Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) has 2 protein isoforms (PPARγ1 and PPARγ2) generated by alternative promoter usage and alternative splicing. However, their functional uniqueness and similarity remain unclear. In the study, we investigated the effects of lentivirus-mediated overexpression of PPARγ1 and PPARγ2 on proliferation, apoptosis, and differentiation of the immortalized chicken preadipocytes. Cell Counting Kit–8 assay showed PPARγ1 and PPARγ2 overexpression markedly suppressed cell proliferation, and fluorescence activated cell sorting analysis showed that PPARγ1 and PPARγ2 overexpression caused cell cycle arrest at G0/G1 phase. Cell death detection ELISA analysis showed both PPARγ1 and PPARγ2 overexpression induced cell apoptosis. Oil red O staining and gene expression analysis showed both PPARγ1 and PPARγ2 overexpression promoted preadipocyte differentiation. In the presence of PPARγ ligand, rosiglitazone, PPARγ2 overexpression was more potent in inducing apoptosis, promoting adipogenesis, and suppressing cell proliferation than PPARγ1 overexpression. We further explored the molecular basis for their functional differences. Reporter gene assay showed that under ligand conditions, PPARγ2 overexpression resulted in 1.68-fold increase in transcription activity compared with PPARγ1. Electrophoretic mobility shift assay showed both PPARγ1 and PPARγ2 could bind to PPAR response element (PPRE) as heterodimer with retinoid X receptor alpha, and by comparison, PPARγ2 had a higher affinity for PPRE than PPARγ1. Reporter gene assay showed expression PPARγ1 and PPARγ2 similarly induced fatty acid synthase and adipocyte fatty acid–binding protein promoter activity but differentially induced lipoprotein lipase and perilipin 1 promoter activities. Coimmunoprecipitation analysis showed that PPARγ1 and PPARγ2 interacted similarly with the coactivators, Tat-interacting protein 60. Taken together, our results demonstrate that PPARγ1 and PPARγ2 differentially regulate preadipocyte proliferation, apoptosis, and differentiation as a result of their distinct and overlapping molecular functions.
Key words: PPARγ isoforms, preadipocyte, proliferation, apoptosis, adipogenesis
Introduction
A fundamental property of preadipocytes is that they are able to constantly proliferate and differentiate into mature adipocytes and therefore maintain the functional plasticity and expansion of adipose tissue (Cawthorn et al., 2012). Peroxisome proliferator-activated receptor γ (PPARγ) is the master regulator of adipogenesis (Cristancho and Lazar, 2011). Peroxisome proliferator-activated receptor γ is a member of the nuclear receptor superfamily of transcription factors and is highly expressed in adipose tissues (Ahmadian et al., 2013; Lee et al., 2019). Peroxisome proliferator-activated receptor γ has 2 protein isoforms PPARγ1 and PPARγ2 that are generated by alternative promoter usage and alternate splicing. PPARγ2 has an additional 28 (human) or 30 (mouse and goat) amino acids, relative to PPARγ1 at the N-terminal, containing the activation function-1 region (Al-Shali et al., 2004; De Sá et al., 2011; Shi et al., 2013).
The differences between the 2 PPARγ isoforms in expression pattern, transcriptional activity, and adipocyte differentiation have been studied in mammals. The isoform PPARγ1 is expressed in adipose and many other tissues, whereas PPARγ2 is predominantly expressed in adipose tissue in humans (Grygiel-Górniak, 2014). The abundance of the PPARγ isoforms in different tissues is directly related to their respective specific functions (Bionaz et al., 2013). The reporter gene assay showed that PPARγ2 displayed 5- to 6-fold greater transcriptional activity than PPARγ1 in rats (Werman et al., 1997). Although PPARγ1 and PPARγ2 are expressed at comparable levels in adipocytes (Zhang et al., 2004), their relative importance in adipogenesis remains an open question.
Our previous study in chickens found that PPARγ2 has an additional 6 amino acids at the N-terminal compared with PPARγ1 (Kui et al., 2015). The functional uniqueness and similarity of the 2 chicken PPARγ isoforms remain unclear. In the present study, using lentivirus-mediated overexpression of PPARγ1 and PPARγ2, we investigated the effects of overexpressing the 2 chicken PPARγ isoforms on preadipocyte proliferation, apoptosis, and differentiation. To further understand the underlying molecular mechanisms, we evaluated their ability to transactivate the PPAR response element (PPRE) and adipogenic gene reporters, bind to PPRE, and interact with the coregulator Tat-interacting protein 60 (Tip60). Our results demonstrate that PPARγ2 is more potent in transcriptional activation and DNA binding than PPARγ1.
Materials and methods
Cell Culture and Treatments
Immortalized chicken preadipocytes (ICP2) were generated in our laboratory (Wang et al., 2017). Immortalized chicken preadipocyte cells were cultured in Dulbecco's Modified Eagle Medium/Ham's F-12 media (Gibco) containing 10% fetal bovine serum (BI, Germany), 100 units/mL penicillin, and 100 μg/mL streptomycin and incubated at 37°C with 5% CO2. Cells were treated with rosiglitazone (20 μmol/L) and 5 μmol/L 9-cis RA or both for 24, 48, and 72 h, and DMSO was used as a negative control. Rosiglitazone and 9-cis RA are PPARγ and retinoid X receptor alpha (RXRα) agonist, respectively. All the reagents for cell treatments were from Sigma-Aldrich. All treatments were performed in triplicate and repeated 3 times.
Plasmid Construction
The thymidine kinase (TK) minimal promoter with 3 copies of the DR1 element (AGGTCAAAGGTCA) at its 5′ end was synthesized by GENEWIZ and cloned into the pGL3-Basic Luciferase Reporter Vector (Promega) to yield the 3xPPRE-TK-Luc reporter construct. The CCAAT/enhancer-binding protein alpha (C/EBPα), fatty acid synthase (FAS), adipocyte fatty acid–binding protein (A-FABP), lipoprotein lipase (LPL), and perilipin 1 (PLIN1) promoter reporters containing the potential PPRE were previously generated by our laboratory (Zhang et al., 2014; Zhang et al., 2019). The full-length PPARγ1 (NM_001001460.1) was amplified from the pooled chicken cDNA using PCR with the following primers: PPARγ1-F (5′-GGAATTCATGGTTGACACAGAAATGCCGT-3′) and PPARγ1-R (5′-CCTCGAGGAGGATAAGAACTACTATCGCC-3′) and introduced into the EcoRI and XhoI sites of a pCMV-HA vector to yield pCMV-HA-PPARγ1. The full-length PPARγ2 (KP736527 and NM_001001460.1) coding region was amplified from the pooled chicken cDNA using PCR with the following primers: PPARγ2-F (5′-GGTCGACCGAGATCTCTCGAGGGAAAAGAGAGATTACAATGGTT-3′) and PPARγ2-R (5′-CATGTCTGGATCCCCGCGGCCGCAAAACATACATTATGTCAGAGGATA-3′) and cloned into the XhoI and NotI sites of the pCMV-HA vector to yield pCMV-HA-PPARγ2. The positions of PPARγ1-F/R and PPARγ2-F/R are show in Supplementary Figure 1. The full-length RXRα (XM_003642291.4) coding region was amplified from the pooled chicken cDNA using PCR with the following primers: RXRα-F (5′-CGGAATTCTGGACACCAAACACTTCCTGCCACT-3′) and RXRα-R (5′-CCTCGAGTTAGATGCAGCAGTGACAGCGAACG-3′) and cloned into the EcoRI and XhoI sites of the pCMV-Myc vector to create pCMV-Myc-RXRα. The full-length CDS of Tip60 (XM_015273024.2) was synthesized by GENEWIZ (Suzhou, China) and cloned into the EcoRI and KpnI sites of the pCMV-Myc vector to yield pCMV-Myc-Tip60.
