Abstract
MicroRNAs (miRNAs) are emerging as critical mediators in tumors, including triple-negative breast cancer (TNBC). The role of miR-518a-3p in TNBC was investigated to identify potential therapeutic target. Data from KM Plotter database (www.kmplot.com) showed that high miR-518a-3p expression was significantly associated with overall survival of patients with TNBC (p = 0.04). The expression of miR-518a-3p was dysregulated in TNBC cells. Functional assays revealed that over-expression of miR-518a-3p inhibited cell invasion and migration of TNBC. Additionally, miR-518a-3p could target TMEM2 (transmembrane protein 2), and decreased protein and mRNA expression of TMEM2 in TNBC cells. Knockdown of TMEM2 suppressed cell invasion and migration through inhibiting phospho (p)-JAK1 (Janus kinase 1) and p-STAT (signal transducer and activator of transcription protein) 1/2. Moreover, over-expression of TMEM2 counteracted the suppressive effect of miR-518a-3p on TNBC invasion and migration through promoting the levels of p-JAK1 and p-STAT1/2. In conclusion, miR-518a-3p negatively regulates the JAK/STAT pathway via targeting TMEM2 and suppresses invasion and migration in TNBC, suggesting that miR-518a-3p may be a potential therapeutic target in TNBC.
Keywords: miR-518a-3p, TMEM2, TNBC, migration, invasion, JAK/STAT
Introduction
Breast cancer is a common female malignant tumor, which is a serious threat to women health worldwide.1 As an independent subtype of breast cancer, triple negative breast cancer (TNBC) accounts for 20% of breast cancer cases and receives increasing attention due to its most aggressive ability among all subtypes of breast cancers.2 TNBC is characterized by triple negative expression of progesterone receptor (PR), estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2).3 Therefore, common therapeutic strategies, including hormonal or trastuzumab-based therapy, could not work for patients with TNBC.4 In addition, TNBC is inclined to metastasis to lungs or brains, thus associating with short overall survival and poor prognosis.5 Current treatment for TNBC shows poor outcome due to high chemoresistance.6 Thus, new potential therapeutic strategies are urgently needed for the treatment of TNBC.
MiRNAs are widely found in eukaryotes and are important key factors that confer negative regulation of target genes.7 MiRNAs are implicated in almost all life processes, including proliferation, development, differentiation and apoptosis.7 Recently, dysregulated miRNAs in TNBC were found to be prognostic biomarkers or therapeutic targets for TNBC.8 For example, miR-138, as a prognostic biomarker for TNBC, could promote tumorigenesis of TNBC.9 MiR-518a-3p was reported to be an anti-tumor effector for colorectal cancer10 and participates in malignant behavior of colon cancer.11 However, the role of miR-518a-3p in TNBC remains elusive.
TMEM2 (transmembrane protein 2), a single-pass transmembrane domain protein, functions as hyaluronidase to degrade hyaluronic acid.12 Hyaluronic acid metabolism has been reported to be involved in neuronal proliferation and migration.13 Recently, TMEM2 has been shown to participate in endoplasmic reticulum homeostasis14 and angiogenesis,15 suggesting that TMEM2 may also play a role in tumor progression. The metastasis-promotive role of TMEM2 in breast cancer has been reported recently,16 and TMEM2 preferential binds with miRNA let-7.17 The present study was conducted to investigate whether miR-518a-3p could suppress TNBC progression through targeting TMEM2.
Material and Methods
Cell Culture
TNBC cells (DA-MB-231, BT-549, MDA-MB-453 and MDA-MB-468) were purchased from ATCC (Manassas, VA, USA), and cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum at 37°C with 5% CO 2.
Cell Transfection
MiR-518a-3p mimics, inhibitor and corresponding negative controls (NC mimic, NC inhibitor), as well as shRNAs targeting TMEM2 (shTMEM2 #1 or #2), were synthesized by GenePharma (Suzhou, China). Full length TMEM2 was constructed and cloned into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). MDA-MB-453 or MDA-MB-468 cells were transfected with miR-518a-3p mimics, inhibitor or corresponding negative controls, shRNAs, or pcDNA-TMEM2 and pcDNA vector via Lipofectamine 3000 (Invitrogen). For the treatment with JAK1 inhibitor, MDA-MB-468 cells were cultured in RPMI 1640 medium containing 100 nM Pyridone 6 (Sigma-Aldrich, St. Louis, MO, USA), and then transfected with miR-518a-3p mimics, NC mimic, pcDNA-TMEM2 or pcDNA vector.
