenotype Is Associated With Proliferation and Differentiation.
In total, 8 of 2761 functional ontology gene sets were enriched with differential expression within the P14 meta-analysis (FDR ˂ .05). All were upregulated in bLRs (Figure 9A, Table S3 in Supplement 2, and Figure S6 in Supplement 1) and predominantly related to neurogenesis, differentiation, and brain development. Within the adult meta-analysis, 2 of the 4 top gene sets enriched with differential expression (FDR ˂ .10) were related to proliferation and development but were upregulated in bHRs. This pattern was confirmed using recent RNA-Seq data (F37/F43) (Table S3 in Supplement 2), ruling out bias toward gene families overrepresented on microarray platforms.
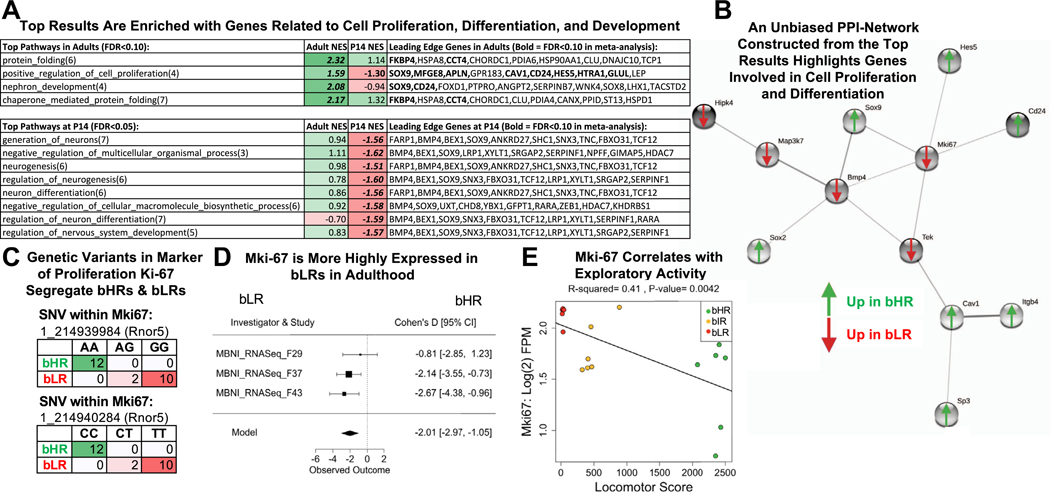
Figure 9.
The top bHR vs. bLR DE results are enriched with genes related to cell proliferation, differentiation, and development, including the canonical Mki67. (A) A table of the top functional ontology gene sets identified as enriched for bHR/bLR DE genes by GSEA (bold = p < .05 in GSEA results; bold + italic = FDR < .05 in GSEA results; NES: positive scores [green] indicate higher expression in bHRs and negative scores [red] indicate higher expression in bLRs). The top 10 leading edge genes for each gene set are shown (bold + italic = FDR < .05 in meta-analysis). These genes have large estimated effect sizes within the meta-analysis and help to drive the enrichment of effects within these gene sets. (B) A PPI network constructed using the top genes from the adult meta-analysis (192 genes with FDR < .10; STRINGdb: confidence setting = .40) had a dominant subnetwork that included Bmp4 and Mki67 as hub genes. Many of these genes are related to cell proliferation and differentiation within the brain. (C) Two genetic variants on Chr 1 within Mki67 fully segregated bHR and bLR rats in our colony (Rnor5 coordinates, Fisher’s exact test: SNV 1_214939984: p = 2.02 × 10−11; SNV 1_214940284: p = 2.02 × 10−11). (D) A forest plot showing that Mki67 was consistently elevated in bLR rats in adulthood (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis; adult: β = 22.01, p = 4.03 × 10−5, FDR = 1.99 × 10−2). (E) Within the behavioral data accompanying the MBNI_RNA-Seq_F37 dataset, Mki67 [units = log(2) FPM] showed a negative relationship with locomotor score (β = 20.000249, R2 = .41, p = .0042). bHR, bred high-responder; bLR, bred low-responder; Chr, chromosome; CI, confidence interval; DE, differentially expressed; FDR, false discovery rate; FPM, fragments per million; GSEA, gene set enrichment analysis; NES, normalized enrichment score; P, postnatal day; PPI, protein–protein interaction; SNV, single nucleotide variant.
A PPI network constructed using the top DE genes (adult meta-analysis; FDR ˂ .10; 192 genes) had a dominant subnetwork highlighting many of these same genes (Figure 9B), including hubs Bmp4 and the canonical Mki67 (marker of proliferation ki-67) (Figure 9C–E). Literature review confirmed the PPI interactions within this subnetwork and their role in proliferation and differentiation in the brain (76–88).
bHR/bLR Phenotype Is Associated With the Dentate Gyrus.
Similarly, when performing GSEA using 69 gene sets custom designed to reflect hippocampal-specific cell types and networks (Table S1 in Supplement 2), we observed an enrichment of differential expression related to the dentate gyrus (DG) (Figure 8E, Figure S7 in Supplement 1, and Table S3 in Supplement 2), the location of neural proliferation within the hippocampus. At P14, bLRs showed an upregulation of genes with enriched expression in the DG versus the cornu ammonis regions (49) (FDR ˂ .05). In adulthood, bHRs showed an upregulation of genes with enriched expression in the ventral DG versus dorsal DG (49) (FDR ˂ .05), including Trhr (FDR ˂ .05).
bHR/bLR Phenotype Is Associated With Hippocampal Coexpression Networks Related to Synaptic Signaling.
Coexpression modules can capture regionally important cell types and functions that remain undocumented in traditional ontology databases (89). We observed an enrichment of bHR/bLR effects within six previously identified hippocampal coexpression modules within the P14 meta-analysis (FDR ˂ .05) and five such modules within the adult meta-analysis (FDR ˂ .05) (Figure 8E, Figure S7 in Supplement 1, and Table S3 in Supplement 2), two of which included genes within implicated chromosomal regions.
The first was a large coexpression module (695 genes) previously identified in the mouse hippocampus (48) (named “lightcyan”), with elevated expression in bLRs relative to bHRs at P14 (FDR ˂ .05) and adulthood (FDR ˂ .10). One of the top DE genes in this module, Etv4 (ETS variant 4) (FDR ˂ .05; bHR > bLR), is a transcription factor required for proper hippocampal dendrite development (90) located within an implicated chromosomal region (Figure S8 in Supplement 1). A PPI network constructed using the DE genes in this module (n = 74; adult p ˂ .05) was enriched with genes related to cell projections, neurons, synapses, and cation binding (FDR ˂ .05).
The second was a small coexpression module (39 genes) previously identified in the mouse hippocampus (48) (named “sienna3”), with elevated expression in bHRs in adulthood (FDR ˂ .05). A PPI network constructed using all 39 genes in this module centered on Trhr and its ligand, Trh (thyrotropin-releasing hormone) (Figure 8G), and included many reward-related signaling molecules, including Cart (CART prepropeptide) (91), Oxt (oxytocin/neurophysin I prepropeptide) (92), and Drd1 (dopamine receptor D1a) (93,94).
