Abstract
Huanglongbing (HLB), caused mainly by ‘Candidatus Liberibacter asiaticus’ (CLas), is the most devastating citrus disease because all commercial species are susceptible. HLB tolerance has been observed in Poncirus trifoliata and their hybrids. A wide-ranging transcriptomic analysis using contrasting genotypes regarding HLB severity was performed to identify the genetic mechanism associated with tolerance to HLB. The genotypes included Citrus sinensis, Citrus sunki, Poncirus trifoliata and three distinct groups of hybrids obtained from crosses between C. sunki and P. trifoliata. According to bacterial titer and symptomatology studies, the hybrids were clustered as susceptible, tolerant and resistant to HLB. In P. trifoliata and resistant hybrids, genes related to specific pathways were differentially expressed, in contrast to C. sinensis, C. sunki and susceptible hybrids, where several pathways were reprogrammed in response to CLas. Notably, a genetic tolerance mechanism was associated with the downregulation of gibberellin (GA) synthesis and the induction of cell wall strengthening. These defense mechanisms were triggered by a class of receptor-related genes and the induction of WRKY transcription factors. These results led us to build a hypothetical model to understand the genetic mechanisms involved in HLB tolerance that can be used as target guidance to develop citrus varieties or rootstocks with potential resistance to HLB.
Subject terms: Biotechnology, Genetics, Molecular biology, Plant sciences
Introduction
Huanglongbing (HLB) or greening has been considered the most devastating citrus disease. HLB is caused by the gram-negative, phloem-limited, α-proteobacterium Candidatus Liberibacter species. The following three Liberibacter species have been associated with HLB: Candidatus Liberibacter asiaticus (CLas), Candidatus Liberibacter americanus (CLam) and Candidatus Liberibacter africanus (CLaf). CLas is the most widespread and is responsible for large economic losses worldwide1,2.
HLB symptoms include blotchy chlorosis, mottling of leaves, yellow shoots, vein corking, stunted growth and small, green, and lopsided fruits with aborted seeds3. HLB symptom development is considered a consequence of a series of molecular, cellular, and physiological disorders in the plant host. The most expressive modifications caused by CLas in the citrus host are alterations in sucrose and starch metabolism, changes of hormone production, biosynthesis of secondary metabolites, phloem function disorders, and source-sink communication4,5.
Poncirus trifoliata is closely related and sexually compatible with the citrus genus, and it shows attenuated HLB symptoms and lower CLas titer, indicating that this genus possibly presents genetic defense mechanism against CLas6,7. Moreover, some citrus hybrids of P. trifoliata have also been reported to present a significant tolerance to HLB7,8; however, it remains unclear which mechanisms are involved in this tolerance. In contrast, all commercial Citrus species are susceptible to CLas infection, and the identification of tolerant genotypes is essential to the maintenance of citrus production2. Studies are still necessary to understand better the differences of genetic responses involved in the susceptibility, tolerance or resistance to such genotypes, aiming to obtain new citrus variety tolerant to HLB by conventional breeding or genetic engineering.
Our study provides a wide-ranging transcriptomic analysis of two CLas-susceptible citrus genotypes (Citrus sinensis and C. sunki), one CLas-tolerant genotype (P. trifoliata), and three pools of hybrids between P. trifoliata and C. sunki, which are classified as susceptible, tolerant, and resistant to HLB. Therefore, this work was the first to study transcriptional reprogramming and to compare the results of a large volume of transcriptomes, including individuals from a population of hybrids infected by CLas, which consequently inherited the susceptible and tolerance genetic mechanisms from their parents.
The results revealed that only a few genes associated with specific pathways were modulated in resistant genotypes to avoid CLas proliferation and plant disease severity. Using the transcriptomic analysis of the hybrid genotypes, we revalidated the mechanisms of susceptibility and tolerance of their parents. Based on the analysis, we built a hypothetical model to explain the genetic mechanism involved in HLB tolerance conferred by P. trifoliata and inherited by its hybrids that could be further used in breeding or biotechnological approaches.
Results
CLas quantification
CLas quantification analysis showed that all plants from C. sinensis, C. sunki, and P. trifoliata were infected by CLas after 240 days of inoculation. From the analysis of the 21 hybrids, nine of them (H68, H106, H109, H113, H142, H156, H154, H161, and H165) were selected for the subsequent steps. The H109, H161, H165, H113, H154, and H146 hybrids were infected, but the H68, H106, and H142 hybrids were negative for the presence of CLas in all biological replicates (Tables 1 and 2).
Table 1.
Detection and quantification of the bacteria by quantitative PCR (qPCR) in Citrus sinensis, C. sunki, Poncirus trifoliata and nine hybrids from an F1 population obtained from the cross between C. sunki and P. trifoliata Raf. cv Rubidoux. Each individual is represented by five repetitions.
| Genotypes | HLB diagnosis (qPCR) days after inoculation | ||||
|---|---|---|---|---|---|
| 30 | 90 | 180 | 240 | 360 | |
| C. sunki | 0/5 | 3/5 | 4/5 | 4/5 | 5/5 |
| C. sinensis | 0/5 | 3/5 | 5/5 | 5/5 | 5/5 |
| P. trifoliata | 0/5 | 0/5 | 0/5 | 3/5 | 3/5 |
| H106 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| H109 | 1/5 | 2/5 | 4/5 | 5/5 | 5/5 |
| H146 | 1/5 | 1/5 | 3/5 | 5/5 | 5/5 |
| H68 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| H161 | 0/5 | 3/5 | 5/5 | 5/5 | 5/5 |
| H142 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| H165 | 0/5 | 3/5 | 5/5 | 5/5 | 5/5 |
| H154 | – | – | 4/5 | 5/5 | 5/5 |
| H113 | – | – | 5/5 | 5/5 | 5/5 |
–: Correspond to samples that were not evaluated due to the absence of leaves.
Table 2.
Candidatus Liberibacter asiaticus (CLas) quantification obtained by comparing the standard curve of the HLB primers with the standard curve of the internal control gene (GAPDH) initiators. The value quantification refers to Log10 of the number of copies of the CLas fragment after 240 days from inoculation in each repetition per genotype included in RNAseq analysis.
