Abstract
The cecal microbiota plays a critical role in energy harvest and nutrient digestion, influencing intestinal health and the performance of chickens. Feed efficiency (FE) is essential for improving economic efficiency and saving social resources in chicken production and may be affected by the cecal microbiota. Therefore, to investigate the composition and functional capacity of cecum microbes related to FE in Xiayan chicken, an indigenous breed in Guangxi province, metagenome sequencing was performed on chicken cecal contents. 173 male and 167 female chickens were divided into high and low FE groups according to the residual feed intake. The cecal microbial genome was extracted and sequenced. The results showed that the genera Bacteroides, Prevotella, and Alistipes were the 3 most abundant in each cecal microbiome. The linear discriminant analysis effect size revealed 6 potential biomarkers in male and 14 in female chickens. Notably, the relative abundance of Lactobacillus in the high FE group was higher than that of the low FE group both in the male and female chickens, and the species Limosilactobacillus oris has a higher score in the high FE group of male chickens. In contrast, some potentially pathogenic microorganisms such as Campylobacter avium in females and Helicobacter pullorum in males were enriched in the low FE group. Predictive functional analysis showed that the high FE group in male chickens had a greater ability of xenobiotics biodegradation and metabolism and signaling molecules and interaction. In addition, the host sex was found to exert effects on the cecal microbial composition and function associated with FE. These results increased our understanding of the cecal microbial composition and identified many potential biomarkers related to FE, which may be used to improve the FE of the chickens.
Key words: Xiayan chicken, feed efficiency, cecal microbiota, metagenome sequencing
Introduction
Owing to the expansion of the human population, the improvement of income level, and urbanization, the requirement for protein and meat is increasing. Low production costs, high feed conversion ratios, and low product prices have contributed to making poultry the meat of choice, both for producers and consumers. Over the next decade, poultry will continue to strengthen its dominant position within the meat complex, accounting for virtually half of all additional meat that will be produced. The feed cost in chicken meat production accounts for a high proportion of the total farming expense, being nearly 70% of the total cost of poultry production (Ong, 2010). Therefore, improvements in the feed efficiency (FE) of the chicken will decrease production costs and reduce the demand of land area than for feed production, while also reducing the environmental impact of broiler production. The residual feed intake (RFI) is defined as the difference between the actual feed intake and predicted requirements based on animal maintenance and growth (Koch et al., 1963). The RFI is superior sensitive and accurate in measuring the FE and is increasingly used in the genetic improvement of the FE in livestock. Besides, the heritability values for the RFI ranged from 0.21 to 0.49 in the previous studies (Do et al., 2014; Yuan et al., 2015; Zhang et al., 2017).
Exploring the microbial community composition has gained a growing interest in breeding animals because this has been allowed to predict the associated metabolites and compositional structure of such communities (Hiergeist et al., 2015). In recent years, due to metagenomic approaches based on high-throughput sequencing methods, the research of the gut microbiota has been made rapid progress (Milani et al., 2013; Mancabelli et al., 2017; Wu et al., 2020). The gastrointestinal tract (GIT) of chickens is the major site for nutrient absorption and food digestion and has a highly diverse and dynamic microbial ecosystem (Yan et al., 2017). Changes of microbial diversity in the GIT have been associated with the differences of breeds (Pandit et al., 2018), sex (Siegerstetter et al., 2017), growth stages (Yan et al., 2017), and intestinal segments (Al-Marzooqi et al., 2020) in chickens, as a result of various environmental and genetic factors. The cecum, which is the main functional section in the distal intestine, plays important roles not only in vitamin and amino acid production but also in the prevention of pathogen colonization (Stanley et al., 2014; Williams et al., 2015). The cecal microbiota was found to be highly related to the FE, which suggested an important role in chicken FE (Yan et al., 2017).
Some indigenous chicken breeds have a higher product quality, productivity, and pathogen resistance, which have been widely reported (Arora et al., 2011; Haunshi et al., 2011; Duah et al., 2020). For example, Xiayan chicken is a famous specialty in Guangxi province and one of the top 10 yellow-feather broilers in China. Xiayan chickens were characterized by the fast growth rate, strong survivability, large size, and tender meat and have been enjoying a high reputation in the broiler market in Guangxi, Guangdong, Hainan, Hong Kong, and Macau. At present, our understanding of the intestinal microbial community of indigenous chickens in China remains limited. Although research on the gut microbiota of poultry is increasing, most of the current information about the intestinal microbiota is still limited to humans (Marchesi et al., 2016; Wu et al., 2020). Moreover, the majority of previous research focused on the male chickens (Stanley et al., 2016) and the adult hens (Yan et al., 2017), whereas cecal microbiota associated with the FE were rarely explored simultaneously in chickens of both sexes. In this experiment, we have compared the microbial community composition in the cecum of Xiayan chickens with different FE by metagenomic sequencing and explored the interactions and relationships between the FE and cecal microbiota.
Materials and methods
Animal Rearing and Management
A total of 340 indigenous Xiayan chickens (63 d old) including 173 males and 167 females from Guangxi Rongxian Zhouyi Breeding Co., Ltd., Rongxian Economic Development Zone, Guangxi were used in this study. The animal works were reviewed and approved by the Animal Care and Use Committee in Guangxi University (approval number GXU2018-058). All experimental chickens were raised in the scientific research base of the College of Animal Science and Technology of Guangxi University. During the whole experiment, all chickens were raised with the same commercial diet and management conditions, and water was freely available. Each chicken was raised in a different cage. One week after the start of feeding, we recorded the total feed consumption and total BW gain of each chicken from 70 d to 90 d of age. Average daily feed intake = total feed consumption/total days; average daily BW gain = total BW gain/total days.
Phenotypic Data and Cecal Sample Collection
The RFI value was calculated using SAS linear simulation fitting function following the model (Koch et al., 1963; Luiting and Urff, 1991) of RFI = ADFI-(b0+b1MBW0.75+b2ADG), where the ADFI represents average daily feed intake; MBW refers to the mean BW; MBW0.75 is the metabolic BW; ADG represents average daily BW gain; b0 is intercepted; b1 and b2 represent partial regression coefficients. The RFI value was used to estimate FE and was negatively associated with FE. We ranked the obtained RFI value, after which the 3 chickens with the highest RFI and the 3 chickens with the lowest RFI were selected from the male and female experimental chickens, respectively. These 12 chickens (3 replications × 2 genders × 2 groups) were slaughtered for collecting the cecal contents at the age of 90 d. All samples were immediately transferred into liquid nitrogen and then stored at −80°C for subsequent metagenomic sequencing.