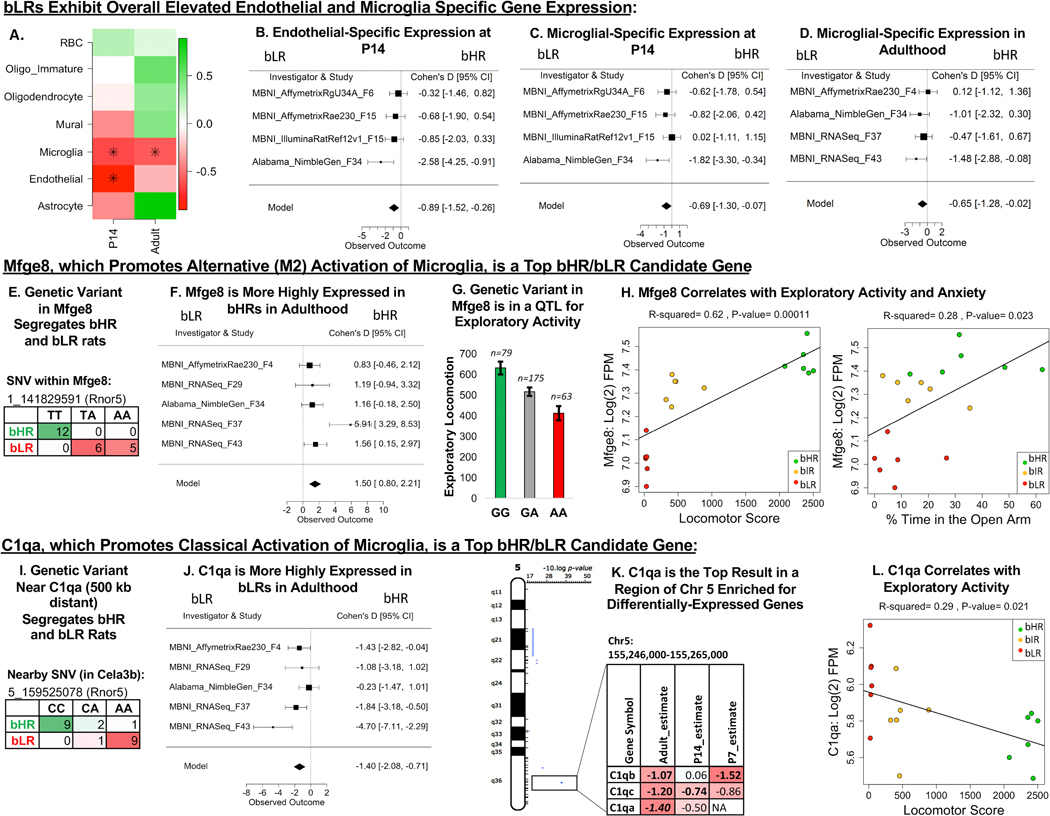
bHR/bLR Phenotype Is Associated With Microglial- and Endothelial-Specific Gene Expression.
To interrogate possible bHR/bLR differences in hippocampal cell type composition, we performed a cell type deconvolution analysis focused on well-characterized nonneuronal categories. bLRs had greater microglial-specific expression than bHRs at P14 and adulthood (Figure 10A, C, D). At P14, bLRs also showed greater endothelial-specific expression (Figure 10B). These effects could reflect differences in cell type composition or activation state. Two of the top DE genes located within implicated chromosomal regions are regulators of microglial state; Mfge8 (milk fat globule-EGF factor 8) (Figure 10E–H) promotes alternative (M2) activation (95), and C1qa (complement component C1q A chain) (Figure 10I–L) promotes classical activation (96). To interrogate less well characterized hippocampal cell types, we compared our meta-analysis results to the new mousebrain.org database (97) and found that the top DE genes (FDR ˂ .10) were highly expressed in a variety of cell types, including neuronal subcategories (Figure S9 in Supplement 1), mirroring the diversity of hippocampal functions implicated in bHR/bLR phenotype.
Figure 10.
Microglial-related gene expression differentiates bHR and bLR rats. (A) A heatmap illustrating the effect of bHR/bLR phenotype on cell type–specific gene expression, which can reflect overall cell type balance (51) or activation (green = upregulated in bHRs; red = upregulated in bLRs; an asterisk (*) indicates p < .05). Note that only well-characterized nonneuronal cell type categories were included in this analysis. (B–D) Forest plots (boxes = Cohen’s d from each study ± 95% CIs; Model = estimated effect size ± 95% CIs provided by the meta-analysis) showing an upregulation in bLRs of endothelial-specific gene expression at P14 (β = –0.89, p = 5.75 × 10−3) (B) and microglial-specific gene expression at P14 (β = –0.69, p = 2.90 × 10−2) (C) and adulthood (β = –0.65, p = 4.47 × 10−2) (D). (E) A genetic variant on Chr 1 in Mfge8 segregates bHR and bLR rats in our colony (Fisher’s exact test: p = 7.53 × 10−8). Mfge8 promotes alternative (M2) activation of microglia. (F) A forest plot illustrating that Mfge8 is more highly expressed in bHRs than in bLRs in the adult meta-analysis in all five adult datasets (β = 1.50, p = 2.70 × 10−5, FDR = 1.73 × 10−2). (G) Mfge8 is located on Chr 1 within a QTL for exploratory locomotor activity. An example of the correlation between genetic variation in this region and behavior is illustrated using the sequencing results from a nearby SNV (Rnor5 coordinates 1_141117448) and exploratory locomotor activity measured in a bHRxbLR F2 intercross (n = 317, adjusted R2 = .061, p = 1.81 × 10−5, FDR = .002). (H) Within the behavioral data accompanying the MBNI_RNASeq_F37 dataset, Mfge8 [units = log(2) FPM] showed a positive relationship with total locomotor score (β = 0.000146, R2 = .62, p = 1.10 × 10−4) as well as the percentage time in the open arms of the EPM (β = 0.00603, R2 = .28, p = 2.30 ×10−2). (I) A genetic variant on Chr 5 near C1qa (500 kb distant) segregates bHR and bLR rats in our colony (Rnor5 coordinates 5_159525078; Fisher’s exact test: p = 1.09 × 10−7). C1qa promotes classical activation of microglia via the complement cascade. (J) A forest plot illustrating that C1qa is more highly expressed in bLRs than in bHRs in all five datasets in the adult meta-analysis (β = –1.40, p = 6.57 × 10−5, FDR = 2.61 × 10−2). (K) C1qa is the top DE gene (FDR < .05) within a segment of Chr 5 enriched for DE genes. The table illustrates the DE genes within this region (estimate = estimated effect size [red/negative = higher expression in bLRs]; bold = p < .05; bold + italic = FDR < .05). (L) Within the behavioral data accompanying the MBNI_RNASeq_F37 dataset, C1qa showed a negative relationship with total locomotor score (C1qa: β = –5.17 × 10−4, R2 = .29, p = 2.09 × 10−2). bHR, bred high-responder; bLR, bred low-responder; Chr, chromosome; CI, confidence interval; EPM, elevated plus maze; FDR, false discovery rate; FPM, fragments per million; P, postnatal day; QTL, quantitative trait locus; RBC, red blood cells; SNV, single nucleotide variant.
DISCUSSION
By selectively breeding rats for more than 16 years, we produced a robust genetic model of the co-occurrence of common internalizing and externalizing behaviors. Such large differences in behavior would be expected to be accompanied by similarly strong differences in gene expression in affective circuitry. By performing a formal meta-analysis across small exploratory datasets, we provide insight into bHR/bLR differences in hippocampal gene expression across development and adulthood. Furthermore, by cross-referencing these results with our concurrent genetic study (25) and previous studies in other bred rats (32–40), we pinpoint strong candidates for mediating the influence of selective breeding on hippocampal function and internalizing/externalizing behavior.
Transcriptional Profiling Converges With Genetic Results to Identify Several Strong Candidates for Contributing to bHR/bLR Behavioral Phenotype
Our exome sequencing study offered a glimpse at the genetic factors contributing to our selectively bred phenotypes (25). However, the implicated chromosomal regions encompass several hundred genes, making their specific effects on gene expression and function difficult to predict. By cross-referencing these findings with our differential expression results, we discovered multiple strong candidates.
Two of these candidates are a compelling proof of principle; Trhr and Ucp2 are located near bHR/bLR fully segregating genetic variants (25) within narrow chromosomal regions highly enriched for DE genes, overlapping QTLs for exploratory activity (25), anxiety (56–58), and stress response (62,63). Trhr was also the top gene within a bHR-upregulated gene set associated with the ventral DG (49), a region important for proliferation and emotional regulation (18), and a hub in a bHR-upregulated hippocampal network containing reward-related signaling molecules. Trhr and Ucp2 have been extensively linked to metabolism and internalizing/externalizing behavior (98–100). Knocking out Ucp2 produces a bLR-like phenotype: higher anxiety-like behavior, lower locomotor activity, and reduced stress resilience (66,67,69,70). Trhr is an important component of the hypothalamic-pituitary-thyroid axis and regulates anxiety and depressive-like behavior (68,71,101).
