Abstract
The research community is in a race to understand the molecular mechanisms of SARS-CoV-2 infection, to repurpose currently available antiviral drugs, and to develop new therapies and vaccines against COVID-19. One major challenge in achieving these goals is the paucity of suitable preclinical animal models. Mice constitute around 70 percent of all of laboratory animal species used in biomedical research. Unfortunately, SARS-CoV-2 only infects mice if they have been genetically modified to express human ACE2. The inherent resistance of wild-type mice to SARS-CoV-2, combined with a wealth of genetic tools that are only available for modifying mice, offer a unique opportunity to create a versatile set of genetically engineered mouse models (GEMMs) useful for COVID-19 research. We propose three broad categories of these models and more than two dozen designs that may be useful for SARS-CoV-2 research and for fighting COVID-19.
EDITORIAL SUMMARY
In this Perspective, the authors discuss strategies for creating a versatile set of 30 genetically engineered mouse models (GEMMs) useful for COVID-19 research. Additionally, they provide the genetic blueprints needed for developing these GEMMs.
Introduction
The research community is in desperate need of animal models for testing therapeutics and vaccines against COVID-19, and for studies aimed at understanding the molecular mechanisms of SARS-CoV-2 infection and pathogenesis. A wide spectrum of laboratory and domestic animal species have been challenged with SARS-CoV-2 in an effort to find suitable models, but only a few, such as hamsters, cats, ferrets, and monkeys are susceptible1–3. Unfortunately, many of these species are neither accessible nor practical for the vast majority of researchers to make rapid progress.
Mice are the most commonly used laboratory animals in biomedical research, and they have helped scientists develop diagnostics, therapeutics, and vaccines for many human diseases. As such, it would have been easy to use mice for COVID-19 research if they were susceptible to SARS-CoV-2 infection, like humans, but that is not the case. One way to render mice susceptible to SARS-CoV-2 is to genetically modify them.
Evaluating the utility of current GEMMs previously developed for SARS-CoV research
Several GEMMs that express the hACE2 gene were created over a decade ago for use in SARS virus studies4–7. Like SARS-CoV, SARS-CoV-2 uses the hACE2 receptor for entry into cells. Therefore some of these transgenic models have been investigated for their suitability for SARS-CoV-2 studies8,9. Repurposing of previously made GEMMs and non-GEMMs models (discussed in the next section), including animals other than mice, for the study of novel coronaviruses has been extensively reviewed by Yuan et al., 202010, and Singh et al., 202011. Even though the existing hACE2 transgenic mice fully support SARS-CoV-2 replication, the pathogenesis does not accurately model the disease course seen in humans12 and they also have other limitations. First, because these models still express the mouse orthologue of ACE2 (mAce2), this will occupy a portion of the cell surface pool of total ACE2, leading to non-physiological amounts of the human ACE2 available for virus interactions. Second, the transgenic expression cassettes in current models are driven by different promoters, and often expressed in different types of cells and at levels that are not similar to endogenous mACE2 expression. Furthermore, constitutively expressing transgenic mouse models are not suited to experiments requiring spatiotemporal regulation of hACE2 expression (i.e., in specific cell types, such as lung epithelial cells, and/or at specific time points). Finally, these models will not be ideal for researchers wanting to simulate comorbid conditions such as obesity, hypertension and diabetes, in which affected tissues will have altered ACE2 expression13,14.
Mouse models developed using non-GEMM approaches
Because the scientific community has such an urgent need for preclinical models, some groups explored non-GEMM strategies that were also previously used for SARS-CoV studies, such as (i) adapting the virus by serial passage through mice so that infectious mutant strains emerge15, and (ii) delivering hACE2-encoding DNA through Adenovirus (AdV) vector systems followed by infection of the mice with SARS-CoV. Both of these strategies were demonstrated recently as proof-of-principle for SARS-CoV-212,16–18. Adaptation of SARS-CoV-2 virus to mice4 did not cause lethality, but the approach seems useful for testing vaccine efficacy12,16. Delivering hACE2 into mice through an AdV vector system demonstrated the utility of the approach for pathogenicity studies, testing vaccines, and therapeutics17,18. Some limitations of the AdV vector approach, however, include the requirement for large quantities of vector and mouse-to-mouse variability in the response to infection. As most mice do not develop severe disease19, they do not develop the extrapulmonary manifestations seen in humans17, and some animals show lung pathology resulting from AdV administration itself18. Direct comparisons of GEMMs versus non-GEMMs show that the GEMMs provide a stable system for testing the ability of vaccines and antivirals to protect against disease, whereas the non-GEMMs offer the flexibility to be used across multiple genetic backgrounds19.
Versatility of GEMMs
While the previously generated transgenic hACE2 mice and the non-GEMM approaches have successfully established that mice can be re-tooled as preclinical models for COVID-19 studies, sophisticated GEMM approaches provide a much more versatile suite of COVID-19 models than can be envisioned for any other species. The natural non-permissiveness of mice to SARS-CoV-2 infection combined with the wide variety of available genetic tools and molecular switches offers a unique opportunity to make this species even more useful for COVID-19 research.
Researchers are now facing a familiar COVID-19 narrative: trying to make sense of a mystifying illness20 affecting not only the respiratory system, but also many other organs, including those of the cardiovascular21, digestive22 and nervous systems23. If a researcher wants to study the pathophysiological and molecular events occurring only in the heart, intestine or brain, it would be difficult to study such events in an animal model in which many other organs are also infected (for example, lung or kidney). If the animal model allowed productive infection of only one type of tissue or cell (cardiac or endothelial or neuronal), and if the model allowed regulation of the dose and timing of infection, it would be very helpful in answering a variety of research questions relevant to cardiac or endothelial or neuronal manifestations. Related to this, a recent report described the expression pattern of ACE2 in human tissues. Although somewhat intriguing, expression of ACE2 seemed to be very low in lung cells while it was detected at high levels in >150 cell types corresponding to 45 tissues24. This observation further highlights the value of the conditional and inducible expression models where researchers working in a wide range of tissue types can use the models to ask specific questions by limiting hACE2 expression and viral infection to only subset of tissue types.
This type of controlled experimental design is possible only in mice because the genetic tools and molecular switches (e.g., Cre-LoxP and Tetracycline-inducible systems) are readily available for multiple cell types and tissue systems. For example, a CRE-activatable hACE2 knock-in mouse model, once created, could be bred to any of the thousands of already available Cre driver lines, thus allowing researchers from a range of biomedical fields, from cardiac physiologists to neuroscientists, to use this model for probing their particular research questions of interest. Additionally, individual gene knockout (KO) mouse models are available for thousands of genes. The molecular pathways of viral infection can be further dissected through classical mouse genetic studies by breeding the COVID-19 models with a number of different gene KO models. Thus, mice can be very useful models in the fight against COVID-19. In this Perspective, we propose concepts for developing more than two dozen GEMMs suitable for a variety of research on COVID-19 and SARS-CoV-2.