Wound Healing and Transwell Assays
Seeded MDA-MB-453 or MDA-MB-468 cells were gently scratched by a pipette tip in the middle of each well 2 days after transfection. Twenty-four hours later, the wound width was imaged and measured under a microscope (Leica, Wetzlar, Germany). For invasion assay, MDA-MB-453 or MDA-MB-468 cells resuspended in serum-free RPMI 1640 medium were seeded into the upper chambers (BD Biosciences, Bedford, MA, USA) pre-coated with matrigel (BD Biosciences). RPMI 1640 with 10% serum was added to the lower chambers. After 24 hours, invasive cells in the lower chambers were fixed with methanol, and then stained with 0.1% crystal violet before imaging under microscope.
Dual-Luciferase Reporter Assay
Targetscan (http://www.targetscan.org/vert_71/) identified the potential binding target of miR-518a-3p as TMEM2. Sequence of TMEM2 3’-UTR or the mutant sequence without miR-518a-3p binding site were subcloned into psiCHECK2 (Promega, Madison, Wisconsin, USA). MDA-MB-468 cells were co-transfected with miR-518a-3p mimics, inhibitor or negative controls and psiCHECK2-wt-TMEM2 or psiCHECK2-mut-TMEM2. After 48 hours, luciferase activities were measured with a Dual-Luciferase Assay Kit (Promega).
qRT-PCR
RNAs or miRNAs from TNBC cells were reverse-transcribed into cDNAs. qRT-PCR was conducted with TB Green Premix Ex Taq (Takara, Dalian, China). GAPDH and U6 were used as endogenous control for mRNA and miRNAs, respectively. Primers used in this study are as follows: GAPDH (F: 5’-ACCACAGTCCATGCCATCAC-3’; R: 5’-TCCACCACCCTGTTGCTGTA-3’), U6 (F: 5’-CTCGCTTCGGCAGCACA-3’; R: 5’-AACGCTTCACGAATTTGCGT-3’), miR-518a-3p (F: 5’-ACAGGCCGGGACAAGTGCAATA-3’; R: 5’-GCTGTCAACGATACGCTACGTAACG-3’), TMEM2 (F: 5’-GGAGATATGCTCCGTCTGACC-3’; R: 5’-CATCTGACTTGCCATACAAGGT-3’).
Western Blotting
Proteins extracted from TNBC cells (30 µg) were separated by SDS-PAGE, and electro-transferred onto a PVDF membrane. Membranes were blocked with 5% skimmed milk powder, and then incubated overnight with primary antibodies: anti-TMEM2 (1:1500, Abcam, Burlingame, CA, USA), anti-JAK1 and anti-p-JAK1 (1:2000, Abcam), anti-STAT1 and anti-p-STAT1 (1:2500, Abcam), anti-STAT2, anti-p-STAT2 and anti-GAPDH (1:3000, Abcam) at 4°C. Following incubation with secondary antibody (1:5000; Abcam), the immunoreactivities were detected by Immobilob ™ Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA).
Statistical Analysis
Data are expressed as mean ± standard deviation. Statistical analyses were performed by GraphPad Prism 5.0 and the difference was determined by one-way analysis of variance (ANOVA) or Student’s t test. Survival rate was determined by Kaplane-Meier method and log-rank test. p < 0.05 was considered as statistically significant.
Results
Expression of miR-518a-3p Is Positively Correlated With Overall Survival of TNBC Patients
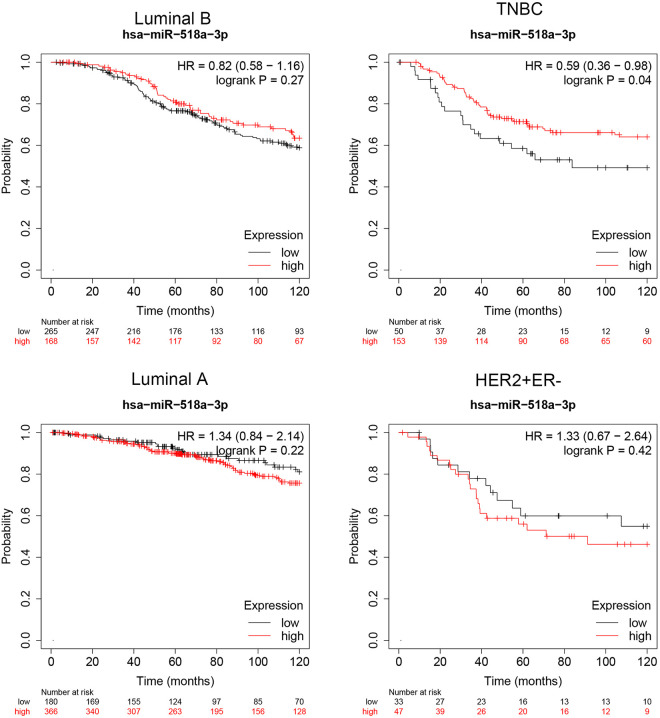
To investigate the correlation between miR-518a-3p expression and TNBC, survival analysis was evaluated using KM Plotter database (www.kmplot.com).18 According to METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) database,19 a total of 203 TNBC, 433 Luminal B, 546 Luminal A and 80 HER2+ER− samples were used for the survival analysis. This database was set up by searching the GEO, EGA, TCGA, and PubMed repositories to identify datasets with published miRNA expression and clinical data. Results revealed that miR-518a-3p expression was not significantly correlated with overall survival of patients with Luminal B (hazard ratio: 0.82; p = 0.27), Luminal A (hazard ratio: 1.34; p = 0.23) and HER2+ER− (hazard ratio: 1.33; p = 0.42) (Figure 1). However, TNBC patients with higher level of miR-518a-3p were found to have better overall survival compared to patients with lower expression of miR-518a-3p (hazard ratio: 0.59; p = 0.04) (Figure 1), suggesting that miR-518a-3p may function as a prognostic biomarker for TNBC.
