Abstract
Introduction
Drug efflux pumps are critical for resistance in Gram-negative organisms, but there are limited data on the role they play in decreased susceptibility to β-lactam/β-lactamase inhibitor combinations. In this study, we aimed to investigate the impact of efflux pump AcrAB on piperacillin–tazobactam (TZP) and ceftolozane–tazobactam (C/T) susceptibility in tigecycline-non-susceptible Klebsiella pneumoniae (TNSKP) strains.
Methods
A tigecycline gradient was used to obtain various TNSKP strains, and in conjunction with the gradient derived strains, a TNSKP clinical strain (TNSKP24) was also included. Minimum inhibitory concentrations (MICs) of antibiotics were determined by the broth microdilution method, and whole-genome sequencing (WGS) was carried out to analyze genomic changes. PCR and sequencing were performed to confirm mutations in ramR, acrR, and the intergenic region of ramR-romA, and qRT-PCR was applied to evaluate levels of gene expression. In-frame acrB knockout and complementation were performed in 3 TNSKP strains.
Results
Two derivatives of K. pneumoniae K2606 (K2606-4 and K2606-16) and TNSKP24 overexpressed efflux pump AcrAB were obtained for further study. The MICs of TZP and C/T exhibited a 4- to 8-fold increase in K2606-4 and K2606-16, respectively, when compared with K2606 (TZP, 2/4 μg/mL; C/T, 0.25/4 μg/mL). Deletion of acrB decreased the MICs of TZP and C/T by 4- to 16-fold in TNSKP24, K2606-4, and K2606-16, respectively, and complementation of acrB increased the MICs of these agents. MICs of clavulanate, sulbactam, and avibactam in the presence of β-lactam compounds did not change after acrB deletion and subsequent introduction of complementation mutants.
Conclusion
This study highlights that decreased susceptibility to TZP and C/T could be caused by the multidrug efflux pump AcrAB in TNSKP strains.
Keywords: drug efflux pump, Enterobacteriaceae, multidrug resistance, β-lactam/β-lactamase inhibitor combinations
Introduction
Over the last decade, Gram-negative bacterial resistance has increased worldwide, and the growing threat of carbapenem-resistant Klebsiella pneumoniae (CRKP) is of great concern. With increased resistance to carbapenems, carbapenem-sparing strategies have been proposed,1,2 and potential alternative agents have been investigated.2,3 β-lactam/β-lactamase inhibitor combinations (BL/BLIs) are considered to be promising candidates for the treatment of mild to moderate infections caused by extended-spectrum-β-lactamase (ESBL)-producing K. pneumoniae.1,3,4 The widely available piperacillin–tazobactam (TZP) and another newly released tazobactam combination, ceftolozane–tazobactam (C/T), both demonstrate excellent in vitro activity against ESBL-producing Gram-negative bacteria, including K. pneumoniae.2,4–8
However, attention should be paid to the development of bacterial resistance to these two tazobactam combinations. Bacterial resistance against TZP is mainly characterized by the following mechanisms used in Enterobacteriaceae: 1) hyper-production of β-lactamases such as TEM-1 in K. pneumoniae,9 and the presence of TEM, SHV, and CTX-M variants in Escherichia coli and K. pneumoniae,7,10,11 and 2) production of β-lactamases that are not readily inhibited by tazobactam (eg, carbapenemases, AmpC β-lactamases, and OXA-group β-lactamases).12 It has been reported that a deficiency of the OmpK36 porin likely results in K. pneumoniae resistance to TZP.13 Another study has also suggested that E. coli may potentially leverage a multidrug efflux pump to promote TZP resistance.14 Lastly, investigations in Pseudomonas aeruginosa have found that an intrinsic AmpC mutation and acquired β-lactamases appear to be the main mechanisms of bacterial resistance to C/T, a new-generation BL/BLI that is tazobactam combined with a novel cephalosporin ceftolozane.15–17 However, it is important to note that the novel tazobactam combination can also be hydrolyzed by carbapenemase enzymes and is considered not to be affected by efflux pumps or porin loss.18 Therefore, the exact role that efflux pumps play in K. pneumoniae susceptibility to TZP and C/T requires further investigation.
Overexpression of efflux pump AcrAB, a member of the resistance-nodulation-division (RND) superfamily, is one of the main mechanisms mediating tigecycline resistance in tigecycline-non-susceptible K. pneumoniae (TNSKP). Upregulation of acrAB can be activated by a global transcriptional activator RamA, and increased expression of ramA occurs mainly because ramR, the gene encoding a local transcriptional repressor of RamA, is mutated.19 While studying tigecycline resistance mechanisms of K. pneumoniae in vitro, we observed that laboratory TNSKP strains simultaneously showed decreased susceptibility to TZP and C/T. This interesting result led us to examine how efflux pump AcrAB affects the susceptibility of clinical and laboratory-derived TNSKP strains to BL/BLIs, including tazobactam, sulbactam, clavulanate, and avibactam combined with their β-lactam partners.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
TNSKP24 is a clinical isolate with a tigecycline MIC 4 μg/mL tested by broth microdilution method as described previously.19 K2606 is a clinical isolate with a tigecycline MIC 2 μg/mL that was randomly selected for tigecycline resistance induction experiment. TNSKP24 and K2606 were obtained from sputum and blood samples, respectively, of two hospitalized patients in Huashan Hospital in 2012. The clinical samples were part of the routine hospital laboratory procedure. In this study, all K. pneumoniae strains used were maintained at −80°C in 20% (vol/vol) glycerol for cryoprotection.
The MICs of tigecycline were determined as described previously20 and interpreted based on the breakpoint for Enterobacteriaceae endorsed by the US Food and Drug Administration (≤2.0 μg/mL is susceptible, 4.0 μg/mL is intermediate, and ≥8.0 μg/mL is resistant). MICs of other antimicrobial agents were also determined by broth microdilution methods and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.21 MIC results were considered significant when there was a ≥ 4-fold difference between the parental strains and their derivatives. E. coli ATCC25922 was used as the quality control strain.
