Abstract
TP53 mutations are the most occurred mutation in HNSCC which might affect the ion channel genes. We aim to investigate the ion channel gene alteration under TP53 mutation and their prognostic implication. The overall mutation status of HNSCC were explored. By screening the TP53-associated ion channel genes (TICGs), an ion channel prognostic signature (ICPS) was established through a series of machine learning algorithms. The ICPS was then evaluated and its clinical significance was explored. 82 TICGs differentially expressed between TP53WT and TP53MUT were screened. Using univariate regression analysis and LASSO regression analysis and multivariate regression analysis, an ICPS containing 7 ion channel genes was established. A series of evaluation was carried out which proved the predictive ability of ICPS. Functional analysis of ICPS revealed that cancer-related pathways were enriched in high-risk group. Next, for clinical application, a nomogram was constructed based on ICPS and other independent clinicopathological factors. TP53 mutation status strongly affects the expression of ion channel genes. The ICPM we have identified is a strong indicator for HNSCC prognosis and could help with patient stratification as well as identification of novel drug targets.
Keywords: head and neck squamous cell carcinoma, TP53, ion channel, BIOMARKER, PROGNOSIS, therapeutic targets
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide, whose occurrence includes oral cavity, oropharynx, larynx and hypopharynx.1 There are 600,000 cases of HNSCC annually, making up of 90% of head and neck cancer.2 The currant acknowledged therapy of HNSCC is to combine surgery, chemotherapy, radiotherapy and biotherapy.3 Despite advances in the means of the treatment of HNSCC, its survival rate remained lower than 50%.4 Stratification of HNSCC patients into high and low risk group upon diagnosis could be a new approach for improvement of the prognosis of HNSCC patients.5,6 Patients with lower risk could receive conventional treatment while higher risk patients will be going through a stricter monitoring and could be assigned to novel clinical trials. This could in turn elevate the benefits of clinical trials,7 and result in a more personalized treatment which could improve the overall clinical outcome of HNSCC patients. Therefore, identifying efficient, sensitive and independent biomarkers to predict prognosis of HNSCC patients is essential.
Ion channel genes encodes pore-forming membrane proteins expressed in every living cell which controls passive influx/efflux of signaling ions into/out of the cell or intracellular organelles thus manipulating ion concentration, membrane potential as well as cell volume.8 Ion channels contributes to virtually all biological processes as well as most of the fundamental biological processes including muscle contraction, secretion of hormones, cell proliferation as well as immune response.9-11 Due to the key role ion channels play in various biological functions, studies have demonstrated that aberrant expression of ion channel genes is crucial to many diseases, including kidney stones caused by malfunction of endocytosis, high blood pressure, insufficient secretion of insulin and cardiac arrhythmias.12 Moreover, ion channels are found to be correlated with the migration and proliferation of cancer cells. Nelson et al found that breast cancer cells that overexpressed gene encoding a subunit of Na+ channel, scn1b, metastasize more than those cancer cells with a normal expression level.13 The potassium channel Kv3.4 is elevated during the tumorigenesis and is found to be a risky factor of head and neck squamous cell carcinoma.14 There are also studies demonstrating the prognosis prediction value of ion channel in breast cancer, lung cancer, and glioma.15-17 Furthermore, these prognostic signatures are also independent of other clinical features. However, few studies concerning the prognostic implications of ion channels have been reported in HNSCC before.
TP53 gene mutation is the most common mutation occurs in HNSCC.18 Its translational product, p53, whose wild type is a known tumor suppressor. WT p53 is a transcription factor, whose function includes the transcription of various genes concerning apoptosis, cell cycle arrest, DNA repair, metabolism as well as senescence.19 In a word, its function makes sure that cells with DNA damage doesn’t duplicate itself via proliferation. Thus, cells with TP53 mutation are easy to spread out the DNA damage and eventually, lead to cancer.18 Growing evidence demonstrated that early stage of TP53 mutations result in a genomic instability, clonal expansion, then progression, leading to HNSCC.20 However, there is no clear picture concerning the expression alterations of ion channel genes under mutations of TP53 gene in HNSCC, and how they might be of clinical significance. The rapid development of next generation sequencing technologies as well as complete clinical follow-up information in the Cancer Genome Atlas (TCGA) database has provided new insights into the construction of signatures, more and more diagnostic/ prognostic cancer biomarkers have been identified.15,21,22 And this advance makes it easier to explore the overall expression alterations of ion channel genes influenced by TP53 mutations. In our study, we systematically investigated the TP53 mutation status in HNSCC, then explored the dysregulated ion channel genes affected by TP53 mutation, which was further passed on to a series of machine learning algorithms for the construction of ion channel prognostic signature (ICPS). Our results demonstrated that ICPS is an independent and robust signature for HNSCC prognostic prediction and we have constructed a nomogram combining the ICPS with several clinical features for clinical use. Besides, drugs known to target the ion channel genes were also revealed. It is important to note that, the ICPS we have identified was influenced by TP53 mutation, which can be used as potential therapeutic targets and might improve the management of HNSCC patients
Materials and Methods
Data Acquisition
The somatic mutation status data (mutect), gene expression data (FPKM, raw count), and clinical follow-up of HNSCC were obtained from The Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.gov/repository). Ion channel genes lists were obtained from the previous 2 studies.15,23 They were integrated and finally we got a list containing 347 ion channel genes (Table S1). Gene-drug interaction relationship were from the Drug Gene Interaction Database (www.dgidb.org). Known drugs that target the given genes were from www.dgidb.org.