We propose the following three broad categories of GEMMs for COVID-19 research. 1) Knocking-in expression cassettes, or point mutations, into the endogenous mouse Ace2 locus. 2) Knocking-in CRE-activatable- or Tetracycline-inducible-hACE2 expression cassettes into safe-harbor loci, by re-engineering the existing reporter or inducer lines. 3) Knocking-in CRE-activatable cassettes into the mouse Ace2 locus. The proposed GEMMs are listed in Table 1; the schematics of the GEMM designs are shown in Figures 1–3.
Table 1.
GEMM designs suitable for COVID-19 and SARS-CoV-2 research
| Model # | Name | Locus/promoter | Gene to express | Expression | Additional features |
|---|---|---|---|---|---|
| Category 1: Knocking-in expression cassettes or point mutations into endogenous mouse Ace2 loci | |||||
| 1 | B6.mAce2KO-hACE2KI | mAce2/ mAce2 | hACE2 | Constitutive | mAce2 gene is inactivated |
| 2 | B6.mAce2KO-hACE2-P2A-EGFPKI | mAce2/ mAce2 | hACE2-P2A-EGFP | Constitutive | mAce2 gene is inactivated, includes a reporter |
| 3 | NSG.mAce2KO-hACE2KI | mAce2/ mAce2 | hACE2 | Constitutive | mAce2 gene is inactivated; immunocompromised mouse strain background, useful for studies involving interaction of human immune system |
| S4 | NSG.mAce2KO-hACE2-P2A-EGFPKI | mAce2/ mAce2 | hACE2-P2A-EGFP | Constitutive | mAce2 gene is inactivated, includes a reporter; immunocompromised mouse strain background, useful for studies involving interaction of human immune system |
| 5 | BALB/c.mAce2KO-hACE2KI | mAce2/ mAce2 | hACE2 | Constitutive | mAce2 gene is inactivated; mouse strain background commonly used for SARS and MERS virus research |
| 6 | BALB/c.mAce2KO-hACE2-P2A-EGFPKI | mAce2/ mAce2 | hACE2-P2A-EGFP | Constitutive | mAce2 gene is inactivated, includes a reporter; mouse strain background commonly used for SARS and MERS virus research |
| 7 | B6.mAce2KI−31K,82M,353K | mAce2/ mAce2 | Partially humanized mAce2 | Constitutive | Enables mACE2 to bind to the SARS-Co-V2 spike protein |
| 8 | NSG.mAce2KI−31K,82M,353K | mAce2/ mAce2 | Partially humanized mAce2 | Constitutive | Enables mACE2 to bind to the SARS-Co-V2 spike protein; immunocompromised mouse strain background, useful for studies involving interaction of human immune system |
| 9 | BALB/c.mAce2KI−31K,82M,353K | mAce2/ mAce2 | Partially humanized mAce2 | Constitutive | Enables mACE2 to bind to the SARS-Co-V2 spike protein; mouse strain background commonly used for SARS and MERS virus research |
| 10 | B6.mAce2KO-hACE2-P2A-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide |
| 11 | NSG.mAce2KO-hACE2-P2A-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; immunocompromised mouse strain background, useful for studies involving interaction of human immune system; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide |
| 12 | BALB/c.mAce2KO-hACE2-P2A-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; mouse strain background commonly used for SARS and MERS virus research; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide |
| 13 | B6.mAce2KO-hACE2-IRES-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; hTMPRSS2 expressed as a separate polypeptide via an IRES |
| 14 | NSG.mAce2KO-hACE2-IRES-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; immunocompromised mouse strain background, useful for studies involving interaction of human immune system; hTMPRSS2 expressed as a separate polypeptide via an IRES |
| 15 | BALB/c.mAce2KO-hACE2-IRES-hTMPRSS2KI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Constitutive | mAce2 gene is inactivated; mouse strain background commonly used for SARS and MERS virus research; hTMPRSS2 expressed as a separate polypeptide via an IRES |
| Category 2: Knocking-in CRE-activatable or tetracycline-inducible expression cassettes into safe-harbor loci by re-engineering existing reporter or inducer mouse lines | |||||
| 16 | ROSA26(ACTB-Lox-hACE2-P2A-tdT-Lox-EGFP) | ROSA26/pCAG | hACE2 | Constitutive | Constitutive expression of hACE2 with reporter capability |
| 17 | ROSA26(ACTB-Lox-hACE2-IRES-tdT-Lox-EGFP) | ROSA26/pCAG | hACE2 | Constitutive | Constitutive expression of hACE2 with reporter capability |
| 18 | ROSA26(ACTB-Lox-tdT-Lox-hACE2-P2A-EGFP) | ROSA26/pCAG | hACE2 | Tissue-specific | CRE-activatable expression of hACE2 with reporter capability. |
| 19 | ROSA26(ACTB-Lox-tdT-Lox-hACE2-IRES-EGFP) | ROSA26/pCAG | hACE2 | Tissue-specific | CRE-activatable expression of hACE2 with reporter capability. |
| 20 | Ai63-TIGRE-TRE-hACE2-P2A-tdT | TIGRE/ TRE | hACE2 | Tetracyclin-inducible | Tetracycline inducible expression of hACE2 with reporter capability |
| 21 | Ai63-TIGRE-TRE-hACE2-IRES-tdT | TIGRE/ TRE | hACE2 | Tetracyclin-inducible | Tetracycline inducible expression of hACE2 with reporter capability |
| Category 3: Knocking-in CRE-activatable cassettes into mouse Ace2 locus | |||||
| 22 | B6.mAce2cKO-hACE2cKI | mAce2/ mAce2 | hACE2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated allowing expression of hACE2 |
| 23 | NSG.mAce2cKO-hACE2cKI | mAce2/ mAce2 | hACE2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated allowing expression of hACE2; immunocompromised mouse strain background, useful for studies involving interaction of human immune system |
| 24 | BALB/c.mAce2cKO-hACE2cKI | mAce2/ mAce2 | hACE2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2; mouse strain background commonly used for SARS and MERS virus research |
| 25 | B6.mAce2cKO-hACE2-P2A-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide |
| 26 | NSG.mAce2cKO-hACE2-P2A-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; immunocompromised mouse strain background, useful for studies involving interaction of human immune system; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide, immunocompromised mouse strain background |
| 27 | BALB/c.mAce2cKO-hACE2-P2A-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; mouse strain background commonly used for SARS and MERS virus research; hTMPRSS2 fused to hACE2 via a self-cleavable P2A peptide |
| 28 | B6.mAce2cKO-hACE2-IRES-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; hTMPRSS2 expressed as a separate polypeptide via an IRES |
| 29 | NSG.mAce2cKO-hACE2-IRES-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; immunocompromised mouse strain background, useful for studies involving interaction of human immune system; hTMPRSS2 expressed as a separate polypeptide via an IRES |
| 30 | BALB/c.mAce2cKO-hACE2-IRES-hTMPRSS2cKI | mAce2/ mAce2 | hACE2, hTMPRSS2 | Tissue-specific expression of hACE2 at physiological levels | mAce2 gene is conditionally inactivated, allowing expression of hACE2 and hTMPRSS2; mouse strain background commonly used for SARS and MERS virus research; hTMPRSS2 expressed as a separate polypeptide via an IRES |
Figure 1. Schematics of COVID-19 GEMM designs, category 1: Knocking-in expression cassettes or point mutation changes into the endogenous mouse Ace2 locus.