RNA Isolation and Quantitative Real-Time RT-PCR
Total RNA was isolated from the cultured cells using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Total RNA was reverse transcribed using the PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). The relative mRNA expression levels were analyzed using the 2−ΔCT method, and TATA box–binding protein was used as an internal control. Experiments were performed in triplicate and repeated 3 times to ensure accuracy. The sequences of the primers are shown in Table 1.
Table 1.
Primers used for quantitative real-time PCR.
| Primer name | Primer sequence |
|---|---|
| LPL | F: ATGTTCATTGATTGGATGGAGGAG |
| R: AAAGGTGGGACCAGCAGGAT | |
| A-FABP | F: ATGTGCGACCAGTTTGT |
| R: TCACCATTGATGCTGATAG | |
| PLIN1 | F: GGGGTGACTGGCGGTTGTA |
| R: GCCGTAGAGGTTGGCGTAG | |
| C/EBPα | F: GCGACATCTGCGAGAACG |
| R: GTACAGCGGGTCGAGCTT | |
| TBP | F: GCGTTTTGCTGCTGTTATTATGAG |
| R: TCCTTGCTGCCAGTCTGGAC |
Abbreviations: A-FABP, adipocyte fatty acid–binding protein; C/EBPα, CCAAT/enhancer binding protein alpha; LPL, lipoprotein lipase; PLIN1, perilipin 1; TBP, TATA box–binding protein.
Immunoprecipitation Assays and Western Blot Analysis
The ICP2 cells were plated at 1 × 106 in 10-cm dishes and cotransfected with PPARγ1 or PPARγ2, RXRα, and Tip60 expression vectors (10 μg) using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. At 24 h after transfection, cells were treated with 20 μmol/L rosiglitazone for 24 h and lysed in 500 μL cell lysis buffer per dish for western and IP buffer (Beyotime, China). The cell lysate was incubated with the anti-HA (ProteinTech Group) at 4°C for 1 h and subsequently incubated with protein A/G beads overnight (Santa Cruz Biotechnology). The beads were washed 4 times with PBS. The whole-cell lysates and immunoprecipitates were subjected to SDS-PAGE. Proteins were transferred onto nitrocellulose membranes (Millipore, MA) and incubated with the following primary antibodies: anti-HA (1:500; Beyotime, China), anti-Myc (1:500; Beyotime China), and anti-caspase3 (1:200; Novus Biologicals). The ECL Plus Detection Kit (HaiGene, China) was used to detect immunoreactive bands. All experiments were repeated 2 times.
Electrophoretic Mobility Shift Assay
Nuclear proteins were isolated from the ICP2 cells transfected with pCMV-HA-PPARγ1 or pCMV-HA-PPARγ2 and pCMV-Myc-RXRα using the NE-PER nuclear extraction kit (Pierce). Binding activity of PPARγ1 or PPARγ2 and RXRα to the indicated probes was performed using the LightShift Chemiluminescent Electrophoretic Mobility Shift Assay (EMSA) Kit (Pierce). The biotin-labeled probes containing PPRE and its corresponding mutant probes were synthesized by Genewiz and are shown in Table 2. The labeled double-stranded probes were added to 3 μL NE-PER nuclear extracts and incubated for 30 min at room temperature. For binding competition experiments, 50- or 100-fold molar excess of unlabeled double-stranded wild-type or mutant probes were added to the binding reactions immediately before the addition of the labeled probes. The DNA–protein complexes were resolved on a 6% native polyacrylamide gel and detected. All experiments were repeated 2 times.
Table 2.
Probes used in EMSA analysis.
| Probe name | Sequences 5′-3′ |
|---|---|
| WT-F | CAAAACTAGGTCAAAGGTCA |
| WT-R | TGACCTTTGACCTAGTTTTG |
| MT-F | CAAAACTAGcaCAAAGcaCA |
| MT-R | TGtgCTTTGtgCTAGTTTTG |
PPAR response element (PPRE) is underlined.
Substitution mutations are presented in lower case italic letter.
Abbreviation: EMSA, electrophoretic mobility shift assay.
Luciferase Reporter Gene Assay
For transactivation assays, ICP2 cells were cotransfected with the 3xPPRE-TK-Luc reporter construct and the indicated expression vectors (pCMV-HA-PPARγ1, pCMV-HA-PPARγ2 or pCMV-Myc-RXRα), along with the pRL-TK vector. After 24 h, the transfected cells were treated with or without rosiglitazone (20 μmol/L) and 9-cis RA (5 μmol/L) for 24 h. For transcriptional regulation analysis, cells were cotransfected with the indicated promoter reporters (C/EBPα, FAS, A-FABP, LPL, or PLIN1 reporters), indicated PPARγ isoform expression vectors (pCMV-HA-PPARγ1 and pCMV-HA-PPARγ2), and indicated the RXRα expression vector (pCMV-Myc-RXRα) along with the pRL-TK Renilla luciferase vector. After 48 h of transfection, the luciferase activity was assessed with the dual-luciferase reporter assay system (Promega). All experiments were performed in triplicate and repeated 3 times. The luciferase activity was normalized to the corresponding Renilla luciferase activity.
Cell Proliferation Assay
The recombinant lentiviruses expressing PPARγ1 (Lenti-PPARγ1), PPARγ2 (Lenti-PPARγ2), and a lentivirus control (Lenti-control) were constructed and packaged by Hanbio (Shanghai, China). Cell proliferation was assessed using the Cell Counting Kit–8 (DOJINDO, Japan). Briefly, ICP2 cells were plated into 48-well plates at 1 × 104 cells/well. After 12 h, the cells were infected with Lenti-PPARγ1 or Lenti-PPARγ2 at 50 multiplicities of infection (MOI) for 24, 48, and 72 h in the absence or presence of rosiglitazone (20 μmol/L). The absorbance was assessed at 450 nm.
Cell Cycle Detection
The cell cycle was analyzed by flow cytometry using the Cell Cycle and Apoptosis Analysis Kit (Beyotime, China). Briefly, ICP2 cells were seeded into 6-well plates at 1 × 106 cells/well and infected with Lenti-PPARγ1 or Lenti-PPARγ2 viruses at 50 MOI in the presence or absence of rosiglitazone (20 μmol/L). After 48 h, the cells were trypsinized and fixed in ice-cold 70% ethanol in PBS at 4°C overnight. The cells were washed with PBS and stained with propidium iodide for 30 min at 37°C in the dark. The cells were then sorted by Fluorescence activated Cell Sorting can analysis (Becton Dickinson, NJ) and analyzed by ModFit LT software (Verity Software House).
Cell Death Detection
Apoptosis assays were determined using Cell Death Detection ELISAplus (Roche, Switzerland). Briefly, ICP2 cells were seeded into 24-well plates at 5 × 104 cells/well and infected with Lenti-PPARγ1 or Lenti-PPARγ2 at 50 MOI for 48 h in the presence or absence of rosiglitazone (20 μmol/L). The cells were lysed directly in lysis buffer for 30 min at 25°C. After centrifugation, the culture supernatant was transferred into a streptavidin-coated microtiter plate and incubated with immunoreagent for 2 h at 25°C with gentle shaking. Each well was rinsed with incubation buffer and incubated with ABTS (The ABTS is the kit content of Cell Death Detection ELISAplus (Roche, Switzerland)) for 15 min. Then, the optical density was measured using an Epoch microplate reader (BioTek Instruments, USA) at 405 nm after adding the ABTS stop solution.
Adipocyte Differentiation and Oil Red O Staining
The ICP2 cells were seeded at 5 × 104 in a 24-well plate and infected with Lenti-PPARγ1, Lenti-PPARγ2, and Lenti-control viruses at 50 MOI with or without rosiglitazone (20 μmol/L). For adipocyte differentiation, ICP2 cells at about 60% confluence were incubated with the differentiation medium containing Dulbecco's Modified Eagle Medium/Ham's F-12 culture medium, 10% fetal bovine serum, 1% antibiotic–antimycotic solution, and 160 μmol/L sodium oleate (Sigma). The differentiation medium was changed every day until the indicated time points.