Differentially Expressed Ion Channel Genes Associated With TP53 Status
489 HNSCC samples were included for differential analysis, which contains 157 TP53 wild and 332 TP53 mutation patients. The R package “edgR” was used to perform differential analysis and gene with FDR < 0.01 were considered as differentially expressed between TP53-wild and TP53-mutaion status. The differentially expressed ion channel genes were selected from the differentially gene list.
Gene-set Enrichment Analysis
To understand how the underlying molecular changes occurred between TP53-wild and mutation status, high risk and low risk groups, GSEA was perform using R package “clusterprofiler.” Annotated gene-set files (h.all.v7.0; c2.cp.kegg.v7.0; c5.bp.v7.0) were chosen as the gene set to refer to. The threshold was set at P < 0.05.
TP53-Acciacted Ion Channel Genes Signature Construction
After excluding samples without clinical information, there were 494 HNSCC patients involved in the construction of the predictive model. The TICGs and HNSCC patients with their clinical parameters were included in the univariate Cox regression analysis for identification of TICGs related to survival. TICGs with a p value lower than 0.05 were candidates for LASSO regression analysis. LASSO regression is commonly used for regression with high-dimensional features. This method is widely used in Cox proportional hazard regression model for survival analysis with high-dimensional data. LASSO Cox regression analysis was performed for the most powerful predictive features selection from candidate TICGs identified before. The most useful survival-related TICGs were used in the next step of analysis. Then, stepwise multivariate Cox regression analysis was performed to develop the risk model. Based on the median score, the patients were separated into 2 groups: high risk and low risk groups. A Kaplan-Meier survival curve was plotted to analyze the difference between the 2 groups. Furthermore, ROC analysis was used to evaluate the efficiency of the predictive model, and AUC values were calculated to validate the performance of the prognostic predictors. To test the robustness of the ICPS signature, bootstrap algorithm was used and we randomly resampled 400 cases for 10,000 times from the HNSCC dataset.
Nomogram Establishment and Estimation
Multivariate Cox regression analysis was used to identify whether ICPS was an independent predictor alone with other clinical features for HNSCC prognosis. Then, coefficients of clinical features and ICPS were integrated for nomogram establishment in order to visualize the predictive ability of several features into a chart. And a calibration curve was plotted to evaluate the predictive ability of the nomogram. This process was conducted by the R package rms. Additionally, ROC curve was plotted to assess the accuracy of the nomogram.
Results and Discussion
Expression Alterations of Ion Channel Genes Under TP53 Mutations
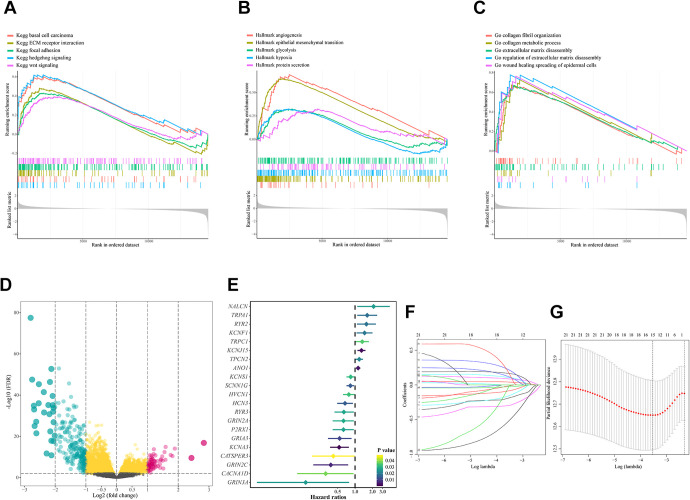
The workflow of the study is presented in Figure 1. We explored the overall mutation status of HNSCC, and TP53 mutation is most commonly occurred (Figure 2a-c). Previous studies have thoroughly investigated the TP53 mutation status on the overall survival as well as drug resistance on HNSCC patients,18 but how they might influence the expression profile of ion channel genes haven’t been explored. Thus, we run a genome wide analysis to identify TP53-associated ion channel genes (TICGs). To find out what biological processes and pathways TP53 mutation were involved in, GESA analysis was performed between TP53WT and TP53MUT, and tumorigenesis and progression related pathways such as epithelial mesenchymal transition, WNT-signaling pathway, glycolysis were enriched in TP53MUT patients (Figure 3a-c). Next, to screen the ion channel genes involved in TP53 mutation, differential analysis was performed, and 82 TICGs were identified (Figure 3d, Table S2).