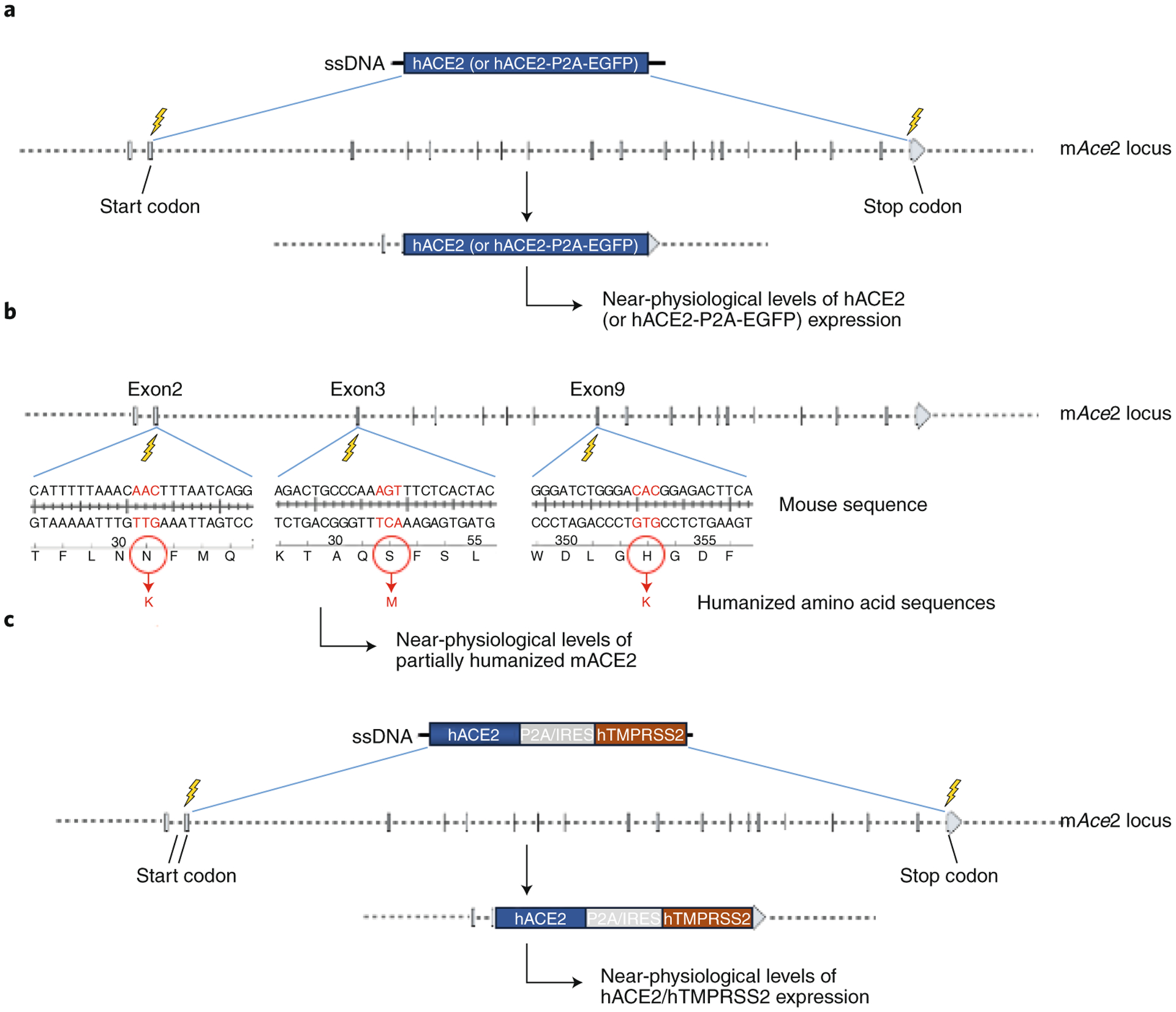
(A) KO:KI design; human ACE2 cDNA (GEMMs #1, #3 and #5) or human ACE2 cDNA fused to P2A-EGFP GEMMs #2, #4 and #6) will be inserted in place of mouse Ace2 by deleting about 48kb of genomic sequence between the start and stop codons of mAce2. (B) Key amino acids KI design (GEMMs #7 to #9); only the key amino acids that differ between mouse and human and are responsible for binding to the spike protein, will be replaced by knocking-in an ssODN donor containing the humanized codons. The locations of amino acids N31K, S82M and H353K are shown. (C) KO:KI designs to co-express hACE2 and hTMPRSS2 (GEMMs #10 to #15). P2A: a self-cleavable viral peptide used for expressing two fusion proteins. IRES: Internal Ribosomal Entry Site.
Figure 3. Schematics of COVID-19 GEMM designs, category 3: Knocking-in CRE-activatable cassettes into the mouse Ace2 locus.