In our exome sequencing study, the variants associated with Trhr and Ucp2 explained a moderate portion of exploratory locomotor behavior [˂10%, w200 locomotor counts (25)]—a magnitude akin to the bHR/bLR difference in locomotor score present in the F1 generation. Altogether, this evidence suggests that bHR/bLR segregating genetic variants drive differential expression of Ucp2 and Trhr in a manner that could meaningfully contribute to bHR/bLR differences in hippocampal function and internalizing/externalizing behavior. Because other novel candidates were associated with variants explaining a similarly promising portion of exploratory behavior in our exome sequencing study, our results offer new avenues for investigation.
Genes Highlighted in the Developmental Meta-analyses Suggest That bHR/bLR Differences in Hippocampal Structure Arise Early in Development
The different propensity of bHR/bLR rats toward externalizing or internalizing behavior is evident at a young age (14,15), paralleling hippocampal development (14). Our meta-analyses encompassed three postnatal ages (P7, P14, and P21) to provide insight into this neurodevelopmental trajectory. These meta-analyses depended on data from earlier generations and older platforms, yet they identified two strong candidates with clear associations with internalizing/externalizing behavior and hippocampal development.
The top P7 result was Ncan, exhibiting a strikingly large effect size (bHR < bLR) as early as generation F6. Ncan is located adjacent to a bHR/bLR segregating variant (25), overlapping a QTL for despair-related behavior (65). As part of the extracellular matrix, Ncan is upregulated during early brain development (102,103) and modulates cell adhesion, migration, and growth factor binding (104). Ncan has been linked to bipolar disorder (102), and knocking out Ncan enhances locomotor activity, risk taking, hedonia, and amphetamine hypersensitivity (103). Therefore, lower levels of Ncan in early development in bHRs could promote externalizing behavior as well as divergent hippocampal development.
The top P14 result was Bmp4, which was elevated in bLRs since the earliest generations in a manner that appeared to persist into adulthood. As a regulator of development, Bmp4 is important for neural induction (105,106), but later it suppresses neurogenesis (107–111) and promotes other cell fates (88,105,112). Bmp4 was an important driver in bHR/bLR-enriched gene sets related to proliferation and differentiation at P14 and was central within a related PPI network constructed from top adult DE genes. In the hippocampus, Bmp signaling promotes dorsal cell type identity and is essential for DG formation (113), matching our results indicating bLR enrichment of DG-related gene expression at P14 and adulthood.
Blocking Bmp signaling produces a bHR-like phenotype, reducing anxiety, fear conditioning, and depressive-like behavior (108,113). Therefore, Bmp4 is a strong candidate for driving long-term hippocampal structural differences capable of producing stable divergence in temperament. However, Bmp4 was not located near a bHR/bLR segregating variant in our exome sequencing study (25), implying that impactful variation is located in a nearby noncoding region or within a gene upstream in Bmp4’s signaling pathway.
Functional Analyses Implicate Hippocampal Proliferation and Differentiation in bHR/bLR Phenotype
A prominent theme in our results were functions related to cell proliferation and differentiation. Indeed, Mki67 itself contained two bHR/bLR segregating genetic variants and was more highly expressed in bLRs in adulthood (FDR < .05), matching the upregulation in bLRs observed histologically in development (14) and maybe adulthood (9,114). These findings confirm that the relationship between internalizing behavior and cell proliferation in our model is not a general stunting of growth-related processes, contrasting with the neurotrophic model of stress-related mood disorders (115).
Many of our top DE genes were also important regulators of cell fate. Bmp4, Sox9 (SRY-box 9), Sox2 (SRY-box 2), Hes5 (Hes family BHLH transcription factor 5), Cd24 (CD24 molecule), and Tek (TEK receptor tyrosine kinase) regulate functions including the developmental progression of neural differentiation, gliogenesis, and endothelial proliferation (76,84,85,87,116–118). Their role in adulthood includes growth and plasticity in response to neural activity and injury (79,119–121). Therefore, these results could explain previous morphological findings indicating that cell differentiation progressed differently in the adult hippocampus in bHRs and bLRs under mild stress (9). Together, these findings raise the interesting possibility that differential expression within neurodevelopmental programming pathways could provide a general mechanism by which environmental stimuli, such as stress and drugs, produce divergent changes in hippocampal structure in bHR/bLR animals.
Functional Analyses Implicate Microglial Activation in bHR/bLR Phenotype
Microglial-specific gene expression was upregulated at both P14 and adulthood in bLRs, suggesting either an increased proportion of microglia cells or microglial activation. Several top candidates were important regulators of microglial function. bLRs had greater expression of C1qa than bHRs since the earliest generations. The C1q genes promote classical microglial activation (96) and mediate phagocytosis-driven synaptic pruning (96,122,123). In contrast, Mfge8 had greater expression in bHRs. Mfge8 is associated with reduced proinflammatory factors (124) as well as alternative (M2) microglial activation (95), playing an important role in phagocytosis (95,125,126). Ucp2 also has anti-inflammatory functions (66,67,69,127,128) and was more highly expressed in bHRs than in bLRs. Both Mfge8 and Ucp2 contain bHR/bLR segregating genetic variants within probable QTLs for exploratory activity (25), suggesting that genetic variation could contribute to their differential expression in the hippocampus.
Together, these results seem to fit proinflammatory theories of internalizing disorders (129). However, we found little evidence of bHR/bLR differences in traditional inflammatory markers, although protein or serological measures could reveal otherwise. Given our other findings, we speculate that bHR/bLR differences in microglial-related expression are likely tied to nonimmune microglial functions, including the regulation of neurogenesis, cell survival (130), and synaptic pruning (123,131) in response to neuronal activity (123). If true, microglial phagocytosis could serve as a multifaceted tool to tailor plasticity during development or in response to environmental stimuli such as stress and drugs of abuse.