Figure 1.
Flowchart of the study. TICGs: TP53-associated ion channel genes; ICPS: ion channel prognostic signature.
Figure 2.
Overview of mutation information in HNSCC of TCGA cohort. (A) Overall distribution of 6 different conversions and as a stacked bar-plot showing fraction of conversions in each sample. Genomic landscape of HNSCC revealed C>T is most frequently conversion type. (B) Distribution of variant classification in TCGA-HNSCC cohort revealed missense mutation is most frequent variant type. (C) Frequency and type of mutations in the top 10 genes in HNSCC. Genes are sorted according to frequency of mutation and TP53 is most mutational gene.
Figure 3.
TP53 associated gene-sets and icon channel genes. (A-C) Significantly enriched gene-sets (KEGG pathway, hallmarks and GO-BP) in TP53-mut HNSCC patients compared with that in TP53-wild patients. (D) Volcano-plot presented differentially expressed genes between TP53 mutation and wild patients. (E) Forest-plot demonstrated the relationship between TP53 associated icon channel genes and HNSCC patients prognosis, which are p < 0.05 in univariate COX analysis. (F-G) LASSO coefficient profiles of 21 TP53-asscociated icon channel genes were related to prognosis. The optimal values of the penalty parameter lambda were determined by 10-fold cross validation and 15 icon channel genes were screened.
Development and Evaluation of the ICPS
To assess the predictive power of the TICGs in prognosis, an ion channel prognostic signature (ICPS) was constructed based on the TICGs. Univariate Cox regression analysis was conducted on TICGs. Setting p < 0.05 as the criteria, 21 prognostic related TICGs was screened out of 82 TICGs (Figure 3e). To avoid overfitting, LASSO regression analysis was conducted on the prognosis related TICGs, and 15 TICGs with non-zero coefficient was chosen with the minimum criteria (Figure 3f-g). Then, these TICGs were passed on to stepwise multivariate Cox regression analysis, from which the ion channel prognostic signature (ICPS) consisted of 7 ion channel genes was finally constructed (Figure 4a). The risk score of every patient was obtained from ICPS (Table S3), which could well distinguish patients in high and low risk group. Risk scores were significantly elevated in TP53MUT group compared with TP53WT (Figure 4b). A higher risk score was related with shorter survival and more death events, and correlation analysis demonstrated that the risk score was significantly negatively related with survival time (Figure 4c-e). To further evaluate the predictive accuracy, ROC analysis was performed and the respective AUC predicting the 1-, 2-, and 3-year survival was 0.694, 0.688 and 0.688 (Figure 4f). Next, to test the accuracy and the uncertainty of the predictive power of the ICPS, bootstrap method was used to calculate the p-value in distinguishing patients between high and low risk group as well as the C-index of the ICPS. The resampling procedure have taken place for 10,000 times, and each of the p-value and C-index was calculated. The median value as well as the 95% confidence interval of the p-value and C-index suggested that the performance of ICPS was rather stable and convincing ( Figure 5a-b).
Figure 4.
Icon channel gene signature construction. (A) Forest-plot demonstrated the 7 TP53-associated icon channel genes consisted of the prognostic signature. (B) Distribution of risk-score between TP53-wild and TP53-mut groups. (C) The distribution of 7-TICG risk score, patients’ survival status and 7 TICGs expression. As the risk score rising, the patients had a shorter survival time, more dead events and the expression value of 7 TICGs ascended or decreased. (D) Analysis of the correlation between risk score and survival time according to TP53 status. (E) The Kaplan–Meier estimates of the OS for high-risk and low-risk patient cohorts grouping by the ICPM. The OS differences between the 2 groups were determined by the 2-sided log-rank test. It can be concluded that higher risk scores are significantly associated with worse OS (P < 0.0001). (F) ROC analysis of sensitivity and specificity for the ICPM in predicting the OS of patients for 1, 2 and 3 years.
Figure 5.
Robust of ICPM and biological function enriched in high-risk group. (A) Histogram of –log10(P) values from the resampling datasets for 10,000 times. There is no one time that p > 0.05. (B) Histogram of C-index values from the resampling datasets for 10,000 times. The mean c-index is 0.66 and sd is 0.02. The red vertical line in A represents -log10(0.05) and the red vertical line in B represents mean c-index. (C-E) GSEA analysis revealed the enriched gene-sets of hallmarks, KEGG pathways and GO-BP in high-risk group.