Selection of Tigecycline-Resistant Mutants by Serial Passage
To explore the potential mechanisms causing reduced susceptibility to BL/BLIs in TNSKP strains, resistance induction with tigecycline gradient was carried out in vitro as described previously with minor modifications.22 Briefly, the parental K2606 strain was statically cultured overnight in 2 mL Muller-Hinton (MH) broth, starting with 0.5× tigecycline MIC of the parental strain. And then, 20 μL of the overnight inoculum was suspended in another 2 mL fresh MH broth containing a two-fold tigecycline concentration of the former, with this process repeated every 24 h until the MH broth reaching a tigecycline concentration of 16× MIC of the original strain after 6 days (ie, reaching 16× MIC of the original strains). The daily subculture (10 μL) was propagated on MH agar plates without antibiotics, and a single colony from each plate was randomly selected and stored at −80°C for further study.
Efflux Pump Inhibition Test
To assess the role of efflux pump in tigecycline MIC alteration, efflux pump inhibition test was performed using Phe-Arg-β-naphthylamide (PAβN) as previously described.23 Briefly, MICs of tigecycline alone or in combination with PAβN (25 μg/mL) were determined in TNSKP24, K2606, and derivatives of K2606.
Whole Genome Sequencing (WGS) and Analysis
To identify the genomic alterations of K2606 after exposure to tigecycline, we sequenced the whole genomes of K2606 and its two derivatives (K2606-4 and K2606-16). Genomic DNA was extracted using Wizard® Genomic DNA Purification Kit (Promega) according to the manufacture’s protocol. Genomes were sequenced using a combination of PacBio RS II Single Molecule Real Time (SMRT) and Illumina sequencing platforms, and the resulting data were used for bioinformatics analysis. Glimmer was used for CDS prediction, tRNA-scan-SE was used for tRNA prediction and Barrnap was used for rRNA prediction. The predicted CDSs were annotated from NR, Swiss-Prot, Pfam, GO, COG and KEGG database using sequence alignment tools such as BLAST, Diamond and HMMER. Briefly, each set of query proteins was aligned with the databases, and annotations of best-matched subjects (e-value <10−5) were obtained for gene annotation.
Genome of K2606 was used as a reference. Variants (SNPs, short indels, and large deletions) of K2606-4 and K2606-16 were called based on the Bowtie2 assembled data using GATK toolkit by Broad Institute. The variants were verified by universal PCR and sequencing.
Universal PCR and Quantitative Real-Time PCR (qRT-PCR)
To detect the presence of mutations in ramR, acrR, and the intergenic region of ramR-romA, universal PCR and sequencing were applied as described previously.19 Genome of K. pneumoniae MGH78578 (tigecycline MIC, 1 μg/mL) was used as a reference for gene mutation analysis. To evaluate the transcriptional expression levels of efflux pump genes acrB and oqxB, and the regulatory genes ramA and rarA in TNSKP strains, qRT-PCR was performed as described previously.19 The normalized relative expression of genes was determined by the 2−ΔΔCT method. A tigecycline-susceptible K. pneumoniae clinical isolate (TSKP1; tigecycline MIC, 0.5 μg/mL) and K2606 were used as a reference isolate for the gene expression analysis of TNSKP24 and the two derivatives of K2606 (K2606-4 and K2606-16), respectively.
Construction of acrB Deletion Mutants
To investigate the role of the efflux pump AcrAB in the susceptibility to BL/BLIs in K. pneumoniae strains, TNSKP24 and two derivatives of K2606 (K2606-4 and K2606-16) were used for acrB gene knockout. Lambda Red recombination and Flp-FRT recombination were used to construct markerless in-frame gene deletion mutants as described previously.24 Finally, 3123-bp of acrB was replaced by an 84-bp segment, and markerless in-frame indel mutants TNSKP24ΔacrB, K2606-4ΔacrB, and K2606-16ΔacrB were obtained, and confirmed by PCR and sequencing. The primers used in this study are listed in Table 1.
Table 1.
Primers Used in This Study
| Primers | Sequence (5′ to 3′) | Product (bp) | Reference |
|---|---|---|---|
| GmF | CGAATTAGCTTCAAAAGCGCTCTGA | 1649 | 24 |
| Gm-R2 | AATTGGGGATCTTGAAGTTCCT | ||
| EBGNHe-5 | CCCGCTAGCGAAAAGATGTTTCGTGAAGC | 1960 | 24 |
| EBGh3-3 | GGGAAGCTTATTATCGTGAGGATGCGTCA | ||
| acrB-UF-F | CGGCAAAGCGAAAGTGGAG | 512 | This study |
| acrB-UF-R | TCAGAGCGCTTTTGAAGCTAATTCGa GAAATTAGGCATGTCTTAACGGC | ||
| acrB-DF-F | AGGAACTTCAAGATCCCCAATTb GAGCATCATTAATCTTCACTCC | 509 | This study |
| acrB-DF-R | GTA TTC GTC CAT AAC GCA TC | ||
| acrB-internal-F | TATCGGCTACGACTGGACC | 317 | This study |
| acrB-internal-R | GCCCTTTGCCCTCTTTCT | ||
| acrB-external-F | TGATTTCCTGCGCCTGAAG | 4247 | This study |
| acrB-external-R | AGCGTTTCG CAG AGAAAG C | ||
| ramR-romA-F | GATGGCGACCACGCTGAA | 305 | This study |
| ramR-romA-R | TATGCCGACTGGGCGAAA | ||
| Hindш-acrB-F | ATGACCATGATTACGCCAAGCTTATGCCTAATTTCTTTATCGATCG | 3192 | This study |
| BamHⅠ-acrB-R | GTACCCCATCGATGGGGGATCCTTAATGATGCTCAACCTGATGGC |
Notes: aReverse-complement to Gm-F; breverse-complement to Gm-R; Gm-F/R for hygromycin B resistance gene hph cassette flanked by the Flp recombinase target sites; EBGNHe-5/EBGh3-3 for the recombination vector pKOBEG; acrB-UF-F/R for 5ʹ upstream flanking of acrB fragment; acrB-DF-F/R for 3ʹ downstream flanking of acrB fragment; acrB-internal-F/R for a fragment inside acrB; acrB-external-F/R for a fragment including UF, DF, and acrB.