Figure 1.
Expression of miR-518a-3p is positively correlated with overall survival of TNBC patients. Correlation between expression and overall survival of Luminal B, Luminal A, HER2+ER− and TNBC patients.
MiR-518a-3p Suppresses Cell Migration and Invasion of TNBC
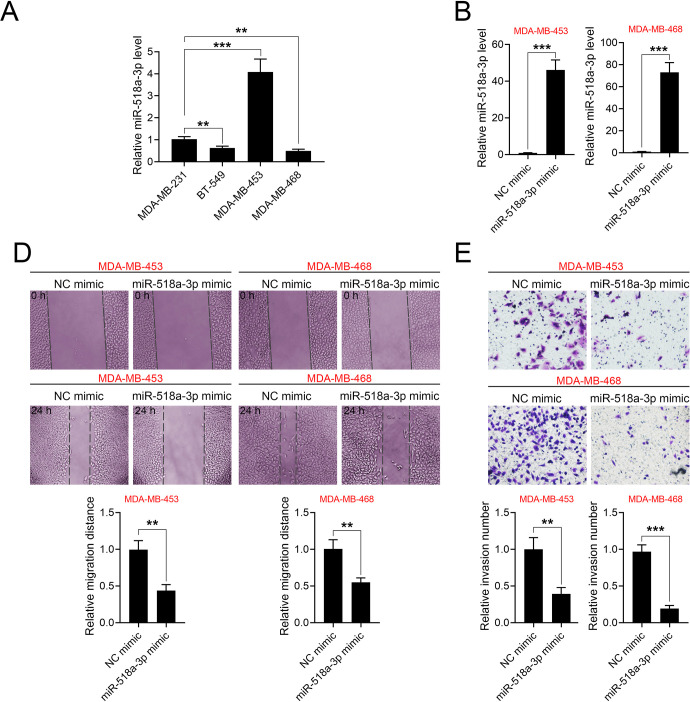
The functional role of miR-518a-3p in TNBC progression was then investigated using in vitro assays. The expression of miR-518-3p in various TNBC cell lines were analyzed by qRT-PCR. Results showed that the expression of miR-518a-3p was the highest in MDA-MB-453 and lowest in MDA-MB-468 (Figure 2A). MDA-MB-453 and MDA-MB-468 were transfected with miR-518a-3p mimics for the subsequent experiments (Figure 2B). Wound healing assays revealed that miR-518a-3p inhibited cell migration (Figure 2C) and cell invasion (Figure 2D) of MDA-MB-453 and MDA-MB-468. Moreover, cell migration (Supplemental Figure S1C and 1D) and invasion (Supplemental Figure S1E) were promoted in cells transfected with miR-518a-3p inhibitor, suggesting that miR-518a-3p might negatively regulate malignant phenotypes of TNBC. Results in Supplemental Figure S1F show that MDA-MB-453 and MDA-MB-468 demonstrated lowest and highest migratory ability, respectively, whereas MDA-MB-231 exhibited higher migratory ability than BT-549.
Figure 2.