Functional Analysis of ICPS
Gene set enrichment analysis (GSEA) was carried out to explore the function of the 7 ion channel genes in ICPS (Figure 5c-e). Cell growth and development related gene sets such as HALLMARK_TGF_BETA_SIGNALING, HALLMARK_MYC_TARGETS_V2, and HALLMARK_DNA_REPAIR were enriched in high-risk group; besides, immune related pathways such as KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY and KEGG_CONPLEMENT_AND_COAGULATION_CASADES were also enriched in high-risk group. However, ion channel genes’ role in these processes remains to be further studied.
ICPS Is an Independent Predictor of the Prognosis of HNSCC
To evaluate the independency of ICPS, univariate Cox regression analysis and multivariate Cox regression analysis were performed on the risk scores of each patient graded by ICPS and several other clinicopathological features including TP53 status, age, margin status, anatomy set, grade, gender, and tumor stage. Both univariate and multivariate regression analysis demonstrated that ICPS is an independent predictor (Figure 6a-b). Subgroup analysis revealed that ICPS could well distinguish patients in both TP53WT and TP53MUT subgroups (Figure 6d-e). Besides, in both univariate and multivariate regression analysis, TP53 status also revealed itself as a strong predictor of prognosis in HNSCC. Thus, we performed survival analysis on TP53WT and TP53MUT group, and overall survival was significantly different between these 2 groups, suggesting the strong relevance of TP53 mutation with overall survival (Figure 6c). Furthermore, we searched for drugs that could be used to target the 7 ion channel genes on http://www.dgidb.org/search_interactions. The results were presented on Table 1.
Figure 6.
ICPM is an independent signature for HNSCC prognosis. (A) Univariate and multivariate regression analysis of the relation between the ICPM and TP53 status regarding prognostic value. (B) Univariate and multivariate regression analysis of the relation between the ICPM and clinicopathological features regarding prognostic value. (C-E) Kaplan-Meier survival analysis according to TP53 status, TP53 mutation subgroup, and TP53 wild-type subgroup.
Table 1.
Gene-Drug Interaction Relationships Were From the Drug Gene Interaction Database (www.dgidb.org).
| Gene | Drug | Interaction types | PMID |
|---|---|---|---|
| ANO1 | cA2 | activator | |
| ANTHRACENE-9-CARBOXYLIC ACID | channel blocker | ||
| METRONIDAZOLE | channel blocker | ||
| FLUFENAMIC ACID | channel blocker | ||
| FLUOXETINE | channel blocker | ||
| CHEMBL23050 | channel blocker | ||
| MIBEFRADIL | channel blocker | ||
| NIFLUMIC ACID | channel blocker | ||
| 5-NITRO-2-PHENYLPROPYLAMINOBENZOIC ACID [NPPB] | channel blocker | ||
| CHEMBL1162149 | channel blocker | ||
| TANNIC ACID | channel blocker | ||
| CROFELEMER | inhibitor antagonist blocker |
19808995 | |
| HCN3 | CILOBRADINE | channel blocker | |
| IVABRADINE | channel blocker | ||
| ZATEBRADINE | channel blocker | ||
| CHEMBL2052019 | channel blocker | ||
| KCNA3 | DALFAMPRIDINE | blocker antagonist |
16472864 |
| CORREOLIDE | channel blocker | ||
| PROCYCLIDINE | channel blocker | ||
| PROMAZINE | channel blocker | ||
| NERISPIRDINE | blocker | ||
| TEDISAMIL | blocker | ||
| GUANIDINE HYDROCHLORIDE | blocker | ||
| GRIA3 | IRAMPANEL | antagonist | |
| CX1739 | positive allosteric modulator | ||
| PF-04958242 | positive allosteric modulator | ||
| Selurampanel | antagonist | ||
| DASOLAMPANEL | antagonist | ||
| BECAMPANEL | antagonist | ||
| MIBAMPATOR | positive allosteric modulator | ||
| MK-8777 | positive allosteric modulator | ||
| ZONAMPANEL | antagonist | ||
| PERAMPANEL | antagonist | ||
| TEZAMPANEL | antagonist | ||
| TOPIRAMATE | antagonist | ||
| FARAMPATOR | positive allosteric modulator | ||
| PHENOBARBITAL | not found | 8786535 | |
| L-GLUTAMATE | not found | 7679210 17139284 17016423 |
|
| TALAMPANEL | not found | 12581229 16861112 17199027 |
|
| ANIRACETAM | not found | 10592235 | |
| TRPA1 | MENTHOL | inducer | 15931068 16829128 12654248 11752352 |
| 1’-ACETOXYCHAVICOL ACETATE | activator | ||
| ACETALDEHYDE | activator | ||
| APOMORPHINE | activator | ||
| ARTEPILLIN | activator | ||
| AURANOFIN | activator | ||
| BENZOQUINONE | activator | ||
