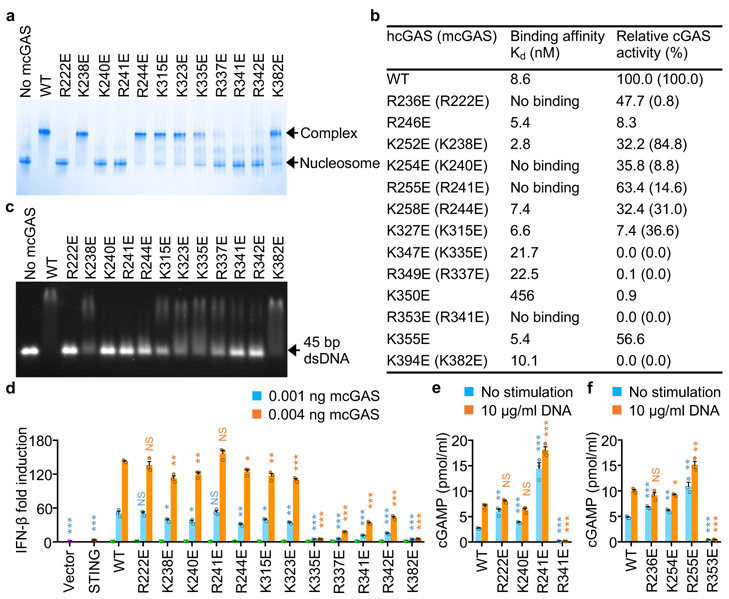
Figure 4. Mutations at the cGAS-nucleosome interface affect nucleosome binding, dsDNA binding, cGAS activity, and cGAS mediated signaling.
a. Polyacrylamide gel electrophoretic mobility shift assay (EMSA) shows that mutations at the cGAS-nucleosome interface affect nucleosome binding by mcGAS. In this assay, mcGAS catalytic domain was mixed with nucleosome at molar ratio of 3:1.
b. Binding affinities of human cGAS mutants to the nucleosome and the relative catalytic activities of human and mouse cGAS mutants.
c. Agarose gel electrophoretic mobility shift assay (EMSA) shows that mutations at the cGAS-nucleosome interface affect dsDNA binding. In this assay, mcGAS catalytic domain was mixed with 45 bp dsDNA at molar ratio of 20:1
d. IFN-β luciferase reporter assays show that mutations of mcGAS affect signaling in HEK 293T cells. Luciferase reporter signals from the cells transfected with 0.004 ng cGAS and 0.4 ng STING are indicated by the orange bars, from cells transfected with 0.001 ng cGAS and 0.4 ng STING by the cyan bars, from cells transfected with 0.004 ng cGAS, 0.4 ng STING, or the vector control by the green, brown and purple bars, respectively.
e. cGAMP levels in HEK 293T cells transfected with mcGAS mutants. The cells were transfected with indicated mcGAS plasmids for 24 hours and stimulated with lipofactamine alone or lipofactamine plus salmon sperm DNA, followed by cGAMP assays from the cell lysates.
f. cGAMP levels in HEK 293T cells transfected with hcGAS mutants with and without DNA stimulation.
In d-f, the data (mean ± SEM) are representative of three independent experiments. Each dot represents a biological replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS-not significant.