Abstract
The liver is the central organ for metabolism and influence the growth and development of the animals. To date, little is known about the microRNA (miRNA) in pigeon livers, particularly in different developmental stages. A comprehensive investigation into miRNA transcriptomes in livers across 3 pigeon developmental stages (1, 14, 28 d old) and an adult stage (2 y old) was performed by small RNA sequencing. We identified 312 known miRNA, 433 conserved miRNA, and 192 novel miRNA in pigeon livers. A set of differentially expressed (DE) miRNA in livers were screened out during pigeon development. This set of miRNA might be involved in hepatospecific phenotype and liver development. A Short Time-series Expression Miner analysis indicated significant expression variations in DE miRNA during liver development of pigeons. These DE miRNA with different expression patterns might play essential roles in response to growth factor, cell morphogenesis, and gland development, etc. Protein–protein interaction network and Molecular Complex Detection analysis identified several vital target genes (e.g., TNRC6B, FRS2, PTCH1, etc.) of DE miRNA, which is closely linked in liver development and enriched in PI3K cascade and regulation of growth. Our results expanded the repertoire of pigeon miRNA and may be of help in better understanding the mechanism of squab's rapid development from the perspective of liver development.
Key words: pigeon, liver, miRNA, small RNA-Seq, development
Introduction
Pigeons (Columba livia) are widely distributed over the whole world and domesticated for thousands of years (Tolba, 2015), which have been domesticated for production of meat, racing, and experimental animals in physiology (Prinzinger and Misovic, 2010), behavior (Ware et al., 2017), and atmospheric environmental monitoring (Cui et al., 2016). As a typical representative of altrices, pigeon squabs have an extremely high growth rate. At the 28th d after hatch, the body weight of pigeons is about 25 times that of newly hatched squabs (Gao et al., 2016). As the main biosynthetic and metabolic organ, the liver has pivotal functions in the synthesis of several vital proteins, the metabolism of various biologically useful materials, the detoxification of toxic substances, and immune defense (Xu et al., 2019). Aside from direct involvement in anabolism, the liver supplies approximately 75% total circulating insulin-like growth factor-I (IGF-I), which is a crucial regulator of skeletal muscle development and could enhance the ability for the muscle to grow (Schwander et al., 1983; Barton, 2006). Animals that cannot express IGF-I exhibit severe growth retardation (Baker et al., 1993). More interestingly, liver indexes of pigeon squabs in early-stage (1- to 7-day-old) have greater values than that of 28- to 35-day-old squabs (Gao et al., 2016), suggesting the liver may have a crucial function in early development stages. Nevertheless, little is known about the mechanism of this phenomenon during squab development.
The liver has been studied mostly biochemically and histopathologically, which consists of different types of cells including hepatocytes, liver sinusoidal endothelial cells, hepatic stellate cells, Kupffer cells, etc. (Chen and Verfaillie, 2014). Genetic studies using gene-targeting technology have identified a number of cytokines, intracellular signaling molecules, and transcription factors involved in liver development and regeneration (Tanimizu and Miyajima, 2007). Specifically, the regulatory network consists of signaling pathways involving fibroblast growth factors (FGF), bone morphogenetic proteins, Wnt, hepatocyte nuclear factors, and CCAAT/enhancer-binding protein (Darlington, 1999; Liu et al., 2017). In addition, as post-transcriptional regulators of gene expression, microRNA (miRNA) are abundant in the liver and exert crucial roles in normal liver development and the fine-tuning of fundamental biological liver processes (Chen and Verfaillie, 2014). For example, miR-122 directly or indirectly stimulates the expression levels of 24 hepatocyte-specific genes (Chen and Verfaillie, 2014) and promotes the hepatic differentiation and maturation (Deng et al., 2014). MiR-27a and -27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation (Ji et al., 2009). MiR-103 and -107 participate in the regulation of insulin sensitivity (Trajkovski et al., 2011). MiR-33 reduces fatty acid degradation by targeting multiple genes involved in fatty acid β-oxidation (Gerin et al., 2010). Furthermore, knockdown of miR-30a would result in defective biliary morphogenesis (Hand et al., 2009). To date, miRNA in liver tissues of some domestic species (e.g., pig (Liu et al., 2017), bovine (Becker et al., 2011), chicken (Hicks et al., 2010)) have been surveyed using high-throughput sequencing. However, miRNA in pigeon liver have not been identified. Here, we aim to explore the miRNA expression profiles in livers and their potential roles during pigeon development. Our findings will be instrumental in understanding the roles of miRNA in pigeon liver development and growth.
Materials and methods
Animal Ethics Statement
All animals used in this study were farmed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004) and approved by the Institutional Animal Care and Use Committee in the College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China, under permit no. DKY-S20174201.