Differentiated ICP2 adipocytes were washed twice with PBS, fixed in 4% formaldehyde for 15 min at room temperature, and rinsed 3 times with distilled water. The cells were stained with Oil red O solution (3:2, 0.6% Oil Red O in isopropanol:water) for 15 min at room temperature, then washed with distilled water. The intracellular lipid droplets were visualized with an optical microscope (Leica). To quantify staining, Oil Red O was extracted from the cells with isopropanol solution, and the optical density was measured at a wavelength of 510 nm. The cell total protein concentration was estimated by the bicinchoninic acid method (Beyotime, China). Intracellular lipid content was normalized against the protein to allow an accurate comparison.
Statistical Analysis
Results are expressed as means ± SD. Comparisons between groups were performed using 2-way ANOVA. Statistical differences were considered significant when P < 0.05.
Results
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Differentially Inhibit Chicken Preadipocyte Proliferation
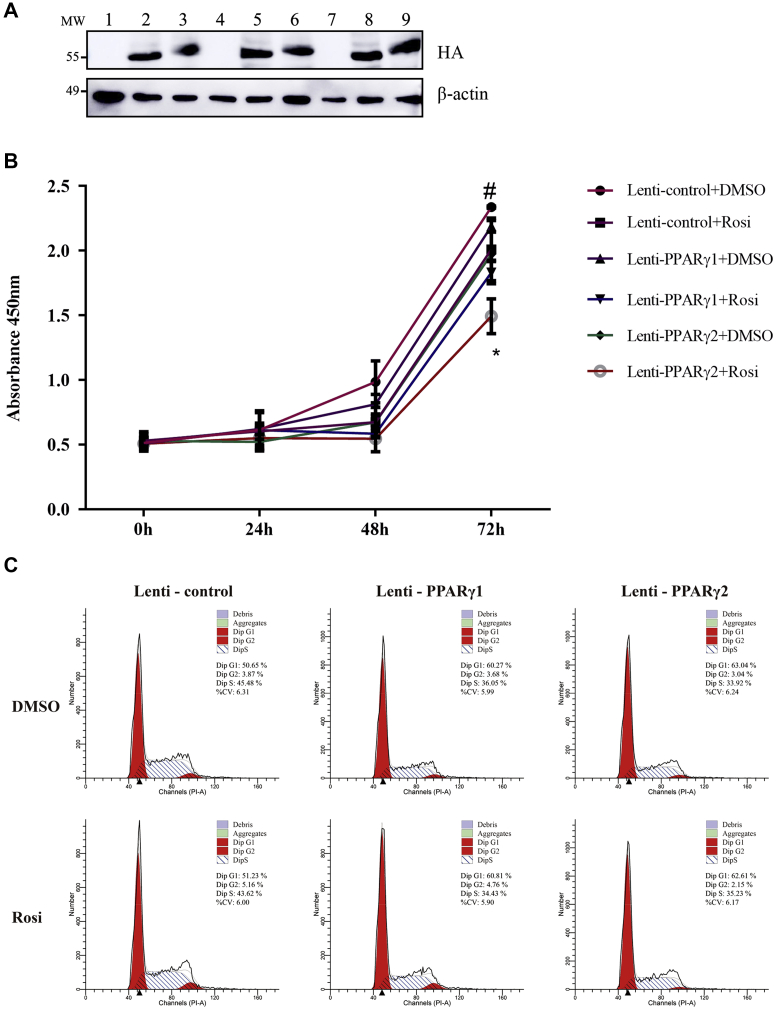
Preadipocyte proliferation and differentiation are 2 vital steps in adipogenesis. To investigate the effects of the two chicken PPARγ isoforms on preadipocyte proliferation, we generated and validated the lentiviruses expressing PPARγ1 and PPARγ2 (Lenti-PPARγ1 and Lenti-PPARγ2) using Western blot assay (Figure 1A). The Cell Counting Kit–8 cell proliferation assay results demonstrated that overexpression of each PPARγ isoform markedly suppressed the proliferation of the ICP2 cells in both the absence and presence of rosiglitazone (Figure 1B). Compared with PPARγ1 overexpression, PPARγ2 overexpression had a more powerful inhibitory effect on proliferation at 72 h (P < 0.05) (Figure 1B). Consistently, further Fluorescence activated Cell Sorting analysis revealed a significant increase in the number of cells accumulating in the G0/G1 phase and a significant decrease in the number of cells accumulating in the S phase in the ICP2 cells infected with either Lenti-PPARγ1 or Lenti-PPARγ2, compared with the cells infected with Lenti-control (P < 0.05) (Figure 1C). Taken together, these results suggest that both PPARγ1 and PPARγ2 inhibit the proliferation of the ICP2 cells, and PPARγ2 has a comparatively stronger antiproliferative effect than PPARγ1.
Figure 1.
Effects of PPARγ1 and PPARγ2 overexpression on the proliferation and cell cycle of ICP2 cells. (A) ICP2 cells were infected with 50 MOI of Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control. PPARγ1 and PPARγ2 protein expression was verified by western blot. Lanes 1, 4, and 7 are the cell lysates harvested from the ICP2 cells infected with Lenti-control at 24, 48, and 72 h after infection, respectively; lanes 2, 5, and 8 are the cell lysates harvested from the ICP2 cells infected with Lenti-PPARγ1 at 24, 48, and 72 h after infection, respectively; and lanes 3, 6, and 9 are the cell lysates harvested from the ICP2 cells infected with Lenti-PPARγ2 at 24, 48, and 72 h after infection, respectively. (B) ICP2 cells were infected with 50 MOI of Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control in the absence or presence of rosiglitazone, and cell proliferation was determined at 0, 24, 48, and 72 h after infection using the CCK-8. (C) ICP2 cells were infected with 50 MOI of Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control, and the cell cycle was evaluated at 48 h after infection using flow cytometry. Statistical significance was determined by two-way ANOVA. Data are reported as means ± SD. ∗P < 0.05, PPARγ2 vs. PPARγ1 in the absence of rosiglitazone. #P < 0.05, PPARγ2 vs. PPARγ1 in the presence of rosiglitazone. Abbreviations: CCK-8, Cell Counting Kit–8; ICP2, immortalized chicken preadipocytes; Lenti-control, lentivirus control; Lenti-PPARγ1, recombinant lentiviruses expressing PPARγ1; Lenti-PPARγ2, recombinant lentiviruses expressing PPARγ2; MOI, multiplicities of infection; PPARγ, peroxisome proliferator-activated receptor γ.