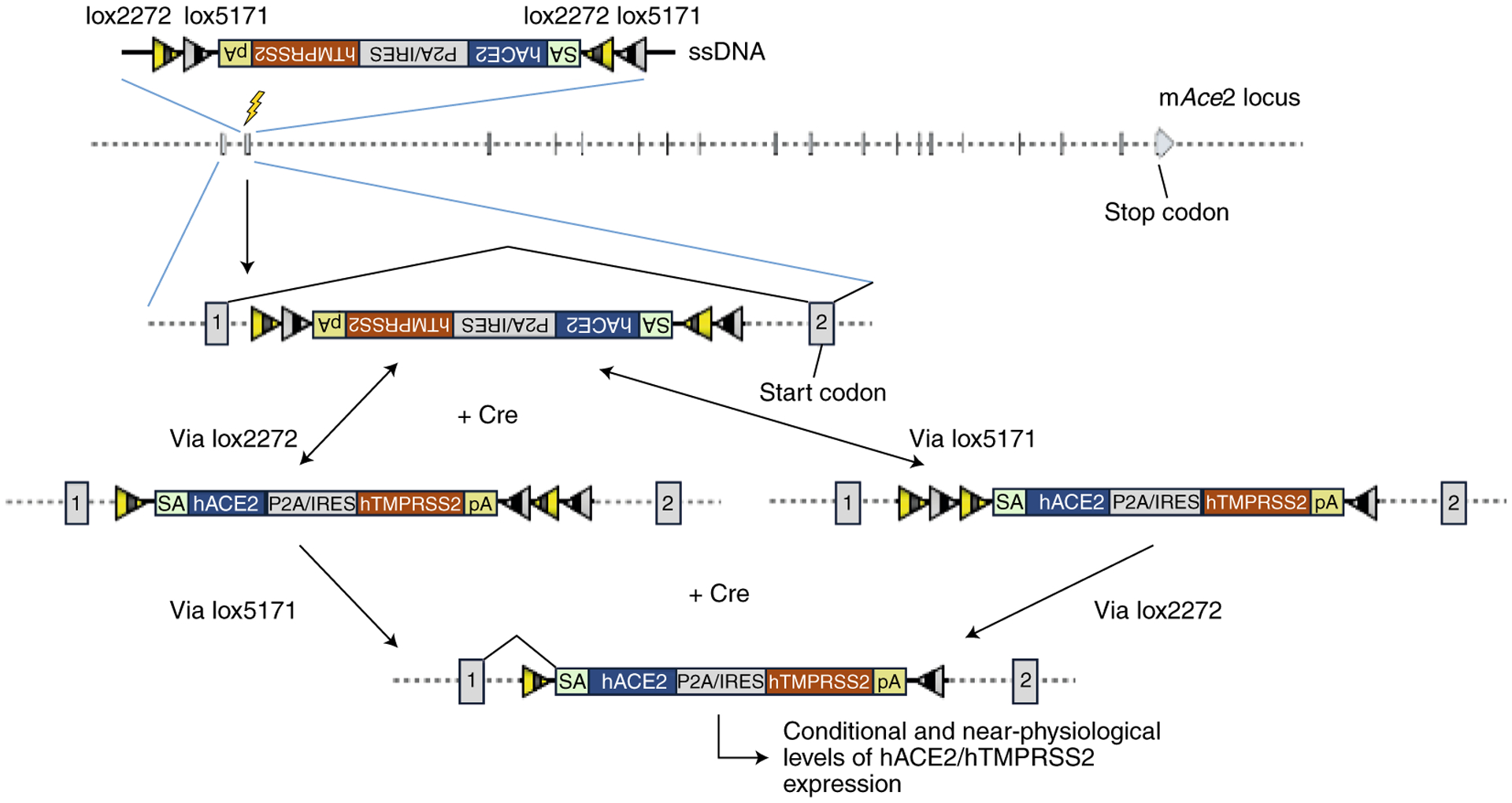
A CRE-invertible KI cassette is inserted in the opposite orientation to the mAce2 locus. Upon CRE-mediated inversion of the cassette through sequential recombinations of lox2272s and lox5171s, the transcript is spliced from exon1 to the inverted KI cassette and downstream transcription is terminated to prevent mAce2 function. This CRE-mediated inversion allows hACE2 expression in models #22 to #24 (not shown), or hACE2 and hTMPRRS2 expression in models #25 to #30. SA: splice acceptor. P2A: a self-cleavable viral peptide used for expressing two fusion proteins. IRES: Internal Ribosomal Entry Site.
GEMM Category 1: Knocking-in expression cassettes or point mutations into endogenous mouse Ace2 loci
One of the ideal GEMMs for COVID-19 research is a Knock-Out:Knock-In (KO:KI) design in which the mouse Ace2 gene is deleted (knocked-out [KO]), and the human ACE2 cDNA is inserted in its place, termed Model #1 herein. Both mouse and human ACE2 genes are located on the X chromosome, and each contains 19 exons. Although all corresponding exons are the same size, the mouse introns are larger, with a total gene size of 49kb and 41kb in mice and humans, respectively. The KO:KI model can be generated by deleting the region between the start and the stop codons of mouse Ace2 (i.e., all coding exons and the introns between) and inserting the 2.4 kb hACE2 cDNA. In the KO:KI design, because the mouse counterpart of ACE2 (mAce2) is deleted, any potential confounding effects on the response to virus infection will not occur, unlike in the hACE2 transgenic mice with random insertions. In this model, the mouse Ace2 promoter is expected to drive human ACE2 cDNA expression at physiological levels, although the effect of deleting the mouse introns cannot be ascertained, as they may contain regulatory regions. While our manuscript was under preparation, a model similar to this design was reported in which the hACE2 cDNA was inserted near the start codon of the mouse Ace225. Although the mAce2 genomic region is still retained in this model, mAce2 is not likely to be expressed because the insertion cassette contains a poly (A) sequence upstream of the mAce2 coding exons. This design also includes additional regulatory elements such as the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), which enhances mRNA stability and translation efficiency and may produce higher than physiological hACE2 protein levels. The ideal model design would be one that can achieve near-physiological levels of hACE2 expression. Such a model design would require precise replacement of all of the 19 mouse exons with the corresponding human exons, leaving the mouse introns, which may contain regulatory sequences, intact. Developing such a model would, however, be quite difficult. One other feature included in the published model25 is a fluorescent reporter (tdTomato [tdT]) expressed as a separate protein via an internal ribosomal entry site (IRES) element, with the reporter serving as a handy tool to mark the hACE2-expressing cells. We propose to include a self-cleaving viral peptide P2A, instead of the IRES element, fused to the C terminus of hACE2, followed by the EGFP coding sequence in model #2.
Ten to fifteen years ago, generating a KO:KI model like the one described above would have required multiple lengthy steps of complex gene targeting in embryonic stem cells, single-cell cloning, and chimera breeding. The process would typically take over a year just to develop a founder mouse, and may be the reason why such a model was not created. In addition, it is possible that when SARS-CoV research declined, scientists were not motivated to invest the necessary time and resources. Now, the situation is quite different; the COVID-19 pandemic has increased the demand for any GEMM available for SARS-CoV-2 infection, even though the existing models are suboptimal for certain COVID-19 research designs. On the bright side, GEMM technologies have improved tremendously during the last decade due to the advent of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology26–35. Thus, more sophisticated GEMMs (e.g., the KO:KI model) can be created within the timespan of a few months.
Additionally, CRISPR tools offer many more technical options than were previously possible36. For example, GEMMs can be created in any strain background. While we propose that models #1 and #2 be made in the C57BL/6 reference mouse strain, which is immunocompetent, we also propose that these design strategies are implemented in an immunocompromised mouse strain background, such as NOD/scid-γcnull (NSG/NOG). NSG mice lack mature T cells, B cells, and natural killer (NK) cells37. These GEMMs (#3 and #4) will be useful in many ways. First, due to the immunosuppressed background, there may be uncontrolled SARS-CoV-2 replication, which would make this strain particularly suitable for screening antiviral drugs. Second, the NSG/NOG strain is very commonly used for transplantation of human hematopoietic stem cells, which leads to the development of a humanized immune system38–41. These models will be suitable for pathogenesis studies involving interactions of the human immune system with the virus and can lead to better understanding of the role of human macrophages permissive for SARS-CoV-2 and mechanisms of human immunopathogenesis42. Third, some COVID-19 patients exhibit cytokine storm syndromes and immunosuppression43, which are the result of active viral replication in an immune deficient host. Therefore, GEMMs allowing SARS-CoV-2 infection in immunocompromised backgrounds can serve as valuable tools for such immunological studies. Lastly, humanization of NSG/NOG mice with hACE2 will provide the ability to study the reaction of human immune cells in viral replication. This can be achieved by creating additional genetic modifications in GEMMs, which will allow more sophisticated immunological studies involving transplanting and infecting human cells into the mice, similar to studies with HIV44,45. Considering that the BALB/c is one of the most commonly used mouse strains used for SARS and MERS virus research46,47, we also propose developing the KO:KI model in the BALB/c background strain (Models #5 and #6). For more information on additional strain considerations see the ‘Future Directions” section.