Complementation with the Wild-Type acrB Gene
To establish the influence of acrB knockout on the susceptibility to TZP and C/T, complementation with the wild-type acrB gene was performed in TNSKP24ΔacrB, K2606-4ΔacrB, and K2606-16ΔacrB. Briefly, the 3147-bp acrB gene of strain TNSKP24 and K2606 was, respectively, amplified using primers Hindш-acrB-F and BamHⅠ-acrB-R (Table 1), digested with Hindш and BamHⅠ, and ligated to vector pHSG396 to generate the recombinant vector (pHSG396acrB). pHSG396acrB was verified by sequencing, and then it was electroporated into competent strains. And the transformants were selected on Luria Bertani (LB) agar plates with 50 μg/mL chloramphenicol and were verified by sequencing. Empty plasmid pHSG396 was used as a negative control.
Results
Susceptibility Profiles of TNSKP24, K2606, and Their Derivatives
The MICs of 18 compounds for K. pneumoniae TNSKP24, K2606, and their derivatives are summarized in Table 2. K2606-4 and K2606-16 were derived from K2606 by gradient exposure at 4× and 16× tigecycline MIC of K2606, respectively. The two derivatives not only exhibited a significant increase in the MICs of tigecycline and ciprofloxacin but also in TZP and C/T when compared with K2606 (Table 2). Interestingly, MICs of other BL/BLIs (sulbactam, clavulanate, and avibactam combinations) tested showed either a marginal increase (2-fold) or none at all (Table 2).
Table 2.
Antimicrobial Susceptibility Patterns of the K. pneumoniae Clinical Parental Strains and Their Derivatives
| Strains | MIC (μg/mL) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | C/T | SAM | CFZ | CXM | CAZ | CCV | CZA | SCF | FEP | MEM | IPM | ATM | AMK | TGC | CIP | CST | |
| TNSKP24a | 256 | 4/4 | 1/4 | 32/16 | >32 | >64 | 0.5 | 0.5/0.25 | 0.25/4 | 16/8 | 8 | ≤0.06 | 0.125 | 2 | 0.5 | 4 | 0.5 | 0.5 |
| TNSKP24ΔacrBb | 256 | 0.25/4 | 0.25/4 | 32/16 | >32 | >64 | 0.5 | 0.5/0.25 | 0.25/4 | 16/8 | 8 | ≤0.06 | 0.125 | 2 | 0.5 | 0.5 | ≤0.06 | 0.5 |
| TNSKP24ΔacrB/pHSG396 | 256 | 0.25/4 | 0.25/4 | 32/16 | >32 | >64 | 0.5 | 0.5/0.25 | 0.25/4 | 16/8 | 8 | ≤0.06 | 0.125 | 2 | 0.5 | 0.5 | ≤0.06 | 0.5 |
| TNSKP24ΔacrB/pHSG396acrB | 256 | 2/4 | 0.5/4 | 32/16 | >32 | >64 | 0.5 | 0.5/0.25 | 0.25/4 | 16/8 | 8 | ≤0.06 | 0.125 | 2 | 0.5 | 2 | 0.5 | 0.5 |
| K2606c | 512 | 2/4 | 0.25/4 | 32/16 | >32 | >64 | 1 | 0.5/0.25 | 0.25/4 | 16/8 | 4 | ≤0.06 | ≤0.06 | 4 | 2 | 2 | 1 | 1 |
| K2606-4d | 512 | 16/4 | 1/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | >128 | 8 | 1 |
| K2606-4ΔacrBe | 256 | 2/4 | 0.25/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 2 | ≤0.06 | 1 |
| K2606-4ΔacrB/pHSG396 | 256 | 2/4 | 0.25/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 2 | ≤0.06 | 1 |
| K2606-4ΔacrB/pHSG396acrB | 512 | 8/4 | 0.5/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 16 | 16 | 1 |
| K2606-16f | 512 | 16/4 | 1/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | >128 | 8 | 1 |
| K2606-16ΔacrBg | 256 | 2/4 | 0.25/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 2 | ≤0.06 | 1 |
| K2606-16ΔacrB/pHSG396 | 256 | 2/4 | 0.25/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 2 | ≤0.06 | 1 |
| K2606-16ΔacrB/pHSG396acrB | 512 | 8/4 | 0.5/4 | 32/16 | >32 | >64 | 2 | 0.5/0.25 | 0.25/4 | 32/16 | 8 | ≤0.06 | ≤0.06 | 8 | 2 | 16 | 16 | 1 |
| ATCC25922 | 2 | 2/4 | 0.125/4 | 4/2 | 4 | 4 | 0.125 | 0.25/0.125 | ≤0.25/4 | 0.25/0.125 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | 2 | 0.125 | ≤0.06 | 0.5 |
Notes: aTNSKP24, a clinical K. pneumoniae strain with tigecycline MIC 4 μg/mL. bTNSKP24ΔacrB, TNSKP24 without acrB. cK2606, a clinical K. pneumoniae strain with tigecycline MIC 2 μg/mL. dK2606-4, K2606 exposure to tigecycline with a concentration 4× MIC of K2606. eK2606-4ΔacrB, K2606-4 without acrB. fK2606-16, K2606 exposure to tigecycline with a concentration 16× MIC of K2606. gK2606-16ΔacrB, K2606-16 without acrB.