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Differentially Enhance Chicken Preadipocyte Apoptosis
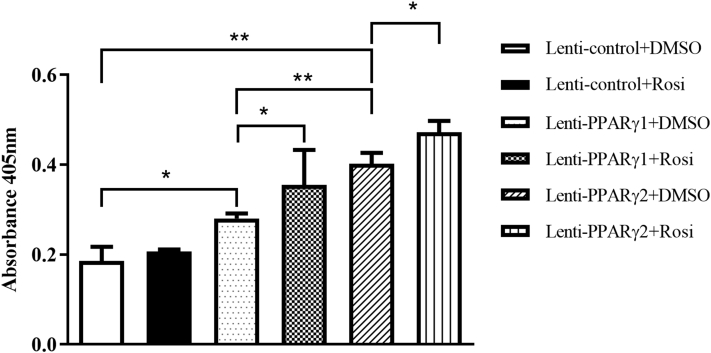
Peroxisome proliferator-activated receptor γ has been shown to play a role in triggering apoptotic pathways, and this activity is depending on cellular signaling (Della-Fera et al., 2001). We investigated the effects of PPARγ1 and PPARγ2 overexpression on the apoptosis of ICP2 cells using cell death detection ELISAplus kits. The results show that in the absence of rosiglitazone, PPARγ1 and PPARγ2 overexpression resulted in 1.51- and 2.17-fold increase in DNA fragmentation, respectively, compared with the Lenti-control (P < 0.05) (Figure 2). In the presence of rosiglitazone, PPARγ1 and PPARγ2 overexpression resulted in 1.71- and 2.28-fold increase in DNA fragmentation, respectively, compared with the Lenti-control (P < 0.05) (Figure 2). Furthermore, in the absence of rosiglitazone, PPARγ2 overexpression resulted in a 1.3-fold increase in DNA fragmentation, compared with PPARγ1 overexpression (P < 0.05), in the presence of rosiglitazone, PPARγ2 overexpression resulted in a 1.4-fold increase in DNA fragmentation, compared with PPARγ1 overexpression (P > 0.05) (Figure 2). These results suggest that PPARγ2 exerts a comparatively greater apoptotic effect on ICP2 cells than PPARγ1.
Figure 2.
Effects of PPARγ1 and PPARγ2 overexpression on apoptosis in ICP2 cells. ICP2 cells were infected with 50 MOI of Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control. In the absence or presence of rosiglitazone, apoptosis was analyzed at 48 h after infection using a cell death detection ELISA kit. All data are representative of 3 independent experiments and shown as the means ± SD. (∗P < 0.05; ∗∗P < 0.01; 2-way ANOVA). Abbreviations: ICP2, immortalized chicken preadipocytes; Lenti-control, lentivirus control; Lenti-PPARγ1, recombinant lentiviruses expressing PPARγ1; Lenti-PPARγ2, recombinant lentiviruses expressing PPARγ2; MOI, multiplicities of infection; PPARγ, peroxisome proliferator-activated receptor γ.
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Differentially Promote Preadipocyte Differentiation
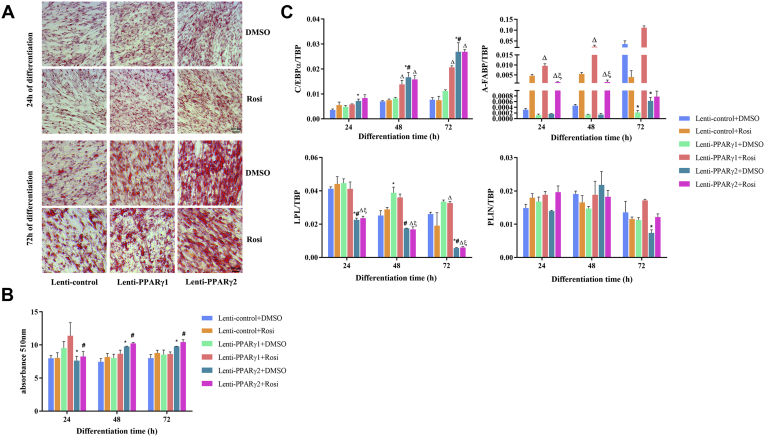
Given that PPARγ is the master regulator of adipocyte differentiation, we investigated the effect of these 2 PPARγ isoforms on the differentiation of ICP2 cells. As shown in Figure 3A, we observed extensive lipid accumulation in the cells infected with either Lenti-PPARγ1 or Lenti-PPARγ2 at 24 and 72 h of differentiation, compared with the Lenti-control–infected cells. In comparison, more lipid accumulation was observed in the cells infected with Lenti-PPARγ1 than those infected with Lenti-PPARγ2 at 24 h of differentiation in both the presence and absence of rosiglitazone (P < 0.05) (Figure 3B). Interestingly, at 48 and 72 h of differentiation, more lipid accumulation was observed in the cells infected with Lenti-PPARγ2 than in those infected with Lenti-PPARγ1 in both the presence and absence of rosiglitazone (P < 0.05) (Figure 3B). Real-time PCR analysis of the adipogenic marker gene expression showed increasing expression levels of A-FABP and C/EBPα from 24 to 72 h after induction of differentiation for Lenti-control, Lenti-PPARγ1, and Lenti-PPARγ2 infections. At 24 h of differentiation, A-FABP and PLIN1 were expressed at higher levels in the cells infected with Lenti-PPARγ1 than in those infected with Lenti-PPARγ2, in both the presence and absence of rosiglitazone, which was consistent with the Oil Red O staining results (P < 0.05) (Figure 3C), whereas, C/EBPα was expressed at a higher level in the cells infected with Lenti-PPARγ2 than in those infected with Lenti-PPARγ1 at 48 and 72 h of differentiation in the absence of rosiglitazone (P < 0.05) (Figure 3C). Collectively, these results suggest that PPARγ1 and PPARγ2 can individually promote chicken preadipocyte differentiation. Overall, PPARγ1 exerted a stronger proadipogenic effect at 24 h of differentiation, whereas PPARγ2 exerted a stronger proadipogenic effect at 48 and 72 h of differentiation.
Figure 3.
Effects of PPARγ1 and PPARγ2 overexpression on the differentiation of ICP2 cells. (A) Oil Red O staining of ICP2 cells infected with Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control were performed at 24 and 72 h of differentiation and (B) Oil Red O quantification at 0, 24, 48, and 72 h ∗P < 0.05, PPARγ2 vs. PPARγ1 in absence of rosiglitazone. #P < 0.05, PPARγ2 vs. PPARγ1 in the presence of rosiglitazone. Statistical significance was determined by 2-way ANOVA. (C) Real-time reverse transcription-polymerase Chain Reaction analysis of adipogenic genes during the differentiation of ICP2 cells infected with Lenti-PPARγ1, Lenti-PPARγ2, or Lenti-control were performed at 24 and 72 h of differentiation, respectively (means ± SD). Statistical significance was determined by two-way ANOVA. ∗P < 0.05 compared with Lenti-control, #P < 0.05, PPARγ2 vs. PPARγ1 in absence of rosiglitazone, respectively. ΔP < 0.05 compared with Lenti-control, ξP < 0.05, PPARγ2 vs. PPARγ1 in presence of rosiglitazone, respectively. Abbreviations: A-FABP, adipocyte fatty acid-binding protein; C/EBPα, CCAAT/enhancer-binding protein alpha; ICP2, immortalized chicken preadipocytes; Lenti-control, lentivirus control; Lenti-PPARγ1, recombinant lentiviruses expressing PPARγ1; Lenti-PPARγ2, recombinant lentiviruses expressing PPARγ2; LPL, lipoprotein lipase; PLIN1, perilipin protein; PPARγ, peroxisome proliferator-activated receptor γ.