As an alternative to the KO:KI design, one could mutate the amino acids of mACE2 into those required for hACE2 binding to the SARS-CoV-2 spike protein. Crystal structures48,49 showed that five residues (K31, E35, D38, M82 and K353) are critical for receptor binding50. Of these, residues K31, M82, and K353 differ in mice. Therefore, changing just these three residues could allow mACE2 to bind effectively to the SARS-CoV-2 spike protein and permit infection of expressing cells. This strategy would retain any regulatory elements within the mouse introns, and it should result in nearly physiological levels of a functional mouse ACE2 receptor. These models are termed #7, #8, and #9 in C57BL/6, NSG/NOG and BALB/c strains, respectively. A similar point mutation knock-in strategy in which two amino acids (288 and 300) of the DPP4 receptor for MERS-CoV were mutated to render mice susceptible to MERS-CoV was reported previously51.
Transmembrane protease serine 2 TMPRSS2 is a protease important in cleaving hACE2 and SARS-CoV-2 spike proteins, enabling efficient viral entry into cells52. It is possible hACE2 expression alone in mice will be insufficient to permit infection and viral pathogenesis at a level similar to humans. However, co-expression of hTMPRSS2 may achieve this level. Models #10 to #15 (and #25 to #30: see below in category 3) are proposed for the co-expression of hTMPRSS2. KI designs for the purpose of this co-expression would involve either fusing TMPRSS2 to the C terminus of the hACE2 KI cassette via a self-cleavable P2A peptide, or expressing TMPRSS2 as a separate polypeptide via an IRES. These designs are similar to models #1, #3, and #5 but contain P2A-hTMPRSS2 for C-terminus fusion (models #10 to #12) or IRES elements (models #13 to #15). In certain cases, fusion of additional peptide sequences (e.g., P2A) may affect protein folding and function, which can be avoided by using IRES elements. However, the gene downstream of the IRES element may not express as efficiently as the gene placed upstream of it. A strategy of co-expressing hACE2 and hTMPRSS2, similar to the ones we proposed here, was also proposed by Soldatov et al., 2020 (ref. 53), although these authors suggested inserting the expression cassettes into the mTMPRSS2 locus instead of the mAce2 locus. Co-expression of hTMPRSS2 may not be necessary in the case of the key amino acid KI mutant models (#7 to #9) because the mACE2 will likely retain its endogenous cleavage site for mouse TMPRSS2 proteins. It is unknown, however, whether mTMPRSS2 will cleave SARS-CoV-2 spike proteins.
GEMM Category 2: Knocking-in CRE-activatable or tetracycline-inducible expression cassettes into safe-harbor loci by re-engineering existing reporter or inducer mouse lines
Models #1 to #15 will express hACE2 alone or in conjunction with hTMPRSS2 in all cells in which the mAce2 promoter is active. There will, however, be many situations in which scientists require GEMMs for answering very specific questions. For example, what molecular events are perturbed when only cardiac, intestinal, or lung epithelial cells in the mouse are infected with SARS-CoV-2? Such a scenario would require a GEMM expressing hACE2 in only one of the respective cell types, and this expression could be switched on or off when needed. Models #1 to #15 do not offer flexibility to control the timing or tissue specificity of expression.
There are sophisticated molecular genetic switches, however, such as Cre-LoxP and tetracycline-inducible systems that can be included in GEMM designs to allow such defined experimental scenarios. Leveraging the vast numbers of the available Cre driver lines created by the mouse genetics community, CRE-activatable KI mice can be bred to those driver lines to remove the stop cassette and to express the insertion cassette. For example, a KI mouse containing a LoxP-Stop-LoxP cassette between the promoter and hACE2 cassette will express hACE2 only after the removal of the stop cassette, and this can be achieved by breeding the KI mouse with a Cre driver line. Similarly, a KI mouse in which a tetracycline response element drives hACE2 expression could be used for turning on or off expression of hACE2 in any desired cell type at any given time point. Numerous tetracycline-inducible models that express the tetracycline transactivator (tTA) or reverse tTA (rtTA) have been generated by the community and can be bred with the KI model to turn on or off hACE2 expression simply by administering/withdrawing doxycycline. The conditional and inducible expression systems described below will also offer a degree of biosafety compared to constitutively expressing models, as they will likely not produce potentially infectious virus particles before the start of the experiments.
GEMMs #16 and #17 offer constitutive expression with a reporter capability, and models #18 and #19 offer CRE-activatable features. These models will contain hACE2 fused to a fluorescent reporter, such as EGFP or tdT, either through a self-cleavable P2A peptide or expressed as a separate protein via an IRES. The four models will be useful for many different scenarios, including constitutive expression, tissue-specific expression or for turning on or off expression of the receptor as needed, simply by breeding them with Cre driver lines (in the case of models #18 and #19), which are available for thousands of promoters. Fluorescent reporters in animal models will offer a useful tool for histological and biochemical experiments. Models #20 and #21 offer tetracycline-inducible features wherein the expression cassettes are placed downstream of tetracycline response elements. As above, these models will contain hACE2 fused to a fluorescent reporter either through a self-cleavable P2A peptide or expressed as a separate protein via IRES. The expression of the transgenes can be achieved in an inducible fashion by breeding the models with tTA/rtTA driver lines and through doxycycline administration.
These KI models (#16 to #21) can be developed by re-engineering the genomes of some of the previously developed GEMMs, using CRISPR approaches. This re-engineering approach will allow the use of available GEMMs harboring proven genetic elements such as LoxP, tetracycline response elements, polyA signals and built-in reporters. For generating models #16 to #19, for example, a reporter mouse model called mT/mG offers a suitable model for re-engineering. The mT/mG double-fluorescent Cre reporter mouse contains a floxed cassette of membrane-targeted tdT (mT) followed by a membrane-targeted green fluorescent protein (mG), targeted into the ROSA26 locus, one of the most commonly used safe harbor sites in the mouse genome. The mT reporter is expressed in every mouse cell. Once the Cre recombinase is introduced via breeding, the mT cassette is deleted, allowing the CAG promoter to express the mG reporter54. This mouse strain is available for purchase from The Jackson Laboratory (JAX Stock No. 007676). To create tetracycline-inducible KI models (#20 and #21), the mouse strain Ai63 can be re-engineered to insert the hACE2 cDNA near the start codon of the tdT sequence, either as a fusion via a P2A peptide or with an IRES. The Ai63 mouse was developed in Dr. Hongkui Zeng’s laboratory at Allen Brain Research Institute in Seattle and expresses tdT under the Tetracycline Response Element (TRE) promoter55.