| BENZYL BROMIDE | activator | ||
| BROMO ACETONE | activator | ||
| PYRIMETHAMINE | activator | ||
| CHLOROPICRIN | activator | ||
| DIBUTYL PHTHALATE | activator | ||
| ISOVELLERAL | activator | ||
| METHYLGLYOXAL | activator | ||
| CHEMBL1608558 | activator | ||
| MORPHANTHRIDINE | activator | ||
| NICOTINE | activator | ||
| 5-NITRO-2-PHENYLPROPYLAMINOBENZOIC ACID [NPPB] | activator | ||
| (-)-OLEOCANTHAL | activator | ||
| PHENACYL CHLORIDE | activator | ||
| CHEMBL1818218 | activator | ||
| SALIRASIB | activator | ||
| THYMOL | activator | ||
| ACROLEIN | activator | ||
| ALLICIN | activator | ||
| CHEMBL184238 | activator | ||
| VERATRIDINE | gating inhibitor | ||
| CHEMBL1086339 | gating inhibitor | ||
| CB-189625 | antagonist | ||
| GRC-17536 | antagonist | ||
| NALCN | PYRIMETHAMINE | channel blocker | |
| COBALT (II) ION | channel blocker | ||
| VERAPAMIL | channel blocker | ||
| KCNJ15 | not found | not found |
Known drugs that target the given genes were from www.dgidb.org.
Construction of Nomogram Based on Combined Features and Its Evaluation
A nomogram was constructed combining risk score of ICPS, gender, anatomic subdivision, TP53 status, margin status, age and tumor stage for clinical application (Figure 7a). To evaluate whether the predicted outcome is similar with the actual outcome, calibration plot was applied and the nomogram performed well (Figure 7b). Next, to compare the predictive performance of ICPS, clinical features, as well as ICPS and clinical features combined together, ROC analysis was performed with the respective C-index being 0.67, 0.69, and 0.73, suggesting the combined model has the best performance (Figure 7c).
Figure 7.
Nomogram construction and validation. (A) Nomogram for predicting the probability of 1-, 2-, and 3-year OS for TCGA-HNSCC patients by integrating ICPM and clinical features. (B) Calibration plot estimated the value of nomogram for predicting the probability of OS at 1, 2, and 3 years. (C) ROC curve analyses of the ICPM, clinical features, and combined nomogram.
Discussion
TP53 mutations are the most common genomic alterations in head and neck squamous cell carcinoma (HNSCC) which was significantly associated worse prognosis and difficulties in treatment.18 TP53WT functions as a tumor suppressor, stabilizing DNA and prevent the onset of cancer, also known as “guardian of the genome.”19 With the loss of function of p53 protein due to mutation of deletion of TP53WT, the control over cell cycle, DNA repair, apoptosis was sabotaged, leading eventually to cancer.24 Recently, the complicated role of ion channel genes in tumorigenesis and prognosis of patients with cancer has been investigated to some extent.15,25,26 Disruptions of ion channel were found to affect cell proliferation and migration in cancer. For example, inhibition of voltage-gated K+ channel can impair the proliferation of cancer cells14,27-29 An ion channel gene based prognostic signature could be used for patient stratification and revealed subtypes for chemotherapy sensitivity in primary glioblastoma.26 In breast cancer, ion channel genes were found to be able to predict the clinical outcome of patients.15 However, few studies have been able to elucidate the expression alterations of ion channel genes under mutation of TP53 and how they might be of clinical significance in prognostic prediction in HNSCC. In our study, the overall mutation status was explored in HNSCC, and expression alterations of ion channel genes under TP53 mutations were systematically explored, from which 82 TP53-associated ion channel genes (TICGs) were identified. Using these TICGs, a series of machine learning algorithms were performed and eventually an ion channel prognostic signature (ICPS) based on 7 ion channel genes were constructed. The predictive value of ICPS was thoroughly investigated and a nomogram was constructed based on ICPS and clinicopathological features. Our study revealed the expression changes of ion channel genes under TP53 mutation and presented an ion channel prognostic signature as well as a nomogram which could be useful for patient classification and may help to enhance the improvement of therapeutic strategy and identify new drug targets for HNSCC.