MiR-518a-3p suppresses cell migration and invasion of TNBC. A, Expression of miR-518-3p in various TNBC cell lines (MDA-MB-231, BT-549, MDA-MB-453 and MDA-MB-468) analyzed by qRT-PCR. B, Transfection efficiencies of miR-518-3p in MDA-MB-453 and MDA-MB-468 cells analyzed by qRT-PCR. C, Effects of miR-518a-3p on cell migration of MDA-MB-453 and MDA-MB-468 cells. D, Effects of miR-518a-3p on cell invasion of MDA-MB-453 and MDA-MB-468 cells. NC mimic = a negative control for miR-518a-3p mimics. **p < 0.01, ***p < 0.001.
Negative Correlation Between miR-518a-3p and TMEM2
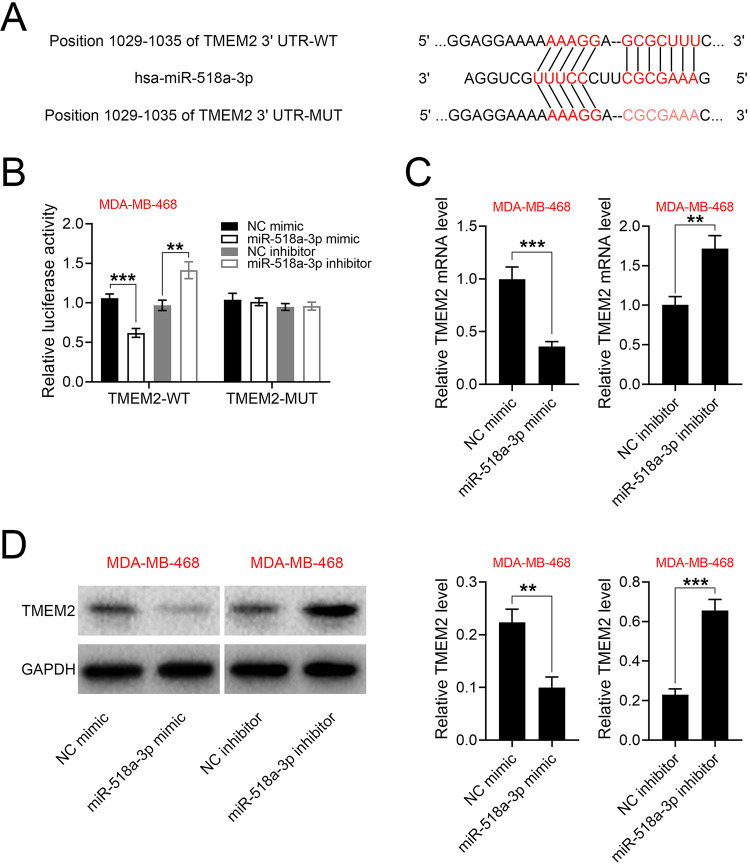
To further validate the promotive role of miR-518a-3p in TNBC, the identification of predicted potential binding targets was performed using Targetscan (http://www.targetscan.org/vert_71/). The analysis result showed that TMEM2 contained putative binding site for miR-518a-3p (Figure 3A). This was further validated by dual-luciferase reporter assays, which showed that the luciferase activity of psiCHECK2-wt-TMEM2 was reduced by miR-518a-3p mimics while enhanced by miR-518a-3p inhibitor (Figure 3B), whereas luciferase activity of psiCHECK2-mut-TMEM2 was not affected by miR-518a-3p mimics or inhibitor (Figure 3B). Furthermore, the mRNA (Figure 3C) and protein (Figure 3D) expression of TMEM2 was reduced in MDA-MB-468 transfected with miR-518a-3p mimics, while up-regulated by miR-518a-3p inhibitor. Collectively, these findings demonstrated that miR-518a-3p directly binds to TMEM2 and inhibits its expression.
Figure 3.
Negative correlation between miR-518a-3p and TMEM2. A, Potential binding site of miR-518a-3p in the 3’UTR of TMEM2. B, Effects of miR-518a-3p on the luciferase activities of psiCHECK2-wt-TMEM2 and psiCHECK2-mut-TMEM2. C, Effects of miR-518a-3p on mRNA expression of TMEM2 in MDA-MB-468. D, Effects of miR-518a-3p on protein expression of TMEM2 in MDA-MB-468. NC inhibitor = a negative control for miR-518a-3p inhibitor. **p < 0.01, ***p < 0.001.