| Genotype | Ct value of GAPDH | Ct value of HLB | Quantification/Log10 number of copies |
|---|---|---|---|
| C. sinensis | 24.24 | 26.48 | 3.48 |
| 22.07 | 24.30 | 4.09 | |
| 20.12 | 31.74 | 2.01 | |
| C. sunki | 18.43 | 24.90 | 3.92 |
| 18.18 | 20.51 | 5.15 | |
| 18.56 | 25.17 | 3.85 | |
| P. trifoliata | 19.99 | 20.31 | 5.20 |
| 19.20 | 21.85 | 4.78 | |
| 18.30 | 25.80 | 3.67 | |
| H109 | 19.16 | 17.95 | 5.86 |
| 18.30 | 20.35 | 5.19 | |
| 19.13 | 20.38 | 5.19 | |
| H161 | 20.32 | 21.12 | 4.98 |
| 19.28 | 18.87 | 5.61 | |
| 19.23 | 18.75 | 5.64 | |
| H165 | 18.98 | 18.85 | 5.61 |
| 19.05 | 22.18 | 4.68 | |
| 18.52 | 18.87 | 5.61 | |
| H113 | 17.67 | 21.93 | 4.75 |
| 19.14 | 26.25 | 3.55 | |
| 19.05 | 20.81 | 5.07 | |
| H154 | 19.47 | 30.79 | 2.28 |
| 19.05 | 19.33 | 5.48 | |
| 19.05 | 20.55 | 5.14 | |
| H146 | 18.78 | 21.33 | 4.92 |
| 18.35 | 22.16 | 4.69 | |
| 18.48 | 25.60 | 3.73 | |
| H68 | 20.28 | Undetermined | 0 |
| 19.39 | Undetermined | 0 | |
| 19.50 | Undetermined | 0 | |
| H106 | 19.96 | Undetermined | 0 |
| 19.86 | Undetermined | 0 | |
| 18.34 | Undetermined | 0 | |
| H142 | 20.93 | Undetermined | 0 |
| 18.59 | Undetermined | 0 | |
| 18.99 | Undetermined | 0 |
Phenotypic analysis
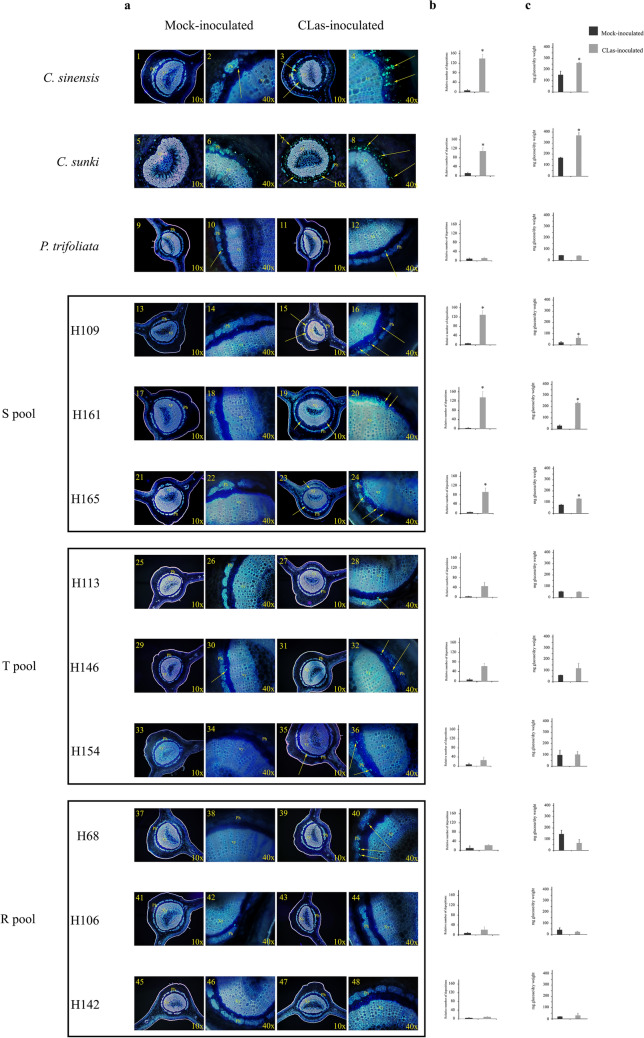
A significant increase in callose deposition was observed for the CLas-infected C. sinensis, C. sunki, H109, H161, and H165 plants compared to the control (Fig. 1). Moreover, P. trifoliata, H113, H154, H146, H68, H106, and H142 showed no difference between the mock and CLas-inoculated plants (Fig. 1). Compared with inoculated and mock-inoculated plants, C. sinensis, C. sunki, and three infected hybrids (H109, H161 and H165) showed a significant difference (p < 0.05) in the amount of starch. In contrast, no significant difference in starch accumulation was observed in P. trifoliata and the other six hybrids (H113, H154, H146, H68, H106, and H142) (Fig. 1).
Figure 1.
Callose deposition. (a) Cross sections of leaf petioles of C. sinensis mock-inoculated (1 and 2) and CLas inoculated (3 and 4), C. sunki mock-inoculated (5 and 6) and CLas inoculated (7 and 8), P. trifoliata mock-inoculated (9 and 10) and CLas inoculated (11 and 12), H109 mock inoculated (13 and 14) and CLas inoculated (15 and 16), H161 mock-inoculated (17 and 18) and CLas inoculated (19 and 20), H165 mock-inoculated (21 and 22) and CLas inoculated (23 and 24), H113 mock-inoculated (25 and 26) and CLas inoculated (27 and 28), H146 mock-inoculated (28 and 30) and CLas inoculated (31 and 32), H154 mock-inoculated (33 and 34) and CLas inoculated (35 and 36), H68 mock-inoculated (37 and 38) and CLas inoculated (39 and 40), H106 mock-inoculated (41 and 42) and CLas inoculated (42 and 44), H142 mock-inoculated (45 and 46) and CLas-inoculated (47 and 48). FL, phloem; Xi, xylem. (b) The bar graph next to the microscopy plates show the callose quantification performed by counting fluorescent spots marked by aniline blue dye. Quantification was performed with tree replicates per genotype, inoculated plants (positive or negative HLB) and mock-inoculated plants. (c) Starch quantification. Individuals were inoculated with CLas (CLas-infected) or mock-inoculated (CLas-free) and collection was performed after 240 days, and quantification was carried by the enzymatic method. Bars represent the standard deviation between 3 biological replicates. * p_value < 0.05 (Mock-inoculated × CLas inoculated).
In general, the visual symptoms were more evident in the susceptible plants, while the visual HLB symptoms were undefined in P. trifoliata and its hybrids. However, according to CLas detection, starch and callose quantification between different treatments, the hybrids were clustered into three distinct groups as follows: Susceptible Pool (S Pool), composed of three different hybrids (H109, H161, and H165) that were diagnosed as HLB-positive and presented elevated starch and callose deposition, similar to that observed for susceptible parental genotypes (Fig. 1); Tolerant Pool (T Pool), composed of three different hybrids (H113, H154, and H146) that were diagnosed as HLB-positive but did not exhibit a significant starch and callose accumulation as observed in susceptible genotypes (Fig. 1); and Resistant Pool (R Pool), composed of three different hybrids (H68, H106, and H142) that were diagnosed as HLB-negative with starch quantification similar to healthy plants (mock-inoculated plants) (Fig. 1).
Transcriptome assembly
To elucidate the different responses to CLas infection, we studied the changes in global transcriptional level in susceptible, tolerant, and resistant genotypes infected by CLas. In this work, 36 cDNA libraries from six different genotypes of either CLas-inoculated or mock-inoculated (control) samples were evaluated. After trimming, 487 million reads were obtained, and 95% of the total was assigned (see Supplementary Table S1). The reads were mapped in 133,976 transcripts on the C. sinensis genome available on http://citrus.hzau.edu.cn/.
HLB-susceptible genotypes, C. sinensis and C. sunki, showed a high number of differentially expressed genes (6141 and 5624 DGEs, respectively) compared with the tolerant parental, P. trifoliata (100 DEGs) (Table 3). A similar pattern was observed between the pool of hybrids. The S Pool showed 708 differentially expressed genes (DEGs), while the R Pool presented only 92 DGEs. The Tolerant Pool (T Pool) showed the highest number of DEGs (2027) among the hybrid pools. Most of these genes were downregulated in HLB-infected plants compared with healthy ones (Table 3).