GEMM Category 3: Knocking-in CRE-activatable cassettes into mouse Ace2 locus
Another advance in developing models for COVID-19 would be to combine the features of knocking-in expression cassettes with conditional potential. In models #22, #23 and #24, CRE-activatable inversion cassettes expressing hACE2, flanked by a combination of mutant LoxP elements (Lox2272 and 5171)56 are inserted into the intron 1 of mouse Ace2. The coding sequences are placed in the opposite orientation to the mAce2 promoter. Upon CRE-mediated recombination, the coding sequences will be inverted, thereby placing them in the correct orientation to be expressed from the mAce2 promoter, enabling the promoter to drive expression of hACE2. In models #25 to #30, the inversion cassettes express both hACE2 and hTMPRSS2 (either fused to P2A, or as separate proteins using an IRES in between). We call this design, used in models #22 to #30, a conditional KI (cKI) strategy. These GEMMs are expected to express near-physiological levels of hACE2 along with the required amounts of hTMPRSS2 upon CRE-mediated recombination. These models will be particularly suitable for research projects that address well-defined questions requiring selective expression of the two key proteins (hACE2 and hTMPRSS2) only in certain cell types of the mouse at a specific time point of interest. While numerous Cre driver lines developed in the C57BL/6 strain (or the ones backcrossed to this strain) are readily available, such strains are not available for the NSG/NOG and BALB/c strains. Considering that Cre driver lines can be generated using robust technologies like Easi-CRISPR30,32 quite easily, such models can be generated as needed very rapidly (~2 months’ time to generating and identifying the G0 founder mice)32.
Details of all the 30 GEMM designs are given in Table 1. The full sequences of the targeting constructs and the guide RNA sequences, and additional notes of the models are given in Supplementary Note 1. The targeting constructs for these models, except the ones containing hTMPRSS2 coding sequences, are ~3 kilobases or shorter in length. These models can be generated using the Easi-CRISPR approach, which uses long single-stranded DNA as a donor for highly efficient insertion of sequences into the mouse genome30,32. The models containing hTMPRSS2 sequences are longer than 4kb and could also be generated using the Easi-CRISPR approach, but the efficiency may be lower and require the injection of more zygotes to achieve the faithful insertion of a full-length cassette. Alternative strategies of using plasmid DNA constructs (circular dsDNA) containing much longer homology arms25,28,57,58 or ssODN mediated knock-in of large cassettes31 can be attempted in the event that the Easi-CRISPR method is not successful in generating the desired models containing hTMPRSS2.
Concluding remarks
Mice are the most commonly used laboratory animals in biomedical research, and they have helped scientists develop diagnostics, therapeutics and vaccines for many human diseases. While the previously generated transgenic hACE2 mice and non-GEMM approaches (such as adaptation of virus to mice and adenovirus mediated delivery of hACE2) have been successfully validated as potential preclinical models for COVID-19 studies, more sophisticated GEMM approaches will render mice much more versatile COVID-19 models than can be envisioned for any other species. The availability of a wide variety of genetic tools and molecular switches available only in mice offers a unique opportunity to make this species useful for COVID-19 research. We described three broad categories of GEMMs and about 30 different model designs, in three important mouse strains that may be useful for SARS-CoV-2 research and for fighting COVID-19. By using robust CRISPR-based strategies such as Easi-CRISPR30,32 and/or i-GONAD33,34, the founder mice for the majority of these models, particularly those involving only insertion of hACE2 and point mutation knock-ins can be generated within ~2 months. The designs involving both hACE2 and hTMPRSS2 insertion are more complex because of the longer insertion sizes. They may require the injection of larger numbers of zygotes to create knock-in models because as the size of the insertion cassettes become longer, the efficiency of full-length cassette insertion becomes lower. Nevertheless, even the simpler models will rapidly provide the opportunity for significant advances. Furthermore, the use of GEMM mice together with SARS-CoV-2 mouse adapted virus12,16 may provide the ideal mouse model for COVID-19.
Future Directions
COVID-19 is a new disease. There are many unknowns concerning SARS-CoV-2, including its pathogenicity. Nevertheless, the research community is making advances at a remarkable speed. At this stage, the proposed GEMMs suitable for COVID-19 and SARS-CoV-2 research are based on the available information. Additional models could include expressing soluble hACE259 along with FURIN protease60, with the latter cleaving and producing mature proteins required for viral entry. A recent study identified polymorphisms in the human ACE2 and TMPRSS2 genes that may explain variability in susceptibility to COVID-1961. These polymorphisms could be engineered into mice for comparison with the proposed GEMMs expressing the standard human genes.
In addition, over the last century, the laboratory mouse has been bred to produce an extensive catalogue of inbred, hybrid and outbred strains, which collectively provide an elegant system suitable for studying many human disease conditions. This wealth of strain resources could be leveraged for SARS-CoV-2 disease research. For example, the GEMM designs proposed here can be generated in additional mouse strains taking into consideration SARS-CoV-2 co-morbidities and human genetic diversity. The New Zealand Obese (NZO) mouse strain could be useful for simulating SARS-CoV-2 co-morbidities62 of obesity and diabetes because of its natural insulin resistance and susceptibility to developing these conditions63. Hybrids of the New Zealand Black (NZB) and New Zealand White (NZW) strains could be considered for simulating autoimmune conditions, as they have been used as models of human systemic lupus erythematosus64. Several inbred and hybrid strains have been tested for their suitability for performing studies of cardiac function65, vascular properties66, hypertension67 and arrythmia68. These demonstrate a wide range of variability in assay performance among genetic backgrounds, however, suggesting that careful considerations should be made in choosing strain backgrounds for a given assay in cardiovascular disease research.
During the last decade, a novel set of strains called Collaborative Cross (CC) strains were also added to the vast array of mouse genetic backgrounds. CC strains are specially designed to overcome the narrow genetic diversity of inbred strains by crossing many inbred mouse lines together69. The complex mix of human genetic diversity is expected to be reflected in the mouse CC strains. These strains have been used for a wide range of studies, including identifying genetic modifiers affecting the susceptibility for the pathogenicity of infectious diseases affecting human lung70. Genome-wide studies have been performed by infecting CC strains with mouse adapted SARS-CoV to identify polymorphic host genes that contribute to pathogenesis, and these have identified genes such as Trim5571 and Ticam272. Related to this, human genome-wide association studies (GWAS), have identified genomic regions that may be important for SARS-CoV-2 susceptibility and severity. Interestingly, preliminary studies have identified a region of chromosome 3p21.31 comprising six genes73,74, one of which is SLC6A20, a gene that encodes a transporter interacting with hACE2.
Incorporating factors like FURIN protease, or polymorphic versions of either ACE2, TMPRSS2 or SLC6A20 similar to our proposed GEMM designs, or producing our proposed GEMMs in additional strain backgrounds including collaborative cross strains can extend the COVID-19 GEMM list from #31 onward and vastly increase opportunities to understand COVID-19. Furthermore, using mouse as the model system allows us to leverage the results of research into over three dozen genes that have already been investigated with respect to coronavirus disease (see Figure 2 of a recent review by LoPresti et al)75. Systematically breeding the 30 knock-in models described here with each of the knockouts for those three dozen genes will serve as a valuable tool to understand more about coronaviruses.
Figure 2. Schematics of COVID-19 GEMM designs, category 2: Knocking-in CRE-activatable or tetracycline-inducible expression cassettes into safe-harbor loci by re-engineering existing reporter or inducer mouse lines.