Knockdown of TMEM2 Suppresses TNBC Invasion and Migration
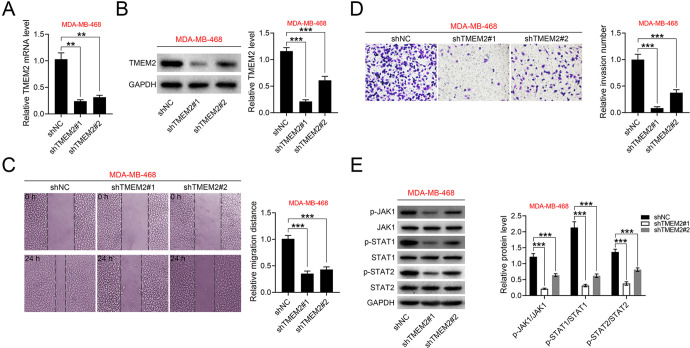
The functional role of TMEM2 in TNBC progression was also investigated in MDA-MB-468 transfected with shRNAs targeting TMEM2 (shTMEM2 #1 or #2) (Figure 4A and 4B). Knockdown of TMEM2 had no significant effect on miR-518a-3p expression (Supplemental Figure S1A). Cell migration (Figure 4C) and invasion (Figure 4D) were inhibited by shTMEM2 #1 or #2, suggesting that TMEM2 might contribute to malignant phenotypes of TNBC. Moreover, the protein expression of JAK1, STAT1 and STAT2 were not affected by shTMEM2 in MDA-MB-468 (Figure 4E). However, the levels of p-JAK1, p-STAT1 and p-STAT2 were reduced in MDA-MB-468 transfected with shTMEM2 (Figure 4E), revealing that the JAK/STAT pathway may be implicated in TMEM2-mediated TNBC.
Figure 4.
Knockdown of TMEM2 suppresses cell migration and invasion of TNBC. A, Transfection efficiencies of shTMEM2 #1 or #2 in MDA-MB-468 analyzed by qRT-PCR. B, Transfection efficiencies of shTMEM2 #1 or #2 in MDA-MB-468 analyzed by western blot. C, Effects of TMEM2 on cell migration of MDA-MB-468 cells. D, Effects of TMEM2 on cell invasion of MDA-MB-468 cells. E, Effects of TMEM2 on protein expression of JAK1, STAT1, STAT2, p-JAK1, p-STAT1 and p-STAT2 in MDA-MB-468 cells. **p < 0.01, ***p < 0.001.
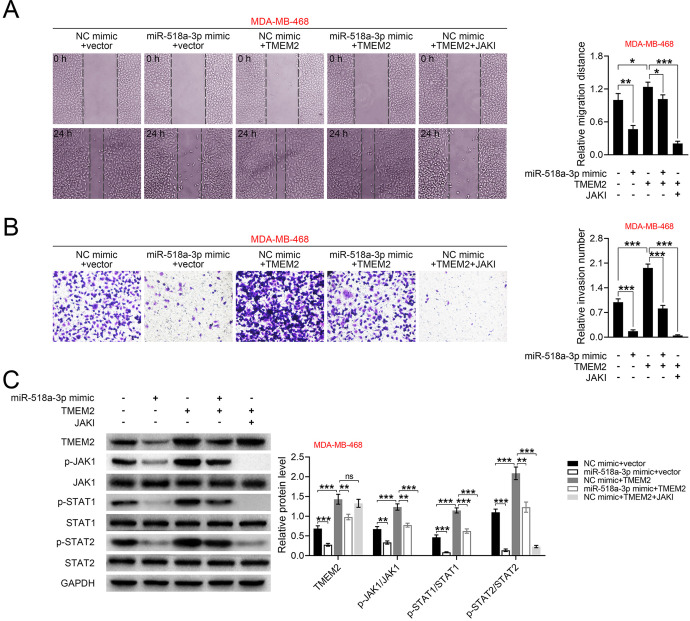
TMEM2 Counteracts the Suppressive Effect of miR-518a-3p on TNBC
MDA-MB-468 was cotransfected with miR-518a-3p mimics and pcDNA-TMEM2 to evaluate the role of miR-518a-3p/TMEM2 axis in TNBC. Results showed that TMEM2 could promote cell migration (Figure 5A) and invasion (Figure 5B). Although transfection with pcDNA-TMEM2 alone had no significant effect on miR-518a-3p expression (Supplemental Figure S1B), cotransfection of miR-518a-3p mimics and pcDNA-TMEM2 resulted in an increased miR-518a-3p level (Supplemental Figure S1B). Cotransfection of MDA-MB-468 with miR-518a-3p mimics and pcDNA-TMEM2 attenuated the suppressive effect of miR-518a-3p on cell migration (Figure 5A) and invasion (Figure 5B). Moreover, malignant phenotypes of TNBC with pcDNA-TMEM2 and JAK inhibitor treatment were also determined, and the result showed that treatment with JAK inhibitor significantly inhibited cell migration (p < 0.001, Figure 5A) and invasion (p < 0.001, Figure 5B). Similarly, over-expression of TMEM2 reversed miR-518a-3p-induced decrease in TMEM2, p-JAK1, p-STAT1 and p-STAT2 levels (Figure 5C), whereas treatment with JAK inhibitor dampened TMEM2 over-expression-induced increase in TMEM2, p-JAK1, p-STAT1 and p-STAT2 (Figure 5C). These results showed that miR-518a-3p negatively regulates the JAK/STAT pathway via targeting TMEM2 to suppress invasion and migration in TNBC.