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Have Differential Transcriptional Activities
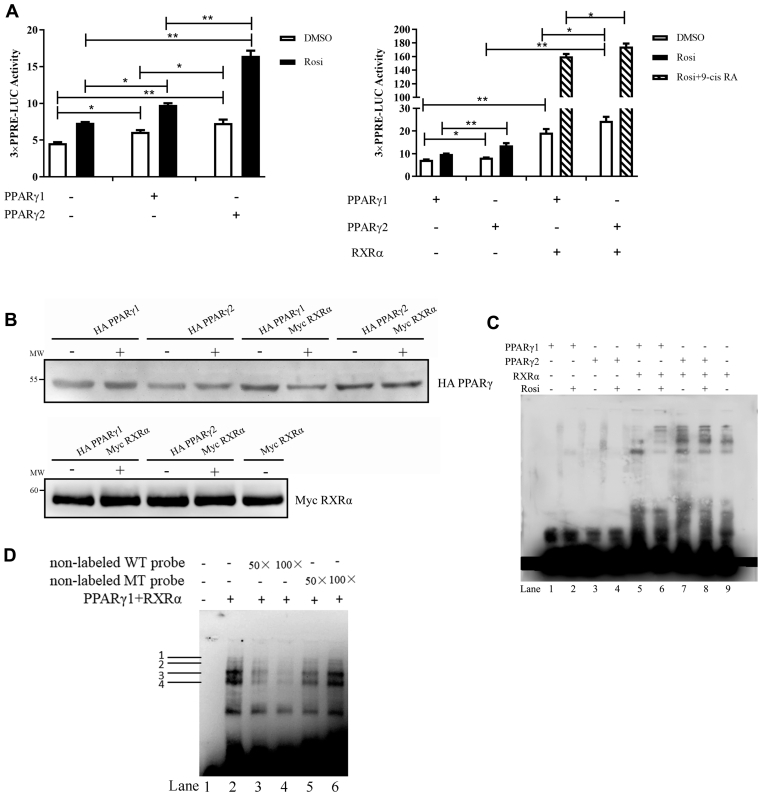
To gain insight into the molecular mechanisms underlying the differential effects of PPARγ1 and PPARγ2, we first evaluated their transcriptional activities in the absence and presence of rosiglitazone. The reporter gene assay showed that, compared with transfection of the pCMV-HA empty vector, transfection of pCMV-HA-PPARγ1 and pCMV-HA-PPARγ2 resulted in a 1.33- and 1.59-fold induction of luciferase activity of the PPRE reporter (3xPPRE-TK-Luc) (P < 0.05), respectively, in the absence of rosiglitazone. However, transfection of pCMV-HA-PPARγ1 and pCMV-HA-PPARγ2 resulted in a 2.13- and 3.58-fold induction of luciferase activity (P < 0.01), respectively, in the presence of rosiglitazone. In comparison, PPARγ2 induced higher luciferase activity than PPARγ1 in both the presence and absence of rosiglitazone (P < 0.05) (Figure 4A), suggesting that PPARγ2 has greater transcriptional activity than PPARγ1.
Figure 4.
Transcriptional activity and DNA binding of PPARγ1 and PPARγ2. (A) ICP2 cells were transfected or cotransfected with indicated expression vectors and 3xPPRE-TK-Luc along with the pRL-TK luciferase vector. After 24 h, transfected cells were treated with or without rosiglitazone (20 μmol/L) or 9-cis RA (5 μmol/L) for 24 h. Luciferase activity was determined at 48 h after transfection. The firefly luciferase activity values were normalized to a Renilla transfection control. Data are expressed as means ± SD. Statistical significance was determined by 2-way ANOVA. ∗P < 0.05, ∗∗P < 0.01. (B) Western blot analysis of PPARγ1, PPARγ2, and RXRα proteins in ICP2 cells transfected with indicated expression vectors in the presence (+) or absence (−) of 20 μmol/L rosiglitazone. (C) EMSA was performed using PPRE probes and the extracted nuclear proteins, PPARγ1 (lanes 1–2) and PPARγ2 (lanes 3–4), PPARγ1 and RXRα (lanes 5–6), PPARγ2 and RXRα (lanes 7–8), RXRα (lane 9). (D) EMSA and competition-EMSA experiment showing the binding of PPARγ1 and RXRα to the PPRE. EMSA using probe alone (lane 1) and incubated with the PPARγ1 and RXRα protein (lane 2). Competition with increasing amounts of unlabeled wild type PPRE probe with PPARγ1 and RXRα (lanes 3–4) and unlabeled mutant type PPRE probe (lanes 5–6). Abbreviations: EMSA, electrophoretic mobility shift assay; ICP2, immortalized chicken preadipocytes; PPARγ, peroxisome proliferator-activated receptor γ; PPRE, PPAR response element.
It is known that PPARγ forms a heterodimer with RXRα to regulate its target genes. To gain further insight into the difference between PPARγ1 and PPARγ2 transcriptional activities, ICP2 cells were cotransfected with either pCMV-HA-PPARγ1 or pCMV-HA-PPARγ2 and pCMV-Myc-RXRα along with the PPRE reporter construct 3xPPRE-TK-Luc, and luciferase activity was determined. The results show that in the absence of the rosiglitazone, cotransfection with PPARγ1 and RXRα resulted in a 2.63-fold increase in luciferase activity compared with transfection with PPARγ1 alone (P < 0.01) (Figure 4A). Furthermore, in the absence of rosiglitazone, cotransfection with PPARγ2 and RXRα resulted in a 2.96-fold increase in luciferase activity compared with transfection with PPARγ2 alone (P < 0.01) (Figure 4A). In the presence of both agonists (rosiglitazone + 9-cis RA), cotransfection with PPARγ1 and RXRα lead to a 21.92-fold increase in luciferase activity compared with transfection with PPARγ1 alone (P < 0.01) (Figure 4A). In the presence of both agonists, cotransfection with PPARγ2 and RXRα resulted in a 21.16-fold increase in luciferase activity compared with transfection with PPARγ2 alone (P < 0.01) (Figure 4A). As shown in Figure 4A, in both the presence and absence of rosiglitazone and 9-cis RA, the PPARγ2/RXRα complex tended to be more potent in inducing luciferase reporter activity than the PPARγ1/RXRα complex (P = 0.12). Altogether, these results suggest that PPARγ2 is a more potent transcriptional activator than PPARγ1 in both the presence and absence of rosiglitazone.
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Have Different DNA Binding Affinities
The isoforms PPARγ1 and PPARγ2 share the same DNA-binding domain but differ at the N-terminal. It has been reported that the variable N-terminal domain of nuclear receptors influences DNA-binding specificity and affinity (Grad et al., 2001; Brodie and McEwan, 2005). We performed EMSA to test whether the 6 additional amino acids at the N-terminal affect DNA binding. In the absence and presence of rosiglitazone, the nuclear-extracted PPARγ1, PPARγ2, and RXRα proteins were confirmed by Western blot analysis (Figure 4B). Equal amounts of these extracted nuclear proteins were used in EMSA. The EMSA assay showed that the binding of PPARγ1 and PPARγ2 to the biotin-labeled PPRE probes did not occur in the absence of RXRα (Figure 4C, lanes 1–4). However, the binding of PPARγ1 and PPARγ2 to the biotin-labeled PPRE probes (4 major bands) was observed in the presence of RXRα (Figure 4C, lanes 6–8). To confirm the specificity of these PPARγ1/2/RXRα-DNA complexes, we performed competition experiments using unlabeled PPRE probes with either a wild-type or mutant PPRE probe as a competitor. The results show that as the concentrations of the unlabeled wild-type PPRE probe increased, the 4 major bands decreased in intensity (Figure 4D, lanes 3–4). As the concentration of the mutant PPRE probe increased, the 4 major bands did not decrease in intensity (Figure 4D, lanes 5–6). Therefore, it can be concluded that the 4 major bands represent the specific PPARγ1/2/RXRα-PPRE complexes. In comparison, the PPARγ2/RXRα heterodimer was capable of forming stronger DNA–protein complexes with the biotin-labeled PPRE probes than the PPARγ1/RXRα heterodimer (Figure 4C), suggesting the PPARγ2/RXRα heterodimer has a stronger binding affinity to PPRE than PPARγ1/RXRα.