The ICPS was constructed base on 7 ion channel genes, including ANO1, TRPA1, HCN3, KCNA3, GRIA3, KCNJ15, NALCN, and ANO1, TRPA1, KCNJ15, NALCN are risky factors for HNSCC prognosis while HCN3, KCNA3, and GRIA3 are protective factors for HNSCC. ANO1 encodes Calcium-activated chloride channel, and was found to be highly expressed in HNSCC as a contributor for tumor progression by inhibiting apoptosis.30 It was further validated in another study that ANO1 was aberrantly expressed in HNSCC patients, and had a significant negative survival impact.31-33 As for TRPA1, it functions as a receptor-activated non-selective cation channel concerning pain detection and possibly cold perception and inner ear function.34 Although it hasn’t been investigated in HNSCC so far, it was found that in human prostate cancer, resveratrol could activate TRPA1, leading to an elevation of intracellular calcium concentration, and enhancing the expression and secretion of growth factors in cancer-associated fibroblast HCN3 encodes a multi-pass membrane protein functioning as a voltage gated cation channel,35 and in neuroblastoma, knockdown of HCN3 increased cell proliferation.36 However, how it influenced the prognosis of HNSCC remains unclear and might require further investigation. KCNA3 encodes a member of potassium voltage gated channel, and it was found that reduced function of KCNA3 in tumor infiltrating lymphocytes might lead to an reduction of immune surveillance in head and neck cancer,37 which corresponded with our finding that KCNA3 was a protective factor in HNSCC. GRIA3 is a receptor for glutamate functioning as ligand-gated ion channel in the central nervous system and plays an important part in excitatory synaptic transmission.35 However, in pancreatic cancer, knock-down of GRIA3 was found to inhibit cell proliferation, migration and enhance apoptosis.38 Similar results were found in none-small cell lung cancer, where GRIA3 promotes cell invasion, metastasis and epithelial-mesenchymal transition.39,40 Nevertheless, it remains unclear how GRIA3 functions as a protective factor in HNSCC. KCNJ15 encodes an integral membrane protein and inward-rectifier type potassium channel.35 Downregulation of KCNJ15 was found in renal cell carcinoma, and its overexpression could significantly inhibit the proliferation, migration and colony formation, as well as arrest cell cycle and induce apoptosis in renal cell carcinoma cells.41 NALCN encodes a voltage-independent, cation-nonselective channel which is permeable to sodium, potassium and calcium ion.42 However, very few studies in cancer have mentioned this gene, only one study concerning pancreatic cancer have found that significant mutation of NALCN was observed in tumor tissues,43 while further studies are needed to reveal the exact mechanism of NALCN in HNSCC.
Additionally, our results revealed that ICPS is an independent prognostic predictor, which stressed the importance of ion channel genes on survival prediction. In order to put ICPS into clinical use, a nomogram was constructed based on ICPS and other independent clinicopathological factors which demonstrated good performance. The nomogram could be used as a scoring system for HNSCC patients which could enhance patient classification and therapeutic management in the future.
Nevertheless, some limitations in our study cannot be overlooked. Since the study was mainly based on methodology, experiment-based researches need to be carried out to reveal the mechanisms and functions behind the 7 ion channel genes. Besides, although we have performed bootstrap to test the robustness of the signature, external datasets are needed for a further validation
Conclusions
In conclusion, we have systematically investigated the aberrant ion channel genes under TP53 mutation in HNSCC, based on which an ion channel prognostic signature was developed. The ICPS was an independent and robust signature based on evaluation analysis. Finally, a nomogram was constructed for clinical use and drugs known to target the 7 ion channel genes were also revealed. These findings may give great assistance in HNSCC prognosis prediction and therapy.
Supplemental Material
Supplemental Material, table_s1 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment
Supplemental Material, table_s2 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment
Supplemental Material, table_s3 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment
Acknowledgments
We appreciate the great help of bioinformatic analysis from Dr. Xiaoqi Zhang and Dr. Lu Xing in West China Stomatology Hospital.