Conclusions and Future Directions
By comparing exome sequencing findings with hippocampal differential expression patterns during development and adulthood across many generations of selective breeding in our bHR/bLR colony, we implicate a diverse and compelling array of genes whose effects may converge to promote internalizing versus externalizing behavior. Owing to our dependence on older platforms and exclusively male rats, we cannot claim to have identified all relevant candidates, nor have we highlighted all promising results in our text. Similarly, by combining results from diverse experimental designs, we may have decreased our statistical power while increasing the generalizability of our results. That said, our results converge with other bred rat models to identify promising candidate genes and implicate two functional pathways that could guide bHRs and bLRs along a divergent developmental trajectory, setting the stage for a widely different reactivity to the environment. These findings inspire new avenues of research (132–134), including cell type–specific morphological analyses and the manipulation of candidate pathways to assess relevance to internalizing/externalizing behavioral and neurological phenotypes.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Commercial Assay Or Kit | Affymetrix Rat Expression Set 230 A Microarray Chip | Affymetrix (now ThermoFisher, Waltham, MA) | ||
| Commercial Assay Or Kit | Illumina RatRef- 12v1 Beadchip | Illumina, San Diego, CA | ||
| Commercial Assay Or Kit | TRIzol reagent | Invitrogen, Calsbad, CA | Catalog #15596018 | |
| Commercial Assay Or Kit | RNeasy RNA purification columns | Qiagen (sold via ThermoFisher, Waltham, MA), Valencia, CA | ||
| Commercial Assay Or Kit | TruSeq library preparation kit | Illumina, San Diego, CA | ||
| Commercial Assay Or Kit | AllPrep DNA RNA miRNA Universal Kit 50 | Qiagen, Valencia, CA | Catalog #: 80224 | |
| Commercial Assay Or Kit | Illumina TruSeq Stranded mRNA Library Preparation kit | Illumina, San Diego, CA | Catalog #s RS-122-2101, RS-122-2102 | |
| Commercial Assay Or Kit | Kapa’s library quantification kit for Illumina Sequencing platforms | Kapa Biosystems,Wilmington MA | Catalog # KK4835 | |
| Commercial Assay Or Kit | QIAshredder kit | Qiagen, Valencia, CA | Catalog #79654 | |
| Commercial Assay Or Kit | RNeasy Mini Kit | Qiagen, Valencia, CA | Catalog #74104 | |
| Commercial Assay Or Kit | iScript cDNA Synthesis Kit | Biorad, Hercules, CA | Catalog #1708891 | |
| Commercial Assay Or Kit | iQTM SYBR® Green Supermix | Biorad, Hercules, CA | ||
| Deposited Data; Public Database | MBNI_AffymetrixRae230_F4 Dataset | Gene Expression Omnibus, ref: this paper | GSE140596 | |
| Deposited Data; Public Database | MBNI_AffymetrixRgU34A_F6 Dataset | Gene Expression Omnibus, ref: PMID: 21864320 | GSE29552 | |
| Deposited Data; Public Database | MBNI_AffymetrixRae230_F15 Dataset | Gene Expression Omnibus, ref: this paper | GSE140595 | |
| Deposited Data; Public Database | MBNI_IlluminaRatRef12v1_F15 Dataset | Gene Expression Omnibus, ref: this paper | GSE140594 | |
| Deposited Data; Public Database | MBNI_RNASeq_F29 Dataset | Gene Expression Omnibus, ref: this paper | GSE140597 | |
| Deposited Data; Public Database | Alabama_Nimblegen_F34 Dataset | Gene Expression Omnibus, ref: PMID: 25791846 | GSE88874 | |
| Deposited Data; Public Database | MBNI_RNASeq_F37 Dataset | Gene Expression Omnibus, ref: this paper | GSE140598 | |
| Deposited Data; Public Database | MBNI_RNASeq_F43 Dataset | Gene Expression Omnibus, ref: this paper | GSE140287 | |
| Deposited Data; Public Database | Flinders Sensitive Line vs. Flinders Resistant Line Affymetrix Rat Expression Set 230 Microarray Dataset | Gene Expression Omnibus, ref: PMID: 20830301 | GSE20388 | |
| Deposited Data; Public Database | bHR vs. bLR Bmp4 qPCR Dataset | FigShare, ref: this paper | https://doi.org/10.1101/774034 | |
| Deposited Data; Public Database | bHR vs. bLR Exome Sequencing Dataset | NCBI Bioproject, https://www.ncbi.nlm.nih.gov/bioproject/, ref: PMID: 31182603 | PRJNA521139 | |
| Deposited Data; Public Database | Rat Genome Database | https://rgd.mcw.edu, ref: PMID: 25355511 | accessed 08/08/2019, RRID:SCR_006444 | |
| Deposited Data; Public Database | GO2MSIG | http://www.bioinformatics.org/go2msig/, ref: PMID: 24884810 | accessed 6/2017, RRID:SCR_018359 | |
| Deposited Data; Public Database | Hipposeq | https://hipposeq.janelia.org, ref: PMID: 27113915 | accessed 05/11/2018, RRID:SCR_015730 | |
| Deposited Data; Public Database | string-db | https://string-db.org/cgi/input.pl, ref: PMID: 25352553 | accessed 04/2018, RRID:SCR_005223 | |
| Deposited Data; Public Database | Interactive web dashboard for viewing bHR vs. bLR differential expression | https://mbni.org/dashboard/huzefak/hrlrMetaAnalysis/, ref: this paper | ||
| Deposited Data; Public Database | Custom chip definition files (.cdf) | http://nmg-r.bioinformatics.nl/NuGO_R.html, http://mbni.org/customcdf/22.0.0/entrezg.download/ , ref: PMID: 16284200 | rae230arnentrezgcdf_19.0.0, rgu34arnentrezgcdr_19.0.0, rat2302rnentrezgcdf_22.0.0 | |
| Deposited Data; Public Database | R package org.Rn.eg.db | http://bioconductor.org/packages/org.Rn.eg.db/ | version 3.4.1, RRID:SCR_018358 | |
| Deposited Data; Public Database | NCBI Genome Remapping Service | https://www.ncbi.nlm.nih.gov/genome/tools/remap | accessed 8/8/2019 | |
| Deposited Data; Public Database | mousebrain.org | mousebrain.org, ref: PMID: 30096314 | accessed 04/09/2018, RRID:SCR_018356 | |
| Deposited Data; Public Database | Pubmed | https://www.ncbi.nlm.nih.gov/pubmed/ | ||
| Organism/Strain | Selectively-bred High Responder rats, species: Rattus Norvegicus, sex: male | Laboratory of Dr. Huda Akil & Dr. Stanley Watson Jr., ref: PMID: 16502134, this paper | ||
| Organism/Strain | Selectively-bred Low Responder rats, species: Rattus Norvegicus, sex: male | Laboratory of Dr. Huda Akil & Dr. Stanley Watson Jr., ref: PMID: 16502134, this paper | ||
| Sequence-Based Reagent | Custom-designed qPCR primers targeting Bmp4 (Rattus Norvegicus) | Laboratory of Dr. Huda Akil & Dr. Stanley Watson Jr., ref: this paper | ACC# NM_012827.2; forward primer: 5’-CCCTGGTCAACTCCGTTAAT-3’, start = 1214; reverse primer: 5’-AACACCACCTTGTCGTACTC-3’, start = 1319 | |
| Sequence-Based Reagent | Custom-designed qPCR primers targeting Gapdh (Rattus Norvegicus) | Laboratory of Dr. Huda Akil & Dr. Stanley Watson Jr., ref: this paper | ACC# NM_017008.4; forward primer: 5’- GTTTGTGATGGGTGTGAACC-3’, start = 459; reverse primer: 5’-TCTTCTGAGTGGCAGTGATG-3’, start = 628 | |
| Software; Algorithm | Noldus Ethovision | Noldus, Leesburg, VA | ||
| Software; Algorithm | R Studio | CRAN: The Comprehensive R Archive Network, https://cran.r-project.org | v.1.0.153 | |
| Software; Algorithm | R | CRAN: The Comprehensive R Archive Network, https://cran.r-project.org | v.3.2.2 | |
| Software; Algorithm | All code used to analyze the genomics data in this paper | Github Repository: https://github.com/isabellie4/PhenotypeProject | ||
| Software; Algorithm | All code used to analyze the genomics data in this paper | Github Repository: https://github.com/hagenaue/bHRbLR_MetaAnalysisProject | ||
| Software; Algorithm | Robust multi-array average (RMA) | ref: PMID: 12582260 | ||
| Software; Algorithm | R package affy | https://www.bioconductor.org/packages/release/bioc/html/affy.html, ref: PMID: 14960456 | version 1.54.0, RRID:SCR_012835 | |
| Software; Algorithm | R package metafor | https://cran.r-project.org/web/packages/metafor/index.html, ref: DOI: 10.18637/jss.v036.i03 | version 2.0-0, RRID:SCR_003450 | |
| Software; Algorithm | R package multtest | https://www.bioconductor.org/packages/release/bioc/html/multtest.html, ref: DOI: 10.1007/0-387-29362-0_15 | version 2.32.0, RRID:SCR_001255 | |
| Software; Algorithm | Positional Gene Enrichment (PGE) Analysis | http://silico.biotoul.fr/pge/ , ref: PMID: 18346969 | RRID:SCR_018360 | |
| Software; Algorithm | R package fgsea | https://bioconductor.org/packages/release/bioc/html/fgsea.html, ref: DOI: 10.1101/060012 | version 1.2.1 | |
| Software; Algorithm | R package BrainInABlender | https://github.com/hagenaue/BrainInABlender, ref: PMID: 30016334 | version 0.0.0.9000 | |
| Software; Algorithm | R package limma | https://bioconductor.org/packages/release/bioc/html/limma.html, ref: PMID: 25605792 | version 3.28.21, RRID:SCR_010943 | |