Table 3.
Number of differentially expressed genes in C. sinensis, C. sunki, P. trifoliata, S Pool, T Pool and R Pool. CLas-infected plants compared with healthy plants.
| Genotypes | Up-regulated | Down-regulated | Total |
|---|---|---|---|
| C. sinensis | 3175 | 2966 | 6141 |
| C. sunki | 3288 | 2336 | 5624 |
| P. trifoliata | 70 | 30 | 100 |
| S Pool | 288 | 420 | 708 |
| T Pool | 939 | 1088 | 2027 |
| R Pool | 63 | 29 | 92 |
| Total | 5812 | 5331 | 14,692 |
The principal component analysis (PCA) using the Bioconductor package (see Supplementary Fig. S1) showed the replicates of the different genotypes in general grouped according to the analyzed condition for C. sunki, C. sinensis, and the susceptible and tolerant hybrids. The resistant groups in fact presented a mixed grouping, which is not surprising if we consider that these hybrids were the ones that showed the fewer number of DEGs. The genotype grouping indicated that the global expression landscape is related more to the different genotypes and not the analyzed condition (infection by CLas). In this case, the identification of genes exclusively differentially expressed in the genotypes considered susceptible, tolerant, or resistant as well as genes that had antagonistic expression between the opposite phenotypes became important to increase our understanding of the different responses.
Differential gene expression analysis
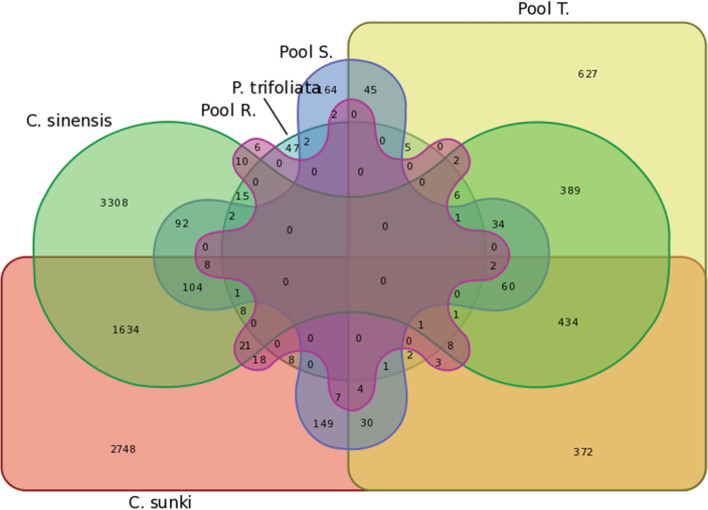
The results are summarized in a Venn diagram (Fig. 2 and Table S2). The susceptible genotypes, C. sinensis and C. sunki, exhibited the highest number of overlapping DEGs (1634), and 88% of these genes presented a similar expression pattern (Fig. 2, Supplementary Table S2), suggesting that a similar gene modulation is caused by CLas infection. In P. trifoliata, 47% of the DGEs were exclusive of this genotype (Fig. 2 and Supplementary Table S2), and 26% of the DGEs were overlapped and showed antagonistic expression compared to susceptible genotypes. Five of the downregulated genes in P. trifoliata were upregulated in both C. sinensis and C. sunki genotype, and one gene was upregulated in the S Pool (see Supplementary Table S3 and S4).
Figure 2.
Venn diagram, considering common and exclusive DGEs of C. sinensis, P. trifoliata, C. sunki, S Pool, T Pool and R Pool.
Among the seven genes upregulated in the R Pool, five were downregulated in C. sinensis, and one gene was downregulated in the T Pool and another one in C. sunki (see Supplementary Table S5). The study of genes with antagonistic expression between susceptible and tolerant and/or resistant genotypes may help to explain possible tolerance mechanisms as well as to identify good targets for plant resistance.
Main processes affected by CLas infection
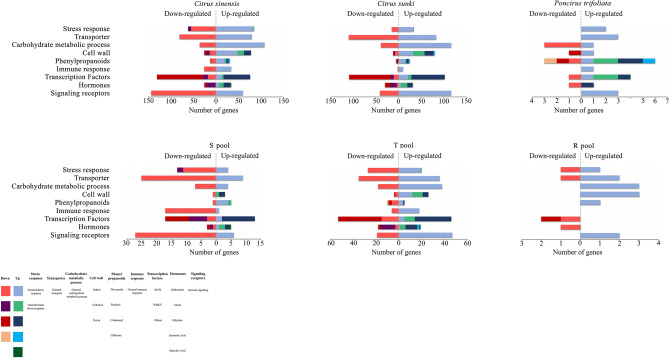
Libraries of DEG functions assigned by Blast2GO9 and Gene ontology (GO)10 analyses helped us better understand the differences in genetic responses involved in susceptibility, tolerance, or resistance (Fig. 3). Susceptible genotypes and tolerant hybrids differentially expressed many genes in comparison to resistant hybrids and P. trifoliata. These different pathways provided valuable information regarding the genetic mechanisms of CLas perception and responses activated in tolerant/resistant and susceptible hosts (Fig. 3).
Figure 3.
C. sinensis, C. sunki, P. trifoliata, S Pool, T Pool and R Pool responses to 240 days of infection by CLas. Genes are classified into nine groups (Stress response, Transporter, Carbohydrate metabolic process, Cell wall, Phenylpropanoids, Immune response, Transcription Factors Hormones and Signaling receptors) according to Blast2GO analysis and based on their expression pattern. The number of down-regulated genes in response to CLas is represented by the bars in reddish tones and upregulated in blue tones. Some bars present subdivisions and the color legend for each pathway is indicating the specific, related gene or specific pathways, which were important to illustrate the proposed tolerance mechanism to HLB.
Differentially expressed genes (DEGs) associated with a specific biological pathway
Signaling receptor
Plant receptors are responsible for the recognition of several external stimuli, including pathogen attack. These transmembrane proteins are directly associated with signaling pathways, which trigger a proper physiological response11. Several types of receptors were regulated in C. sinensis, C. sunki, the S Pool, and the T Pool, and most of them were downregulated in those genotypes (Fig. 3). In P. trifoliata and the R Pool only a few receptors were differentially expressed, and most of them were induced (Fig. 3). These receptors included G-type lectin S-receptor-like, cysteine-rich receptor kinase, and serine/threonine-protein kinase, which were upregulated in P. trifoliata, and leucine-rich repeat transmembrane kinase and leucine-rich repeat receptor-like protein kinase, which were induced in the R Pool (see Supplementary Table S6). Therefore, our results suggests that downregulation of receptors may be associated with susceptible response to CLas.
Hormones
Genes associated with auxin and ethylene pathways were barely or not affected in P. trifoliata and the R Pool, whereas many auxin and ethylene-related genes were differentially expressed in C. sunki, C. sinensis, the T Pool, and the S Pool under CLas infection. Interestingly, no important changes in the transcriptional profiles of genes related to SA and JA biosynthesis were found (Fig. 3). In addition, CLas induced key genes involved with gibberellin (GA) degradation in tolerant and resistant genotypes, while the related GA synthesis genes were downregulated. In P. trifoliata, the gibberellin-induced gene was one of the top three downregulated DEGs (log2 fold change = − 10) (see Supplementary Table S6). The opposite pattern was observed in CLas-susceptible genotypes, in which an induction of genes involved with GA synthesis and downregulation of GA degradation was observed. Thus, these findings suggests that GA plays an important role in CLas-citrus interactions, affecting plant physiology and consequently HLB symptoms.
