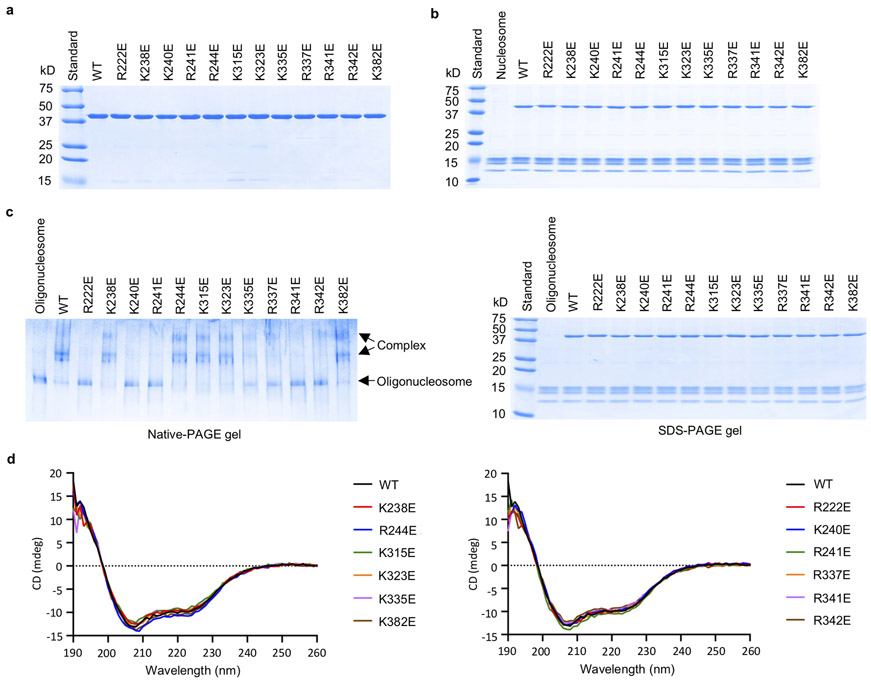
Extended Data Figure 7. Characterization of mcGAS catalytic domain mutants and oligonucleosome binding studies.
a. SDS-PAGE analysis of mouse cGAS catalytic domain mutants used for the gel shift assays and enzyme activity assays.
b. SDS-PAGE analysis of mcGAS and nucleosome mixture samples used for the gel shift assay.
c. Polyacrylamide gel electrophoretic mobility shift assay (EMSA) shows that mutations at the cGAS-nucleosome interface affect oligonucleosome binding by cGAS. In these samples, mcGAS was mixed with oligonucleosome at molar ratio of 6:1. The samples used for the binding studies were analyzed by SDS-PAGE (right panel).
d. Circular dichroism of mouse cGAS catalytic domain and its mutants used for gel shift assays and enzyme activity assays. cGAS mutants that have strong binding to nucleosomes are shown by the spectra on the left. cGAS mutants that have weak or no binding to nucleosomes are shown by the spectra on the right.