| Software; Algorithm | R package preprocessCore | https://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html, ref: S8 | version 1.34.0 | |
| Software; Algorithm | Tophat2 | https://ccb.jhu.edu/software/tophat/index.shtml, ref: PMID: 23618408 | RRID:SCR_013035 | |
| Software; Algorithm | Bowtie2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml, ref: PMID: 22388286 | RRID:SCR_005476 | |
| Software; Algorithm | Cufflinks | http://cole-trapnell-lab.github.io/cufflinks/, ref: PMID: 22383036 | RRID:SCR_014597 | |
| Software; Algorithm | CuffDiff | http://cole-trapnell-lab.github.io/cufflinks/, ref: PMID: 22383036 | RRID:SCR_001647 | |
| Software; Algorithm | Cuffmerge | https://www.genepattern.org/modules/docs/Cuffmerge, ref: PMID: 22383036 | RRID:SCR_015688 | |
| Software; Algorithm | SubRead aligner | http://subread.sourceforge.net, ref: PMID: 23558742 | RRID:SCR_009803 | |
| Software; Algorithm | featureCounts | http://subread.sourceforge.net, ref: PMID: 24227677 | RRID:SCR_012919 | |
| Software; Algorithm | R package compute.es | https://cran.r-project.org/web/packages/compute.es/index.html | version 0.2-2 | |
| Software; Algorithm | R package GEOQuery | https://www.bioconductor.org/packages/release/bioc/html/GEOquery.html, ref: PMID: 17496320 | version 2.42.0, RRID:SCR_000146 | |
| Other | The Genomic Services Laboratory at HudsonAlpha | (now called HudsonAlpha Discovery), Huntsville, AL, https://gsl.hudsonalpha.org/index | ||
| Other | University of Michigan DNA Sequencing Core | Ann Arbor, MI, https://seqcore.brcf.med.umich.edu |
ACKNOWLEDGMENTS AND DISCLOSURES
The work that was performed at the Molecular Behavioral Neuroscience Institute was supported by grants from the Hope for Depression Research Foundation (RGA No. DTF Phase II [D] [principal investigator, Bruce S. McEwen, now deceased]), Pritzker Neuropsychiatric Disorders Research Consortium [to HA and SJW], National Institute on Drug Abuse (Grant Nos. 5P01 DA021633-02 and 5RO1 DA013386 [to HA]), Office of Naval Research (Grant Nos. N00014-12-1-0366, N00014-09-1-0598, and N00014-02-1-0879 [to HA]), National Institute of Mental Health (NIMH) Conte Center (Grant No, L99MH60398 [to HA]), and NIMH Program Project (Grant No. P01MH42251-01 [to HA]). The University of Alabama study was funded by the NIMH (Grant No. 4R00MH085859-02 [to SMC]). Research training for IAB was supported by the Undergraduate Research Opportunity Program at the University of Michigan.
We acknowledge everyone involved in the studies included in the meta-analysis: James Stewart, Angela Koelsch, Sharon Burke, Mary Hoverstein, Jennifer Fitzpatrick, Hui Li, Fei Li, and Amy Tang. We also acknowledge the University of Michigan Advanced Genomics Core and the HudsonAlpha Institute for Biotechnology Genomics Services Lab. Finally, we thank everyone who provided feedback on the manuscript: Dr. Shelly Flagel, Alek Pankonin, Pouya Mandi Gholami, Dr. Aram Parsegian, and Leah Johnson, as well as Dr. Albert Fernandez Teruel, whose careful review of our manuscript inspired the cross-comparison of our results with other bred models.
This article was published as a preprint on bioRxiv: doi: https://doi.org/10.1101/774034.
MHH, RCT, FM, HA, and SJW are members of the Pritzker Neuropsychiatric Disorders Research Consortium, which is supported by Pritzker Neuropsychiatric Disorders Research Fund. A shared intellectual property agreement exists between the academic and philanthropic entities of the consortium. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2020.05.024.
Contributor Information
Isabelle A. Birt, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
Megan H. Hagenauer, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
Sarah M. Clinton, School of Neuroscience, Virginia Tech, Blacksburg, Virginia
Cigdem Aydin, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
Peter Blandino, Jr., Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
John D.H. Stead, Department of Neuroscience, Carleton University, Ottawa, Ontario, Canada
Kathryn L. Hilde, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
Fan Meng, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
Robert C. Thompson, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
Huzefa Khalil, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
Alex Stefanov, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
Pamela Maras, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
Zhifeng Zhou, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Bethesda, Maryland..
Elaine K. Hebda-Bauer, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
David Goldman, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Bethesda, Maryland..
Stanley J. Watson, Jr., Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan
Huda Akil, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan.
REFERENCES
- 1.Krueger RF, Markon KE (2006): Reinterpreting comorbidity: A modelbased approach to understanding and classifying psychopathology. Annu Rev Clin Psychol 2:111–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smoller JW (2016): The genetics of stress-related disorders: PTSD, depression, and anxiety disorders. Neuropsychopharmacology 41:297–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. (2018): Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aydin C, Frohmader K, Akil H (2015): Revealing a latent variable: Individual differences in affective response to repeated injections. Behav Neurosci 129:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhury S, Aurbach EL, Sharma V, Blandino P, Turner CA, Watson SJ, Akil H (2014): FGF2 is a target and a trigger of epigenetic mechanisms associated with differences in emotionality: Partnership with H3K9me3. Proc Natl Acad Sci U S A 111:11834–11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinton SM, Vazquez DM, Kabbaj M, Kabbaj M-H, Watson SJ, Akil H (2007): Individual differences in novelty-seeking and emotional reactivity correlate with variation in maternal behavior. Horm Behav 51:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flagel SB, Robinson TE, Clark JJ, Clinton SM, Watson SJ, Seeman P, et al. (2010): An animal model of genetic vulnerability to behavioral disinhibition and responsiveness to reward-related cues: Implications for addiction. Neuropsychopharmacology 35:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flagel SB, Waselus M, Clinton SM, Watson SJ, Akil H (2014): Antecedents and consequences of drug abuse in rats selectively bred for high and low response to novelty. Neuropharmacology 76(pt B):425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez JA, Clinton SM, Turner CA, Watson SJ, Akil H (2009): A new role for FGF2 as an endogenous inhibitor of anxiety. J Neurosci 29:6379–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prater KE, Aurbach EL, Larcinese HK, Smith TN, Turner CA, Blandino P, et al. (2017): Selectively bred rats provide a unique model of vulnerability to PTSD-like behavior and respond differentially to FGF2 augmentation early in life. Neuropsychopharmacology 42:1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stead JDH, Clinton S, Neal C, Schneider J, Jama A, Miller S, et al. (2006): Selective breeding for divergence in novelty-seeking traits: Heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet 36:697–712. [DOI] [PubMed] [Google Scholar]
- 12.Stedenfeld KA, Clinton SM, Kerman IA, Akil H, Watson SJ, Sved AF (2011): Novelty-seeking behavior predicts vulnerability in a rodent model of depression. Physiol Behav 103:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner CA, Clinton SM, Thompson RC, Watson SJ, Akil H (2011): Fibroblast growth factor-2 (FGF2) augmentation early in life alters hippocampal development and rescues the anxiety phenotype in vulnerable animals. Proc Natl Acad Sci U S A 108:8021–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinton SM, Stead JDH, Miller S, Watson SJ, Akil H (2011): Developmental underpinnings of differences in rodent novelty-seeking and emotional reactivity. Eur J Neurosci 34:994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner CA, Hagenauer MH, Aurbach EL, Maras PM, Fournier CL, Blandino P, et al. (2019): Effects of early-life FGF2 on ultrasonic vocalizations (USVs) and the mu-opioid receptor in male Sprague-Dawley rats selectively-bred for differences in their response to novelty. Brain Res 1715:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JL, Kagan J, Reznick JS, Corley R (1992): The heritability of inhibited and uninhibited behavior: A twin study. Dev Psychol 28:1030–1037. [Google Scholar]