Preparation of Experimental Animals and Tissues
White King pigeons and squabs were purchased from the Feng Mao pigeon breeding farm (Mianyang, China). Parent White King pigeons (8 males and 8 females at 2 y old) and 48 squabs (mixed sex) were grouped into 4 stages (16 replicates for each stage, the age of 1 d, 14 d, 28 d, and 2 y old). After weighing the body weight, pigeons were anesthetized with ether and subsequently euthanized. Next, the whole liver of each pigeon was separated and weighed. The liver tissues from the same anatomical location were collected from pigeons representing each of the 4 stages. Subsequently, the samples for small RNA sequencing were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
RNA Extraction and High-throughput Sequencing
Total RNA was respectively extracted from frozen liver samples collected from male pigeons with TRIzol reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's protocol. The quantity of total RNA was assessed using an Agilent 2100 Bioanalyzer with Agilent RNA 6000 nano Reagents Part 1 kit (Agilent Technologies). Each development stage has 3 replicates that came from the 3 pigeons. The total RNA of each replicate was individually used for library construction. For each library, small RNA ranging from 10 to 45 nt in length was purified by polyacrylamide gel electrophoresis and ligated using proprietary adapters. The modified small RNA was then reverse-transcribed to cDNA and amplified by PCR. Finally, libraries were sequenced by an Illumina HiSeq 2500 platform and generated 50 bp single-end reads.
Identification and Differential Expression Analysis of miRNA
The bioinformatics pipeline for miRNA discovery and profiling was carried out as previously described, with some improvements (Ma et al., 2018). To identify the pigeon miRNA, the initial sequence was subjected to a series of stringent filters (such as removing low-quality reads, repeated sequences, and adapter sequences), and the output was called clean data. Filtered sequences were then mapped to the pigeon reference genome (C. livia, ColLiv2, GenBank assembly accession: GCA_001887795.1) (https://www.ncbi.nlm.nih.gov/assembly/GCA _001887795.1) with stringent criteria (0 mismatches for full length) using Bowtie software. Next, mappable reads were extended in the reference genome as predicted miRNA precursors. Only candidate precursors that perfectly matched to known pigeon (C. livia) mature miRNA annotated by miRBase (Release 22.0) (Kozomara et al., 2019) were identified as known pigeon miRNA. Subsequently, to identify the conserved miRNA, we performed alignments between remaining candidate precursors and seed sequences of mature miRNA from chicken (Gallus gallus), the zebra finch (Taeniopygia guttata) and other mammals, allowing no mismatch. Novel miRNA were further predicted using the miRDeep2 core algorithm (Friedlander et al., 2008). edgeR was used for differential expression analysis among different stages in the OmicShare tools (www.omicshare.com/tools). A unique miRNA was considered to be differentially expressed (DE) when |log2(fold change) |>1 and false discovery rate < 0.05.
qRT-PCR
MicroRNA was reverse-transcribed respectively using Mir-X miRNA First-Strand Synthesis Kit (Takara, Otsu, Japan) following the manufacturer's recommendations. The qRT-PCR assays were performed on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using SYBR Premix Ex Taq II (Takara, Dalian, China) in accordance with the manufacturer's protocols. The qRT-PCR was carried out on 3 biological replicates. Relative miRNA levels were normalized against U6 snRNA and calculated using the 2−ΔΔCt method. Information on the primer sequences used is available in Supplementary Table 1.
STEM Analysis
A time-series analysis of the miRNA expression data was performed using Short Time-series Expression Miner software (STEM) http://www.cs.cmu.edu/∼jernst/stem/(Ernst and Bar-Joseph, 2006). By using STEM analysis, each miRNA was assigned to the model profile most closely matched to its time series based on the correlation coefficient. Short Time-series Expression Miner software was run using the log normalize data option, with all other settings set to the defaults.
Prediction and Functional Annotation of miRNA Target Genes
The TargetScan algorithm (http://www.targetscan.org) was applied to predict the target genes of DE miRNA (Agarwal et al., 2015). Enrichment analyses for the target genes of DE miRNA in different expression profiles were performed using Metascape (http://metascape.org/gp/index.html) with the default parameters set (Zhou et al., 2019).
Protein–Protein Interaction Network Analysis of miRNA Targets
Targets of miRNA were used to construct protein–protein interaction (PPI) networks by using the Metascape tool with default parameters. Subsequently, using the Molecular Complex Detection (MCODE) algorithm, densely connected network components were first identified. Enrichment analysis was applied to each MCODE component independently, and the 3 best-scoring (by P-value) terms were retained as the functional description of the resulting modules.
Construction of the miRNA–mRNA Regulatory Network
MicroRNA interacting with mRNA of seed genes identified from the PPI network were predicted using TargetScan (http://www.targetscan.org). Omicshares tool (https://www.omicshare.com/tools/) was used for the construction of the miRNA–mRNA regulatory network.
Statistical Analysis
Phenotypic data were analyzed and compared for statistically significant differences using ANOVA in R. Results were considered statistically significant for P-values < 0.05.