Authors’ Note: The RNA-seq data used to support the findings of this study have been deposited in the TCGA project repository (https://portal.gdc.cancer.gov/).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Dongdong Tong  https://orcid.org/0000-0002-5498-8582
https://orcid.org/0000-0002-5498-8582
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2. Feng Z, Xu Q, Chen W. Epigenetic and genetic alterations-based molecular classification of head and neck cancer. Expert Rev Mol Diagn. 2012;12(3):279–290. doi:10.1586/erm.12.19 [DOI] [PubMed] [Google Scholar]
- 3. Zhang L, Zhang W, Wang YF, et al. Dual induction of apoptotic and autophagic cell death by targeting survivin in head neck squamous cell carcinoma. Cell Death Dis. 2015;6(5):e1771 doi:10.1038/cddis.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 5. Xing L, Zhang X, Guo M, Zhang X, Liu F. Application of machine learning in developing a novelty five-pseudogene signature to predict prognosis of head and neck squamous cell carcinoma: a new aspect of “Junk Genes” in biomedical practice. DNA Cell Biol. 2020;39(4):709–723. doi:10.1089/dna.2019.5272 [DOI] [PubMed] [Google Scholar]
- 6. Xing L, Zhang X, Zhang X, Tong D. Expression scoring of a small-nucleolar-RNA signature identified by machine learning serves as a prognostic predictor for head and neck cancer. J Cell Physiol. 2020;235(11):8071–8084. doi:10.1002/jcp.29462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo W, Zhu L, Yu M, Zhu R, Chen Q, Wang Q. A five-DNA methylation signature act as a novel prognostic biomarker in patients with ovarian serous cystadenocarcinoma. Clin Epigenetics. 2018;10(1):142 doi:10.1186/s13148-018-0574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kondratskyi A, Kondratska K, Skryma R, Klionsky DJ, Prevarskaya N. Ion channels in the regulation of autophagy. Autophagy. 2018;14(1):3–21. doi:10.1080/15548627.2017.1384887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Camerino DC, Desaphy JF, Tricarico D, Pierno S, Liantonio A. Therapeutic approaches to ion channel diseases. Adv Genet. 2008;64:81–145. doi:10.1016/S0065-2660(08)00804-3 [DOI] [PubMed] [Google Scholar]
- 10. Fiske JL, Fomin VP, Brown ML, Duncan RL, Sikes RA. Voltage-sensitive ion channels and cancer. Cancer Metastasis Rev. 2006;25(3):493–500. doi:10.1007/s10555-006-9017-z [DOI] [PubMed] [Google Scholar]
- 11. Roger S, Potier M, Vandier C, Besson P, Le Guennec JY. Voltage-gated sodium channels: new targets in cancer therapy? Curr Pharm Des. 2006;12(28):3681–3695. [DOI] [PubMed] [Google Scholar]
- 12. Jentsch TJ, Hubner CA, Fuhrmann JC. Ion channels: function unravelled by dysfunction. Nat Cell Biol. 2004;6(11):1039–1047. doi:10.1038/ncb1104-1039 [DOI] [PubMed] [Google Scholar]
- 13. Nelson M, Millican-Slater R, Forrest LC, Brackenbury WJ. The sodium channel beta1 subunit mediates outgrowth of neurite-like processes on breast cancer cells and promotes tumour growth and metastasis. Int J Cancer. 2014;135(10):2338–2351. doi:10.1002/ijc.28890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Menendez ST, Rodrigo JP, Allonca E, et al. Expression and clinical significance of the Kv3.4 potassium channel subunit in the development and progression of head and neck squamous cell carcinomas. J Pathol. 2010;221(4):402–410. doi:10.1002/path.2722 [DOI] [PubMed] [Google Scholar]
- 15. Ko JH, Ko EA, Gu W, Lim I, Bang H, Zhou T. Expression profiling of ion channel genes predicts clinical outcome in breast cancer. Mol Cancer. 2013;12(1):106 doi:10.1186/1476-4598-12-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ko JH, Gu W, Lim I, Bang H, Ko EA, Zhou T. Ion channel gene expression in lung adenocarcinoma: potential role in prognosis and diagnosis. PLoS One. 2014;9(1):e86569 doi:10.1371/journal.pone.0086569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang R, Gurguis CI, Gu W, et al. Ion channel gene expression predicts survival in glioma patients. Sci Rep. 2015;5:11593 doi:10.1038/srep11593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou G, Liu Z, Myers JN. TP53 Mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response. J Cell Biochem. 2016;117(12):2682–2692. doi:10.1002/jcb.25592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893 doi:10.1101/cshperspect.a000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. el-Naggar AK, Lai S, Luna MA, et al. Sequential p53 mutation analysis of pre-invasive and invasive head and neck squamous carcinoma. Int J Cancer. 1995;64(3):196–201. doi:10.1002/ijc.2910640309 [DOI] [PubMed] [Google Scholar]