DNA Extraction and Metagenomic Sequencing
We used the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Germany) to extract cecal microbial genome DNA according to the manufacturer's instructions. The agarose gel electrophoresis was used to evaluate the DNA quality and integrity, and DNA concentration was measured using Thermo Scientific NanoDrop 2000 spectrophotometers.
All 12 metagenomic DNA samples were sequenced on the Illumina platform by using 150 bp paired-end sequencing. Sequence reads were treated to remove low-quality reads, trim the read sequences, and remove the host genome sequences. Specifically, reads with low-quality bases (quality value ≤ 38) greater than 40 bp were removed. Reads with N bases greater than 10 bp were removed, and the reads which shared the overlap above 15 bp with adapter were removed. Host genomic sequences were removed (Karlsson et al., 2012, 2013b) by bowtie2 (v2.2.4). The remaining clean reads were assembled using SOAPdenovo (v2.04) (Luo et al., 2012) to gain Scaftigs. Bowtie2 was used to align the clean data of each sample to Scaftigs and collect unmapped reads for another SOAPdenovo assembly with the same parameters. After combining the unused reads of each sample, SOAPdenovo undergo mixed assembly with the same parameters as above. The mixed assembled Scaffolds were broken from the N junction to get new Scaftigs (≥500 bp). MetaGeneMark (v2.10) (Zhu et al., 2010) was performed for predicting the open reading frames according to Scaftigs (≥ 500 bp) from single-sample assembly and mixed-sample assembly. The redundancy of the predicted open reading frames (>100 nt) was eliminated using CD-HIT (v4.5.8) (Li and Godzik, 2006) to obtain a nonredundant initial gene catalog (genes). The clean data from each sample were mapped to the initial gene catalog (genes) using bowtie2 and get the number of reads. The gene in which the number of reads ≤ 2 (Li et al., 2014) were filtered out and the final gene catalog (unigenes) used for subsequent analysis were obtained.
Abundance Analysis and Taxonomy Annotation
The abundance information of each gene in each sample was counted based on the length of the gene and the number of mapped reads. The formula is as follows: where r refers to the number of reads mapped to the genes and L is the gene's length (Qin et al., 2010; Karlsson et al., 2012). The abundance of each gene in each sample was used to analyze the number of differential genes between groups. The results were visualized by the Venn figure.
The sequences of bacteria, fungi, archaea, and viruses are all extracted from the RefSeq nonredundant proteins database (accessed 2 January 2018) of the NCBI. DIAMOND software (Buchfink et al., 2015) (V0.9.9) was used to blast the unigenes to the sequences of the microbiome database. The aligned result (e value ≤ the smallest e value ∗ 10) (Oh et al., 2014) was chosen and the LCA algorithm was performed to make sure the species annotation information of sequences. The table containing the number of genes and the abundance information of each sample in each taxonomy hierarchy (kingdom, phylum, class, order, family, genus, species) was obtained based on the LCA annotation result. The abundance of species in one sample equals the sum of the gene abundance annotated for the species (Qin et al., 2010; Karlsson et al., 2012, 2013b). To measure the differences of cecal microbiota composition between samples from different FE groups, the vegan package was used to calculate the Bray–Curtis distance matrixes. The results were visualized by principal coordinate analysis and ggplot2 using R (v3.6.1). The nonparametric Kruskal–Wallis sum-rank test was used to detect different species between different FE groups, and the linear discriminant analysis effect size (LEfSe) analysis was used to reduce dimensionality and evaluate the impact of different species to obtain the biomarkers with significant differences between groups (Segata et al., 2011).
Functional Annotation Analysis
DIAMOND software (v0.9.9.110) was used to blast the unigenes against the functional databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (v01012018) and the Carbohydrate-Active enZYmes (CAZy) database (v04072015) with parameters of blast -e 1e-5. The annotation information of KEGG Orthology (KO) from the KEGG database was acquired based on the relative abundance profile. The differential function pathway between different FE groups was demined by STAMP software (P < 0.05).
Statistical Analysis
In our research, SAS (v9.2) was used to perform statistical analysis and significance analysis on the data.
Results
Phenotypic Values of Chicken RFI
The feed conversion ratio, RFI, daily BW gain, and daily feed intake from 70 to 90 d of age were recorded separately in all 340 experimental chickens and ranked according to the RFI value. In this study, we selected 3 chickens with the highest RFI (low FE) and 3 chickens with the lowest RFI (high FE) among male and female chickens, respectively (Supplementary Table 1). The RFI value of the group with a high FE was significantly lower than that of the group with a lower FE (low FE) (P < 0.01), either in male or female experimental chickens. The daily feed intake and daily BW gain were significantly different between the high FE and low FE groups (P < 0.01) in female chickens; no differences were observed in male chickens (Table 1).
Table 1.
Feed efficiency and phenotype data for female and male chickens with high and low residual feed intake.
| Parameter | Males |
Females |
||
|---|---|---|---|---|
| HFE | LFE | HFE | LFE | |
| Daily feed intake(g) | 75.30 ± 10.58 | 95.85 ± 6.73 | 65.63 ± 4.82A | 82.95 ± 1.69B |
| Daily BW gain(g) | 27.50 ± 4.58 | 22.00 ± 2.05 | 21.83 ± 0.95A | 18.17 ± 0.29B |
| FCR (g/g) | 2.75 ± 0.21A | 4.37 ± 0.20B | 3.01 ± 0.15A | 4.57 ± 0.09B |
| RFI (g) | −13.02 ± 0.92A | 13.56 ± 1.47B | −11.79 ± 2.55A | 11.89 ± 1.62B |
A,BDifferent capital letters indicate that the difference is statistically significant (P < 0.01). HFE and LFE denote high feed efficiency and low feed efficiency, respectively.
Abbreviations: FCR, feed conversion ratio; RFI, residual feed intake.
Community Composition of Chicken Cecal Microbe
To assess the cecal microbial composition at the species level, cecal contents of all 12 experimental chickens were collected, and metagenomic sequencing was performed. We obtained a total of 80,994.61 Mbp of clean reads after quality control. To obtain Scaftigs >500 bp, the sequence reads were de novo assembled. A total of 2.456 million Scaftigs with an average length of 1,496 bp and an average N50 length of 1,925 bp were produced after subsequent assembly. After removing redundancies, a total of 1,096,825 genes remained, which constituted the final gene catalog (unigenes) for subsequent analyses (Supplementary Table 2). The Venn diagrams indicated that 92,109 and 148,540 genes were unique in the high and low FE groups of male chickens, respectively. At the same time, 80,377 and 169,651 genes are unique in the high and low FE groups of female chickens, respectively (Supplementary Figure 1).