Results
Phenotypic Measurements
In this study, we investigated body weight, liver weight, and liver index changes during pigeon development. As shown in Figure 1A, body weight progressively increased from the time squabs were 1-day-old until the pigeons reached the age of 2-y-old. However, liver weight and liver indexes exhibited first-increase and then-decrease tendency with the development of the squabs. Particularly, both liver weight and liver index in 14-day-old and 28-day-old squabs were conspicuously higher than those of 2-y-old pigeons.
Figure 1.
Body weight, liver weight, and liver index changes during development of pigeons. (A) Body weight, (B) liver weight, (C) liver index. (n = 16).
Summary of Deep-sequencing Data
To identify miRNA and investigate miRNA expression changes during the development of pigeon squabs, 12 total RNA samples were used to construct small RNA libraries. Libraries were sequenced using Illumina HiSeq 2500 platform and 50 bp single-end reads were generated. All sequence data have been submitted to the NCBI Gene Expression Omnibus with the accession number GSE149501. In total, we obtained 176.64 million raw reads (Supplementary Table 2). The adapter sequences, contamination, and low-quality reads were then trimmed, and the remaining reads were considered high-quality clean reads. The proportion of high-quality clean reads ranged from 76.18 to 93.00% in the 12 libraries. The length distribution analysis (Supplementary Figure 1) showed that most (57.89∼ 80.22%) of the small RNAs in the 12 libraries were approximately 21–24 nt in length, which is the typical length of Dicer-processed products.
MicroRNA Transcriptome Profiles During Liver Development
A total of 937 mature miRNA derived from 641 pre-miRNA were identified in pigeon livers across 3 development stages (1 d, 14 d, and 28 d) and an adult stage (2 y) (Table 1). These miRNA candidates were classified into 3 types: pigeon known miRNA, conserved miRNA, and putative novel miRNA. There are 312 pigeon known miRNA corresponded to 172 pigeon known pre-miRNA (Supplementary Table 3). Of the pigeon conserved miRNA, 433 corresponded to 332 other 2 avian species (G. gallus and T. guttata) and mammalian species pre-miRNA (Supplementary Table 4), whereas 192 putative novel miRNA corresponded to 137 candidate pre-miRNA (Supplementary Table 5).
Table 1.
Pigeon miRNA identified in 12 sRNA libraries.
| Group (number of pre-miRNA/miRNA) | 1 d | 14 d | 28 d | 2 y | Total |
|---|---|---|---|---|---|
| Known miRNA | 162/293 | 165/298 | 153/274 | 153/275 | 172/312 |
| Conserved miRNA | 168/217 | 221/285 | 127/155 | 109/143 | 332/433 |
| Putative novel miRNA | 72/101 | 89/128 | 61/86 | 48/66 | 137/192 |
To uncover the possible roles of miRNA in pigeon livers during development, we ranked the miRNA by expression level (Figure 2). The miRNA expression profile was different across the 4 different age groups. Of note, the expression abundance of the top 10 unique miRNA accounts for 69.68–84.58% of the total counts in each stage. In addition, the set of the top 10 unique miRNA over the 4 stages came to a total of 15 unique miRNA. Of these miRNA, 7 miRNA (cli-miR-22-3p, cli-miR-26-5p, cli-miR-30a-5p, cli-miR-30 d-5p, cli-miR-30e-5p, cli-miR-143-3p, and cli-miR-148a-3p) overlapped in all 4 stages.
Figure 2.
Top 10 unique miRNA with the highest expression levels in pigeon livers over the 4 stages from 1-day-old to 2-y-old. A plot of the unique miRNA vs. their total copy number % of all unique miRNA for each library. The 7 miRNA that are present in the top 10 miRNA in all libraries are connected by lines.
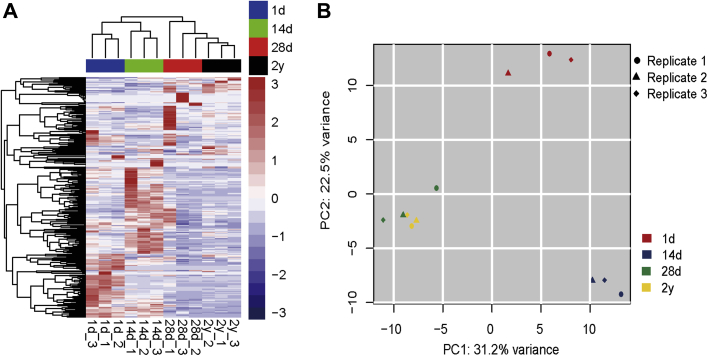
Subsequently, we conducted a hierarchical clustering analysis and principal component analysis based on the miRNA expression profiles. As shown in Figure 3A, a clustering of miRNA expression profiles in pigeon livers mainly based on developmental stage and time. The younger squabs (1 d and 14 d) were tightly clustered into a subgroup, and separated from 28-day-old and 2-year-old pigeons. It suggested that the development stage contributes to the different miRNA transcriptome of pigeon livers. This set of dissimilarities was confirmed by principal component analysis (Figure 3B).