Transcription factors
Plant responses to pathogen attack require large-scale transcriptional reprogramming. P. trifoliata showed only five transcription factor (TF)-related genes modulated by CLas infection. Only the MYB TF was downregulated. The other four TFs were upregulated, including two WRKY TFs (Fig. 3). The resistant hybrids suppressed the expression of another class of transcription factor, the SCL domain (see Supplementary Table S6). In contrast, hundreds of TF genes showed changes at the transcription level in C. sinensis, C. sunki, the S Pool and the T Pool (Fig. 3). In this context, the large number of TFs affected in these genotypes may be directly related to the regulation of genes responsive to HLB infection. Of note, several WRKY TFs were identified in C. sinensis and C. sunki, and most of them were repressed in CLas-infected plants (Fig. 3). Therefore, these results indicated that the increase in transcription of WRKY TFs in P. trifoliata is associated with the genetic defense mechanism involved with HLB tolerance.
Defense-related genes
Defense-related genes are directly related to processes or production of compounds able to inhibit pathogen reproduction or to make further infection more difficult12. In particular, one defense-related gene, endochitinase B, was differentially expressed and highly upregulated in resistant hybrids (see Supplementary Table S6). Endochitinases have previously been reported as important bactericides, and some of them have ability to cleave peptidoglycan chains, promoting bacterial cell lysis13. Other defense-related genes were differentially expressed in susceptible plants by CLas. Among them, regions encoding lipid transfer, molecular factors that help the innate immune system of plants, and small lipid-transfer proteins can inhibit fungal growth and pathogenic bacteria14. Genes encoding these proteins were differentially expressed in C. sinensis, C. sunki, and the S Pool (see Supplementary Table S6 and Fig. S2). These results indicated the activation of defense pathways in response to CLas infection in susceptible genotypes.
CDR1 also represents an important defense related gene in Poncirus and Poncirus-hybrids15. CDR1 showed high expression in all the Poncirus hybrids, including the S pool, but it was only induced in the R pool. Therefore, even though it could be associated with resistance, high CDR1 constitutive expression level seems not to be sufficient to lead to the resistance phenotype.
Secondary metabolism and cell wall composition
Secondary metabolites often play an important role in many physiological responses, such as growth, photosynthesis, reproduction, and plant defenses against pathogens16. The most upregulated genes in P. trifoliata included a variety of phenylpropanoids and lignin-related genes, such as caffeic acid O-methyltransferase, chalcone synthase, feruloyl ortho-hydroxylase 1, hydroxycinnamoyl transferase and laccase precursor (see Supplementary Fig. S2). In our study, the laccase precursor gene, whose protein catalyzes lignin and its derivatives17, was exclusive and highly induced in CLas-infected P. trifoliata (see Supplementary Table S6).
Pectin hydrolysis occurs frequently in response to bacterial infection18. Just one pectin degradation-related gene was differentially expressed (downregulated) in P. trifoliata (Fig. 3 and Supplementary Table S6). Many genes involved in pectin synthesis and degradation were differentially expressed in C. sinensis and C. sunki. Pectin methyltransferases are enzymes that induce pectin modification. In C. sinensis and C. sunki under stress caused by CLas infection, the pectin methyltransferase 1 gene was upregulated (see Supplementary Fig. S2).
A larger number of DEGs involved in cellulose synthesis showed mRNA levels altered in susceptible genotypes; however, P. trifoliata and the R Pool did not exhibit differentially expressed regions encoding cellulose (see Supplementary Table S6).
These results demonstrated that the cell wall is highly affected in susceptible plants even at 240 days after CLas inoculation. At the same time, genes involved in cell strengthening proved to be important in P. trifoliata.
Phloem-related genes
It is already known that callose deposition and phloem proteins (PP2) act as a physical barrier, attempting to block systemic spread of CLas; however, they also likely cause phloem disorders19. The current study identified DEGs coding phloem proteins that had altered expression induced by CLas in C. sinensis, C. sunki, the S Pool, and T Pool. Although P. trifoliata did not present callose-induced phloem blockage (Fig. 1), we observed modulation of PP2-B15 in response to CLas with ninefold higher expression than the control (see Supplementary Table S6). That result suggests that P. trifoliata modulates phloem genes in response to CLas without over-deposition of callose, consequently not causing important phloem function disorders. Anatomical divergences between P. trifoliata and Citrus may represent an important feature to avoid collapse of the sieve tube elements20.
As shown by our phenotypic data, only susceptible plants had affected callose deposition. Different callose synthases were differentially expressed in the susceptible plants, whereas those genes were absent in P. trifoliata and the R Pool inoculated with CLas (see Supplementary Table S7).
Interestingly, genes encoding sieve element occlusion c (SEOc) and d (SEOd), which are part of a protein family that encodes specialized crystalloid phloem proteins21, were largely upregulated in all susceptible plants under study. Some of these genes were also upregulated in tolerant hybrids (see Supplementary Table S6 and S7).
Carbohydrate metabolism
Carbohydrate metabolism was the biological function most affected by HLB (Fig. 3). In the presence of CLas, susceptible genotypes overexpressed genes involved with starch synthesis and suppressed genes that encode enzymes for starch degradation (see Supplementary Table S8 and S9). This phenomenon was not observed for the tolerant and resistant genotypes. Several DEGs involved in the metabolism of starch were identified in C. sinensis, C. sunki, and the T Pool, especially in the former two (see Supplementary Table S6). Genes encoding ADP-glucose pyrophosphorylase and starch branching enzyme II, which participate in the synthesis of starch and starch granules, were upregulated in C. sinensis and C. sunki (see Supplementary Table S6 and S8). Beta and alpha-amylase, important enzymes for normal degradation of the starch in plants22, also had their genes expression modulated in both susceptible plants (C. sinensis and C. sunki) and the T Pool (see Supplementary Table S9). Corroborating our phenotypic data (Fig. 1), resistant and tolerant genotypes did not exhibit altered expression of the main genes involved in synthesis of starch (see Supplementary Table S6). While the R Pool had only beta-amylase-encoding gene upregulated, P. trifoliata did not have any DEGs related to synthesis and degradation of starch (see Supplementary Table S6).
Transporters
The transport of substances was also one of the main biological functions affected by CLas. The transcription levels of genes related to transporters were overwhelmingly altered by CLas infection in all genotypes and hybrids (Fig. 3). In general, susceptible plants had the greatest number of transport-related genes affected by CLas (Fig. 3). The R Pool showed few DEGs related to transport function, including ABC transporter family, phosphate transporter (PHO1-2), and amino acid transmembrane transport (Supplementary Table 2). Zinc transporter (ZIP1 and ZIP8) genes were differentially expressed in C. sinensis, C. sunki, and the T Pool (see Supplementary Table S6). Most transport family genes affected by CLas infection were involved with transport of sugars, amino acids, and ions (see Supplementary Table S6). When comparing the transporter-related DEGs in the tolerant genotypes, P. trifoliata, and the T Pool, we observed different responses among them. The T Pool exhibited 73 differentially expressed transporter-related genes. The parental P. trifoliata showed only 4 differentially expressed transporter-related genes, among which potassium transporter was exclusively differentially expressed in P. trifoliata (Fig. 3 and Supplementary Table S6).