Abbreviations: PIP, piperacillin; TZP, piperacillin–tazobactam; C/T, ceftolozane–tazobactam; SAM, ampicillin-sulbactam; CFZ, cefazolin; CXM, cefuroxime; CAZ, ceftazidime; SCF, cefoperazone-sulbactam; CCV, ceftazidime-clavulanate; CZA, ceftazidime-avibactam; FEP, cefepime; MEM, meropenem; IPM, imipenem; ATM, aztreonam; AMK, amikacin; TGC, tigecycline; CIP, ciprofloxacin; CST, colistin.
The tigecycline MIC of TNSKP24 was 4 μg/mL and decreased to 1 μg/mL in the presence of 25 μg/mL PAβN. Additionally, the MICs of K2606-4 and K2606-16 were >128 μg/mL, and both were reduced to 16 μg/mL in the presence of 25 μg/mL PAβN. On the other hand, the tigecycline MIC of K2606 was not significantly decreased (2 μg/mL to 1μg/mL) in the presence of PAβN.
Genomic Changes of K2606 After Exposure to Tigecycline
We obtained complete genome sequences and summarized all mutations of K2606-4 and K2606-16 (Table 3, Figure S1). When compared with K2606, nonsynonymous mutations occurred in five genes (K2606-4, n=3; K2606-16, n=3). Among the five genes, only one (tet(A), encoding tetracycline efflux pump) exhibited a mutation (S251A) in both of the derivatives (Table 3, Figure S1A). Of the four remaining genes with nonsynonymous mutations, two were mutated in each of the derivatives. dksA, a gene encoding a transcriptional regulator, and rpsJ, a gene encoding ribosomal protein RpsJ, were found to be mutated in K2606-4 and K2606-16, respectively. The other two genes encode hypothetical proteins (Table 3, Figure S1B, C). It is important to note that genes encoding β-lactamases and their corresponding promoters did not exhibit mutations in the two derivatives of K2606.
Table 3.
Genomic Changes of K2606 After Exposure to Tigecycline
| Location | Mutation | Position | Mutation Type | Information | Annotation | Gene Description |
|---|---|---|---|---|---|---|
| SNPa in both K2606-4 and K2606-16 | ||||||
| Chrb | T843830G | Intergenic of gene0838 and gene0839 | - | - | fimB;- | type 1 fimbriae regulatory protein; hypothetical protein |
| Chr | A1127404G, A1127631G A1127769G, A1128074G |
Intergenic of gene1117 and gene1123 | - | - | csrA;- | carbon storage regulator; phospholipid-binding domain protein |
| Chr | C3872385 T | Intergenic of gene3810 and gene3811 | - | - |
romA; ramR |
beta-lactamase domain protein; TetR family transcriptional regulator |
| pAc | A90269C | Internal of gene0095 | Nonsynonymous | c.T751G p.S251A |
tetA | tetracycline efflux MFS transporter |
| SNP in K2606-4 only | ||||||
| Chr | G303519A | Intergenic of gene0279 and gene0278 | - | - | tusA;- | tRNA 2-thiouridine (34) synthase; hypothetical protein |
| Chr | G586814C | Intergenic of gene0574 and gene0575 | - | -;- | hypothetical protein; hypothetical protein |
|
| Chr | G4324175T | Internal of gene4263 | Nonsynonymous | c.G386T, p.R129L | dksA | transcriptional regulator |
| Chr | C4350054A | Internal of gene4291 | Nonsynonymous | c.G59T, p.C20F | - | hypothetical protein |
| pBd | C125391G | Intergenic of gene0112 and gene0113 | - | -;higA | hypothetical protein; transcriptional regulator |
|
| SNP in K2606-16 only | ||||||
| Chr | C427705A | gene0395 | Nonsynonymous | c. C116A, p.P39Q |
rpsJ | ribosome protein RpsJ |
| Chr | C2049592G | Intergenic of gene1998 and gene1999 | - | - | -;- | GNAT family acetyltransferase; transcriptional regulator |
| Chr | A4052373C | Internal of gene3986 | Nonsynonymous | c. A101C, p.Y34S, |
- | hypothetical protein |
| pDe | C29141G | Intergenic of gene0036 and gene0037 | - | - | -;- | hypothetical protein; hypothetical protein |
| fINDEL in K2606-4 | ||||||
| Chr | GCTATACCAAA deleted between 586,798 and 586,808 | Intergenic of gene0572 and gene0573 | Deletion | - | glxK;- | glycerate 2-kinase; hypothetical protein |
Notes: aSNP, single nucleotide polymorphism; bChr, chromosome; cpA, plasmid A; dpB, plasmid B; epD, plasmid D; fINDEL, insertion-deletion.
Multiple intergenic point mutations were detected in K2606-4 (n=9) and K2606-16 (n=8) (Table 3). Of these mutations, six (T843830G, A1127404G, A1127631G, A1127769G, A1128074G, and C3872385T) were concurrent in both of the two strains (Table 3, Figure S1A). These six mutations occurred up or downstream of six distinct genes, and four of the genes (fimB, csrA, gene1123, and ramR) were associated with bacterial virulence, metabolism, signal transduction, and resistance, respectively. Five derivative specific intergenic point mutations, three in K2606-4 and two in K2606-16, were also noted (Table 3, Figure S1B and S1C). Lastly, only one 11-bp fragment deletion was detected in K2606-4, and none were found in K2606-16 (Table 3, Figure S1D).