Figure 3.
MicroRNA transcriptome profiles in pigeon livers across the 4 different age groups. (A) Hierarchical clustering analysis for the normalized expression of miRNA among 12 miRNA libraries. (B) Principal component analysis (PCA) of miRNA across all 12 libraries.
Differentially Expressed miRNA During Liver Development Stages
To screen the DE miRNA among pigeon livers during development, differential expression analysis by taking |log2(fold change) |>1 and false discovery rate <0.05 as criteria was performed after removing miRNA of less than 10 count reads. We totally identified 177 miRNA that were DE during the liver development (Figure 4A, Supplementary Table 6), accounting for 18.89% of total identified miRNA in pigeon livers. Specifically, 6 contrasts were generated (1 d vs. 14 d, 1 d vs. 28 d, 1 d vs. 2 y, 14 d vs. 28 d, 14 d vs. 2 y, 28 d vs. 2 y), and screened out 48, 40, 92, 54, 117, and 2 DE miRNA (Figure 4B), respectively.
Figure 4.
Differentially expressed miRNA in pigeon livers during development. (A) An upset plot of the intersections between 4 different age groups. (B) Volcano plot of the DE miRNA between 2 different age groups. The x-axis indicates the difference in expression level on a log2 (fold change). The y-axis represents the corresponding false discovery rate on a negative log 10(FDR).
To confirm the small RNA-Seq results, we selected 6 miRNA (miR-29a-3p, miR-143-3p, miR-215-5p, miR-29 b-3p, miR-363-3p, miR-1388-5p) to conduct qPCR assay using 3 independent samples. The results were in line with our sequencing result (Pearson r = 0.915 ± 0.065, Figure 5), which highlighted the reliability of our sequencing data.
Figure 5.
Validation of the sequencing data using qPCR. Six randomly selected DE miRNA in pigeon livers were validated by real-time qPCR (n = 3). The Pearson product–moment correlation coefficient (r) was calculated using R.
Distinct miRNA Expression Patterns During Liver Development
Differentially expressed miRNA expression patterns in pigeon livers during development were assessed using the STEM algorithm. The STEM clustering tool assigned each miRNA to the model profile that most closely matched its temporal expression profile. Three significant model profiles (profiles 0, 7, and 13) from the 20 distinct expression patterns were generated (Figure 6 and Supplementary Table 7). Profiles 0, 7, and 13 comprised 20, 46, and 12 miRNA, respectively. The expression level of miRNA in expression pattern 0 decreased across all 4 stages, whereas other miRNA in expression patterns 7 and 13 exhibited a decreasing tendency with fluctuation.
Figure 6.
The miRNA expression analysis by STEM clustering. MicroRNA are organized into different clusters based on expression patterns using STEM software. Line plots (left panels) and box plots (right panels) are used to show fold changes (log2 scale) and expression levels (log2RPM), respectively. Representative miRNA of each cluster are listed at the right. In each line plot, one representative miRNA is highlighted in red. Abbreviations: FC, fold change; RPM, reads per million reads; STEM, Short Time-series Expression Miner.
Enrichment Analysis of Target Genes of miRNA in Different Profiles
Given miRNA function presents a dose-dependent manner (Carlsbecker et al., 2010), only the relatively more abundant miRNA (>1,000 read counts) were carried out the prediction of potential target genes. As depicted in Figure 7, the target genes of miRNA in profile 0 were enriched in developmental growth (GO: 0048589), cellular response to growth factor stimulus (GO: 0071363), heart development (GO: 0007507), etc. The target genes of miRNA in profile 7 are mainly involved in response to growth factor (GO: 0070848), cellular response to growth factor stimulus (GO: 0071363), cell morphogenesis involved in differentiation (GO: 0000904), etc., whereas the target genes of miRNA in profile 13 were enriched in cell–cell adhesion via plasma-membrane adhesion molecules (GO: 0098742), regulation of neuron differentiation (GO: 0045664), synapse organization (GO: 0050808), etc. (Supplementary Table 8). From these results, we found that some enriched GO categories were overlapped by 3 different profiles. Therefore, we analyzed intersection target genes of miRNA in different profiles. The miRNA in aforementioned 3 different profiles (profiles 0, 7, and 13) shared 197 intersection target genes, which mainly enriched in gland development (GO: 0048732), cell part morphogenesis (GO: 0032990), developmental growth (GO: 0048589), etc. (Figures 8A–8C, Supplementary Table 9). In addition, we also performed a meta-enrichment analysis based on 3 target gene lists, which suggested that target genes from 3 profiles mainly enriched in developmental growth (GO: 0048589), tissue morphogenesis (GO: 0048729), response to growth factor (GO: 0070848), Wnt signaling pathway (GO: 0070848), etc. (Figures 9A–9C).