We determined the taxonomic composition of cecal microbiota by blasting the unigenes to the NCBI RefSeq nonredundant proteins database. The bacteroidetes, firmicutes, proteobacteria, and actinobacteria were the most prevalent phyla in all 12 samples, accounting for >77% of the cecal microbial populations (Table 2). Nevertheless, at the genus level, the predominant genera in experimental chickens were different between the high and low FE group (Table 2 and Supplementary Figure 2). As can be seen from Table 1, the Lactobacillus of the low FE group was 1.41 and 2.31% lower than those of the high FE group in male and female chickens, respectively.
Table 2.
Relative abundance of the dominant phyla and genera in the cecum of the high and low FE groups of male and female chickens.
| Phylum (%) | Male |
Female |
Genus (%) | Male |
Female |
||||
|---|---|---|---|---|---|---|---|---|---|
| HFE | LFE | HFE | LFE | HFE | LFE | HFE | LFE | ||
| Bacteroidetes | 55.08 | 58.03 | 48.23 | 49.86 | Bacteroides | 22.10 | 24.41 | 18.39 | 21.54 |
| Firmicutes | 20.89 | 19.32 | 22.93 | 25.98 | Prevotella | 7.52 | 7.02 | 4.30 | 4.74 |
| Proteobacteria | 4.48 | 3.43 | 4.74 | 4.08 | Alistipes | 4.70 | 4.41 | 8.01 | 6.18 |
| Actinobacteria | 1.65 | 1.06 | 1.71 | 2.18 | Muribaculum | 2.29 | 3.42 | 1.42 | 1.46 |
| Verrucomicrobia | 0.32 | 0.35 | 0.99 | 0.64 | Lactobacillus | 2.35 | 0.94 | 3.43 | 1.12 |
| Spirochaetes | 0.29 | 0.47 | 0.94 | 0.25 | Megamonas | 1.14 | 0.81 | 0.83 | 1.94 |
| Euryarchaeota | 0.38 | 0.87 | 1.08 | 0.81 | Blautia | 1.42 | 1.68 | 1.37 | 2.54 |
| Fusobacteria | 0.05 | 0.15 | 0.64 | 0.11 | Lachnoclostridium | 1.55 | 1.34 | 1.80 | 2.15 |
| Synergistetes | 0.83 | 0.99 | 0.91 | 1.18 | Parabacteroides | 2.14 | 1.96 | 1.21 | 1.75 |
| Elusimicrobia | 0.3 | 0.08 | 0.24 | 0.25 | Mediterranea | 2.04 | 2.42 | 1.82 | 1.55 |
| Others | 15.72 | 15.25 | 17.57 | 14.67 | Others | 52.74 | 51.60 | 57.42 | 55.03 |
HFE and LFE denote high feed efficiency and low feed efficiency, respectively.
Similarities of the Cecal Microbial Communities
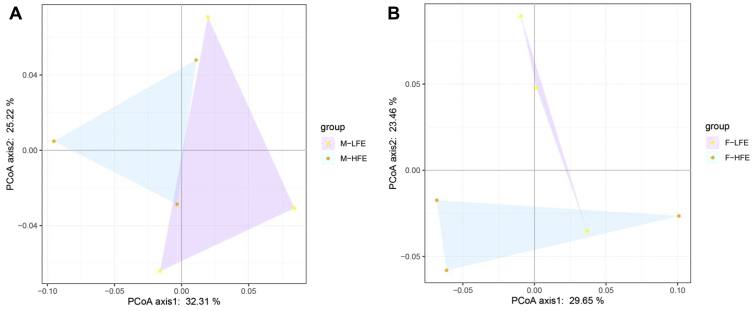
To explore the differences in the cecal microbiota composition between high FE and low FE groups, we calculated the Bray-Curtis distance between high FE and low FE group of male chickens and high FE and low FE group of female chickens at the species level. The distance matrix was visualized by principal coordinate analysis (Figure 1). As can be seen from Figure 1, the samples could be clustered by FE, which was consistent with the grouping results.
Figure 1.
Principal coordinate analysis (PCoA) of the cecal microbiota based on the Bray-Curtis distance at the species level. (A) PCoA between high FE and low FE of male chickens. (B) PCoA between high FE and low FE of female chickens. Abbreviation: FE, feed efficiency.
Comparison of Chicken Cecal Microbiota in High and Low FE Groups
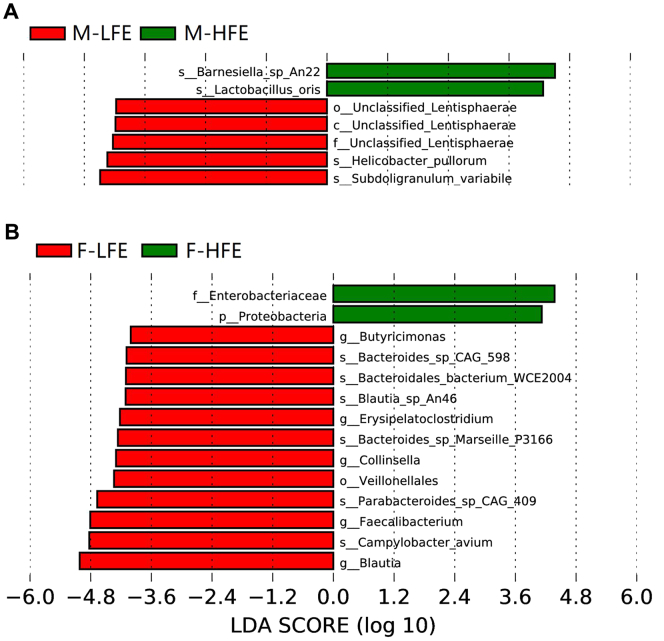
An LEfSe analysis was performed to identify any significant differences in the relative abundances of microbial taxa between chickens with different FE group, which could be used as biomarkers. In total, 22 biomarkers were identified with linear discriminant analysis scores >4 (Supplementary Table 3). In the microbial population of male chickens, Barnesiella sp An22 and Limosilactobacillus oris in the high FE group and Bacteroides sp An322, Subdoligranulum variabile, and Helicobacter pullorum in the low FE group were characteristic of the respective FE groups (Figure 2A). The Enterobacteriaceae and Proteobacteria in the high FE group and Blautia, Campylobacter avium, and Faecalibacterium in the low FE group could be considered as a potential biomarker in the microbial population of female chickens (Figure 2B).
Figure 2.
Linear discriminant analysis (LDA) effect size (LEfSe) analysis of cecal microbiota. The same analysis was performed in the (A) male chickens, and (B) female chickens to compare the microbial communities in the cecum between high FE and low FE groups. The LDA plots indicate species that can be used as biomarkers. Abbreviation: FE, feed efficiency.