Mutations of ramR, acrR, the Intergenic Region of ramR-romA, and Increased Transcriptional Level of acrB, oqxB, ramA, and rarA
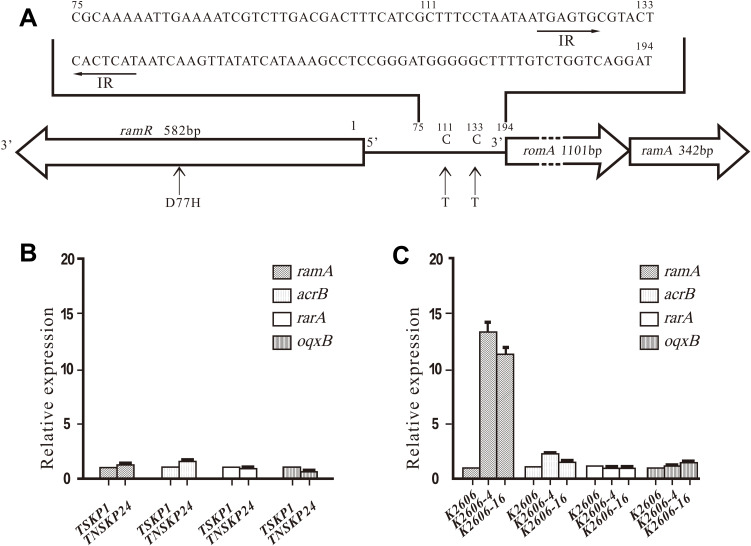
No mutations were detected in ramR, acrR, and intergenic regions of ramR-romA in TNSKP24 when compared with K. pneumoniae MGH78578. Also, no mutations were detected in acrR in K2606, K2606-4, and K2606-16. Interestingly, two mutations that were found in the K2606 parent strain persisted in its derivatives. A missense mutation (D77H) in ramR and a point mutation (C111T) in the intergenic region of ramR-romA were detected in K2606 and its two derivatives (K2606-4 and K2606-16) when compared with MGH78578. However, a novel point mutation (C133T, mentioned above C3872385T) was found in K2606-4 and K2606-16 (Figure 1A) but was absent in K2606.
Figure 1.
Gene mutation and expression in TNSKP24, K2606 K2606-4, and K2606-16. (A) Mutations in ramR and the intergenic region of ramR-romA in K. pneumoniaeK2606 and its two derivatives when compared with K. pneumoniae MGH78578 (GenBank: CP000647.1). The arrow segments in the intergenic region of romR-romA are the inverted repeat (IR) sequences recognized by the RamR protein (24). The numbering system is based on the “t” prior to romA’s start codon ATG as the 194. Missense mutation (D77H) in ramR, and point mutation (C111T) in the intergenic region of ramR-romA was detected in K2606, K2606-4, and K2606-16, and an additional novel point mutation (C133T) was found between the palindromic repeats in the 2 derivatives. (B) and (C) Transcriptional expression (mean±standard deviation) of ramA, acrB, rarA, and oqxB in TNSKP24, K2606 K2606-4, and K2606-16.
The TNSKP24 clinical strain showed slightly altered expression levels of acrB, oqxB, ramA, and rarA when compared with TSKP1 (Figure 1B). K2606-4 and K2606-16 possessed relatively high transcriptional expression levels of ramA (13- and 11-fold), while those of acrB, rarA and oqxB had little to no increase (Figure 1C).
Antimicrobial Susceptibility Patterns of acrB Deletion and Complementation Mutants
The efflux pump gene acrB was deleted in-frame in three strains (TNSKP24, K2606-4, and K2606-16), and the corresponding mutants were designated as TNSKP24ΔacrB, K2606-4ΔacrB, and K2606-16ΔacrB, respectively. The TZP MICs of TNSKP24ΔacrB, K2606-4ΔacrB, and K2606-16ΔacrB were 0.25/4 μg/mL, 2/4 μg/mL, and 2/4 μg/mL (Table 2), respectively, which were reduced remarkably (16-fold or 8-fold) compared with the corresponding original strains. Similarly, the C/T MICs of the three knockout mutants were all 0.25/4 μg/mL and were also significantly reduced (4-fold) compared with the original strains. The MICs of piperacillin and other BL/BLIs tested, however, were only marginally reduced and in some cases, exhibited no reduction at all. These results suggest that functional AcrAB contributes to reduced TZP and C/T susceptibility in these strains.
As expected, the MICs of tigecycline and ciprofloxacin were also significantly decreased among the acrB knockout mutants compared with the corresponding original strains (Table 2), suggesting that AcrAB can efficiently pump out tigecycline and ciprofloxacin as well. On the other hand, the MICs of certain cephalosporins, carbapenems, aztreonam, aminoglycosides, and colistin were unaffected, indicating that these compounds are likely not good substrates of AcrAB.
Complementation with wild-type acrB was successfully constructed in TNSKP24ΔacrB, K2606-4ΔacrB, and K2606-16ΔacrB (designated as TNSKP24ΔacrB/pHSG396acrB, K2606-4ΔacrB/pHSG396acrB, and K2606-16ΔacrB/pHSG396acrB, respectively). The complementation of acrB increased the MICs of TZP, C/T, tigecycline, and ciprofloxacin, confirming that acrB plays a role in decreasing susceptibilities to these agents in TNSKP (Table 2).
Discussion
Multidrug efflux pumps, especially RND family transporters such as AcrAB, are critical for antibiotic resistance in Gram-negative bacteria, including the intrinsic and acquired resistance to structurally diverse agents. Identifying the actual role of efflux pumps in β-lactam susceptibility, however, is difficult due to complicated resistance backgrounds in clinical isolates. In this study, one clinical and two laboratory-derived tigecycline-non-susceptible K. pneumoniae strains simultaneously showed decreased susceptibility to two tazobactam-containing compounds, TZP and C/T. A study by Nicolas-Chanoine et al suggested that piperacillin, ceftolozane, and tazobactam may be substrates of AcrAB.25 Tam et al confirmed that AcrB can bind and extrude carboxylated β-lactams.26 To this end, our study recapitulates results from the aforementioned studies and concludes that the mechanism underlying MIC changes in TZP and C/T is the extrusion of carboxylated β-lactams by AcrAB in K. pneumoniae. Very recently, a study by Suzuki et al indicated that AcrAB also contributes to TZP non-susceptibility in E. coli.27 Although piperacillin and ceftazidime are considered substrates of AcrAB,25,28 our study showed that the MICs of these substances exhibited marginal change, if at all, in acrB deletion mutants of TNSKP24, K2606-4, and K2606-16, and the minimal changes in MICs may be due to β-lactamase production in these strains (blaSHV-168 and blaCTX-M-14 existed in TNSKP24; blaSHV-27 and blaCTX-M-14 detected in K2606, K2606-4, and K2606-16, data not shown).