Figure 7.
Function enrichment analysis of target genes of miRNA (read counts >1,000) in different profiles (Profile 0, 7 and 13). The x-axis indicates –log (P-value).The y-axis indicates functional categories.
Figure 8.
Function enrichment analysis of shared target genes in the 3 profiles (Profile 0, 7, and 13). (A) Venn diagram revealed that 197 common target genes existed in the 3 profiles. (B) The network of enriched terms is colored by cluster-ID, and nodes that share the same cluster are typically close to each other. (C) Bar plot of enriched terms across common target genes in the 3 profiles, colored by P values.
Figure 9.
Meta-enrichment analysis summary for target genes of miRNA (read counts >1,000) in the 3 profiles (Profile 0, 7, and 13). (A) Overlap among target gene lists, where purple curves link identical genes, and blue curves link genes belong to the same enriched ontology term. The inner circle represents gene lists, where hits are arranged along the arc. Genes hit multiple lists are colored in dark orange, and genes unique to a list are shown in light orange. (B) The network of enriched terms is colored by cluster ID, and nodes that share the same cluster are typically close to each other. (C) Heatmap of top 20 enriched terms across 3 target gene lists corresponding 3 profiles, colored by P-values.
Protein–Protein Interaction Network and MCODE Enrichment Analysis
Protein–protein interaction network was generated using Metascape and visualized with Cytoscape 3.4.0 (Figure 10A). In total, 98 nodes and 157 PPI relationships were obtained. The MCODE method was applied to identify closely related proteins from the PPI network. The MCODE algorithm subclustered PPI network into 4 subclusters containing 13 genes from which 3 genes, TNRC6B, FRS2, and PTCH1, were defined as seed genes (Figure 10B, Supplementary Table 10). The Gene Ontology (GO) enrichment analysis was applied to each MCODE network to assign “meanings” to the network component. The enriched terms of module 1 included nuclear-transcribed mRNA poly(A) tail shortening, positive regulation of mRNA catabolic process, nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay, etc. The enriched categories for module 2 included PI3K cascade, insulin receptor substrate (IRS)-mediated signaling, IRS-related events triggered by IGF1R, etc. Module 3 included regulation of growth (Supplementary Table 11).
Figure 10.
Protein–protein interaction (PPI) networks and miRNA–mRNA regulatory networks. (A) A PPI network. (B) PPI MCODE component. Red, blue, green, and violet colors indicate modules 1, 2, 3, and 4, respectively. (C) miRNA–mRNA regulatory networks. White nodes denote seed genes, and orange nodes denote miRNA; Gray edges denote miRNA–mRNA interaction relationship.
MicroRNA-Target Regulatory Network
To screen out the miRNA having binding sites to seed genes (TNRC6B, FRS2, and PTCH1) from the 3 different profiles, we performed target gene prediction by using TargetScan and constructed the mRNA–miRNA network. As shown in Figure 10C, 10 miRNA (miR-107-3p, miR-15c-5p, miR-92-3p, miR-103-3p, miR-18a-5p, miR-148a-5p, miR-16a-5p, miR-20 b-5p, miR-9-5p, miR-10 b-5p) target TNRC6B. Two miRNA (miR-18a-5p, miR-20 b-5p) target FRS2, and 3 miRNA (miR-16a-5p, let-7d-3p, miR-15c-5p) target PTCH1. Of these, cli-miR-20 b-5p can antagonize both TNRC6B and FRS2, and cli-miR-16a-5p can antagonize both TNRC6B and PTCH1, whereas cli-miR-15c-5p targets TNRC6B and PTCH1.
Discussion
The liver is a key metabolic organ as well as a predominant source of IGF-I (Schwander et al., 1983), exerting vital functions in the normal growth and development of animals. In the present study, the liver indexes of squabs are higher than those of adult pigeons. It implies that the liver has pivotal functions during the early development of pigeons. Accordingly, the present study is focused on the identification of miRNA in pigeon livers during development and the exploration of their potential function. As a class of gene expression regulatory factors, miRNA expressed in the liver tissues (Hicks et al., 2010; Becker et al., 2011; Liu et al., 2017), while it has not been reported in pigeons. In the present study, we used deep sequencing to identify the miRNA and their expression levels in pigeon livers. We totally identified 937 mature miRNA in pigeon livers across 3 development stages and an adult stage, and DE miRNA were determined. We found some of these miRNA that had been previously reported to be associated with liver development and regeneration, for example, miR-21, miR-148, miR-26, let-7 family, miR-22, miR-19 b, miR-106, miR-23, miR-29, miR-30, miR-145, etc. (Liu et al., 2010; Finch et al., 2014). Among these miRNA, miR-148a-3p exhibited the highest abundance in livers on day 1 and day 28, whereas miR-22-3p and miR-26-5p are the richest miRNA at 14-day-old and 2-y-old, respectively. This is basically consistent with the reported studies that miR-148a is the most abundant miRNA in chicken hepatocytes (Wang et al., 2014) and porcine livers of different breeds (Li et al., 2012). Gailhouste et al. found miR-148a promoted the hepatospecific phenotype (Gailhouste et al., 2013). By contrast, miR-26a and miR-22 regulated mouse hepatocyte proliferation during liver regeneration (Zhou et al., 2012) and participating in hepatic steatosis (Hu et al., 2020), respectively. These results indicated that miR-148a, miR-22-3p, and miR-26-5p might affect pigeon liver development through promoting the hepatospecific phenotype and regulating the hepatocyte proliferation and metabolism.