Comparison of the Functionality of Chicken Cecal Microbiome
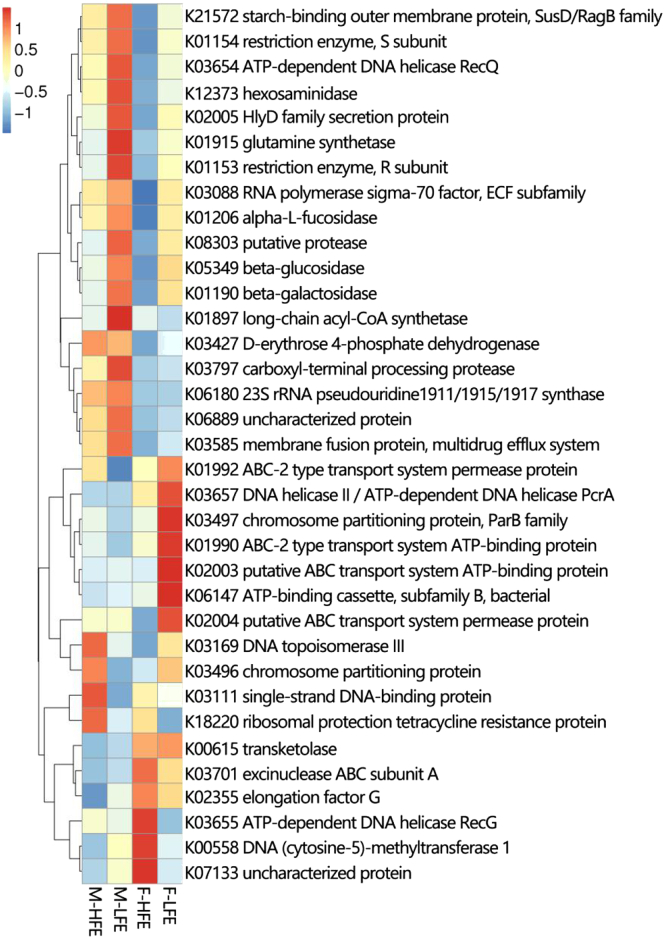
To investigate the functional capacity of cecal microbiota related to chicken FE, unigenes were annotated based on the KEGG and CAZy databases. First, the unigenes were aligned to the KEGG gene database, and a total of 5,407 KOs were obtained. Based on the functional annotations and abundance information of KO in the KEGG gene database, we selected the KOs of the top 35 and hierarchically clustered from the functional difference (Figure 3 and Supplementary Table 4). These KOs enriched in male chickens with high FE were associated with genetic information processing (KO3169, KO3111, K18220, KO3496). These KOs associated with amino acid metabolism (KO1915), fatty acid metabolism (KO1897), amino sugar and nucleotide sugar metabolism (K12373), and genetic information processing (KO1153, KO3797) were more abundant in male chickens with low FE. In the female chickens, these KOs abundant in high FE were associated with genetic information processing (KO0558, KO3655, KO3701). Notably, those KOs associated with energy transportation (KO6147, KO2003, KO1990, and KO2004) were abundant in low FE. The presence of these KOs may reduce the FE of female chickens, as Virkel et al. pointed out in 2019 that ATP-binding cassette transporting protein may influence the bioavailability in domestic animals (Virkel et al., 2019).
Figure 3.
Heat map of the top 35 predicted functions by KEGG. The X-axis shows the group IDs; M-HFE, M-LFE, F-HFE, and F-LFE denote the high feed efficiency group in male chickens, the low feed efficiency group in male chickens, the high feed efficiency group in female chickens, and the low feed efficiency group in female chickens, respectively. The Y-axis refers to the KEGG orthologues. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
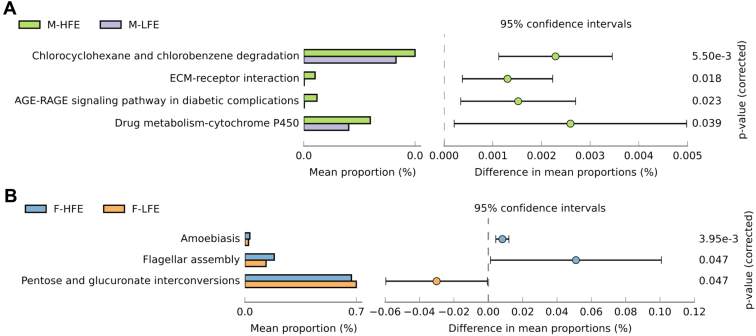
We investigated pathways of significant differences between the high FE and low FE group with P < 0.05. We identified 2 kinds of xenobiotics biodegradation and metabolism such as chlorocyclohexane and chlorobenzene degradation and drug metabolism—cytochrome P450 (CYP), and signaling molecules and interaction, and agarose gel electrophoresis–receptor for advanced glycation end products signaling pathway in diabetic complications (Figure 4A). These function terms were enriched in the high FE group of male chickens. In the female chickens, amoebiasis and cell motility in high FE group and carbohydrate metabolism in low FE group were abundant (Figure 4B).
Figure 4.
Differential KEGG function pathways between high feed efficiency group and low feed efficiency group. (A) The KEGG pathways detected in male chickens. (B) The KEGG pathways detected in female chickens. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Subsequently, we further aligned the sequences of unigenes to protein sequences in the CAZy database and classified the sequences into 6 enzyme classes. Glycoside hydrolases and glycosyltransferases were the 2 classes enriched the most in all samples (Supplementary Figure 3). The function pathways of significant differences between the high FE and low FE groups were identified. At the enzyme family level, we observed the differential pathway associated with the carbohydrate metabolism in the high FE group was less than the low FE group in male chickens. Whereas in the female chickens, 6 differential pathways associated with the carbohydrate metabolism in the high FE group and 2 pathways in the low FE group were identified (Supplementary Figure 4).
Discussion
The gut microbiota in chickens and mammals (McCormack et al., 2017) play important roles in the nutrient digestion, harvesting of ingested energy, and regulation of intestinal function, and its variations affected the metabolism (Stanley et al., 2013) and immune functions (Derrien et al., 2011) of the host. The finding that differentially abundant microbes were detected in the cecum might have implied a crucial role for cecal microbiota in FE (Yan et al., 2017). In this experiment, we explored the cecal microbial composition of Xiayan chickens and systematically estimated the interactions and relationships between cecal microbiota and FE with metagenomic sequencing data.