C/T is a second-generation BL/BLI that was recently approved by the US Food and Drug Administration.6 The emergence of C/T resistance is rare and mainly reported in P. aeruginosa due to AmpC structural modification.15–17 However, our study is the first to show that the overexpression of efflux pump AcrAB contributes to decreased susceptibility to C/T in K. pneumoniae.
After K2606 exposure to tigecycline, the derivatives acquired tigecycline resistance expectedly. Surprisingly, tigecycline MICs were extremely high (>128 μg/mL) in the two derivatives (K2606-4 and K2606-16). The high MICs were likely a result of the synergistic effects of an efflux pump TetA mutation and efflux pump AcrAB overexpression, and the synergic effect of efflux pump TetA and RND transporters on conferring higher tigecycline resistance has been observed in Acinetobacter baumannii recently.29 However, tigecycline MIC returned to basal level when acrB was deleted in K2606-4 and K2606-16, suggesting AcrAB plays a critical role in mediating tigecycline resistance. Moreover, when wild-type acrB was introduced into K2606-4ΔacrB and K2606-16ΔacrB, tigecycline MICs increased to 16 μg/mL in the mutants, indicating that other factors may be contributing to high tigecycline MICs (>128 μg/mL) in the parental strains. Du et al reported that tet(A) mutation can confer a relatively high-level of tigecycline resistance (32 μg/mL) in vivo in K. pneumoniae strain,30 but further study is needed to elucidate how tet(A) mutation and synergy between efflux pumps TetA and AcrAB affect tigecycline resistance in K. pneumoniae.
Next, we observed transcriptional expression levels of acrB in the clinical strain, TNSKP24, and the two laboratory-derived strains. Although the transcriptional expression levels of acrB were not markedly increased, MICs of TZP and C/T were significantly decreased in the corresponding acrB deletion mutants, suggesting that overexpression of efflux pump AcrAB contributes to reduced susceptibility to these two tazobactam combinations. Complementation with the acrB gene partially restored the MICs of TZP and C/T. As a result, this finding supports the notion that AcrAB functions to alter the MICs of the two antibiotics. Although the MICs were only partially restored, this may be due, in part, to the insufficient restoration of acrB expression as well as other factors.
RamR has been shown to indirectly affect transcriptional levels of acrAB in K. pneumoniae. When RamR binds the palindromic repeats in the intergenic region of ramR-romA, romA and ramA are transcriptionally repressed.31 Additionally, another study showed that increased expression of ramA can up-regulate the expression of acrAB.19 In our study, two derivatives of K2606 exhibited a novel point mutation in the palindromic repeats of ramR-romA after exposure to tigecycline, and this likely contributed to the up-regulation of ramA in the two strains. Previously, we also identified that a 12-bp deletion in the intergenic region of ramR-romA caused the elevated expression of ramA and acrB which also conferred tigecycline resistance in vivo in K. pneumoniae.32 In summary, these results indicate that mutations in the intergenic region of ramR-romA may help promote tigecycline resistance by upregulating expression of acrB in K. pneumoniae.
The present study shows that tigecycline and ciprofloxacin can be efficiently expelled by efflux pump AcrAB. However, carbapenems, certain cephalosporins, aztreonam, and aminoglycosides cannot be removed by the pump, and this is consistent with what has been shown in previous studies.19,28,33,34 Previous studies have also reported that efflux played a primary role in resistance to cefepime in P. aeruginosa.35,36 Consistent with our study, however, the studies also found that the efflux pump did not contribute to cefepime resistance in K. pneumoniae strains.36 It is also worth mentioning that the net contribution of AcrAB to the MICs of β-lactams was likely masked by the presence of various β-lactamases, especially in clinical isolates. Overall, our study supports the notion that efflux pump AcrAB can significantly reduce the susceptibility of K. pneumoniae strains to tazobactam-containing compounds.
Our study also showed that the MICs of TZP and C/T were still in the susceptible range in TNSKP24, K2606-4, and K2606-16. In many cases, however, TZP resistance can be promoted in vitro when exposed to E. coli, which may leverage a multidrug efflux pump system to help augment resistance.14 The efflux pump AcrAB may reduce the BL/BLIs susceptibility only below the clinical breakpoint, and resistance may only become obvious when reduction in membrane permeability and production of β-lactamases occur simultaneously. For instance, high-level ceftazidime-avibactam resistance can be mediated in vitro by a combination of increased β-lactamase expression, loss of functional outer membrane proteins, and activated efflux in K. pneumoniae.37
Juan et al reported that prior fluoroquinolone use was an independent risk factor that promoted K. pneumoniae non-susceptibility to tigecycline.38 The underlying mechanism may be due to the overexpression of AcrAB, which mediates non-susceptibility to both fluoroquinolone and tigecycline. AcrAB is widely distributed in K. pneumoniae and helps mediate cross-resistance to various antimicrobials. Due to the aforementioned mechanisms, the simultaneous or sequential use of various AcrAB substrates requires caution and should be avoided if possible.
It is important to note that this study has a few limitations. First, the two parental clinical strains, TNSKP24 and K2606, are both β-lactamase producers. β-lactamases have the potential to mask the role of efflux pumps, including AcrAB, and thus cause the effect of efflux to be underestimated in clinical strains. Despite this setback, the present study clearly supports that efflux pump AcrAB plays a significant role in reducing TZP and C/T susceptibilities in K. pneumoniae. Secondly, although the present study supports tazobactam as a likely substrate of AcrAB, it was not confirmed to be one in K. pneumoniae, and this could be partially attributed to β-lactamases produced by our strains. Irrespective of this limitation, the conclusion that efflux pump AcrAB contributes to reducing the susceptibilities of the two tazobactam combinations as a whole is still strongly supported. Lastly, the present study mainly evaluated AcrAB contribution via acrB deletion and complementation in TNSKP clinical and laboratory strains, but the impact of β-lactamase and other mutated genes (such as tet(A)) was not confirmed by gene knockout. The MICs of TZP and C/T were still significantly reduced in acrB-deletion mutants, which strongly suggests acrB plays an important role in bacterial susceptibility to the two tazobactam combinations. In order to address the limitations presented in this work, further study is needed to clarify the synergistic effects of efflux pump AcrAB and various other proteins, such as efflux pump TetA and β-lactamases, on BL/BLIs in K. pneumoniae strains.