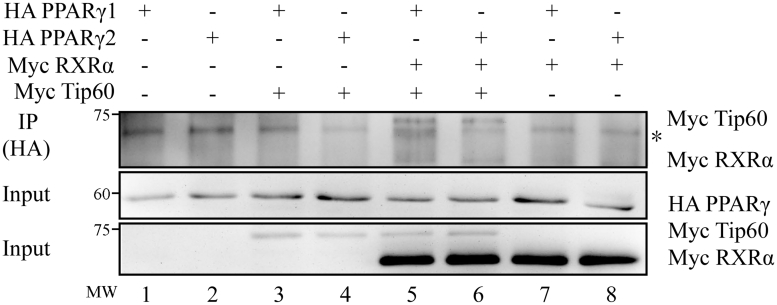
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Interact Similarly With Tip60
Peroxisome proliferator-activated receptor γ functions by interacting with various transcriptional coregulators, coactivators, and corepressors.Tat-interacting protein 60, a member of the MYST family of acetyltransferases, is a coactivator of PPARγ (van Beekum et al., 2007). A previous study showed that Tip60 can interact with the activation function-1 domain of PPARγ (van Beekum et al., 2007). To understand the mechanisms underlying the functional differences between PPARγ1 and PPARγ2, we also investigated the differences between the 2 PPARγ isoforms in their interaction with Tip60. Coimmunoprecipitation experiments were performed with an anti-HA tag antibody, and the precipitated proteins were subjected to Western blot analysis with the anti-Myc antibody. As shown in Figure 5, as expected, PPARγ1 and PPARγ2 interacted similarly with RXRα (lanes 5–8, Figure 5). In the absence of RXRα (lanes 3–4, Figure 5), neither PPARγ1 nor PPARγ2 interacted with Tip60. But in the presence of RXRα (lanes 5–6, Figure 5), PPARγ1 and PPARγ2 interacted similarly with Tip60. These data indicate that RXRα is required for the interaction of PPARγ and Tip60, and PPARγ1 and PPARγ2 have comparable binding affinities for RXRα and Tip60.
Figure 5.
Coimmunoprecipitation (co-IP) analysis of interaction between PPARγ isoforms and Tip60. Co-immunoprecipitation analysis of the PPARγ isoforms and Tip60. ICP2 cells were transfected with the indicated expression vectors (pCMV-HA-PPARγ1, pCMV-HA-PPARγ2, pCMV-Myc-Tip60, and pCMV-Myc-RXRα). Cells were harvested and lysed in western and IP buffer, and Tip60 and RXRα were immunoprecipitated (IP) using anti-HA. The precipitated proteins were subjected to Western blot analysis to detect Myc-RXRα and Myc-Tip60 using Anti-Myc. An aspecific band is indicated (∗). Abbreviations: ICP2, immortalized chicken preadipocytes; PPARγ, peroxisome proliferator-activated receptor γ; Tip60, Tat-interacting protein 60.
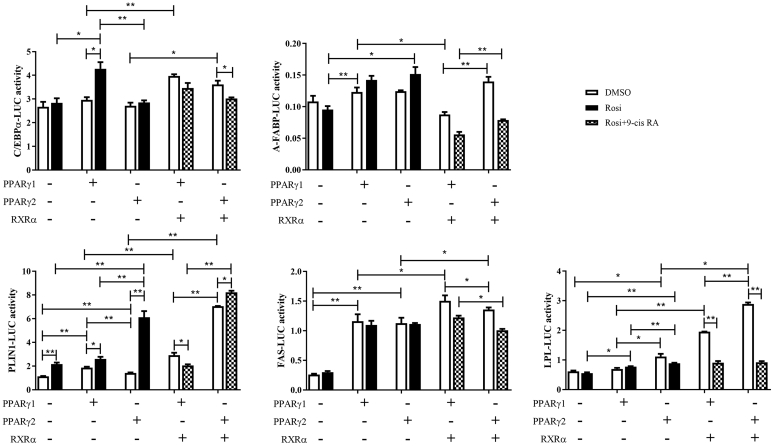
Peroxisome Proliferator-Activated Receptor γ1 and PPARγ2 Exert Overlapping and Distinct Regulatory Roles
The adipogenic genes, LPL, PLIN1, A-FABP, C/EBPα, and FAS, are known target genes of PPARγ. We further compared the regulation of these adipogenic genes by PPARγ1 and PPARγ2. The reporter gene assays showed that both PPARγ1 and PPARγ2 markedly enhanced the promoter activity of PLIN1 and FAS in the absence of rosiglitazone and enhanced the promoter activity of LPL, PLIN1, and A-FABP in the presence of rosiglitazone (Figure 6, P < 0.05). In comparison, PPARγ2 was more potent in activating LPL-promoter activity than PPARγ1 in the presence of rosiglitazone (Figure 6, P < 0.05), and PPARγ2, but not PPARγ1, could activate LPL-promoter activity in the absence of rosiglitazone (Figure 6, P < 0.05). In contrast, PPARγ1, but not PPARγ2, could activate C/EBPα-promoter activity in the presence of rosiglitazone. Cotransfection of any one of the PPARγ isoform genes and RXRα resulted in a synergistic increase in the promoter activity of LPL, PLIN1, C/EBPα, and FAS in the absence of the agonists (rosiglitazone + 9-cis RA) (Figure 6). Comparatively, cotransfection with PPARγ2 and RXRα was more potent in activating the promoter activity of LPL, PLIN1, and A-FABP than cotransfection with PPARγ1 and RXRα (Figure 6, P < 0.05). Interestingly, the cotransfection of either of the 2 PPARγ isoform genes and RXRα markedly repressed the promoter activity of LPL, A-FABP, and FAS in the presence of both agonists (rosiglitazone + 9-cis RA) (P < 0.05). These results demonstrate that PPARγ1 and PPARγ2 exert overlapping and distinct regulatory roles in the regulation of the adipogenic genes tested, which might explain the differential effects of overexpressing PPARγ1 and PPARγ2 during adipocyte differentiation (Figure 3).
Figure 6.
Effects of PPARγ1 and PPARγ2 overexpression on the promoter activity of adipogenic marker genes. ICP2 cells were transfected or co-transfected with indicated reporter constructs and expression vectors (PPARγ1, PPARγ2, and RXRα) along with the pRL-TK luciferase vector. At 24 h after transfection, the cells were treated with or without rosiglitazone (20 μmol/L) and 9-cisRA (5 μmol/L) for 24 h. Luciferase activity was determined 48 h after transfection. The firefly luciferase activity values were normalized to a Renilla transfection control. Data are reported as means ± SD. (∗P < 0.05, ∗∗P < 0.01; two-way ANOVA). Abbreviations: A-FABP, adipocyte fatty acid-binding protein; C/EBPα, CCAAT/enhancer-binding protein alpha; FAS, fatty acid synthase; ICP2, immortalized chicken preadipocytes; LPL, lipoprotein lipase; PLIN1, perilipin protein; PPARγ, peroxisome proliferator-activated receptor γ.
Discussion
The function and regulation of PPARγ in adipose tissue have been well documented in the mouse. However, the functional differences between the PPARγ isoforms are controversial. Several reports have shown that PPARγ1 and PPARγ2 can individually stimulate adipocyte differentiation in the mouse (Mueller et al., 2002; Zhang et al., 2004; Li et al., 2016), whereas other studies have reported that PPARγ2, but not PPARγ1, induces adipogenesis in the mouse (Delin et al., 2002). In vivo studies indicated that PPARγ1 and PPARγ2 can drive adipose tissue development, but PPARγ2 plays the dominant role in adipogenesis in mouse (Zhang et al., 2004). In the present study, we demonstrated that PPARγ1 and PPARγ2 differentially regulate chicken preadipocyte proliferation, apoptosis, and differentiation.
In agreement with the previous study (Zhang et al., 2004), we found that both PPARγ1 and PPARγ2 promoted adipocyte differentiation, as demonstrated by Oil red O staining and mRNA expression analysis of adipogenic marker genes. Of the 2 isoforms, PPARγ2 exerts a stronger adipogenic effect than PPARγ1 (Figure 3). In addition, we unexpectedly found that PPARγ2 overexpression decreased the expression of A-FABP and LPL during adipocyte differentiation (P < 0.01; Figure 3C). The marked discrepancies between the results of Oil red O staining and quantitative real-time reverse transcription-polymerase chain reaction analysis may be owing to the negative feedback loop between PPARγ and A-FABP during adipocyte differentiation (Garin-Shkolnik et al., 2014). Our results provide compelling evidence that the 2 PPARγ isoforms have different abilities to promote adipocyte differentiation.