- 21. Liu G, Zheng J, Zhuang L, et al. A Prognostic 5-lncRNA expression signature for head and neck squamous cell carcinoma. Sci Rep. 2018;8(1):15250 doi:10.1038/s41598-018-33642-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu F, Xing L, Zhang X, Zhang X. A four-pseudogene classifier identified by machine learning serves as a novel prognostic marker for survival of osteosarcoma. Genes (Basel). 2019;10(6):414 doi:10.3390/genes10060414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. D’Arcangelo D, Scatozza F, Giampietri C, Marchetti P, Facchiano F, Facchiano A. Ion channel expression in human melanoma samples: in silico identification and experimental validation of molecular targets. Cancers (Basel). 2019;11(4):446 doi:10.3390/cancers11040446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi:10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- 25. Lastraioli E, Iorio J, Arcangeli A. Ion channel expression as promising cancer biomarker. Biochim Biophys Acta. 2015;1848(10 pt B):2685–2702. doi:10.1016/j.bbamem.2014.12.016 [DOI] [PubMed] [Google Scholar]
- 26. Wang HY, Li JY, Liu X, et al. A three ion channel genes-based signature predicts prognosis of primary glioblastoma patients and reveals a chemotherapy sensitive subtype. Oncotarget. 2016;7(46):74895–74903. doi:10.18632/oncotarget.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abdul M, Hoosein N. Voltage-gated potassium ion channels in colon cancer. Oncol Rep. 2002;9(5):961–964. [PubMed] [Google Scholar]
- 28. Chang KW, Yuan TC, Fang KP, et al. The increase of voltage-gated potassium channel Kv3.4 mRNA expression in oral squamous cell carcinoma. J Oral Pathol Med. 2003;32(10):606–611. doi:10.1034/j.1600-0714.2003.00197.x [DOI] [PubMed] [Google Scholar]
- 29. Fraser SP, Grimes JA, Djamgoz MB. Effects of voltage-gated ion channel modulators on rat prostatic cancer cell proliferation: comparison of strongly and weakly metastatic cell lines. Prostate. 2000;44(1):61–76. doi:10.1002/1097-0045(20000615)44:1<61:: aid-pros9>3.0.co;2-3 [DOI] [PubMed] [Google Scholar]
- 30. Godse NR, Khan N, Yochum ZA, et al. TMEM16A/ANO1 inhibits apoptosis via downregulation of bim expression. Clin Cancer Res. 2017;23(23):7324–7332. doi:10.1158/1078-0432.CCR-17-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perez Sayans M, Chamorro Petronacci CM, Lorenzo Pouso AI, et al. Comprehensive genomic review of TCGA head and neck squamous cell carcinomas (HNSCC). J Clin Med. 2019;8(11):1896 doi:10.3390/jcm8111896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reddy RB, Khora SS, Suresh A. Molecular prognosticators in clinically and pathologically distinct cohorts of head and neck squamous cell carcinoma—a meta-analysis approach. PLoS One. 2019;14(7):e0218989 doi:10.1371/journal.pone.0218989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hermida-Prado F, Menendez ST, Albornoz-Afanasiev P, et al. Distinctive expression and amplification of genes at 11q13 in relation to HPV status with impact on survival in head and neck cancer patients. J Clin Med. 2018;7(12):501 doi:10.3390/jcm7120501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moparthi L, Survery S, Kreir M, et al. Human TRPA1 is intrinsically cold- and chemosensitive with and without its N-terminal ankyrin repeat domain. Proc Natl Acad Sci U S A. 2014;111(47):16901–16906. doi:10.1073/pnas.1412689111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi:10.1002/cpbi.5 [DOI] [PubMed] [Google Scholar]
- 36. Tang W, Dong K, Li K, Dong R, Zheng S. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1alpha and p53 pathways. Sci Rep. 2016;6:36268 doi:10.1038/srep36268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chimote AA, Hajdu P, Sfyris AM, et al. Kv1.3 channels mark functionally competent CD8+ tumor-infiltrating lymphocytes in head and neck cancer. Cancer Res. 2017;77(1):53–61. doi:10.1158/0008-5472.CAN-16-2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ripka S, Riedel J, Neesse A, et al. Glutamate receptor GRIA3--target of CUX1 and mediator of tumor progression in pancreatic cancer. Neoplasia. 2010;12(8):659–667. doi:10.1593/neo.10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei CH, Wu G, Cai Q, et al. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol. 2017;10(1):125 doi:10.1186/s13045-017-0493-0 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Wei C, Zhang R, Cai Q, et al. MicroRNA-330-3p promotes brain metastasis and epithelial-mesenchymal transition via GRIA3 in non-small cell lung cancer. Aging (Albany NY). 2019;11(17):6734–6761. doi:10.18632/aging.102201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Wang H, Ni B, et al. Loss of KCNJ15 expression promotes malignant phenotypes and correlates with poor prognosis in renal carcinoma. Cancer Manag Res. 2019;11:1211–1220. doi:10.2147/CMAR.S184368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129(2):371–383. doi:10.1016/j.cell.2007.02.041 [DOI] [PubMed] [Google Scholar]
- 43. Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi:10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, table_s1 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment
Supplemental Material, table_s2 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment
Supplemental Material, table_s3 for TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma by Jing Sun, Xijiao Yu, Lande Xue, Shu Li, Jianxia Li, Dongdong Tong and Yi Du in Technology in Cancer Research & Treatment