To minimize the influence of genetics, diet, and external factors, all experimental chickens were selected with similar genetic backgrounds and the same management, environmental, and dietary conditions. However, FE evaluated by the RFI still had great differences in experimental chickens, which indicated that the difference of gut microbiota might result in this phenomenon. At the phylum level, bacteroidetes, firmicutes, and proteobacteria were the most abundant phyla in cecal microbiota of chickens, which were consistent with previous reports (Lee et al., 2017; Dong et al., 2019; Shi et al., 2019). A recent study has shown that the abundance of bacteroidetes in the intestinal microbiota of the high FE pigs was lower than that of low FE pigs (Quan et al., 2020). In this experiment, we found the same result in chickens. Turnbaugh et al. (2006) point out that firmicutes play an important role in energy extraction in mice (Turnbaugh et al., 2006) and Yang et al. (2016) argued that the fatness might be improved when firmicutes increases in pigs' intestine (Yang et al., 2016). The results of these studies may suggest that the predominant phyla keep balance, and the stability of gut function can be ensured. However, compared with previous results, the composition and relative abundance of cecal-dominant microbes were different at the genus level (Pandit et al., 2018; Shi et al., 2019). Because the breed, sex, or feed of experimental animals between each study were different, the highly alterable microbial community composition may be affected by a variety of factors at a lower taxonomic level. For example, in the study by Shi et al. (2019), Bacteroides, Rikenellaceae_RC9, and Faecalibacterium were the 3 most abundant genera in laying hens (Shi et al., 2019). In Tamil Nadu, Bacteroides, unclassified Bacteria, and Alistipes were the 3 dominant genera in Ross 308 chicken breeds/lines, and unclassified Clostridiales, Clostridium, and Faecalibacterium presented higher proportions in commercial Cobb 400 in the study by Pandit et al. (2018) (Pandit et al., 2018).
Notably, the relative abundance of Lactobacillus in high FE groups was higher than the low FE groups, both in the male and female chickens (Table 2). We speculated that Lactobacillus may have a potential link to the FE. The LEfSe confirmed our hypothesis. That analysis indicated that the species Lactobacillus oris in the high FE group of male chickens could be considered as a potential biomarker. Yan et al. (2017) point out that Lactobacillus was one of the differentially abundant taxa and was highly related to the host FE (Yan et al., 2017). Studies have shown that Lactobacilli generally undergo 2 metabolic pathways: 1) converting glucose to lactic acid, which is called homofermentation, and 2) transforming glucose to lactic acid, acetic acid, ethanol, and CO2, which is called heterofermentation (Forte et al., 2018). These metabolites decrease the pH of the intestinal lumen to put potential pathogenic bacteria in an unfavorable environment (Menconi et al., 2011). The presence of Lactobacillus can effectively increase some beneficial bacteria and decrease potentially harmful bacteria to maintain the stability of microbial microbiota in the GIT (Forte et al., 2016). Wang et al. (2017) point out that Lactobacillus johnsonii BS15 reduces fat deposition and promotes the growth performance of chickens (Wang et al., 2017). Besides, Lactobacilli are often used as the formulation of prebiotics and probiotics that enhance the intestinal health for improved the colonization resistance to gut pathogens such as Campylobacter and Salmonella and the host performance (Kobierecka et al., 2017; Muyyarikkandy and Amalaradjou, 2017; Saint-Cyr et al., 2017; Khan and Chousalkar, 2020; Khan et al., 2020).
Campylobacter avium that could be considered as a potential biomarker was observed in the low FE group of the female chickens. Campylobacteriosis is a primary food-borne zoonosis all over the world (Carron et al., 2018). Some investigators have proven that poultry can serve as a natural host for Campylobacter species and a reservoir during dissemination (Torralbo et al., 2014). Besides, broilers are often colonized by Campylobacter (Garcia-Sanchez et al., 2018), and previous studies have reported that the colonization of Campylobacter jejuni in broiler chickens has a medium impact on the composition of intestinal microbiota (Kaakoush et al., 2014; Sofka et al., 2015). Furthermore, in the low FE group of the male chickens, H. pullorum has a higher score with LEfSe analysis. Helicobacter pullorum is a putative enterohepatic pathogen that has been associated with hepatobiliary and gastrointestinal diseases in chickens and humans (Sirianni et al., 2013). Another report showed that the chicken was infected with Helicobacter with no symptoms during infection, but the cecum of sacrificed chickens had mild lesions (Ceelen et al., 2007). The analysis by Pineda-Quiroga (2018) showed that lower abundance H. pullorum in the cecum was beneficial to the growth of broiler weight (Pineda-Quiroga et al., 2018). And the reduction of this bacterium also minimizes the risk of being transmitted to humans by chicken product consumption (Borges et al., 2015). Both C. avium and H. pullorum were associated with the inflammatory response. Therefore, we hypothesize that the colonization of these bacteria in cecum may reduce the FE of chickens.
Notably, a number of species under the Bacteroides genus such as Bacteroides sp An322, Bacteroides sp Marseille_P3166, and Bacteroides sp CAG_598 were observed. Bacteroides were gram-negative, spore-free, obligate anaerobic small bacilli. Bacteroides can cause endogenous infections when the organism's immune function is disordered or the microbiota is imbalanced. Previous studies have reported that the bacteroidales S24 7 group was more abundant in the low FE group of pigs (He et al., 2019). Ivarsson et al. (2014) pointed out that the abundance of Bacteroides–Prevotella–Porphyromonas in pigs was positively associated with the capacity of fermenting nonstarch polysaccharides to short-chain fatty acids (Ivarsson et al., 2014), and the short-chain fatty acids were linked to promoting human obesity (Cho et al., 2012). As is known to all, fat deposition decreases the FE of pigs (Martinsen et al., 2015). Taken together, we speculate that these bacteria may lead to a decrease in FE by promoting the development of host fatness. The results of functional annotation that was used to predict the function pathway associated with FE confirmed our hypothesis. At the enzyme family level, we observed the differential pathway associated with the carbohydrate metabolism in the low FE group was higher than that of the high FE group in male chickens. Besides, the KEGG pathway of the metabolism of monosaccharides was enriched in low FE group of female chickens; the same result was observed in pigs (Yang et al., 2017). A previous study showed that gut microbiota related to obesity had a higher capacity for energy extraction (Turnbaugh et al., 2006).
Consist with the previous study, the pathway of the flagellar assembly was enriched in the high FE group of female chickens (Tan et al., 2017), which suggests that the growth environment for microorganisms in the high FE group was better than that of the low FE group. In addition, the pathway associated with xenobiotics biodegradation and metabolism was abundant in the high FE group of male chickens. The previous report showed the toxic effects of hexabromocyclododecane on mammals, and chlorocyclohexane and chlorobenzene degradation and drug metabolism–CYP were associated with the degradation of hexabromocyclododecane (Wang et al., 2019). The CYP in the intestinal mucosa serves as a main metabolic barrier against orally ingested xenobiotics (Obach et al., 2001). In addition, CYP1A, CYP3A, and CYP2H subfamilies play a vital role in hepatic drug metabolism in chickens (Ourlin et al., 2000). Interestingly, consistent with the previous finding (Lee et al., 2017), the FE-associated gut microbes and pathway were diverse in male and female chickens, suggesting that the host gender had a significant effect on gut microbiota in chickens.