In this study, we screened out 177 DE miRNA during pigeon liver development. Of these, 78 DE miRNA were significantly clustered in 3 expression profiles. GO and Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that the target genes of DE miRNA (read counts >1,000) in different profiles were involved in response to growth factor, cellular response to growth factor stimulus, etc. Previous studies found that at least 3 growth factors, EGF, TGF alpha, and HGF, are complete mitogens for hepatocytes in culture and stimulators of liver growth (Fausto and Webber, 1993). Furthermore, the liver is an important source of IGF-1 mediating many of the growth-promoting actions of GH (Norbeck et al., 2007). In our study, we found cli-miR-454-3p was predicted to target pigeon IGF-I mRNA, whereas several DE miRNA (e.g., cli-miR-16a-5p, cli-miR-133a-3p, cli-miR-9-5p, cli-let-7d-3p, cli-miR-15c-5p, cli-let-7f-5p, etc.) were predicted to target pigeon IGF-I receptor mRNA. More intriguingly, some vital target genes identified by PPI and MCODE analysis are mainly enriched in IRS-related events triggered by IGF1R, PI3K cascade, and regulation of growth. These findings imply that DE miRNA in squab livers are highly implicated in liver development and pigeon growth by targeting growth factor–related genes and signaling pathways. Besides, the liver is the biggest gland in an organism, while the overlapped target genes of the DE miRNA (read counts >1,000) in profiles were enriched in gland development, which is the most significantly enriched term for these target genes. Accordingly, combining the enrichment results of target genes of DE miRNA in different profiles, it can be concluded that these miRNA may participate in pigeon development at the cellular, organ, and organismal levels.
By means of gene–gene network reconstruction analysis, several key target genes of DE miRNA in different profiles were identified during the pigeon development, including trinucleotide repeat–containing 6 B (TNRC6B), fibroblast growth factor receptor substrate 2 (FRS2), patched 1 (Ptch1), etc. These target genes are responsible for the regulation of liver development in pigeons via multiple signaling pathways. TNRC6B, the predicted target gene of at least 10 DE miRNA in this study, is localized to mRNA-degrading cytoplasmic P bodies and is functionally required to mediate miRNA-guided mRNA cleavage (Meister et al., 2005). Alterations in TNRC6B gene expression affected cell proliferation and cell adhesion (Murakami et al., 2013). Considering that cell proliferation and cell adhesion were the critical processes during development (Ekblom et al., 1986), it was worthwhile further to explore the potential roles of TNRC6B in pigeon development. Fibroblast growth factor signal pathway controls liver specification and regulates the metabolism of lipids, cholesterol, and bile acids. Fibroblast growth factor signaling also promotes hepatocyte proliferation (Tsai et al., 2013). For example, FGF8 and FGF10 function as paracrine signals in embryonic liver development, and FGF21 regulates glucose and lipid metabolism in white adipose tissue (Itoh et al., 2016). FRS2 is the main mediator of signaling in the FGF pathway. Meanwhile, FRS2 also functions as a molecular sensor integrating external regulatory signals into the FGF pathway (Zhou et al., 2009). Some downregulated DE miRNA (e.g., cli-miR-20 b-5p and cli-miR-18a-5p) have binding sites in FRS2 mRNA, suggesting that these DE miRNA might be implicated in pigeon liver development via FGF signaling pathway. Hedgehog (Hh) signaling is highly linked with the development and homeostasis of various organs. Yoshikazu Hirose et al. demonstrated that Hh signal pathway modulates the hepatoblast proliferation, and further suggest that this pathway needs to be shut off for hepatic differentiation of hepatoblasts to proceed normally (Hirose et al., 2009). Ptch1, as another predicted key target gene of DE miRNA (e.g., cli-miR-16a-5p and cli-miR-15c-5p), is a negative regulatory factor of the Hh signaling pathway (Yu et al., 2015). Therefore, some miRNA have a chance to regulate hepatoblast proliferation and differentiation by targeting Ptch1 to act on Hh signaling pathway during pigeon liver development.