Discussion
The hybrids evaluated in this work and the parents, Citrus sunki and P. trifoliata, were classified as susceptible, tolerant, or resistant according to bacterial presence, callose deposition, and starch accumulation (Fig. 1). RNAseq data indicated that the genotypes responded differently under CLas infection, which was confirmed by RT-qPCR analysis. Overall, the genes showed similar patterns in the RNAseq and RT-qPCR data, but some divergent values were found, which was similar to other transcriptome studies when the results of different techniques were compared23.
Our findings indicated that few genes were differentially expressed according to RNAseq analysis of the tolerant and resistant plants. In contrast, RNAseq analysis of susceptible plants showed transcription modulation of many genes. Resistant and tolerant plants have a tendency to respond more rapidly and vigorously to a pathogen than susceptible plants12. It is possible that the resistant hybrids have an early response to CLas presence. Early molecular interactions are well-known mechanisms in plant-pathogen interactions24–26. Nevertheless, to verify that the genetic responses were due to CLas infection and to avoid false positives, the samples for transcriptomic analysis were collected 8 months after CLas infection.
P. trifoliata showed upregulation of receptor-related genes, which presented an efficient recognition of CLas and possibly an effective signaling and activation of defense response against CLas. The reprogramming of defense signaling pathways has previously been reported as a critical element of the early response to CLas in tolerant genotypes27, such as P. trifoliata. Previous studies have also highlighted the induction of phenylpropanoid-related genes as a molecular mechanism of HLB tolerance5. Lignin-related genes and several phenylpropanoids were strongly upregulated in P. trifoliata transcriptome (Supplementary Table S6). As reorganization of plant growth and development are critical to maximize plant survival under stress28, cell wall reinforcement is a tolerance mechanism of P. trifoliata against CLas. When comparing P. trifoliata and resistant hybrids, we observed a distinct transcriptional response to CLas (Fig. 2). However, all replicates of the resistant hybrids did not present any detection of CLas, even after almost 1 year of the experiment (Tables 1 and 2), and probably for this reason, they exhibited few DEGs in RNAseq. Interestingly, the exclusive DEGs of the R Pool, formed by the CLas-negative hybrids, may be linked with genes and mechanisms capable of eliminating the bacteria from the plant, such as endochitinase B. Plant endochitinases cleave peptidoglycan chains, thereby promoting bacterial cell lysis13.
CLas infection is erratic and unpredictable, and even susceptible plants can escape from infection. Until almost 1 year, all plant replicates classified as resistant did not present CLas titer (Tables 1 and 2). Therefore, until that moment, we considered that those plants were resistant to CLas infection and that a mechanism was utilized to avoid spreading the disease.
In the transcriptome of tolerant genotypes, downregulation of GA synthesis genes and upregulation of genes involved with GA degradation were observed, and the opposite behavior was observed in the susceptible genotypes (induction of GA synthesis and repression of GA degradation). In addition, we observed upregulation of several auxin-induced genes and repression of auxin responsive factors (Supplementary Table S6). It is known that the GA pathway presents cross-talk with auxin and ethylene hormones, which are plant growth regulators that also have been associated with plant defense and microbial pathogenesis29,30. The present study showed that these regulators were strongly differentially expressed in the tolerant plants by CLas. It has been reported that auxin induces GA biosynthesis and suppresses GA degradation through modulation of several transcription factors and transporters31,32. In citrus-pathogen interactions, crosstalk between auxin and GA has also been reported. Inhibition of GA synthesis promotes inhibition of auxin-induced transcription, consequently reducing symptoms in the citrus-Xanthomonas citri interaction33.
The plant tolerance mechanism is better explained by the interaction of GA and the salicylic acid (SA) hormone. The GA pathway is considered a hormone modulator of the SA signaling backbone during plant responses to pathogens34–36. In Arabidopsis thaliana, Alonso-Ramírez et al. (2009)36 showed that GAs and the overexpression of GA-responsive genes increase not only the endogenous levels of SA but also the expression of ics1 and npr1 genes involved in SA biosynthesis and action, respectively. However, SA-related genes were almost not modulated in the present study, which might be due to the high SA level in the evaluated stage, resulting in the expression of SA synthesis-related genes no longer being necessary as shown by Oliveira et al., 201920. Moreover, it is known that SA accumulation and downstream signaling events are important components of both pathogen‐associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI)37,38 through increasing the expression of WRKY transcription factors. Many WRKY TFs were induced in the tolerant genotypes and affected in the susceptible plants (Fig. 5). WRKY TFs have been considered key regulators of plant defense against many pathogens, including CLas27. The function of some WRKY genes remains unexplored, but in some crop species, specific WRKYs promote tolerance or even resistance to biotic and abiotic stresses27. Thus, the induction of WRKY TFs may also be related to the activation of genes involved with the tolerance mechanism. For example, in P. trifoliata, the WRKY transcription factor 14–1 was induced, and its orthologue in Arabidopsis (known as WRKY22) is an essential component of MAPK-mediated plant defense responses against pathogens. MAPKs are associated with one of the earliest signaling events after plant sensing of PAMPs and pathogen effectors.
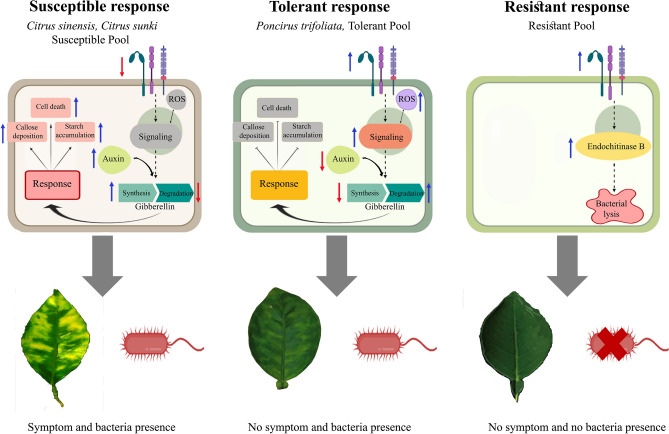
Figure 5.
Model of interaction between CLas and Citrus plants. Susceptible plants, the downregulation of signaling receptors promotes a late recognition of CLas infection and consequently, no proper signaling is activated. Auxin-related genes positively modulate the gibberellin synthesis, which activates response mechanisms to CLas infection, such as callose and PP2 deposition and impaired substances transport. Interference on substance transport along with callose deposition causes phloem dysfunction resulting in starch accumulation on photosynthetic tissues. Starch accumulation promotes thylakoid rupture and chlorophyll degradation culminating in HLB classical symptoms. Tolerant plants, the induction of signaling receptors causes a fast and efficient defense response modulated by suppression of auxin pathway and induction of GA degradation. The suppression of these pathways prevents the events that lead to the phloem dysfunction (callose deposition, starch accumulation and transport alteration) and activates defense response through the synthesis of phenylpropanoids and cell wall-strengthened related genes. This transcriptional reprograming is efficient to impair the development of symptoms. Resistant genotypes, a possibly early and fast defense may occur in response to CLas, since low numbers of the genes are modulated after 240 days post inoculation. Nonetheless, this response is related to induction of signaling receptors and upregulation of Endochitinase B, which might be associated with bacterial cell lysis (Created with BioRender.com).