Figure 5.
TMEM2 counteracts the suppressive effect of miR-518a-3p on TNBC. A, Effects of miR-518a-3p, TMEM2 and JAK inhibitor on cell migration of MDA-MB-468 cells. B, Effects of miR-518a-3p, TMEM2 and JAK inhibitor on cell invasion of MDA-MB-468 cells. C, Effects of miR-518a-3p, TMEM2 and JAK inhibitor on protein expression of TMEM2, JAK1, STAT1, STAT2, p-JAK1, p-STAT1 and p-STAT2 in MDA-MB-468 cells. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Due to its extremely aggressive ability and poor prognosis, TNBC is considered incurable with high mortality rates.9 Thus, novel potential therapeutic targets or prognostic biomarkers are a dire need for the treatment of TNBC. MiRNAs, with the ability to regulate tumor progression, have been regarded as critical biomarkers or therapeutic targets of TNBC.20 Considering the anti-tumor effect of miR-518a-3p against colon cancer, this study was conducted to investigate the role of miR-518a-3p in TNBC.
Molecular characterization in breast cancer provides predictive information and critical prognostic for the treatment of the patients.21 Luminal A or B represent hormone-receptor positive while HER2 negative, and less aggressive than TNBC.22 Data from KM Plotter database showed that high miR-518a-3p expression was not significantly correlated with overall survival of Luminal A or B, HER2+ER− breast cancer patients, but significantly associated with TNBC patients, suggesting that miR-518a-3p might be related to malignant phenotypes of TNBC. Considering that metastasis is one of the most important causes that leads to relapse and eventually death of TNBC patients,23 the role of miR-518a-3p in metastasis of TNBC was then evaluated.
MiR-518a-3p inhibits colorectal cancer cell growth through targeting NF-κB-inducing kinase,10 and knockdown of miR-518a-3p promotes colon cancer cell migration and invasion via up-regulation of murine double minute 2.11 Our results confirmed the anti-tumor effects of miR-518a-3p against migration and invasion of TNBC, revealing its potential clinical application in TNBC. Moreover, TMEM2 was validated as a target of miR-518a-3p in TNBC. Although TMEM2 has been shown to be a downstream target of miRNA let-7,17 miRNAs involve in TNBC progression have not been reported to date. Transmembrane proteins, including transmembrane 4 L6 family member 124 or TMEM88,25 play an oncogenic role in TNBC progression. TMEM2, transcriptionally regulated by SOX4, could promote metastasis of breast cancer.16 Results from this study also indicated that knockdown of TMEM2 suppressed TNBC invasion and migration, and over-expression of TMEM2 counteracted the suppressive effect of miR-518a-3p on TNBC invasion and migration.
Previous study has shown that TMEM2 could promote the activation of JAK/STAT pathway during hepatitis B virus infection.26 Here, phosphorylation of JAK1, STAT1 and STAT2 was inhibited by knockdown of TMEM2 in TNBC, suggesting that miR-518a-3p/TMEM2 axis may suppress TNBC progression through inactivation of the JAK/STAT pathway. STAT functions as a transcription factor that regulates gene expression involved in cell proliferation, differentiation and apoptosis. Moreover, the JAK/STAT pathway plays an important role in carcinogenesis.27 Overactivation of JAK/STAT signaling promotes tumorigenesis and metastasis,28 whereas its inhibition has been widely used in clinical treatment for TNBC.29-31 Results in this study revealed that inhibition of JAK by Pyridone 6 significantly suppressed TMEM2 over-expression-promoted cell migration and invasion of TNBC cells. However, how TMEM2 regulates phosphorylation of JAK/STAT remains elusive. In addition, chemoresistance is also considered a leading cause of relapse and death in TNBC patients.22 MiR-770 could suppress chemoresistance of TNBC through modification of tumor-associated macrophages.32 Considering that JAK/STAT signaling is involved in macrophage-mediated therapeutic resistance via regulation of protumorigenic factors,33 miR-518a-3p/TMEM2/JAK/STAT axis may participate in chemoresistance of TNBC, which requires further elucidation. The clinical application of miR-518a-3p in TNBC should be investigated in future study. Moreover, following Dicer cleavage of the stem loop of primary miRNA, miR-518a with the 5p and 3p strands are produced. According to previous studies,34-36 miR-518a-5p is also implicated in tumor progression. However, the association of miR-518a-5p and TNBC progression needs further investigation.