- 17.Turner CA, Flagel SB, Blandino P, Watson SJ, Akil H (2017): Utilizing a unique animal model to better understand human temperament. Curr Opin Behav Sci 14:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanselow MS, Dong H-W (2010): Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray JA (1982): The Neuropsychology of Anxiety: An Enquiry Into the Functions of the Septo-hippocampal System. New York: Clarendon/Oxford University Press. [Google Scholar]
- 20.Papez JW (1937): A proposed mechanism of emotion. Arch Neurol Psychiatry 38:725–743. [Google Scholar]
- 21.Campbell S, Macqueen G (2004): The role of the hippocampus in the pathophysiology of major depression. J Psychiatry Neurosci 29:417–426. [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach JL, McEwen BS (1972): Rat brain binds adrenal steroid hormone: Radioautography of hippocampus with corticosterone. Science 175:1133–1136. [DOI] [PubMed] [Google Scholar]
- 23.Clinton S, Miller S, Watson SJ, Akil H (2008): Prenatal stress does not alter innate novelty-seeking behavioral traits, but differentially affects individual differences in neuroendocrine stress responsivity. Psychoneuroendocrinology 33:162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabbaj M, Devine DP, Savage VR, Akil H (2000): Neurobiological correlates of individual differences in novelty-seeking behavior in the rat: Differential expression of stress-related molecules. J Neurosci 20:6983–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Blandino P, Yuan Q, Shen P-H, Hodgkinson CA, Virkkunen M, et al. (2019): Exploratory locomotion, a predictor of addiction vulnerability, is oligogenic in rats selected for this phenotype. Proc Natl Acad Sci U S A 116:13107–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aurbach EL, Inui EG, Turner CA, Hagenauer MH, Prater KE, Li JZ, et al. (2015): Fibroblast growth factor 9 is a novel modulator of negative affect. Proc Natl Acad Sci U S A 112:11953–11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isgor C, Kabbaj M, Akil H, Watson SJ (2004): Delayed effects of chronic variable stress during peripubertal-juvenile period on hippocampal morphology and on cognitive and stress axis functions in rats. Hippocampus 14:636–648. [DOI] [PubMed] [Google Scholar]
- 28.Cohen JL, Glover ME, Pugh PC, Fant AD, Simmons RK, Akil H, et al. (2015): Maternal style selectively shapes amygdalar development and social behavior in rats genetically prone to high anxiety. Dev Neurosci 37:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003): Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. [DOI] [PubMed] [Google Scholar]
- 30.Viechtbauer W (2010): Conducting meta-analyses in R with the metafor package. J Stat Softw 36. 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 31.Pollard KS, Dudoit S, van der Laan MJ (2005): Multiple testing procedures: The multtest package and applications to genomics In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer, 249–271. [Google Scholar]
- 32.Meckes JK, Lim PH, Wert SL, Luo W, Gacek SA, Platt D, et al. (2018): Brain region-specific expression of genes mapped within quantitative trait loci for behavioral responsiveness to acute stress in Fisher 344 and Wistar Kyoto male rats. PLoS One 13:e194293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrus BM, Blizinsky K, Vedell PT, Dennis K, Shukla PK, Schaffer DJ, et al. (2012): Gene expression patterns in the hippocampus and amygdala of endogenous depression and chronic stress models. Mol Psychiatry 17:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabariego M, Morón I, Gómez MJ, Donaire R, Tobeña A, Fernández-Teruel A, et al. (2013): Incentive loss and hippocampal gene expression in inbred Roman high- (RHA-I) and Roman low- (RLA-I) avoidance rats. Behav Brain Res 257:62–70. [DOI] [PubMed] [Google Scholar]
- 35.Díaz-Morán S, Palència M, Mont-Cardona C, Cañete T, Blázquez G, Martínez-Membrives E, et al. (2013): Gene expression in hippocampus as a function of differential trait anxiety levels in genetically heterogeneous NIH-HS rats. Behav Brain Res 257:129–139. [DOI] [PubMed] [Google Scholar]
- 36.Raghavan NS, Chen H, Schipma M, Luo W, Chung S, Wang L, Redei EE (2017): Prepubertal ovariectomy exaggerates adult affective behaviors and alters the hippocampal transcriptome in a genetic rat model of depression. Front Endocrinol (Lausanne) 8:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garafola CS, Henn FA (2014): A change in hippocampal protocadherin gamma expression in a learned helpless rat. Brain Res 1593:55–64. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, Amstein T, Shen J, Brush FR, Gershenfeld HK (2005): Molecular correlates of emotional learning using genetically selected rat lines. Genes Brain Behav 4:99–109. [DOI] [PubMed] [Google Scholar]
- 39.Wilhelm CJ, Choi D, Huckans M, Manthe L, Loftis JM (2013): Adipocytokine signaling is altered in Flinders sensitive line rats, and adiponectin correlates in humans with some symptoms of depression. Pharmacol Biochem Behav 103:643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaveri E, Kelly F, Mallei A, Harris K, Taylor A, Reid J, et al. (2010): Expression profiling of a genetic animal model of depression reveals novel molecular pathways underlying depressive-like behaviours. PLoS One 5:e12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoyama M, De Pons J, Hayman GT, Laulederkind SJF, Liu W, Nigam R, et al. (2015): The Rat Genome Database 2015: Genomic, phenotypic and environmental variations and disease. Nucleic Acids Res 43:D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Preter K, Barriot R, Speleman F, Vndesompele J, Moreau Y (2008): Positional gene enrichment analysis of gene sets for high-resolution identification of overrepresented chromosomal regions. Nucleic Acids Res 36:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. (2003): PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. [DOI] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. (2005): Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sergushichev A (2016): An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 10.1101/060012. [DOI] [Google Scholar]
- 46.Powell JAC (2014): GO2MSIG, an automated GO based multispecies gene set generator for gene set enrichment analysis. BMC Bioinformatics 15:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson MR, Shkura K, Langley SR, Delahaye-Duriez A, Srivastava P, Hill WD, et al. (2016): Systems genetics identifies a convergent gene network for cognition and neurodevelopmental disease. Nat Neurosci 19:223–232. [DOI] [PubMed] [Google Scholar]
- 48.Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, et al. (2011): Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol 5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cembrowski MS, Wang L, Sugino K, Shields BC, Spruston N (2016): Hipposeq: A comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5:e14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. (2015): STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagenauer MH, Schulmann A, Li JZ, Vawter MP, Walsh DM, Thompson RC, et al. (2018): Inference of cell type content from human brain transcriptomic datasets illuminates the effects of age, manner of death, dissection, and psychiatric diagnosis. PLoS One 13:e200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD (2001): Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 53.Yuan JS, Reed A, Chen F, Stewart CN (2006): Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calvo N, Cecchi M, Kabbaj M, Watson SJ, Akil H (2011): Differential effects of social defeat in rats with high and low locomotor response to novelty. Neuroscience 183:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabbaj M, Evans S, Watson SJ, Akil H (2004): The search for the neurobiological basis of vulnerability to drug abuse: Using microarrays to investigate the role of stress and individual differences. Neuropharmacology 47(suppl 1):111–122. [DOI] [PubMed] [Google Scholar]