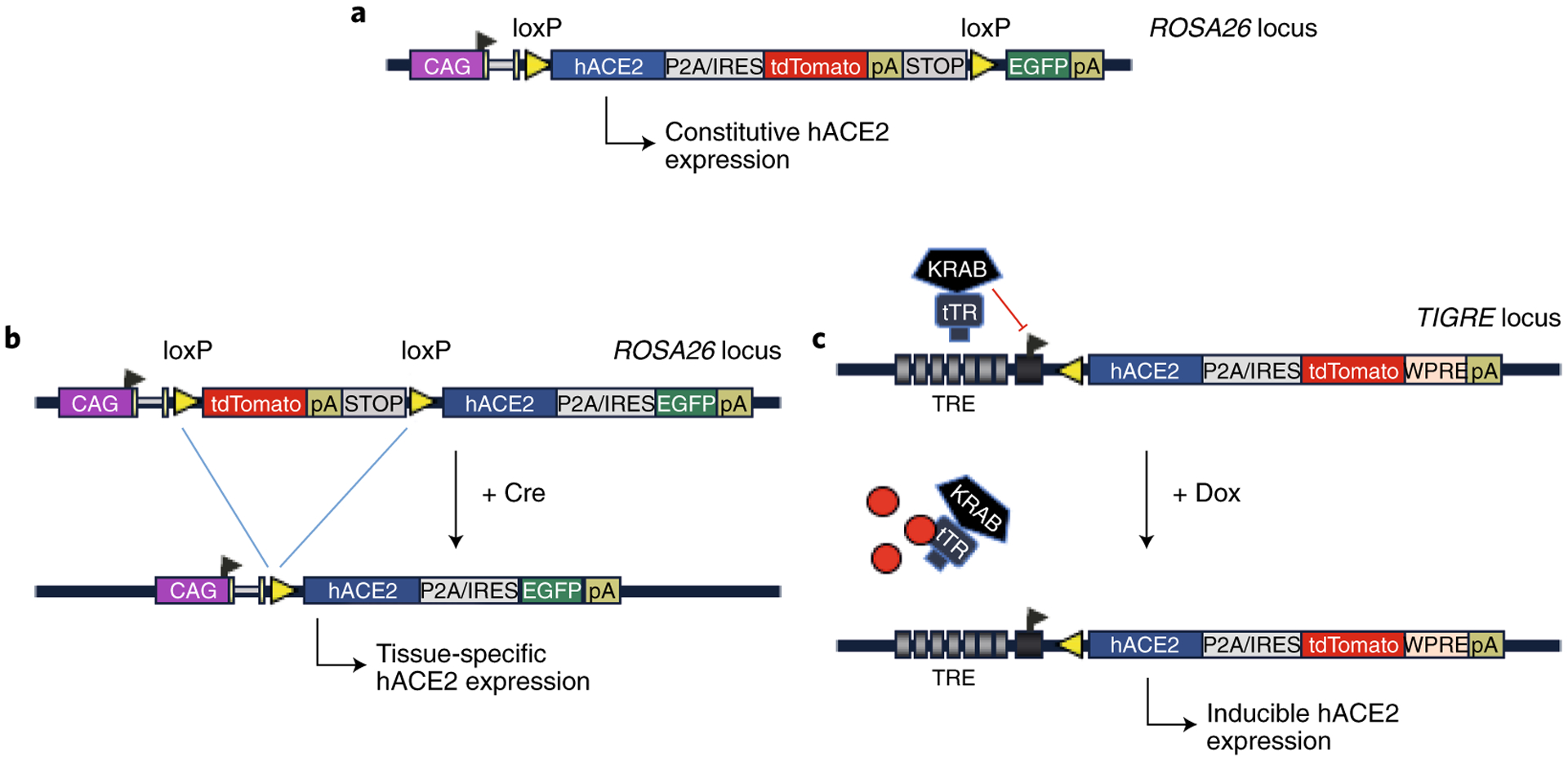
(A) Constitutive (B) CRE-activatable and (C) tetracycline-inducible designs (GEMMs #16 to #21). These models can be generated by re-engineering existing reporter and tetracycline-inducible models. P2A: a self-cleavable viral peptide used for expressing two fusion proteins. IRES: Internal Ribosomal Entry Site.
The list of 30 GEMMs proposed herein offers a foundation for the research community to generate the first set of small animal models for COVID-19 and SARS-CoV-2 research. These are expected to prove valuable preclinical models for evaluating COVID-19 therapeutics and vaccines, which may continue to be important for studying the next coronavirus that may jump from animals to people.
Supplementary Material
Supplementary Note 1. Sequences of the ssDNA constructs and the guide RNAs with additional details of the 30 proposed GEMMs. This file contains the genetic blueprints of the model designs, additional rationale and notes relevant to each model as well as the technical information needed for developing the models.
Acknowledgements:
We thank Hongkui Zeng, Bosiljka Tasic, and Tanya Daigle (Allen Institute for Brain Science) for advice and discussions on the design of re-engineered alleles. We thank Melody A. Montgomery and Douglas D. Meigs (University of Nebraska Medical Center) for professional editing. C.B.G is funded by NIH grants R35HG010719, R21GM129559, R21AI143394, and R21DA046831. S.L.M is funded by NIH grants R01DC011819, R01DC014470. M.O is funded by JSPS (16KK0189) for the Promotion of Joint International Research (Fostering Joint International Research)
Footnotes
Competing interests: University of Nebraska Medical Center has filed a provisional patent on the proposed mouse model designs (inventors: C.B.G, R.M.Q and M.O).