Moreover, the tolerant and susceptible genotypes had changes in the level of transcription of many callose synthases and phloem protein (PP2) genes in response to CLas infection (Supplementary Table S6 and S7). In addition, all susceptible plants showed induction of a class of genes that includes the SEOc gene (Supplementary Table S7). This class of genes has been reported to encode P-protein subunits21. Overexpression of these genes could increase callose and PP2 protein synthesis in the citrus phloem sieve elements. Callose and PP2 accumulation is a crucial factor of phloem blockage in CLas-infected plants19,39,40. Phloem blockage causes disturbance of photoassimilate flows from source organs (leaves) to sink organs (roots), resulting in starch accumulation in the leaves as observed in this work and in previous studies41.
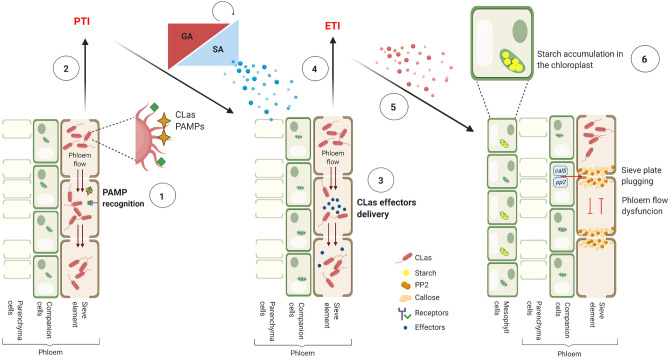
Based on the knowledge of CLas-susceptible plant interaction that culminates in HLB symptoms, a zig-zag model as illustrated previously by Jones & Dang (2006)42 was adapted to explain such genetic molecular response to CLas (Fig. 4). During the beginning of infection, receptors from citrus plants detect the CLas PAMPs, which triggers a PTI response, resulting in the production of GA and SA as well as in the induction of several downstream genes (asymptomatic stage). In a second phase, CLas delivers effectors, such as Las531543 and others44, which interfere with PTI or enable pathogen nutrition and dispersal, resulting in effector-triggered susceptibility (ETS). In phase 3, effectors activate an ETI and an amplified version of PTI leading to induction of callose synthases and pp2 gene expression that results in callose and PP2 accumulation. Therefore, callose and PP2 accumulation and the consequent anatomical alterations of the sieve pores may lead to hypersensitive cell death (HR) of the infected plants, which spatially isolate the CLas to reduce their colonizing ability via the phloem19,40.
Figure 4.
Defense response of susceptible genotypes against CLas. In the phase 1 of this model, citrus plants receptors detect the CLas PAMPs. In phase 2, a PAMP-triggered immunity (PTI) response is initiated, resulting in the production of gibberellic acid (GA), salicylic acid (SA) and the SA-dependent gene expression activation (in blue). In phase 3, CLas deliver effectors leading in effector-triggered susceptibility (ETS). In phase 4, effectors are recognized by plants proteins, activating effector-triggered immunity (ETI). In phase 5, ETI triggers a series of genetic events (in red), including the induction of calloses synthases and pp2 expression. This exaggerated response could be considered as hypersensitive cell death (HR), since the attempt to isolate spatially the CLas leading to callose and PP2 accumulation, that cause phloem dysfunctions. The phase 6 represents the starch accumulation in the mesophyll chloroplasts(Created with BioRender.com).
To describe the genetic mechanisms potentially involved in a susceptible, tolerant, and resistant interaction with CLas based on the data obtained in this study, we built a hypothetical model (Fig. 5). The model shows that in the susceptible plants (Fig. 5), auxin-related genes positively modulate GA synthesis, which activates response mechanisms to CLas infection, such as callose deposition, PP2 deposition, phloem dysfunction, and impaired flow transport. The impaired flow results in starch accumulation on mesophyll chloroplasts, which promotes thylakoid rupture and chlorophyll degradation, culminating in HLB typical symptoms. In the tolerant plants, including P. trifoliata (Fig. 5), the induction of signaling receptors cause a fast and efficient defense response modulated by suppression of the auxin pathway and induction of GA degradation. The suppression of these pathways prevents the events that lead to phloem dysfunction (callose deposition, starch accumulation, and transport alteration), and it activates the defense response through the synthesis of phenylpropanoids and cell wall strengthened-related genes. This transcriptional reprograming is efficient to impair the development of symptoms. In the resistant genotypes (Fig. 5), a potentially early and rapid defense may occur in response to CLas because only a few genes were differentially expressed after 240 days after inoculation. However, this response is related to induction of signaling receptors and upregulation of endochitinase B, which is associated with bacterial cell lysis.
Both hypothetical models showed that there are many pathways acting in citrus defense against CLas infection. The data acquired in this study can help to generate citrus varieties of scions or rootstocks with potential resistance to HLB based on citrus conventional breeding programs or biotechnological approaches, including the development of transgenic or cisgenic lines as well as genome editing and host-induced gene silencing.
Materials and methods
Plant material
C. sinensis, C. sunki, P. trifoliata, and 21 hybrids obtained from a controlled cross between Citrus sunki ex Tan (female parent and susceptible to HLB) and Poncirus trifoliata Raf. cv Rubidoux (male parent and tolerant to HLB) were used in the analysis. C. sinensis was included because it is one of most important citrus scions in the world, and it can also be considered an internal control of the experiment considering that C. sinensis is characterized as a species highly susceptible to HLB40. The experimental design was completely randomized and consisted of five biological replicates for each inoculated genotype (CLas-infected budwoods) and mock-inoculated genotype (health budwoods). Plants were propagated using buds that were grafted onto rootstocks of Rangpur lime (C. limonia Osb.). At the end of 6 months, the plant scions were grafted using two CLas-infected buds obtained from C. sinensis (L.) Osbeck cv Pera. All plants were kept in a greenhouse at Centro de Citricultura Sylvio Moreira of the Agronomic Institute (IAC), SP with an average temperature of 25 °C for 12 months. The starch content and callose deposition were estimated only in the genotypes selected for the further analysis (C. sinensis, C. sunki, P. trifoliata, and 15 hybrids obtained from crosses between C. sunki and P. trifoliata). Leaves from inoculated and mock-inoculated plants from all evaluated genotypes were collected with three biological replicates of each genotype after 8 months of CLas infection.
CLas quantification
CLas presence and HLB symptoms were evaluated according to previously described methodology40. Briefly, 30, 90, 180, 240, and 360 days after inoculation, to confirm HLB infection, leaves above the inoculation point were collected and tested by qPCR using 16S ribosomal DNA primer sets and FAM/Iowa Black FQ label probe (IDT Inc., Coralville, IA) probes as described by Li et al. (2006)45. Citrus GAPDH (glyceraldehyde 3-phosphate dehydrogenase F: GGAAGGTCAAGATCGGAATCAA; R: CGTCCCTCTGCAAGATGACTCT) was used as the reference gene. Values above 34 Ct were considered negative for CLas infection7. After 240 days of CLas inoculations, the bacterial titer was evaluated according to Boava et al. (2015)7 by qPCR using a standard curve with serial dilutions of 16S ribosomal DNA (rDNA) cloning into pGEM-T vector (PROMEGA).