Currently, research on the intestinal microbiota of indigenous breed chickens in China, especially local breeds with special genetic traits, is scarce. The number of indigenous breed chickens in China is huge, and almost every indigenous breed chicken has its outstanding traits such as low abdominal fat rate, low-temperature resistance, and low-hypoxia resistance. These traits may be related to gut microbial composition and function. The Tibetan chicken lives in a high altitude and hypoxic environment and has the characteristics of strong resistance to hypoxia and roughage. Comparing the cecal microbiota of free-range Tibetan chickens with large-scale chickens, the researchers found that the cecal microbial composition and abundance were different (Zhou et al., 2016). This difference may be related to the excellent characteristics of Tibetan chickens. Therefore, research on the intestinal microbiota of Chinese indigenous breed chickens should be strengthened to enrich and improve information on the gut microbial composition and function of indigenous chicken.
Conclusion
In conclusion, we have revealed the compositional differences within the cecal microbiota associated with FE in Xiayan chicken, suggesting a potential association between cecal microbiota and FE. Meanwhile, we identified a total of 22 potential biomarkers associated with FE, beneficial bacteria including Lactobacillus and Limosilactobacillus oris, and harmful bacteria such as C. avium and H. pullorum in female and male chickens, respectively. The present study increased our understanding of the cecal microbial composition and identified many potential biomarkers related to FE, which could help improve the FE of Xiayan chickens.
Acknowledgments
This work was supported by the Science and Technology Major Project of Guangxi (GK AA17204027).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.09.076.
Disclosures
The authors declare no conflict of interest.
Supplementary data
References
- Al-Marzooqi W., Al-Maskari Z.A.S., Al-Kharousi K., Johnson E.H., El Tahir Y. Diversity of intestinal bacterial microbiota of indigenous and commercial strains of chickens using 16S rDNA-based analysis. Animals (Basel) 2020;10:391. doi: 10.3390/ani10030391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G., Mishra S.K., Nautiyal B., Pratap S.O., Gupta A., Beura C.K., Singh D.P. Genetics of hyperpigmentation associated with the Fibromelanosis gene (Fm) and analysis of growth and meat quality traits in crosses of native Indian Kadaknath chickens and non-indigenous breeds. Br. Poult. Sci. 2011;52:675–685. doi: 10.1080/00071668.2011.635637. [DOI] [PubMed] [Google Scholar]
- Borges V., Santos A., Correia C.B., Saraiva M., Menard A., Vieira L., Sampaio D.A., Pinheiro M., Gomes J.P., Oleastro M. Helicobacter pullorum isolated from fresh chicken meat: antibiotic resistance and genomic traits of an emerging foodborne pathogen. Appl. Environ. Microbiol. 2015;81:8155–8163. doi: 10.1128/AEM.02394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Carron M., Chang Y.M., Momanyi K., Akoko J., Kiiru J., Bettridge J., Chaloner G., Rushton J., O'Brien S., Williams N., Fevre E.M., Hasler B. Campylobacter, a zoonotic pathogen of global importance: Prevalence and risk factors in the fast-evolving chicken meat system of Nairobi, Kenya. Plos Negl. Trop. Dis. 2018;12:e0006658. doi: 10.1371/journal.pntd.0006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceelen L.M., Decostere A., Chiers K., Ducatelle R., Maes D., Haesebrouck F. Pathogenesis of Helicobacter pullorum infections in broilers. Int. J. Food Microbiol. 2007;116:207–213. doi: 10.1016/j.ijfoodmicro.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Cho I., Yamanishi S., Cox L., Methe B.A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., Li H., Alekseyenko A.V., Blaser M.J. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Van Baarlen P., Hooiveld G., Norin E., Muller M., de Vos W.M. Modulation of mucosal immune response, Tolerance, and Proliferation in mice colonized by the Mucin-Degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do D.N., Ostersen T., Strathe A.B., Mark T., Jensen J., Kadarmideen H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014;15:27. doi: 10.1186/1471-2156-15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hu B., Wan W., Gong Y., Feng Y. Effects of husbandry systems and Chinese indigenous chicken strain on cecum microbial diversity. Asian-Australas J. Anim. Sci. 2019;33:1610–1616. doi: 10.5713/ajas.19.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duah K.K., Essuman E.K., Boadu V.G., Olympio O.S., Akwetey W. Comparative study of indigenous chickens on the basis of their health and performance. Poult. Sci. 2020;99:2286–2292. doi: 10.1016/j.psj.2019.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte C., Acuti G., Manuali E., Casagrande Proietti P., Pavone S., Trabalza-Marinucci M., Moscati L., Onofri A., Lorenzetti C., Franciosini M.P. Effects of two different probiotics on microflora, morphology, and morphometry of gut in organic laying hens. Poult. Sci. 2016;95:2528–2535. doi: 10.3382/ps/pew164. [DOI] [PubMed] [Google Scholar]
- Forte C., Manuali E., Abbate Y., Papa P., Vieceli L., Tentellini M., Trabalza-Marinucci M., Moscati L. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens. Poult. Sci. 2018;97:930–936. doi: 10.3382/ps/pex396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez L., Melero B., Diez A.M., Jaime I., Rovira J. Characterization of Campylobacter species in Spanish retail from different fresh chicken products and their antimicrobial resistance. Food Microbiol. 2018;76:457–465. doi: 10.1016/j.fm.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Haunshi S., Niranjan M., Shanmugam M., Padhi M.K., Reddy M.R., Sunitha R., Rajkumar U., Panda A.K. Characterization of two Indian native chicken breeds for production, egg and semen quality, and welfare traits. Poult. Sci. 2011;90:314–320. doi: 10.3382/ps.2010-01013. [DOI] [PubMed] [Google Scholar]
- He B., Li T., Wang W., Gao H., Bai Y., Zhang S., Zang J., Li D., Wang J. Metabolic characteristics and nutrient utilization in high-feed-efficiency pigs selected using different feed conversion ratio models. Sci. China Life Sci. 2019;62:959–970. doi: 10.1007/s11427-018-9372-6. [DOI] [PubMed] [Google Scholar]
- Hiergeist A., Glasner J., Reischl U., Gessner A. Analyses of intestinal microbiota: culture versus sequencing. ILAR J. 2015;56:228–240. doi: 10.1093/ilar/ilv017. [DOI] [PubMed] [Google Scholar]