References
- 1.Shi J et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 368, 1016–1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockx B et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science eabb7314 (2020) doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sia SF et al. Pathogenesis and transmission of SARS-CoV-2 virus in golden Syrian hamsters. https://www.researchsquare.com/article/rs-20774/v1 (2020) doi: 10.21203/rs.3.rs-20774/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X-H et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med 57, 450–459 (2007). [PubMed] [Google Scholar]
- 5.McCray PB et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng C-TK et al. Severe Acute Respiratory Syndrome Coronavirus Infection of Mice Transgenic for the Human Angiotensin-Converting Enzyme 2 Virus Receptor. J. Virol 81, 1162–1173 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menachery VD et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci 113, 3048–3053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao L et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature (2020) doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 9.Jiang R-D et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell S009286742030622X (2020) doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan L, Tang Q, Cheng T & Xia N Animal models for emerging coronavirus: progress and new insights. Emerg. Microbes Infect 9, 949–961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A et al. A Comprehensive Review of Animal Models for Coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol. Sin 35, 290–304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinnon KH et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. http://biorxiv.org/lookup/doi/10.1101/2020.05.06.081497 (2020) doi: 10.1101/2020.05.06.081497. [DOI] [Google Scholar]
- 13.Winkler ES et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol (2020) doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Zhou W, Yang L & You R Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res 157, 104833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts A et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu H et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science eabc4730 (2020) doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell S0092867420307418 (2020) doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan AO et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell S009286742030742X (2020) doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rathnasinghe R et al. Comparison of Transgenic and Adenovirus hACE2 Mouse Models for SARS-CoV-2 Infection. http://biorxiv.org/lookup/doi/10.1101/2020.07.06.190066 (2020) doi: 10.1101/2020.07.06.190066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couzin-Frankel J From ‘brain fog’ to heart damage, COVID-19’s lingering problems alarm scientists. Science (2020) doi: 10.1126/science.abe1147. [DOI] [Google Scholar]
- 21.Lindner D et al. Association of Cardiac Infection With SARS-CoV-2 in Confirmed COVID-19 Autopsy Cases. JAMA Cardiol. (2020) doi: 10.1001/jamacardio.2020.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parasa S et al. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw. Open 3, e2011335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koralnik IJ & Tyler KL COVID ‐19: A Global Threat to the Nervous System. Ann. Neurol 88, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hikmet F et al. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol 16, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun S-H et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 28, 124–133.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen B et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23, 720–723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms DW et al. Mouse Genome Editing Using the CRISPR/Cas System. Curr. Protoc. Hum. Genet 83, 15.7.1–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aida T et al. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 16, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura H CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci Rep 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quadros RM et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimi K ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura H, Quadros RM, Gurumurthy CB & Ohtsuka M Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc 13, 195–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtsuka M et al. i-GONAD: a robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 19, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurumurthy CB et al. Creation of CRISPR-based germline-genome-engineered mice without ex vivo handling of zygotes by i-GONAD. Nat. Protoc 14, 2452–2482 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Gurumurthy CB & Lloyd KCK Generating mouse models for biomedical research: technological advances. Dis. Model. Mech 12, dmm029462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurumurthy CB et al. CRISPR/Cas9 and the Paradigm Shift in Mouse Genome Manipulation Technologies in Genome Editing (ed. Turksen K) 65–77 (Springer International Publishing, 2016). [Google Scholar]
- 37.Shultz LD et al. Human Lymphoid and Myeloid Cell Development in NOD/LtSz- scid IL2R γ null Mice Engrafted with Mobilized Human Hemopoietic Stem Cells. J. Immunol 174, 6477–6489 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Ito M et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100, 3175–3182 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Saito Y et al. The in vivo development of human T cells from CD34(+) cells in the murine thymic environment. Int. Immunol 14, 1113–1124 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa F et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood 106, 1565–1573 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorantla S, Poluektova L & Gendelman HE Rodent models for HIV-associated neurocognitive disorders. Trends Neurosci. 35, 197–208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merad M & Martin JC Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol (2020) doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta P et al. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet 395, 1033–1034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dagur RS et al. Human-like NSG mouse glycoproteins sialylation pattern changes the phenotype of human lymphocytes and sensitivity to HIV-1 infection. BMC Immunol. 20, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dash PK et al. Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat. Commun 10, 2753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts A et al. Aged BALB/c Mice as a Model for Increased Severity of Severe Acute Respiratory Syndrome in Elderly Humans. J. Virol 79, 5833–5838 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J et al. Cellular Immune Responses to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection in Senescent BALB/c Mice: CD4+ T Cells Are Important in Control of SARS-CoV Infection. J. Virol 84, 1289–1301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, Li W, Farzan M & Harrison SC Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Wan Y, Shang J, Graham R, Baric RS & Li F Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol 94, e00127–20, /jvi/94/7/JVI.00127–20.atom (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luan J, Lu Y, Jin X & Zhang L Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun 526, 165–169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leist SR & Cockrell AS Genetically Engineering a Susceptible Mouse Model for MERS-CoV-Induced Acute Respiratory Distress Syndrome in MERS Coronavirus (ed. Vijay R) vol. 2099 137–159 (Springer; US, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soldatov VO, Kubekina MV, Silaeva Y. Yu., Bruter AV & Deykin AV On the way from SARS-CoV-sensitive mice to murine COVID-19 model. Res. Results Pharmacol 6, 1–7 (2020). [Google Scholar]
- 54.Muzumdar MD, Tasic B, Miyamichi K, Li L & Luo L A global double-fluorescent Cre reporter mouse. Genes. N. Y. N 2000 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Daigle TL et al. A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robles-Oteiza C et al. Recombinase-based conditional and reversible gene regulation via XTR alleles. Nat. Commun 6, 8783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abe T, Inoue K, Furuta Y & Kiyonari H Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes. Cell Rep. 31, 107653 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Yoshimi K et al. Combi-CRISPR: combination of NHEJ and HDR provides efficient and precise plasmid-based knock-ins in mice and rats. Hum. Genet (2020) doi: 10.1007/s00439-020-02198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monteil V et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181, 905–913.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersen KG, Rambaut A, Lipkin WI, Holmes EC & Garry RF The proximal origin of SARS-CoV-2. Nat. Med 26, 450–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou Y et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 18, 216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson S et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 323, 2052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veroni MC, Proietto J & Larkins RG Evolution of Insulin Resistance in New Zealand Obese Mice. Diabetes 40, 1480–1487 (1991). [DOI] [PubMed] [Google Scholar]
- 64.Vyse TJ et al. Genetic linkage of IgG autoantibody production in relation to lupus nephritis in New Zealand hybrid mice. J. Clin. Invest 98, 1762–1772 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnabei MS, Palpant NJ & Metzger JM Influence of genetic background on ex vivo and in vivo cardiac function in several commonly used inbred mouse strains. Physiol. Genomics 42A, 103–113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steppan J et al. Commonly used mouse strains have distinct vascular properties. Hypertens. Res (2020) doi: 10.1038/s41440-020-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao W et al. Differential Expression of Hypertensive Phenotypes in BXD Mouse Strains in Response to Angiotensin II. Am. J. Hypertens 31, 108–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jelinek M, Wallach C, Ehmke H & Schwoerer AP Genetic background dominates the susceptibility to ventricular arrhythmias in a murine model of β-adrenergic stimulation. Sci. Rep 8, 2312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Threadgill DW, Miller DR, Churchill GA & de Villena FP-M The Collaborative Cross: A Recombinant Inbred Mouse Population for the Systems Genetic Era. ILAR J. 52, 24–31 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Lorè NI et al. Collaborative Cross Mice Yield Genetic Modifiers for Pseudomonas aeruginosa Infection in Human Lung Disease. mBio 11, e00097–20, /mbio/11/2/mBio.00097–20.atom (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gralinski LE et al. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLOS Genet. 11, e1005504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gralinski LE et al. Allelic Variation in the Toll-Like Receptor Adaptor Protein Ticam2 Contributes to SARS-Coronavirus Pathogenesis in Mice. G3amp58 GenesGenomesGenetics 7, 1653–1663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellinghaus D et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med NEJMoa2020283 (2020) doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The COVID-19 Host Genetics Initiative. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet 28, 715–718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.LoPresti M, Beck DB, Duggal P, Cummings DAT & Solomon BD The Role of Host Genetic Factors in Coronavirus Susceptibility: Review of Animal and Systematic Review of Human Literature. Am. J. Hum. Genet S0002929720302755 (2020) doi: 10.1016/j.ajhg.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note 1. Sequences of the ssDNA constructs and the guide RNAs with additional details of the 30 proposed GEMMs. This file contains the genetic blueprints of the model designs, additional rationale and notes relevant to each model as well as the technical information needed for developing the models.