In addition, consistent with the previous proliferation study in mouse NIH-3T3 cells (Altiok et al., 1997), we demonstrated that both PPARγ1 and PPARγ2 exert antiproliferative effects (Figures 1B, 1C). Comparatively, our results show that PPARγ2 had a stronger antiproliferative effect on ICP2 cells than PPARγ1 (Figure 1A). Our results differ from those of Altiok et al. who showed that both PPARγ1 and PPARγ2 activation by pioglitazone induced a similar growth arrest (Altiok et al., 1997). This discrepancy could be explained by several reasons. First, different cell lines were used to assay cell proliferation in our study compared with the previous study; we used the immortal chicken cell line (ICP2), but Altiok et al. used the mouse cell line, NIH-3T3. Second, different PPARγ ligands were used in our study, we used the exogenous PPARγ ligand rosiglitazone, but Altiok et al. used pioglitazone. Third, different cell proliferation assays were used between the 2 studies, we used a Cell Counting Kit–8 cell proliferation assay, whereas Altiok et al. used a BrdU cell proliferation assay.
Activation of PPARγ has been shown to stimulate apoptosis in a variety of cell types (Harris and Phipps, 2001; Elrod and Sun, 2008; Xiao et al., 2010); however, the differential effects of PPARγ isoforms on apoptosis has not been explored. To this end, we tested the effect of PPARγ1 and PPARγ2 overexpression on the apoptosis of ICP2 cells. The ELISA results show that PPARγ1 and PPARγ2 overexpression induced mild apoptosis in ICP2 cells, and PPARγ2 overexpression comparatively exerted a higher apoptotic effect than PPARγ1 (Figure 2). However, Western blot analysis showed no obvious difference in the protein levels of cleaved caspase-3, a marker of apoptosis, between PPARγ1 and PPARγ2 overexpression (data not shown). This contradictory result may be because of their mild apoptotic effect or the lower sensitivity of the Western blot.
Chicken PPARγ1 and PPARγ2 differ only in the N-terminal domain (A/B domain) containing the activation function-1–transactivation domain. Our results show that PPARγ2 displayed a stronger transcriptional activity than PPARγ1 (Figure 4A), which is consistent with the published data in mice (Vidal-Puig et al., 1996). The difference in transcription activity between PPARγ1 and PPARγ2 is obviously due to differences in the A/B domain, which is involved in transcriptional activation (Brunmeir and Xu, 2018). The A/B domain has been shown to provide protein phosphorylation sites and physically interacts with other receptor domains or regulatory proteins (Chandra et al., 2008; Frkic et al., 2018). For example, MAPK phosphorylates the A/B-domain of PPARγ and inhibits its transactivation (Armoni et al., 2015), and a coactivator, thyroid hormone receptor–associated protein 220, contributed to the difference in transcription activity between the 2 PPARγ isoforms (Mueller et al., 2002; Bugge et al., 2009). In the present study, PPARγ1 and PPARγ2 interacted similarly with Tip60 (Figure 5), but we cannot exclude the possibility that they interact differentially with other coregulators, such as cAMP response element-binding protein–binding protein, p300 (Koppen and Kalkhoven, 2010), DRIP205/thyroid hormone receptor–associated protein 220 (Mueller et al., 2002), PPARγ coactivator 2 (Koppen and Kalkhoven, 2010), and tribbles homolog 3 (Takahashi et al., 2008).
The isoforms PPARγ1 and PPARγ2 share the DNA-binding domain and ligand-binding domain but differ in the A/B domain. The A/B domain is known to physically and functionally cooperate with the ligand-binding domain (Wärnmark et al., 2003; Khorasanizadeh and Rastinejad, 2016), and ligand-binding domain cooperates with the DNA-binding domain to enhance DNA binding (Chandra et al., 2008). The results of the present study show that PPARγ2 had a higher binding affinity for PPRE than PPARγ1, which is presumably because of differences in the intramolecular interactions between these functional domains (Figure 4C). In addition, the differential regulation of adipogenic genes by PPARγ1 and PPARγ2 (Figure 6) may be caused by differences in their transcription activation and DNA binding, alone or in combination.
Interestingly, our results show that PPARγ1 had a stronger proadipogenic effect at 24 h of differentiation, but PPARγ2 had a stronger proadipogenic effect at 48 and 72 h of differentiation (Figure 3B). One possible explanation for this phenomenon is that these two PPARγ isoforms are involved at different stages of the differentiation and differentially regulate target adipogenic genes. In agreement with our prediction, PPARγ1 is expressed during the early stages of the differentiation of mouse 3T3-L1 preadipocytes, whereas PPARγ2 is mainly expressed during the later stages of the differentiation (Lee and Ge, 2014).
In the present study, our results show that in the presence of both agonists (rosiglitazone + 9-cis RA), the PPARγ isoforms/RXRα heterodimer decreased LPL-promoter luciferase activity more than in the absence of both agonists (Figure 6), which is inconsistent with a previous study (Schoonjans et al., 1996). This discrepancy may be explained by two possible reasons. First, 9-cis RA treatment could induce inactivation of the RXRα-PPARγ heterodimers by suppressing the level of RXRα (Sagara et al., 2013). Second, binding of 9-cis RA to RXRα may alter the distinct conformation of the receptor heterodimer (Vivat-Hannah et al., 2003).
We cannot rule out the possibility that the endogenous PPARγ may interfere with our results. To gain a better understanding of these 2 chicken PPARγ isoforms, it is worth generating PPARγ gene knockout ICP2 cells using CRISPR/Cas9 technology and investigating the functional and molecular differences between the 2 PPARγ isoforms in the PPARγ knockout cells.
In conclusion, we demonstrated that PPARγ1 and PPARγ2 differentially regulated preadipocyte proliferation, apoptosis, and differentiation as a result of their distinct and overlapping molecular functions.
Acknowledgments
This work was supported by the China National Natural Science Foundation Grant (No.31572392) and the China Agriculture Research System (No. CARS-41).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.09.086.
Contributor Information
Hui Li, Email: lihui@neau.edu.cn.
Ning Wang, Email: wangning@neau.edu.cn.
Disclosures
The authors declare no conflicts of interest.