- 56.Albert FW, Carlborg O, Plyusnina I, Besnier F, Hedwig D, Lautenschläger S, et al. (2009): Genetic architecture of tameness in a rat model of animal domestication. Genetics 182:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terenina-Rigaldie E, Moisan M-P, Colas A, Beaugé F, Shah KV, Jones BC, Mormède P (2003): Genetics of behaviour: Phenotypic and molecular study of rats derived from high- and low-alcohol consuming lines. Pharmacogenetics 13:543–554. [DOI] [PubMed] [Google Scholar]
- 58.Conti LH, Jirout M, Breen L, Vanella JJ, Schork NJ, Printz MP (2004): Identification of quantitative trait loci for anxiety and locomotion phenotypes in rat recombinant inbred strains. Behav Genet 34:93–103. [DOI] [PubMed] [Google Scholar]
- 59.Baum AE, Solberg LC, Churchill GA, Ahmadiyeh N, Takahashi JS, Redei EE (2006): Test- and behavior-specific genetic factors affect WKY hypoactivity in tests of emotionality. Behav Brain Res 169:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johannesson M, Lopez-Aumatell R, Stridh P, Diez M, Tuncel J, Blázquez G, et al. (2009): A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: The NIH heterogeneous stock. Genome Res 19:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Teruel A, Escorihuela RM, Gray JA, Aguilar R, Gil L, Giménez-Llort L, et al. (2002): A quantitative trait locus influencing anxiety in the laboratory rat. Genome Res 12:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW, et al. (2006): Genetic analysis of the stress-responsive adrenocortical axis. Physiol Genomics 27:362–369. [DOI] [PubMed] [Google Scholar]
- 63.Yamazato M, Ohya Y, Nakamoto M, Sakima A, Tagawa T, Harada Y, et al. (2006): Sympathetic hyperreactivity to air-jet stress in the chromosome 1 blood pressure quantitative trait locus congenic rats. Am J Physiol Regul Integr Comp Physiol 290:R709–R714. [DOI] [PubMed] [Google Scholar]
- 64.Llamas B, Contesse V, Guyonnet-Duperat V, Vaudry H, Mormède P, Moisan M-P (2005): QTL mapping for traits associated with stress neuroendocrine reactivity in rats. Mamm Genome 16:505–515. [DOI] [PubMed] [Google Scholar]
- 65.Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW, et al. (2004): Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm Genome 15:648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du R-H, Wu F-F, Lu M, Shu X-D, Ding J-H, Wu G, Hu G (2016): Uncoupling protein 2 modulation of the NLRP3 inflammasome in astrocytes and its implications in depression. Redox Biol 9:178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gimsa U, Kanitz E, Otten W, Aheng C, Tuchscherer M, Ricquier D, et al. (2011): Alterations in anxiety-like behavior following knockout of the uncoupling protein 2 (ucp2) gene in mice. Life Sci 89:677–684. [DOI] [PubMed] [Google Scholar]
- 68.Pekary AE, Sattin A, Lloyd RL (2015): Ketamine modulates TRH and TRH-like peptide turnover in brain and peripheral tissues of male rats. Peptides 69:66–76. [DOI] [PubMed] [Google Scholar]
- 69.Sun X-L, Liu Y, Dai T, Ding J-H, Hu G (2011): Uncoupling protein 2 knockout exacerbates depression-like behaviors in mice via enhancing inflammatory response. Neuroscience 192:507–514. [DOI] [PubMed] [Google Scholar]
- 70.Wang D, Zhai X, Chen P, Yang M, Zhao J, Dong J, Liu H (2014): Hippocampal UCP2 is essential for cognition and resistance to anxiety but not required for the benefits of exercise. Neuroscience 277:36–44. [DOI] [PubMed] [Google Scholar]
- 71.Zeng H, Schimpf BA, Rohde AD, Pavlova MN, Gragerov A, Bergmann JE (2007): Thyrotropin-releasing hormone receptor 1-eficient mice display increased depression and anxiety-like behavior. Mol Endocrinol 21:2795–2804. [DOI] [PubMed] [Google Scholar]
- 72.Hoover AN, Wynkoop A, Zeng H, Jia J, Niswander LA, Liu A (2008): C2cd3 is required for cilia formation and hedgehog signaling in mouse. Development 135:4049–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao B, Harada Y, Kawakami K, Nabika T (2011): A 1.8-Mbp fragment on chromosome 1 affects sympathetic response to stress: Evaluation in reciprocal congenic strains between strokeprone spontaneously hypertensive rat and Wistar-Kyoto rat. J Hypertens 29:257–265. [DOI] [PubMed] [Google Scholar]
- 74.Klimes I, Weston K, Gasperíková D, Kovács P, Kvetnanský R, Jezová D, et al. (2005): Mapping of genetic determinants of the sympathoneural response to stress. Physiol Genomics 20:183–187. [DOI] [PubMed] [Google Scholar]
- 75.Potenza MN, Brodkin ES, Joe B, Luo X, Remmers EF, Wilder RL, et al. (2004): Genomic regions controlling corticosterone levels in rats. Biol Psychiatry 55:634–641. [DOI] [PubMed] [Google Scholar]
- 76.Belvindrah R, Rougon G, Chazal G (2002): Increased neurogenesis in adult mCD24-deficient mice. J Neurosci 22:3594–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cerezo A, Guadamillas MC, Goetz JG, Sánchez-Perales S, Klein E, Assoian RK, del Pozo MA (2009): The absence of caveolin-1 increases proliferation and anchorage-independent growth by a Rac-dependent, Erk-independent mechanism. Mol Cell Biol 29:5046–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galach M (2014): Molecular mechanisms involved in epithelial differentiation of human induced pluripotent stem cells [dissertation] Available at: 10.11588/heidok.00017870. Accessed May 15, 2018. [DOI]
- 79.Head BP, Peart JN, Panneerselvam M, Yokoyama T, Pearn ML, Niesman IR, et al. (2010): Loss of caveolin-1 accelerates neurodegeneration and aging. PLoS One 5:e15697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hossain MB, Shifat R, Li J, Luo X, Hess KR, Rivera-Molina Y, et al. (2017): TIE2 associates with caveolae and regulates caveolin-1 to promote their nuclear translocation. Mol Cell Biol 37:e142–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, et al. (2014): The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis 1:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X-L, Liu L, Li D-D, He Y-P, Guo L-H, Sun L-P, et al. (2017): Integrin b4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Sci Rep 7:40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu C, Goswami M, Talley J, Chesser-Martinez PL, Lou C-H, Sater AK (2012): TAK1 promotes BMP4/Smad1 signaling via inhibition of erk MAPK: A new link in the FGF/BMP regulatory network. Differentiation 83:210–219. [DOI] [PubMed] [Google Scholar]
- 84.Panaliappan TK, Wittmann W, Jidigam VK, Mercurio S, Bertolini JA, Sghari S, et al. (2018): Sox2 is required for olfactory pit formation and olfactory neurogenesis through BMP restriction and Hes5 upregulation. Development 145:dev153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poché RA, Furuta Y, Chaboissier M-C, Schedl A, Behringer RR (2008): Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J Comp Neurol 510:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shakiba N, White CA, Lipsitz YY, Yachie-Kinoshita A, Tonge PD, Hussein SMI, et al. (2015): CD24 tracks divergent pluripotent states in mouse and human cells. Nat Commun 6:7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M (2003): The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suzuki Y, Montagne K, Nishihara A, Watabe T, Miyazono K (2008): BMPs promote proliferation and migration of endothelial cells via stimulation of VEGF-A/VEGFR2 and angiopoietin-1/Tie2 signalling. J Biochem 143:199–206. [DOI] [PubMed] [Google Scholar]
- 89.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH (2008): Functional organization of the transcriptome in human brain. Nat Neurosci 11:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fontanet PA, Ríos AS, Alsina FC, Paratcha G, Ledda F (2018): Pea3 transcription factors, Etv4 and Etv5, are required for proper hippocampal dendrite development and plasticity. Cereb Cortex 28:236–249. [DOI] [PubMed] [Google Scholar]
- 91.Jaworski JN, Jones DC (2006): The role of CART in the reward/reinforcing properties of psychostimulants. Peptides 27:1993–2004. [DOI] [PubMed] [Google Scholar]
- 92.Young LJ, Wang Z (2004): The neurobiology of pair bonding. Nat Neurosci 7:1048–1054. [DOI] [PubMed] [Google Scholar]
- 93.Higa KK, Young JW, Ji B, Nichols DE, Geyer MA, Zhou X (2017): Striatal dopamine D1 receptor suppression impairs reward-associative learning. Behav Brain Res 323:100–110. [DOI] [PubMed] [Google Scholar]
- 94.Milienne-Petiot M, Groenink L, Minassian A, Young JW (2017): Blockade of dopamine D1-family receptors attenuates the mania-like hyperactive, risk-preferring, and high motivation behavioral profile of mice with low dopamine transporter levels. J Psychopharmacol (Oxford) 31:1334–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spittau B, Rilka J, Steinfath E, Zöller T, Krieglstein K (2015): TGFβ1 increases microglia-mediated engulfment of apoptotic cells via upregulation of the milk fat globule-EGF factor 8. Glia 63:142–153. [DOI] [PubMed] [Google Scholar]