Conclusions
Efflux pump AcrAB can remarkably reduce susceptibilities to two tazobactam-containing combinations (piperacillin–tazobactam and ceftolozane–tazobactam) in TNSKP strains. This study also highlights how the often overlooked and underestimated role of efflux is critical to resistance in K. pneumoniae clinical strains. In addition, simultaneous decreases in susceptibility to various other antimicrobials could be caused by efflux pump AcrAB. In turn, this could impact the combination or sequential use of these agents which would ultimately have serious clinical implications. To this end, it is important to be cognizant of the far-reaching impacts that efflux pumps have on bacterial susceptibility, especially as K. pneumoniae drug resistance continues to increase.
Acknowledgments
We thank Yohei Doi for his critical review of the manuscript. This study was supported by the funding from the National Natural Science Foundation of China (NO. 81703567 to J. Li, NO. 81603166 to Z. Sheng, and NO. 81473250 and 81773785 to M. Wang), the Shanghai Three-Year Plan of the Key Subjects Construction in Public Health-Infectious Diseases and Pathogenic Microorganism (NO. 15GWZK0102 to Q. Xie), Shanghai Municipal Key Clinical Specialty (Infectious disease, YW20190002 to Q. Xie), and partially funded by the National Science and Technology Key Project on Major Infectious Diseases (NO. 2017ZX10302201-004-005 to Z. Sheng).
Abbreviations
TZP, piperacillin–tazobactam; C/T, ceftolozane–tazobactam; TNSKP, tigecycline-non-susceptible Klebsiella pneumoniae; MIC, minimum inhibitory concentration; WGS, whole-genome sequencing; CRKP, carbapenem-resistant Klebsiella pneumoniae; BL/BLIs, β-lactam/β-lactamase inhibitor combinations; ESBL, extended-spectrum-β-lactamase; RND, resistance-nodulation-division; CLSI, Clinical and Laboratory Standards Institute; MH, Muller-Hinton; PAβN, Phe-Arg-β-naphthylamide; SMRT, Single Molecule Real Time; qRT-PCR, quantitative real-time PCR; TSKP, tigecycline-susceptible K. pneumoniae clinical isolate; LB, Luria Bertani.
Data Sharing Statement
The complete genome sequence of K2606 is available at US National Center for Biotechnology Information (NCBI) website under the following GenBank accession numbers: CP047633 for chromosome, CP047634 for plasmid A, CP047635 for plasmid B, CP047636 for plasmid C, CP047637 for plasmid D, and CP047638 for plasmid E.
Supplemental material is available online in Figure S1.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bassetti M, Peghin M, Pecori D. The management of multidrug-resistant Enterobacteriaceae. Curr Opin Infect Dis. 2016;29(6):583–594. doi: 10.1097/QCO.0000000000000314 [DOI] [PubMed] [Google Scholar]
- 2.Harris PNA, Tambyah PA, Paterson DL. β-lactam and β-lactamase inhibitor combinations in the treatment of extended-spectrum β-lactamase producing Enterobacteriaceae: time for a reappraisal in the era of few antibiotic options? Lancet Infect Dis. 2015;15(4):475–485. doi: 10.1016/S1473-3099(14)70950-8 [DOI] [PubMed] [Google Scholar]
- 3.Tamma PD, Rodriguez-Bano J. The use of noncarbapenem β-lactams for the treatment of extended-spectrum β-lactamase infections. Clin Infect Dis. 2017;64(7):972–980. doi: 10.1093/cid/cix034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris PNA, Tambyah PA, Lye DC, et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance. JAMA. 2018;320(10):984–994. doi: 10.1001/jama.2018.12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu F-P, Guo Y, Zhu D-M, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–14. doi: 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 6.Popejoy MW, Paterson DL, Cloutier D, et al. Efficacy of ceftolozane/tazobactam against urinary tract and intra-abdominal infections caused by ESBL-producing Escherichia coli and Klebsiella pneumoniae: a pooled analysis of Phase 3 clinical trials. J Antimicrob Chemother. 2017;72(1):268–272. doi: 10.1093/jac/dkw374 [DOI] [PubMed] [Google Scholar]
- 7.Shen Z, Ding B, Bi Y, et al. CTX-M-190, a novel beta-lactamase resistant to tazobactam and sulbactam, identified in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2016;61(1):e01848–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Duin D, Bonomo RA, Saravolatz LD. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis. 2016;63(2):234–241. doi: 10.1093/cid/ciw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhan S, et al. Conjugative Resistance to Tazobactam plus piperacillin Among Extended-spectrum Beta-lactamase-producing Nosocomial Klebsiella pneumoniae. Scand J Infect Dis 2001;33(7):512–515. doi: 10.1080/00365540110026520 [DOI] [PubMed] [Google Scholar]
- 10.Dubois V, Poirel L, Arpin C, et al. SHV-49, a novel inhibitor-resistant β-lactamase in a clinical isolate of Klebsiella pneumoniae. Antimicrob AgentsChemother. 2004;48(11):4466–4469. doi: 10.1128/AAC.48.11.4466-4469.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou K, Tao Y, Han L, et al. Piperacillin-tazobactam (TZP) resistance in Escherichia coli due to hyperproduction of TEM-1 β-lactamase mediated by the promoter Pa/Pb. Fron Microbiol. 2019;10:833. doi: 10.3389/fmicb.2019.00833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canton R, Morosini MI, Martin OM, et al. IRT and CMT β-lactamases and inhibitor resistance. Clin Microbiol Infect. 2008;14(Suppl 1):53–62. doi: 10.1111/j.1469-0691.2007.01849.x [DOI] [PubMed] [Google Scholar]
- 13.Wang XD, Cai JC, Zhou HW, et al. Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated β-lactamase production and OmpK36 porin deficiency. J Med Microbiol. 2009;58(9):1196–1202. doi: 10.1099/jmm.0.008094-0 [DOI] [PubMed] [Google Scholar]