In conclusion, we demonstrated that miR-518a-3p is positively correlated with overall survival of TNBC patients and inhibits the metastasis of TNBC through targeting TMEM2. Conversely, over-expression of TMEM2 counteracts the suppressive effect of miR-518a-3p on TNBC progression through phosphorylation of JAK1, STAT1 and STAT2. In vivo experiments should be performed to demonstrate that newly validated miR-518a-3p/TMEM2/JAK/STAT axis provides a potential therapeutic target for the treatment of TNBC.
Supplemental Material
Supplemental Material, fig.s1 for miR-518a-3p Suppresses Triple-Negative Breast Cancer Invasion and Migration Through Regulation of TMEM2 by Lin Gan, Huachao Yang, Zhifeng Xiong, Zailiang Yang, Ting Wang and Gang Lyu in Technology in Cancer Research & Treatment
Abbreviations
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- JAK1
Janus kinase 1
- miRNAs
microRNAs
- PR
progesterone receptor
- STAT
signal transducer and activator of transcription protein
- shRNAs
short hairpin RNAs
- TNBC
triple-negative breast cancer
- TMEM2
transmembrane protein 2
Footnotes
Authors’ Note: LG and GL designed the study, supervised the data collection, analyzed the data, HY and ZX interpreted the data and prepare the manuscript for publication, ZY and TW supervised the data collection, analyzed the data and reviewed the draft of the manuscript. All authors have read and approved the manuscript. All data generated or analyzed during this study are included in this published article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the 2018 Performance Incentive Guidance Project of Chongqing Science and Technology Bureau (Grant No. cstc2018jxj1130063).
ORCID iD: Gang Lyu  https://orcid.org/0000-0003-0773-5432
https://orcid.org/0000-0003-0773-5432
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sung-Im D, Shin H-C, Kim HS. Clinical significance of MET receptor protein and mRNA expression in invasive breast cancer. Eur J Gynaecol Oncol. 2020;41(2):240–245. [Google Scholar]
- 2. Zeng Y, JM X, Wang B, et al. Mechanism for ginsenoside Rh2-induced apoptosis of triple-negative breast cancer MDA-MB-231 cells. Clin Exp Obstet Gynecol. 2020;47(1):99–104. [Google Scholar]
- 3. Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8(3):235–244. [DOI] [PubMed] [Google Scholar]
- 4. Bilici A, Arslan C, Altundag K. Promising therapeutic options in triple-negative breast cancer. J BUON. 2012;17(2):209–222. [PubMed] [Google Scholar]
- 5. Gucalp A, Traina TA. Triple-negative breast cancer: adjuvant therapeutic options. Chemothe Res Pract. 2011;2011:696208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andergassen U, Kolbl AC, Mumm JN, Mahner S, Jeschke U. Triple-negative breast cancer: new therapeutic options via signalling transduction cascades. Oncol Rep. 2017;37(5):3055–3060. [DOI] [PubMed] [Google Scholar]
- 7. Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA). Front Biosci. 2008;13:2216–2230. [DOI] [PubMed] [Google Scholar]
- 8. D’Ippolito E, Iorio MV. MicroRNAs and triple negative breast cancer. Int J Mol Sci. 2013;14(11):22202–22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nama S, Muhuri M, Di Pascale F, et al. MicroRNA-138 is a prognostic biomarker for triple-negative breast cancer and promotes tumorigenesis via TUSC2 repression. Sci Rep. 2019;9(1):12718–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qu LL, He L, Zhao X, Xu W. Downregulation of miR-518a-3p activates the NIK-dependent NF-κB pathway in colorectal cancer. Int J Mol Med. 2015;35(5):1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun Y, Cao B, Zhou J. Roles of DANCR/microRNA-518a-3p/MDMA ceRNA network in the growth and malignant behaviors of colon cancer cells. BMC Cancer. 2020;20(1):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamaguchi Y, Yamamoto H, Tobisawa Y, Irie F. TMEM2: a missing link in hyaluronan catabolism identified? Matrix Biol. 2018;78-79:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherman LS, Matsumoto S, Su W, Srivastava T, Back SA. Hyaluronan synthesis, catabolism, and signaling in neurodegenerative diseases. Int J Cell Biol. 2015;2015:368584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schinzel RT, Higuchi-Sanabria R, Shalem O, et al. The hyaluronidase, TMEM2, promotes ER homeostasis and longevity independent of the UPR(Er). Cell. 2019;179(6):1306–1318 e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Angelis JE, Lagendijk AK, Chen H, et al. Tmem2 regulates embryonic Vegf signaling by controlling hyaluronic acid turnover. Develop Cell. 2017;40(2):123–136. [DOI] [PubMed] [Google Scholar]