- Ivarsson E., Roos S., Liu H.Y., Lindberg J.E. Fermentable non-starch polysaccharides increases the abundance of Bacteroides-Prevotella-Porphyromonas in ileal microbial community of growing pigs. Animal. 2014;8:1777–1787. doi: 10.1017/S1751731114001827. [DOI] [PubMed] [Google Scholar]
- Kaakoush N.O., Sodhi N., Chenu J.W., Cox J.M., Riordan S.M., Mitchell H.M. The interplay between Campylobacter and Helicobacter species and other gastrointestinal microbiota of commercial broiler chickens. Gut Pathog. 2014;6:18. doi: 10.1186/1757-4749-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Fak F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D., Backhed F., Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Tremaroli V., Nookaew I., Bergstrom G., Behre C.J., Fagerberg B., Nielsen J., Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Khan S., Chousalkar K.K. Short-term feeding of probiotics and synbiotics modulates caecal microbiota during Salmonella Typhimurium infection but does not reduce shedding and invasion in chickens. Appl. Microbiol. Biot. 2020;104:319–334. doi: 10.1007/s00253-019-10220-7. [DOI] [PubMed] [Google Scholar]
- Khan S., Moore R.J., Stanley D., Chousalkar K.K. Gut microbiota of laying hens and its manipulation with prebiotics and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 2020;86:e00600-20. doi: 10.1128/AEM.00600-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobierecka P.A., Wyszynska A.K., Aleksandrzak-Piekarczyk T., Kuczkowski M., Tuzimek A., Piotrowska W., Gorecki A., Adamska I., Wieliczko A., Bardowski J., Jagusztyn-Krynicka E.K. In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. Microbiologyopen. 2017;6:e00512. doi: 10.1002/mbo3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R.M., Swiger L.A., Chambers D., Gregory K.E. Efficiency of feed Use in Beef Cattle. J. Anim. Sci. 1963;22:486–494. [Google Scholar]
- Lee K.C., Kil D.Y., Sul W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017;55:939–945. doi: 10.1007/s12275-017-7202-0. [DOI] [PubMed] [Google Scholar]
- Li J.H., Jia H.J., Cai X.H., Zhong H.Z., Feng Q., Sunagawa S., Arumugam M., Kultima J.R., Prifti E., Nielsen T., Juncker A.S., Manichanh C., Chen B., Zhang W.W., Levenez F., Wang J., Xu X., Xiao L., Liang S.S., Zhang D.Y., Zhang Z.X., Chen W.N., Zhao H.L., Al-Aama J.Y., Edris S., Yang H.M., Wang J., Hansen T., Nielsen H.B., Brunak S., Kristiansen K., Guarner F., Pedersen O., Dore J., Ehrlich S.D., Bork P., Wang J., Consortium M. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Luiting P., Urff E.M. Optimization of a model to estimate residual feed consumption in the laying hen. Livestock Prod. Sci. 1991;27:321–338. [Google Scholar]
- Luo R.B., Liu B.H., Xie Y.L., Li Z.Y., Huang W.H., Yuan J.Y., He G.Z., Chen Y.X., Pan Q., Liu Y.J., Tang J.B., Wu G.X., Zhang H., Shi Y.J., Liu Y., Yu C., Wang B., Lu Y., Han C.L., Cheung D.W., Yiu S.M., Peng S.L., Zhu X.Q., Liu G.M., Liao X.K., Li Y.R., Yang H.M., Wang J., Lam T.W., Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancabelli L., Milani C., Lugli G.A., Turroni F., Mangifesta M., Viappiani A., Ticinesi A., Nouvenne A., Meschi T., van Sinderen D., Ventura M. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci. Rep. 2017;7:9879. doi: 10.1038/s41598-017-10663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J.R., Adams D.H., Fava F., Hermes G.D., Hirschfield G.M., Hold G., Quraishi M.N., Kinross J., Smidt H., Tuohy K.M., Thomas L.V., Zoetendal E.G., Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen K.H., Odegard J., Olsen D., Meuwissen T.H. Genetic variation in efficiency to deposit fat and lean meat in Norwegian Landrace and Duroc pigs. J. Anim. Sci. 2015;93:3794–3800. doi: 10.2527/jas.2015-9174. [DOI] [PubMed] [Google Scholar]
- McCormack U.M., Curiao T., Buzoianu S.G., Prieto M.L., Ryan T., Varley P., Crispie F., Magowan E., Metzler-Zebeli B.U., Berry D., O'Sullivan O., Cotter P.D., Gardiner G.E., Lawlor P.G. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00380-17. e00380–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi A., Wolfenden A.D., Shivaramaiah S., Terraes J.C., Urbano T., Kuttel J., Kremer C., Hargis B.M., Tellez G. Effect of lactic acid bacteria probiotic culture for the treatment of Salmonella enterica serovar Heidelberg in neonatal broiler chickens and Turkey poults. Poult. Sci. 2011;90:561–565. doi: 10.3382/ps.2010-01220. [DOI] [PubMed] [Google Scholar]
- Milani C., Hevia A., Foroni E., Duranti S., Turroni F., Lugli G.A., Sanchez B., Martin R., Gueimonde M., van Sinderen D., Margolles A., Ventura M. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One. 2013;8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyyarikkandy M.S., Amalaradjou M.A. Lactobacillus bulgaricus, Lactobacillus rhamnosus and Lactobacillus paracasei Attenuate Salmonella Enteritidis, Salmonella Heidelberg and Salmonella Typhimurium colonization and Virulence gene expression in vitro. Int. J. Mol. Sci. 2017;18:2381. doi: 10.3390/ijms18112381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obach R.S., Zhang Q.Y., Dunbar D., Kaminsky L.S. Metabolic characterization of the major human small intestinal cytochrome p450s. Drug Metab. Dispos. 2001;29:347–352. [PubMed] [Google Scholar]
- Oh J., Byrd A.L., Deming C., Conlan S., Kong H.H., Segre J.A., Progra N.C.S. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.E. Whole proteomes as internal standards in quantitative proteomics. Genome Med. 2010;2:49. doi: 10.1186/gm170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourlin J.C., Baader M., Fraser D., Halpert J.R., Meyer U.A. Cloning and functional expression of a first inducible avian cytochrome P450 of the CYP3A subfamily (CYP3A37) Arch. Biochem. Biophys. 2000;373:375–384. doi: 10.1006/abbi.1999.1566. [DOI] [PubMed] [Google Scholar]
- Pandit R.J., Hinsu A.T., Patel N.V., Koringa P.G., Jakhesara S.J., Thakkar J.R., Shah T.M., Limon G., Psifidi A., Guitian J., Hume D.A., Tomley F.M., Rank D.N., Raman M., Tirumurugaan K.G., Blake D.P., Joshi C.G. Microbial diversity and community composition of caecal microbiota in commercial and indigenous Indian chickens determined using 16s rDNA amplicon sequencing. Microbiome. 2018;6:115. doi: 10.1186/s40168-018-0501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Quiroga C., Camarinha-Silva A., Borda-Molina D., Atxaerandio R., Ruiz R., Garcia-Rodriguez A. Feeding broilers with dry whey powder and whey protein concentrate affected productive performance, ileal digestibility of nutrients and cecal microbiota community. Animal. 2018;12:692–700. doi: 10.1017/S1751731117002208. [DOI] [PubMed] [Google Scholar]