- 14.Dos Santos KV, Diniz CG, de Castro Veloso L, et al. Proteomic analysis of Escherichia coli with experimentally induced resistance to piperacillin/tazobactam. Res Microbiol. 2010;161(4):268–275. doi: 10.1016/j.resmic.2010.03.006 [DOI] [PubMed] [Google Scholar]
- 15.Cabot G, Bruchmann S, Mulet X, et al. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother. 2014;58(6):3091–3099. doi: 10.1128/AAC.02462-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraile-Ribot PA, Cabot G, Mulet X, et al. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother. 2018;73(3):658–663. doi: 10.1093/jac/dkx424 [DOI] [PubMed] [Google Scholar]
- 17.MacVane SH, Pandey R, Steed LL, et al. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother. 2017;61(12):e01183–17. doi: 10.1128/AAC.01183-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cluck D, Lewis P, Stayer B, et al. Ceftolozane–tazobactam: a new-generation cephalosporin. Am J Health Syst Pharm. 2015;72(24):2135–2146. doi: 10.2146/ajhp150049 [DOI] [PubMed] [Google Scholar]
- 19.Sheng ZK, Hu F, Wang W, et al. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2014;58(11):6982–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford PA, Petersen PJ, Young M, et al. Tigecycline MIC testing by broth dilution requires use of fresh medium or addition of the biocatalytic oxygen-reducing reagent oxyrase to standardize the test method. Antimicrob Agents Chemother. 2005;49(9):3903–3909. doi: 10.1128/AAC.49.9.3903-3909.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.In C. Performance standards for antimicrobial susceptibility testing. Wayne, PA: Clinial and Laboratory Standards Institute; 2018. [Google Scholar]
- 22.Hornsey M, Ellington MJ, Doumith M, et al. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J Antimicrob Chemother. 2010;65(3):479–482. doi: 10.1093/jac/dkp475 [DOI] [PubMed] [Google Scholar]
- 23.Xu Q, Jiang J, Zhu Z, et al. Efflux pumps AcrAB and OqxAB contribute to nitrofurantoin resistance in an uropathogenic Klebsiella pneumoniae isolate. Int J Antimicrob Agents. 2019;54(2):223–227. doi: 10.1016/j.ijantimicag.2019.06.004 [DOI] [PubMed] [Google Scholar]
- 24.Bi D, Jiang X, Sheng Z-K, et al. Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J Antimicrob Chemother. 2015;70(10):2770–2774. doi: 10.1093/jac/dkv204 [DOI] [PubMed] [Google Scholar]
- 25.Nicolas-Chanoine M-H, Mayer N, Guyot K, et al. Interplay between membrane permeability and enzymatic barrier leads to antibiotic-dependent resistance in Klebsiella pneumoniae. Frontiers in Microbiology. 2018;9:1422. doi: 10.3389/fmicb.2018.01422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam H-K, Malviya VN, Foong W-E, et al. Binding and transport of carboxylated drugs by the multidrug transporter AcrB. J Mol Biol. 2020;432(4):861–877. doi: 10.1016/j.jmb.2019.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y, Sato T, Fukushima Y, et al. Contribution of β-lactamase and efflux pump overproduction to tazobactam-piperacillin resistance in clinical isolates of Escherichia coli. Int J Antimicro Agents. 2020;55(4):105919. doi: 10.1016/j.ijantimicag.2020.105919 [DOI] [PubMed] [Google Scholar]
- 28.Mazzariol A, Cornaglia G, Nikaido H. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob Agents Chemother. 2000;44(5):1387–1390. doi: 10.1128/AAC.44.5.1387-1390.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foong WE, Wilhelm J, Tam H-K, et al. Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J Antimicrob Chemother. 2020;75(5):1135–1139. doi: 10.1093/jac/dkaa015 [DOI] [PubMed] [Google Scholar]
- 30.Du X, He F, Shi Q, et al. The rapid emergence of tigecycline resistance in <sub>blaKPC–2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front Microbiol. 2018;9:648. doi: 10.3389/fmicb.2018.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Majumdar S, Yu J, Fookes M, et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 2015;11(1):e1004627. doi: 10.1371/journal.ppat.1004627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye M, Ding B, Qian H, et al. In vivo development of tigecycline resistance in Klebsiella pneumoniae owing to deletion of the ramR ribosomal binding site. Int J Antimicrob Agents. 2017;50(4):523–528. doi: 10.1016/j.ijantimicag.2017.04.024 [DOI] [PubMed] [Google Scholar]
- 33.Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli J Bacteriol. 2005;187(6):1923–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido H, Basina M, Nguyen V, et al. Multidrug efflux pump AcrAB of Salmonella typhimuriumexcretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180(17):4686–4692. doi: 10.1128/JB.180.17.4686-4692.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hocquet D, Nordmann P, El Garch F, et al. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicro Agents Chemother. 2006;50(4):1347–1351. doi: 10.1128/AAC.50.4.1347-1351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laudy AE, Osinska P, Namyslowska A, et al. Modification of the susceptibility of Gram-negative rods producing ESβLS to β-lactams by the efflux phenomenon. PLoS One. 2015;10(3):e0119997. doi: 10.1371/journal.pone.0119997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson K, Hemarajata P, Sun D, et al. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother. 2017;61(10):e00989–17. doi: 10.1128/AAC.00989-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juan CH, Huang YW, Lin YT, et al. Risk factors, outcomes, and mechanisms of tigecycline-nonsusceptible Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother. 2016;60(12):7357–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]