- 16. Lee H, Goodarzi H, Tavazoie SF, Alarcón CR. TMEM2 is a SOX4-regulated gene that mediates metastatic migration and invasion in breast cancer. Cancer Res. 2016;76(17):4994–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Werfel S, Leierseder S, Ruprecht B, Kuster B, Engelhardt S. Preferential microRNA targeting revealed by in vivo competitive binding and differential argonaute immunoprecipitation. Nucleic Acids Res. 2017;45(17):10218–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. [DOI] [PubMed] [Google Scholar]
- 19. Hwang K-T, Kim BH, Oh S, et al. Prognostic role of KRAS mRNA expression in breast cancer. J Breast Cancer. 2019;22(4):548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugita B, Gill M, Mahajan A, et al. Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget. 2016;7(48):79274–79291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vohra P, Buelow B, Chen Y-Y, et al. Estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 expression in breast cancer FNA cell blocks and paired histologic specimens: a large retrospective study. Cancer Cytopathol. 2016;124(11):828–835. [DOI] [PubMed] [Google Scholar]
- 22. Hon JD, Singh B, Sahin A, et al. Breast cancer molecular subtypes: from TNBC to QNBC. Am J Cancer Res. 2016;6(9):1864–1872. [PMC free article] [PubMed] [Google Scholar]
- 23. Parvani JG, Davuluri G, Wendt MK, et al. Deptor enhances triple-negative breast cancer metastasis and chemoresistance through coupling to survivin expression. Neoplasia. 2015;17(3):317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan C, Liu N, Zheng D, Du J, Wang K. MicroRNA-206 inhibits metastasis of triple-negative breast cancer by targeting transmembrane 4 L6 family member 1. Cancer Manag Res. 2019;11:6755–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu X, Zhang X, Zhang Y, Jiang G, Mao X, Jin F. Cytosolic TMEM88 promotes triple-negative breast cancer by interacting with Dvl. Oncotarget. 2015;6(28):25034–25045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu X, Xie C, Li Y-m, et al. TMEM2 inhibits hepatitis B virus infection in HepG2 and HepG2.2.15 cells by activating the JAK–STAT signaling pathway. Cell Death Dis. 2016;7(6):e2239–e2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xin P, Xu X, Deng C, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol. 2020;80:106210. [DOI] [PubMed] [Google Scholar]
- 29. Stover DG, Gil Del Alcazar CR, Brock J, et al. Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast cancer. 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng T, Cao W, Shen W, et al. Arctigenin inhibits STAT3 and exhibits anticancer potential in human triple-negative breast cancer therapy. Oncotarget. 2017;8(1):329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen M, Pockaj B, Andreozzi M, et al. JAK2 and PD-L1 amplification enhance the dynamic expression of PD-L1 in triple-negative breast cancer. Clin Breast Cancer. 2018;18(5):e1205–e1215. [DOI] [PubMed] [Google Scholar]
- 32. Li Y, Liang Y, Sang Y, et al. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Irey EA, Lassiter CM, Brady NJ, et al. JAK/STAT inhibition in macrophages promotes therapeutic resistance by inducing expression of protumorigenic factors. Proc Natl Acad Sci U S A. 2019;116(25):12442–12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rubie C, Kruse B, Frick VO, et al. Chemokine receptor CCR6 expression is regulated by miR-518a-5p in colorectal cancer cells. J Transl Med. 2014;12(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhan L, Mu Z, Yang M, Zhang T, Li H, Qian L. Elevation of circ-PITX1 upregulates interleukin 17 receptor D expression via sponging miR-518a-5p and facilitates cell progression in glioma. J Cell Biochem. 2019;120(10):16495–16502. [DOI] [PubMed] [Google Scholar]
- 36. Zhang N, Jin Y, Hu Q, et al. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J Ovarian Res. 2020;13(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, fig.s1 for miR-518a-3p Suppresses Triple-Negative Breast Cancer Invasion and Migration Through Regulation of TMEM2 by Lin Gan, Huachao Yang, Zhifeng Xiong, Zailiang Yang, Ting Wang and Gang Lyu in Technology in Cancer Research & Treatment