Supplementary data
References
- Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M., Evans R.M. PPARγ signaling and metabolism: the good, the bad and the future. Nat. Med. 2013;19:557. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shali K., Cao H., Knoers N., Hermus A.R., Tack C.J., Hegele R.A. A single-base mutation in the peroxisome proliferator-activated receptor γ4 promoter associated with altered in vitro expression and partial lipodystrophy. J. Clin. Endocrinol. Metab. 2004;89:5655–5660. doi: 10.1210/jc.2004-0280. [DOI] [PubMed] [Google Scholar]
- Altiok S., Xu M., Spiegelman B.M. PPARγ induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armoni M., Harel C., Karnieli E. PPARγ gene expression is autoregulated in primary adipocytes: ligand, sumoylation, and isoform specificity. Horm. Metab. Res. 2015;47:89–96. doi: 10.1055/s-0034-1394463. [DOI] [PubMed] [Google Scholar]
- Bionaz M., Chen S., Khan M.J., Loor J.J. Functional role of PPARs in ruminants: potential targets for fine-tuning metabolism during growth and lactation. PPAR Res. 2013;2013:684159. doi: 10.1155/2013/684159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie J., McEwan I.J. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. J. Mol. Endocrinol. 2005;34:603–615. doi: 10.1677/jme.1.01723. [DOI] [PubMed] [Google Scholar]
- Brunmeir R., Xu F. Functional regulation of PPARs through post-translational modifications. Int. J. Mol. Sci. 2018;19:1738. doi: 10.3390/ijms19061738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A., Grontved L., Aagaard M.M., Borup R., Mandrup S. The PPARγ2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol. Endocrinol. 2009;23:794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthorn W.P., Scheller E.L., MacDougald O.A. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J. Lipid Res. 2012;53:227–246. doi: 10.1194/jlr.R021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T.P., Rastinejad F. Structure of the intact PPAR-γ–RXR-α nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sá P.M., Richard A.J., Hang H., Stephens J.M. Transcriptional regulation of adipogenesis. Compr. Physiol. 2011;7:635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- Delin R., Collingwood T.N., Rebar E.J., Wolffe A.P., Camp H.S. PPARgamma knockdown by engineered transcription factors: exogenous PPARgamma2 but not PPARgamma1 reactivates adipogenesis. Genes Dev. 2002;16:27. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Fera M.A., Qian H., Baile C.A. Adipocyte apoptosis in the regulation of body fat mass by leptin. Diabetes Obes. Metab. 2001;3:299–310. doi: 10.1046/j.1463-1326.2001.00112.x. [DOI] [PubMed] [Google Scholar]
- Elrod H.A., Sun S.-Y. PPARγ and apoptosis in cancer. PPAR Res. 2008;2008:704165. doi: 10.1155/2008/704165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frkic R.L., Marshall A.C., Blayo A.-L., Pukala T.L., Kamenecka T.M., Griffin P.R., Bruning J.B. PPARγ in complex with an antagonist and inverse agonist: a tumble and trap mechanism of the activation helix. iScience. 2018;5:69–79. doi: 10.1016/j.isci.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin-Shkolnik T., Rudich A., Hotamisligil G.S., Rubinstein M. FABP4 attenuates PPARγ and adipogenesis and is inversely correlated with PPARγ in adipose tissues. Diabetes. 2014;63:900–911. doi: 10.2337/db13-0436. [DOI] [PubMed] [Google Scholar]
- Grad J.M., Lyons L.S., Robins D.M., Burnstein K.L. The androgen receptor (AR) amino-terminus imposes androgen-specific regulation of AR gene expression via an exonic enhancer. Endocrinology. 2001;142:1107–1116. doi: 10.1210/endo.142.3.8049. [DOI] [PubMed] [Google Scholar]
- Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications-a review. Nutr. J. 2014;13:17. doi: 10.1186/1475-2891-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.G., Phipps R.P. The nuclear receptor PPAR gamma is expressed by mouse T lymphocytes and PPAR gamma agonists induce apoptosis. Eur. J. Immunol. 2001;31:1098–1105. doi: 10.1002/1521-4141(200104)31:4<1098::aid-immu1098>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S., Rastinejad F. Visualizing the architectures and interactions of nuclear receptors. Endocrinology. 2016;157:4212–4221. doi: 10.1210/en.2016-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen A., Kalkhoven E. Brown vs white adipocytes: the PPARγ coregulator story. FEBS Lett. 2010;584:3250–3259. doi: 10.1016/j.febslet.2010.06.035. [DOI] [PubMed] [Google Scholar]
- Kui D., Yingning S., Xiaofei Z., Tianmu Z., Wenjian Z., Jiyang Z., Guihua W., Shouzhi W., Li L., Hui L. Identification and characterization of transcript variants of chicken peroxisome proliferator-activated receptor gamma. Poult. Sci. 2015;94:2516–2527. doi: 10.3382/ps/pev229. [DOI] [PubMed] [Google Scholar]
- Lee J.-E., Ge K. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 2014;4:29. doi: 10.1186/2045-3701-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-E., Schmidt H., Lai B., Ge K. Transcriptional and epigenomic regulation of adipogenesis. Mol. Cell Biol. 2019;39:e00601–e00618. doi: 10.1128/MCB.00601-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang F., Zhang X., Xue C., Namwanje M., Fan L., Reilly M.P., Hu F., Qiang L. Distinct functions of PPARγ isoforms in regulating adipocyte plasticity. Biochem. Biophys. Res. Commun. 2016;481:132–138. doi: 10.1016/j.bbrc.2016.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller E., Drori S., Aiyer A., Yie J., Sarraf P., Chen H., Hauser S., Rosen E.D., Ge K., Roeder R.G. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms. J. Biol. Chem. 2002;277:41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- Sagara C., Takahashi K., Kagechika H., Takahashi N. Molecular mechanism of 9-cis-retinoic acid inhibition of adipogenesis in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2013;433:102–107. doi: 10.1016/j.bbrc.2013.02.057. [DOI] [PubMed] [Google Scholar]
- Schoonjans K., Peinado-Onsurbe J., Lefebvre A.-M., Heyman R.A., Briggs M., Deeb S., Staels B., Auwerx J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- Shi H., Luo J., Yao D., Zhu J., Xu H., Shi H., Loor J.J. Peroxisome proliferator-activated receptor-γ stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. J. Dairy Sci. 2013;96:7844–7853. doi: 10.3168/jds.2013-7105. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Ohoka N., Hayashi H., Sato R. TRB3 suppresses adipocyte differentiation by negatively regulating PPARγ transcriptional activity. J. Lipid Res. 2008;49:880–892. doi: 10.1194/jlr.M700545-JLR200. [DOI] [PubMed] [Google Scholar]
- van Beekum O., Brenkman A.B., Grøntved L., Hamers N., van den Broek N.J., Berger R., Mandrup S., Kalkhoven E. The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisome proliferator-activated receptor γ. Endocrinology. 2007;149:1840–1849. doi: 10.1210/en.2007-0977. [DOI] [PubMed] [Google Scholar]
- Vidal-Puig A., Jimenez-Liñan M., Lowell B.B., Hamann A., Hu E., Spiegelman B., Flier J.S., Moller D.E. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Invest. 1996;97:2553–2561. doi: 10.1172/JCI118703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivat-Hannah V., Bourguet W., Gottardis M., Gronemeyer H. Separation of retinoid X receptor homo-and heterodimerization functions. Mol. Cell Biol. 2003;23:7678–7688. doi: 10.1128/MCB.23.21.7678-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang T., Wu C., Wang S., Wang Y., Li H., Wang N. Immortalization of chicken preadipocytes by retroviral transduction of chicken TERT and TR. PLoS One. 2017;12:e0177348. doi: 10.1371/journal.pone.0177348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wärnmark A., Treuter E., Wright A.P., Gustafsson J.-A. k. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol. Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- Werman A., Hollenberg A., Solanes G., Bjørbæk C., Vidal-Puig A.J., Flier J.S. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor γ (PPARγ) differential activity of PPARγ1 and-2 isoforms and influence of insulin. J. Biol. Chem. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Yuan T., Yao W., Liao K. 3T3-L1 adipocyte apoptosis induced by thiazolidinediones is peroxisome proliferator-activated receptor-γ-dependent and mediated by the caspase-3-dependent apoptotic pathway. FEBS J. 2010;277:687–696. doi: 10.1111/j.1742-4658.2009.07514.x. [DOI] [PubMed] [Google Scholar]
- Zhang X., Cheng B., Liu C., Du Z., Zhang H., Wang N., Wu M., Li Y., Cao Z., Li H. A novel regulator of preadipocyte differentiation, transcription factor TCF21, functions partially through promoting LPL expression. Front. Physiol. 2019;10:458. doi: 10.3389/fphys.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E. Selective disruption of PPARγ2 impairs the development of adipose tissue and insulin sensitivity. Proc. Natl. Acad. Sci. U S A. 2004;101:10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-W., Wu C.-Y., Li H., Wang N. Expression and functional analyses of Krüppel-like factor 3 in chicken adipose tissue. Biosci. Biotechnol. Biochem. 2014;78:614–623. doi: 10.1080/09168451.2014.896735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.