- Qin J.J., Li R.Q., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J.H., Xu J.M., Li S.C., Li D.F., Cao J.J., Wang B., Liang H.Q., Zheng H.S., Xie Y.L., Tap J., Lepage P., Bertalan M., Batto J.M., Hansen T., Le Paslier D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H.M., Yu C., Li S.T., Jian M., Zhou Y., Li Y.R., Zhang X.Q., Li S.G., Qin N., Yang H.M., Wang J., Brunak S., Dore J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S.D., Wang J., Consortium M. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–U70. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Wu Z., Ye Y., Peng L., Wu J., Ruan D., Qiu Y., Ding R., Wang X., Zheng E., Cai G., Huang W., Yang J. Metagenomic characterization of intestinal regions in pigs with contrasting feed efficiency. Front Microbiol. 2020;11:32. doi: 10.3389/fmicb.2020.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Cyr M.J., Haddad N., Taminiau B., Poezevara T., Quesne S., Amelot M., Daube G., Chemaly M., Dousset X., Guyard-Nicodeme M. Use of the potential probiotic strain Lactobacillus salivarius SMXD51 to control Campylobacter jejuni in broilers. Int. J. Food Microbiol. 2017;247:9–17. doi: 10.1016/j.ijfoodmicro.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Qi Z., Gu B., Cheng B., Tu J., Song X., Shao Y., Liu H., Qi K., Li S. Analysis of high-throughput sequencing for cecal microbiota diversity and function in hens under different rearing systems. 3 Biotech. 2019;9:438. doi: 10.1007/s13205-019-1970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegerstetter S.C., Schmitz-Esser S., Magowan E., Wetzels S.U., Zebeli Q., Lawlor P.G., O'Connell N.E., Metzler-Zebeli B.U. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS One. 2017;12:e0187766. doi: 10.1371/journal.pone.0187766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirianni A., Kaakoush N.O., Raftery M.J., Mitchell H.M. The pathogenic potential of Helicobacter pullorum: possible role for the type VI secretion system. Helicobacter. 2013;18:102–111. doi: 10.1111/hel.12009. [DOI] [PubMed] [Google Scholar]
- Sofka D., Pfeifer A., Gleiss B., Paulsen P., Hilbert F. Changes within the intestinal flora of broilers by colonisation with Campylobacter jejuni. Berl Munch Tierarztl. 2015;128:104–110. [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Denman S.E., Haring V.R., Crowley T.M., Hughes R.J., Moore R.J. Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet. Microbiol. 2013;164:85–92. doi: 10.1016/j.vetmic.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Stanley D., Hughes R.J., Geier M.S., Moore R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. 2016;7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl. Microbiol. Biot. 2014;98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- Tan Z., Yang T., Wang Y., Xing K., Zhang F., Zhao X., Ao H., Chen S., Liu J., Wang C. Metagenomic analysis of cecal microbiome identified microbiota and functional Capacities associated with feed efficiency in Landrace Finishing pigs. Front Microbiol. 2017;8:1546. doi: 10.3389/fmicb.2017.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torralbo A., Borge C., Allepuz A., Garcia-Bocanegra I., Sheppard S.K., Perea A., Carbonero A. Prevalence and risk factors of Campylobacter infection in broiler flocks from southern Spain. Prev. Vet. Med. 2014;114:106–113. doi: 10.1016/j.prevetmed.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Virkel G., Ballent M., Lanusse C., Lifschitz A. Role of ABC Transporters in Veterinary Medicine: pharmaco-Toxicological Implications. Curr. Med. Chem. 2019;26:1251–1269. doi: 10.2174/0929867325666180201094730. [DOI] [PubMed] [Google Scholar]
- Wang R., Lin C.Y., Chen S.H., Lo K.J., Liu C.T., Chou T.H., Shih Y.H. Using high-throughput transcriptome sequencing to investigate the biotransformation mechanism of hexabromocyclododecane with Rhodopseudomonas palustris in water. Sci. Total Environ. 2019;692:249–258. doi: 10.1016/j.scitotenv.2019.07.140. [DOI] [PubMed] [Google Scholar]
- Wang H., Ni X., Qing X., Zeng D., Luo M., Liu L., Li G., Pan K., Jing B. Live probiotic Lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving Lipid metabolism, intestinal development, and gut microflora in broilers. Front Microbiol. 2017;8:1073. doi: 10.3389/fmicb.2017.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Milner J., Boudreau M.D., Gokulan K., Cerniglia C.E., Khare S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology. 2015;9:279–289. doi: 10.3109/17435390.2014.921346. [DOI] [PubMed] [Google Scholar]
- Wu I.W., Gao S.S., Chou H.C., Yang H.Y., Chang L.C., Kuo Y.L., Dinh M.C.V., Chung W.H., Yang C.W., Lai H.C., Hsieh W.P., Su S.C. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. 2020;10:5398–5411. doi: 10.7150/thno.41725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Sun C., Yuan J., Yang N. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci. Rep. 2017;7:45308. doi: 10.1038/srep45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Huang X., Fang S., He M., Zhao Y., Wu Z., Yang M., Zhang Z., Chen C., Huang L. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front Microbiol. 2017;8:1555. doi: 10.3389/fmicb.2017.01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Huang X., Fang S., Xin W., Huang L., Chen C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016;6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Dou T., Ma M., Yi G., Chen S., Qu L., Shen M., Qu L., Wang K., Yang N. Genetic parameters of feed efficiency traits in laying period of chickens. Poult. Sci. 2015;94:1470–1475. doi: 10.3382/ps/pev122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu Z.Q., Luo Y.Y., Zhang H.B., Gao N., He J.L., Ji C.L., Zhang D.X., Li J.Q., Zhang X.Q. Whole genomic prediction of growth and carcass traits in a Chinese quality chicken population. J. Anim. Sci. 2017;95:72–80. doi: 10.2527/jas.2016.0823. [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang X., Yang C., Ma B., Lei C., Xu C., Zhang A., Yang X., Xiong Q., Zhang P., Men S., Xiang R., Wang H. Cecal microbiota of Tibetan Chickens from five geographic regions were determined by 16S rRNA sequencing. Microbiologyopen. 2016;5:753–762. doi: 10.1002/mbo3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.H., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.