- 96.Kettenmann H, Kirchhoff F, Verkhratsky A (2013): Microglia: New roles for the synaptic stripper. Neuron 77:10–18. [DOI] [PubMed] [Google Scholar]
- 97.Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, et al. (2018): Molecular architecture of the mouse nervous system. Cell 174:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fischer S, Ehlert U (2018): Hypothalamic-pituitary-thyroid (HPT) axis functioning in anxiety disorders: A systematic review. Depress Anxiety 35:98–110. [DOI] [PubMed] [Google Scholar]
- 99.Maes M, Vandewoude M, Maes L, Schotte C, Cosyns P (1989): A revised interpretation of the TRH test results in female depressed patients: I. TSH responses: Effects of severity of illness, thyroid hormones, monoamines, age, sex hormonal, corticosteroid and nutritional state. J Affect Disord 16:203–213. [DOI] [PubMed] [Google Scholar]
- 100.Lanni A, Moreno M, Lombardi A, Goglia F (2003): Thyroid hormone and uncoupling proteins. FEBS Lett 543:5–10. [DOI] [PubMed] [Google Scholar]
- 101.Choi J, Kim J, Kim T-K, Park J-Y, Lee J-E, Kim H, et al. (2015): TRH and TRH receptor system in the basolateral amygdala mediate stress-induced depression-like behaviors. Neuropharmacology 97:346–356. [DOI] [PubMed] [Google Scholar]
- 102.Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. (2011): Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet 88:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miró X, Meier S, Dreisow ML, Frank J, Strohmaier J, Breuer R, et al. (2012): Studies in humans and mice implicate neurocan in the etiology of mania. Am J Psychiatry 169:982–990. [DOI] [PubMed] [Google Scholar]
- 104.Rauch U, Feng K, Zhou XH (2001): Neurocan: A brain chondroitin sulfate proteoglycan. Cell Mol Life Sci 58:1842–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cole AE, Murray SS, Xiao J (2016): Bone morphogenetic protein 4 signalling in neural stem and progenitor cells during development and after injury. Stem Cells Int 2016:9260592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harland R (2000): Neural induction. Curr Opin Genet Dev 10:357–362. [DOI] [PubMed] [Google Scholar]
- 107.Bond AM, Bhalala OG, Kessler JA (2012): The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev Neurobiol 72:1068–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brooker SM, Gobeske KT, Chen J, Peng C-Y, Kessler JA (2017): Hippocampal bone morphogenetic protein signaling mediates behavioral effects of antidepressant treatment. Mol Psychiatry 22:910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gobeske KT, Das S, Bonaguidi MA, Weiss C, Radulovic J, Disterhoft JF, Kessler JA (2009): BMP signaling mediates effects of exercise on hippocampal neurogenesis and cognition in mice. PLoS One 4:e7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meyers EA, Gobeske KT, Bond AM, Jarrett JC, Peng C-Y, Kessler JA (2016): Increased bone morphogenetic protein signaling contributes to age-related declines in neurogenesis and cognition. Neurobiol Aging 38:164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, et al. (2010): Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7:78–89. [DOI] [PubMed] [Google Scholar]
- 112.Gomes WA, Mehler MF, Kessler JA (2003): Transgenic over-expression of BMP4 increases astroglial and decreases oligodendroglial lineage commitment. Dev Biol 255:164–177. [DOI] [PubMed] [Google Scholar]
- 113.Caronia G, Wilcoxon J, Feldman P, Grove EA (2010): Bone morphogenetic protein signaling in the developing telencephalon controls formation of the hippocampal dentate gyrus and modifies fear-related behavior. J Neurosci 30:6291–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.García-Fuster MJ, Perez JA, Clinton SM, Watson SJ, Akil H (2010): Impact of cocaine on adult hippocampal neurogenesis in an animal model of differential propensity to drug abuse. Eur J Neurosci 31:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duman RS, Monteggia LM (2006): A neurotrophic model for stress-related mood disorders. Biol Psychiatry 59:1116–1127. [DOI] [PubMed] [Google Scholar]
- 116.Bansod S, Kageyama R, Ohtsuka T (2017): Hes5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development. Development 144:3156–3167. [DOI] [PubMed] [Google Scholar]
- 117.Sakai D, Suzuki T, Osumi N, Wakamatsu Y (2006): Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development 133:1323–1333. [DOI] [PubMed] [Google Scholar]
- 118.Sun W, Cornwell A, Li J, Peng S, Osorio MJ, Aalling N, et al. (2017): SOX9 is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. J Neurosci 37:4493–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ming G-L, Song H (2011): Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 70:687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fuller ML, DeChant AK, Rothstein B, Caprariello A, Wang R, Hall AK, Miller RH (2007): Bone morphogenetic proteins promote gliosis in demyelinating spinal cord lesions. Ann Neurol 62:288–300. [DOI] [PubMed] [Google Scholar]
- 121.Yan H, Zhu X, Xie J, Zhao Y, Liu X (2016): β-Amyloid increases neurocan expression through regulating Sox9 in astrocytes: A potential relationship between Sox9 and chondroitin sulfate proteoglycans in Alzheimer’s disease. Brain Res 1646:377–383. [DOI] [PubMed] [Google Scholar]
- 122.Schafer DP, Stevens B (2015): Microglia function in central nervous system development and plasticity. Cold Spring Harb Perspect Biol 7:a20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu Y, Dissing-Olesen L, MacVicar BA, Stevens B (2015): Microglia: Dynamic mediators of synapse development and plasticity. Trends Immunol 36:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Albus E, Sinningen K, Winzer M, Thiele S, Baschant U, Hannemann A, et al. (2016): Milk fat globule–epidermal growth factor 8 (MFG-E8) is a novel anti-inflammatory factor in rheumatoid arthritis in mice and humans. J Bone Miner Res 31:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fricker M, Neher JJ, Zhao J-W, Théry C, Tolkovsky AM, Brown GC (2012): MFG-E8 mediates primary phagocytosis of viable neurons during neuroinflammation. J Neurosci 32:2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fuller AD, Van Eldik LJ (2008): MFG-E8 regulates microglial phagocytosis of apoptotic neurons. J Neuroimmune Pharmacol 3:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vogler S, Pahnke J, Rousset S, Ricquier D, Moch H, Miroux B, Ibrahim SM (2006): Uncoupling protein 2 has protective function during experimental autoimmune encephalomyelitis. Am J Pathol 168:1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, et al. (2000): Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet 26:435–439. [DOI] [PubMed] [Google Scholar]
- 129.Jo WK, Zhang Y, Emrich HM, Dietrich DE (2015): Glia in the cytokine-mediated onset of depression: Fine tuning the immune response. Front Cell Neurosci 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sato K (2015): Effects of microglia on neurogenesis. Glia 63:1394–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mastellos DC (2014): Complement emerges as a masterful regulator of CNS homeostasis, neural synaptic plasticity and cognitive function. Exp Neurol 261:469–474. [DOI] [PubMed] [Google Scholar]
- 132.Maras PM, Daugherty J, Hebda-Bauer E, Watson SJ, Akil H (2018): Minocycline treatment reduces anxiety- and depressive-like behaviors in a genetic model of internalizing mood disorders Presented at Neuroscience 2018, November 3–7, San Diego, California Neuroscience Meeting Planner Program No. 234.03. [Google Scholar]
- 133.Hilde KL, Hagenauer MH, Stefanov AV, Birt I, Hebda-Bauer E, Watson SJ, Akil H (2018): Mechanisms of hippocampal development: Molecular and cellular phenotypes in the selectively bred high-responder/low-responder model of affective disorders Presented at Neuroscience 2018, November 3–7, San Diego, California Neuroscience Meeting Planner Program No. 233.04. [Google Scholar]
- 134.O’Connor AM, Pardo T, Birt I, Hagenauer MH, Maras PM, Prater KE, et al. (2018): The hippocampal glycome contributes to behavioural phenotype in a novel rodent model of mood disorders Presented at Neuroscience 2018, November 3–7, San Diego, California Neuroscience Meeting Planner Program No. 233.01. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.