In summary, we totally identified 312 known miRNA, 433 conserved miRNA, and 192 novel miRNA in pigeon livers across 3 developmental stages and an adult stage using small RNA sequencing. These miRNA might play pivotal roles in response to growth factor, cell morphogenesis, gland development, and regulate liver development and pigeon growth by targeting growth factor–related genes and signaling pathways. Our results expanded the repertoire of pigeon miRNA and may be of help in better understanding the mechanism of squab's rapid development from the perspective of liver development.
Disclosures
The authors declare that there are no conflicts of interest relevant to this work.
Acknowledgments
This work was supported by grants from the National Key R & D Program of China (2018YFD0500403), the National Natural Science Foundation of China (31872335).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.09.039.
Contributor Information
Xun Wang, Email: xunwang@sicau.edu.cn.
Mingzhou Li, Email: Mingzhou.li@sicau.edu.cn.
Supplementary data
References
- Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J., Liu J.P., Robertson E.J., Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- Barton E.R. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl. Physiol. Nutr. Metab. 2006;31:791–797. doi: 10.1139/h06-054. [DOI] [PubMed] [Google Scholar]
- Becker C., Riedmaier I., Reiter M., Tichopad A., Pfaffl M.W., Meyer H.H. Changes in the miRNA profile under the influence of anabolic steroids in bovine liver. Analyst. 2011;136:1204–1209. doi: 10.1039/c0an00703j. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J.Y., Roberts C.J., Dettmer J., Lehesranta S., Zhou J., Lindgren O., Moreno-Risueno M.A., Vaten A., Thitamadee S., Campilho A., Sebastian J., Bowman J.L., Helariutta Y., Benfey P.N. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Verfaillie C.M. MicroRNAs: the fine modulators of liver development and function. Liver Int. 2014;34:976–990. doi: 10.1111/liv.12496. [DOI] [PubMed] [Google Scholar]
- Cui J., Halbrook R.S., Zang S.Y., You J. Use of homing pigeons as biomonitors of atmospheric metal concentrations in Beijing and Guangzhou, China. Ecotoxicology. 2016;25:439–446. doi: 10.1007/s10646-015-1601-y. [DOI] [PubMed] [Google Scholar]
- Darlington G.J. Molecular mechanisms of liver development and differentiation. Curr. Opin. Cell Biol. 1999;11:678–682. doi: 10.1016/s0955-0674(99)00035-6. [DOI] [PubMed] [Google Scholar]
- Deng X.G., Qiu R.L., Wu Y.H., Li Z.X., Xie P., Zhang J., Zhou J.J., Zeng L.X., Tang J., Maharjan A., Deng J.M. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver Int. 2014;34:281–295. doi: 10.1111/liv.12239. [DOI] [PubMed] [Google Scholar]
- Ekblom P., Vestweber D., Kemler R. Cell-matrix interactions and cell adhesion during development. Annu. Rev. Cell Biol. 1986;2:27–47. doi: 10.1146/annurev.cb.02.110186.000331. [DOI] [PubMed] [Google Scholar]
- Ernst J., Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N., Webber E.M. Control of liver growth. Crit. Rev. Eukaryot. Gene Expr. 1993;3:117–135. [PubMed] [Google Scholar]
- Finch M.L., Marquardt J.U., Yeoh G.C., Callus B.A. Regulation of microRNAs and their role in liver development, regeneration and disease. Int. J. Biochem. Cell Biol. 2014;54:288–303. doi: 10.1016/j.biocel.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Friedlander M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Gailhouste L., Gomez-Santos L., Hagiwara K., Hatada I., Kitagawa N., Kawaharada K., Thirion M., Kosaka N., Takahashi R.U., Shibata T., Miyajima A., Ochiya T. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- Gao C.Q., Yang J.X., Chen M.X., Yan H.C., Wang X.Q. Growth curves and age-related changes in carcass characteristics, organs, serum parameters, and intestinal transporter gene expression in domestic pigeon (Columba livia) Poult. Sci. 2016;95:867–877. doi: 10.3382/ps/pev443. [DOI] [PubMed] [Google Scholar]
- Gerin I., Clerbaux L.A., Haumont O., Lanthier N., Das A.K., Burant C.F., Leclercq I.A., MacDougald O.A., Bommer G.T. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand N.J., Master Z.R., Eauclaire S.F., Weinblatt D.E., Matthews R.P., Friedman J.R. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.A., Trakooljul N., Liu H.C. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol. Genomics. 2010;41:185–193. doi: 10.1152/physiolgenomics.00156.2009. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Itoh T., Miyajima A. Hedgehog signal activation coordinates proliferation and differentiation of fetal liver progenitor cells. Exp. Cell Res. 2009;315:2648–2657. doi: 10.1016/j.yexcr.2009.06.018. [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu H.X., Jena P.K., Sheng L., Ali M.R., Wan Y.Y. miR-22 inhibition reduces hepatic steatosis via FGF21 and FGFR1 induction. JHEP Rep. 2020;2:100093. doi: 10.1016/j.jhepr.2020.100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N., Nakayama Y., Konishi M. Roles of FGFs as paracrine or Endocrine signals in liver development, Health, and disease. Front. Cell Dev. Biol. 2016;4:30. doi: 10.3389/fcell.