Phenotypic analysis
Starch and callose quantification of CLas-inoculated and mock-inoculated plants was performed after 240 days of infection. Callose quantification was performed following the methodology reported previously40. Leaf petioles were fixed in FAA solution (50 mL of formaldehyde, 50 mL of glacial acetic acid, and 900 mL of 70% ethanol) for 72 h and then kept in 70% ethanol. Transversal sections of 10 μm were generated using an automatic slide microtome (Leica SM2010R). The sections were stained with blue aniline, and the stained samples were examined on an Olympus BX61 fluorescence microscope using 355–375 nm excitation filter, 400-nm dichromatic mirror, and 435–490 nm emission filter. Callose quantification was performed by counting fluorescent spots in the total phloem area in 10 fields of view for each sample. The starch measurement was performed using leaves dried in an oven at 60 °C for 48 h and ground. Starch content was estimated by enzymatic analysis using 10 mg of dried leaves according to46. Absorbance was measured in 96-well microtiter plates using a Microplate Reader (Model 3550–BIO-RAD) at 490 nm. A standard curve was performed using a glucose solution (SIGMA) at concentrations of 0, 2.5, 5.0, 7.5, and 10 μg/mL.
According to starch, callose, and CLas quantification, the genotypes were classified as susceptible, tolerant, and resistant (see supplementary Fig. S3).
RNA extraction and sequencing (RNA-seq)
Leaves from three biological replicates of the three genotypes (C. sinensis, C. sunki, and P. trifoliata) and the three hybrid pools (S Pool: H109, H161, and H165; T Pool: H113, H154, and H146; and R Pool: H68, H106, and H142), either CLas-infected inoculated or mock-inoculated plants, were collected for transcriptomic analysis after 240 days of infection. It is difficult to establish the ideal time for studying the first responses and stages of infection because it is difficult to confirm that the plant tissue is colonized by bacteria. Thus, to verify that the genetic responses were due to CLas infection, we performed RNA-seq analysis at 8 months. Total RNA was isolated with the MasterPure Plant RNA Purification Kit (EPICENTRE Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. A total of 10 µg of RNA from each sample was sent for sequencing at the Centro de Genômica Funcional in Centro de Biotecnologia Agricola in ESALQ/USP (http://www.esalq.usp.br/genomicafuncional/). RNA-seq was performed using the Illumina HiSeq 2500 platform. All procedures were performed according to Illumina’s protocols. RNA-seq was performed in triplicate with a total of 36 samples.
Data analysis
The quality of obtained fragments from the sequencing was verified using CLC Genomics Workbench v.6 program (CLC BIO) software (https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/). The sequences were trimmed using the trimmomatic tool47 and mapped on the v 2.0 C. sinensis genome (http://citrus.hzau.edu.cn/) using the STAR-2.5.2b program48. The R subread package was used for counting. DEGs between the control and CLas-infected plants were established using the DESeq in Bioconductor package49 using an adjusted p-value of 0.005 and FDR threshold of 0.05. Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/) were used to identify common and unique DEGs among the analyzed genotypes. We used Blast2Go9 for functional categorization, and the DEGs were annotated by Gene Ontology (GO) using default parameters10.
Real time PCR (RT-qPCR) validation
To ensure reproducibility of the biological phenomenon observed by transcriptomic analysis, we performed a second experiment with other plants following the same design used for RNA-seq. We sampled one hybrid of each pool to represent the susceptible, tolerant, and resistant pools. We used only one hybrid from each pool because it represents the hybrids that comprise each pool regarding CLas infection behavior. Total RNA was extracted using the protocol described by Chang et al. (1993)50. Traces of genomic DNA were eliminated using the DNase RNase-Free Ket (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. cDNAs were synthesized from 1.0 μg of total RNA using Superscript III (200 U/μL) (INVITROGEN) with an oligo (dT) primer (dT12-18, INVITROGEN) according to the manufacturer’s instructions.
Ten genes that showed the opposite expression profile between the genotypes with different responses were selected, including chalcone synthase, lipid transfer, cytochrome P450, gibberellin-regulated 9, sieve element occlusion c, cinnamoyl-reductase, pectin methylesterase 1, starch branching enzyme II, PRR response regulator, and choline transporter like-protein 2 (see Supplementary Table S10). Primers were designed using Primer3Plus51, and the Primer-BLAST tool52 was used to check the specificity of the primers. Two endogenous genes, GAPDH and FBOX, were used for normalization of the data. Relative gene expression was calculated with the 2−ΔΔCt method53.
Supplementary information
Acknowledgements
This study was supported by INCT-Citros (FAPESP 2014/50880-0 and CNPq 465440/2014-2); PhD fellowships to ISP (FAPESP 2015/13971-0) and MC (FAPESP 2016/22133-0); and postdoc fellowships to DMG (CNPq 103228/2018-7) and LMG (FAPESP 2019/01901-8).
Author contributions
M.C.Y. and M.A.M. planned and supervised the study. M.C. and I.S.P. contributed to the design and execution of the experiments detailed. M.C. and M.A.T. performed the functional genomic and bioinformatics data analyses. M.C., L.P.B., L.M.G., and D.M.G. conducted and evaluated plant growth. M.C. and I.S.P. drafted the manuscript. M.C.Y., L.M.G., D.M.G., M.A.T., A.A.D.S., and M.A.M. provided intellectual input. All authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77840-2.
References
- 1.Bové JM, Ayres AJ. Etiology of three recent diseases of citrus in São Paulo State: sudden death, variegated chlorosis and huanglongbing. IUBMB Life. 2007;59:346–354. doi: 10.1080/15216540701299326. [DOI] [PubMed] [Google Scholar]
- 2.Munir S, et al. Huanglongbing control: perhaps the end of the beginning. Microb. Ecol. 2018;76:192–204. doi: 10.1007/s00248-017-1123-7. [DOI] [PubMed] [Google Scholar]
- 3.Johnson EG, Wu J, Bright DB, Graham JH. Association of ‘Candidatus Liberibacter asiaticus’ root infection, but not phloem plugging with root loss on huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathol. 2014;63:290–298. doi: 10.1111/ppa.12109. [DOI] [Google Scholar]
- 4.Albrecht U, Bowman KD. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 2012;185–186:118–130. doi: 10.1016/j.plantsci.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Balan B, Ibáñez AM, Dandekar AM, Caruso T, Martinelli F. Identifying host molecular features strongly linked with responses to huanglongbing disease in citrus leaves. Front. Plant Sci. 2018;9:1–13. doi: 10.3389/fpls.2018.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folimonova SY, Robertson CJ, Garnsey SM, Gowda S, Dawson WO. Examination of the responses of different genotypes of citrus to huanglongbing (Citrus Greening) under different conditions. Phytopathology. 2009;99:1346–1354. doi: 10.1094/PHYTO-99-12-1346. [DOI] [PubMed] [Google Scholar]
- 7.Boava LP, Sagawa CHD, Cristofani-Yaly M, Machado MA. Incidence of ‘Candidatus Liberibacter asiaticus’-infected plants among citrandarins as rootstock and scion under field conditions. Phytopathology. 2015;105:518–524. doi: 10.1094/PHYTO-08-14-0211-R. [DOI] [PubMed] [Google Scholar]
- 8.Albrecht U, Bowman KD. Tolerance of the trifoliate citrus hybrid US-897 (Citrus reticulate Blanco × poncirus trifoliata L. Raf.) to huanglongbing. HortScience. 2011;46:16–22. doi: 10.21273/HORTSCI.46.1.16. [DOI] [Google Scholar]
- 9.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 10.Consortium, T. G. O Gene ontology : tool for the unification of biology. Nature. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villena J, Kitazawa H, Van Wees SCM, Pieterse CMJ, Takahashi H. Receptors and signaling pathways for recognition of bacteria in livestock and crops: prospects for beneficial microbes in healthy growth strategies. Front. Immunol. 2018;9:2223. doi: 10.3389/fimmu.2018.02223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP. Disease resistance mechanisms in plants. Genes (Basel). 2018;9:339. doi: 10.3390/genes9070339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Medeiros SC, Monteiro-Júnior JE, Passos Sales GW, Grangeiro TB, Pinto Nogueira NA. Chitinases as antibacterial proteins: a systematic review. J. Young Pharm. 2018;10:144–148. doi: 10.5530/jyp.2018.10.33. [DOI] [Google Scholar]
- 14.Finkina EI, Melnikova DN, Bogdanov IV, Ovchinnikova TV. Lipid transfer proteins as components of the plant innate immune system: structure, functions, and applications. Acta Nat. 2016;8:47–61. doi: 10.32607/20758251-2016-8-2-47-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawat N, et al. Genome resequencing and transcriptome profiling reveal structural diversity and expression patterns of constitutive disease resistance genes in Huanglongbing-tolerant Poncirus trifoliata and its hybrids. Hortic. Res. 2017;4:1–8. doi: 10.1038/hortres.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartwal A, Mall R, Lohani P, Guru SK, Arora S. Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. J. Plant Growth Regul. 2013;32:216–232. doi: 10.1007/s00344-012-9272-x. [DOI] [Google Scholar]
- 17.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009;344:1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Granato L, Galdeano D, D’Alessandre N, Breton M, Machado M. Callose synthase family genes plays an important role in the Citrus defense response to Candidatus Liberibacter asiaticus. Eur. J. Plant Pathol. 2019;155:25–38. doi: 10.1007/s10658-019-01747-6. [DOI] [Google Scholar]
- 20.Oliveira TS, et al. Genetic analysis of salicylic acid-mediated defenses responses and histopathology in the huanglongbing pathosystem. Citrus Res. Technol. 2019;40:1–13. doi: 10.4322/crt.18419. [DOI] [Google Scholar]
- 21.Ernst AM, et al. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1980–E1989. doi: 10.1073/pnas.1202999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd JR, Kossmann J, Ritte G. Leaf starch degradation comes out of the shadows. Trends Plant Sci. 2005;10:130–137. doi: 10.1016/j.tplants.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zhou L, Yu X, Stover E, Luo F. Transcriptome profiling of huanglongbing ( HLB ) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 2016;7:1–13. doi: 10.3389/fpls.2016.00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, et al. Genome-wide identification, classification and expression analysis in fungal–plant interactions of cutinase gene family and functional analysis of a putative ClCUT7 in Curvularia lunata. Mol. Genet. Genom. 2016;291:1105–1115. doi: 10.1007/s00438-016-1168-1. [DOI] [PubMed] [Google Scholar]
- 25.Shen Y, et al. The early response during the interaction of fungal phytopathogen and host plant. Open Biol. 2017;7:170057. doi: 10.1098/rsob.170057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nirmala J, et al. Concerted action of two avirulent spore effectors activates reaction to Puccinia graminis 1 (rpg1)-mediated cereal stem rust resistance. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14676–14681. doi: 10.1073/pnas.1111771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Q, et al. Reprogramming of a defense signaling pathway in rough lemon and sweet orange is a critical element of the early response to Candidatus Liberibacter asiaticus. Hortic. Res. 2017;4:1–15. doi: 10.1038/hortres.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iglesias MJ, Terrile MC, Bartoli CG, D’Ippólito S, Casalongué CA. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 2010;74:215–222. doi: 10.1007/s11103-010-9667-7. [DOI] [PubMed] [Google Scholar]
- 29.Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than Just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 31.Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B. Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007;52:499–511. doi: 10.1111/j.1365-313X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- 32.Richter R, Behringer C, Zourelidou M, Schwechheimer C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13192–13197. doi: 10.1073/pnas.1304250110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cernadas RA, Benedetti CE. Role of auxin and gibberellin in citrus canker development and in the transcriptional control of cell-wall remodeling genes modulated by Xanthomonas axonopodis pv. citri. Plant Sci. 2009;177:190–195. doi: 10.1016/j.plantsci.2009.05.006. [DOI] [Google Scholar]
- 34.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 35.De Bruyne L, Höfte M, De Vleesschauwer D. Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant. 2014;7:943–959. doi: 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- 36.Alonso-Ramírez A, et al. Cross-talk between gibberellins and salicylic acid in early stress responses in Arabidopsis thaliana seeds. Plant Signal. Behav. 2009;4:750–751. doi: 10.4161/psb.4.8.9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, et al. ‘ Candidatus Liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant-Microbe Interact. 2017;30:620–630. doi: 10.1094/MPMI-12-16-0257-R. [DOI] [PubMed] [Google Scholar]
- 38.An C, Mou Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 39.Xie B, Hong Z. Unplugging the callose plug from sieve pores. Plant Signal. Behav. 2011;6:491–493. doi: 10.4161/psb.6.4.14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boava LP, Cristofani-Yaly M, Machado MA. Physiologic, anatomic, and gene expression changes in citrus sunki, poncirus trifoliata, and their hybrids after ‘candidatus liberibacter asiaticus’ infection. Phytopathology. 2017;107:590–599. doi: 10.1094/PHYTO-02-16-0077-R. [DOI] [PubMed] [Google Scholar]
- 41.Etxeberria E, Gonzalez P, Achor D, Albrigo G. Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol. Mol. Plant Pathol. 2009;74:76–83. doi: 10.1016/j.pmpp.2009.09.004. [DOI] [Google Scholar]
- 42.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 43.Pitino M, Allen V, Duan Y. LasΔ5315 effector induces extreme starch accumulation and chlorosis asCa. liberibacter asiaticus infection in nicotiana benthamiana. Front. Plant Sci. 2018;9:1–11. doi: 10.3389/fpls.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granato, L. M. et al. ‘Candidatus Liberibacter asiaticus’ putative effectors : in silico analysis and gene expression in citrus leaves displaying distinct huanglongbing symptoms. Trop. Plant Pathol. **(2020).
- 45.Li W, Hartung JS, Levy L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods. 2006;66:104–115. doi: 10.1016/j.mimet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Amaral L, Gaspar M, Costa P, Aidar M, Buckeridge M. Novo método enzimático rápido e sensível de extração e dosagem de amido em materiais vegetais. Hoehnea. 2007;34:425–431. doi: 10.1590/S2236-89062007000400001. [DOI] [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Wolfgang H. Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. Genome Biol. 2010;11:4310–4315. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PubMed] [Google Scholar]
- 50.Chang S, Puryear J, Cairney J. A Simple and Efficient Method for Isolating RNA from Pine Trees. Plant Mol. Biol. Report. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 51.Untergasser A, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:1–12. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye J, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.