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Zhang J., Huang G., Qian J., Wang X., Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sun Q., Jia Y., Cong R., Ni Y., Yang X., Jiang Z., Zhao R. Coordinated miRNA/mRNA expression profiles for understanding breed-specific metabolic characters of liver between Erhualian and large white pigs. PLoS One. 2012;7:e38716. doi: 10.1371/journal.pone.0038716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Fan J., Zeng W., Zhou Y., Ingvarsson S., Chen H. Quantitative analysis of miRNA expression in several developmental stages of human livers. Hepatol. Res. 2010;40:813–822. doi: 10.1111/j.1872-034X.2010.00683.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Jin L., Lou P., Gu Y., Li M., Li X. Dynamic microRNAome profiles in the developing porcine liver. Biosci. Biotechnol. Biochem. 2017;81:127–134. doi: 10.1080/09168451.2016.1240602. [DOI] [PubMed] [Google Scholar]
- Ma Y., Feng S., Wang X., Qazi I.H., Long K., Luo Y., Li G., Ning C., Wang Y., Hu S., Xiao J., Li X., Lan D., Hu Y., Tang Q., Ma J., Jin L., Jiang A., Li M. Exploration of exosomal microRNA expression profiles in pigeon 'Milk' during the lactation period. BMC Genomics. 2018;19:828. doi: 10.1186/s12864-018-5201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Tamori A., Itami S., Tanahashi T., Toyoda H., Tanaka M., Wu W., Brojigin N., Kaneoka Y., Maeda A., Kumada T., Kawada N., Kubo S., Kuroda M. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer. 2013;13:99. doi: 10.1186/1471-2407-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbeck L.A., Kittilson J.D., Sheridan M.A. Resolving the growth-promoting and metabolic effects of growth hormone: differential regulation of GH-IGF-I system components. Gen. Comp. Endocrinol. 2007;151:332–341. doi: 10.1016/j.ygcen.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Prinzinger R., Misovic A. Age-correlation of blood values in the Rock pigeon (Columba livia) Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2010;156:351–356. doi: 10.1016/j.cbpa.2009.07.028. [DOI] [PubMed] [Google Scholar]
- Schwander J.C., Hauri C., Zapf J., Froesch E.R. Synthesis and secretion of insulin-like growth factor and its binding protein by the perfused rat liver: dependence on growth hormone status. Endocrinology. 1983;113:297–305. doi: 10.1210/endo-113-1-297. [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Miyajima A. Molecular mechanism of liver development and regeneration. Int. Rev. Cytol. 2007;259:1–48. doi: 10.1016/S0074-7696(06)59001-1. [DOI] [PubMed] [Google Scholar]
- Tolba A.R. Gross anatomical study on the hepatic portal vein tributaries in the common domestic pigeon "Columba Livia Domestica". Inter. J. Vet. Sci. 2015;4:63–68. [Google Scholar]
- Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- Tsai S.M., Liu D.W., Wang W.P. Fibroblast growth factor (Fgf) signaling pathway regulates liver homeostasis in zebrafish. Transgenic Res. 2013;22:301–314. doi: 10.1007/s11248-012-9636-9. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang L., Wang H., Shao F., Yu J., Jiang H., Han Y., Gong D., Gu Z. Growth hormone-regulated mRNAs and miRNAs in chicken hepatocytes. PLoS One. 2014;9:e112896. doi: 10.1371/journal.pone.0112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware E.L.R., Saunders D.R., Troje N.F. Social interactivity in pigeon courtship behavior. Curr. Zool. 2017;63:85–95. doi: 10.1093/cz/zow066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E., Zhang L., Yang H., Shen L., Feng Y., Ren M., Xiao Y. Transcriptome profiling of the liver among the prenatal and postnatal stages in chickens. Poult. Sci. 2019;98:7030–7040. doi: 10.3382/ps/pez434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Lu Z., Chen B., Wu X., Dong P., Zheng J. Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis by attenuating DNMT1-mediated Patched1 methylation. J. Cell. Mol. Med. 2015;19:2617–2632. doi: 10.1111/jcmm.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ju W., Wang D., Wu L., Zhu X., Guo Z., He X. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One. 2012;7:e33577. doi: 10.1371/journal.pone.0033577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Feng X., Wu Y., Benge J., Zhang Z., Chen Z. FGF-receptor substrate 2 functions as a molecular sensor integrating external regulatory signals into the FGF pathway. Cell Res. 2009;19:1165–1177. doi: 10.1038/cr.2009.95. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.