Abstract
The calcium-calmodulin–dependent protein kinase kinase-2 (CaMKK2) is a key regulator of cellular and whole-body energy metabolism. It is known to be activated by increases in intracellular Ca2+, but the mechanisms by which it is inactivated are less clear. CaMKK2 inhibition protects against prostate cancer, hepatocellular carcinoma, and metabolic derangements induced by a high-fat diet; therefore, elucidating the intracellular mechanisms that inactivate CaMKK2 has important therapeutic implications. Here we show that stimulation of cAMP-dependent protein kinase A (PKA) signaling in cells inactivates CaMKK2 by phosphorylation of three conserved serine residues. PKA-dependent phosphorylation of Ser495 directly impairs calcium-calmodulin activation, whereas phosphorylation of Ser100 and Ser511 mediate recruitment of 14-3-3 adaptor proteins that hold CaMKK2 in the inactivated state by preventing dephosphorylation of phospho-Ser495. We also report the crystal structure of 14-3-3ζ bound to a synthetic diphosphorylated peptide that reveals how the canonical (Ser511) and noncanonical (Ser100) 14-3-3 consensus sites on CaMKK2 cooperate to bind 14-3-3 proteins. Our findings provide detailed molecular insights into how cAMP-PKA signaling inactivates CaMKK2 and reveals a pathway to inhibit CaMKK2 with potential for treating human diseases.
Keywords: Ca2+-calmodulin–dependent protein kinase kinase-2 (CaMKK2), calmodulin (CaM), cyclic AMP (cAMP), protein kinase A (PKA), inhibition mechanism, adaptor protein, 14-3-3, Ca2+-calmodulin–dependent protein kinase (CaMK), 14-3-3 protein, Ca2+, calmodulin, CaMKK2, cAMP, PKA
The calcium ion (Ca2+) is a dynamic second messenger that relays signals from ligand-activated receptors and voltage-stimulated ion channels at the cell membrane to regulate a wide array of physiological functions (1). A key transducer of Ca2+ signaling is calmodulin (CaM), a ubiquitous Ca2+-binding protein that regulates the activity of numerous downstream effectors in response to elevations in intracellular Ca2+ (2). Important to the actions of the Ca2+-CaM complex is the Ca2+-CaM–dependent protein kinase kinase-2 (CaMKK2), which is the core component of a phosphorylation signaling pathway that regulates appetite and whole-body energy metabolism (3). CaMKK2 inhibition protects against prostate cancer development, hepatocellular carcinoma, high-fat diet–induced obesity, glucose intolerance, and insulin resistance (4–6); therefore, identifying the intracellular mechanisms that inactivate CaMKK2 has important implications for understanding and treating human diseases.
CaMKK2 regulation involves a complex interplay between allosteric activation by Ca2+-CaM and multisite phosphorylation. It has a modular structure composed of an internal catalytic domain and a regulatory module made up of overlapping autoinhibitory and CaM-binding sequences, flanked by N- and C-terminal sequences of unknown function (7). The autoinhibitory sequence obstructs the catalytic site by an intrasteric mechanism that is relieved by Ca2+-CaM binding, allowing for maximal kinase activity (8). In human CaMKK2, activation by Ca2+-CaM induces Thr85 autophosphorylation, which creates a molecular memory of the Ca2+ signal that keeps CaMKK2 in the activated state after the stimulus has diminished (9, 10). Once activated, CaMKK2 triggers the downstream actions of the Ca2+-CaM–dependent protein kinases 1 and 4 (CaMK1 and CaMK4) and the AMP-activated protein kinase (AMPK) signaling pathways (11–13). CaMKK2 is also regulated by Ca2+-CaM–independent mechanisms involving hierarchical phosphorylation of sequential serine residues (Ser129, Ser133, and Ser137) in the S3-node, a regulatory sequence located N-terminal to the catalytic domain. Phosphorylation of Ser137 by proline-directed kinases primes for sequential phosphorylation of Ser133 and Ser129 by glycogen synthase kinase-3, which inhibits CaMKK2 basal activity (14).
CaMKK2 knockout mice are protected against high-fat diet–induced weight gain, insulin resistance, and glucose intolerance (4). Likewise, deletion of the RIIα regulatory subunit of cAMP-dependent protein kinase A (PKA) in mice (which results in activation of the PKA catalytic subunit) causes a similar phenotype (15, 16); therefore, the physiological regulation of whole-body energy metabolism by PKA may involve some inhibitory cross-talk with the CaMKK2 pathway. Consistent with this idea, it was reported recently that CaMKK2 is inactivated by cAMP-dependent PKA in cell-free assays (17). However, it is unclear to what extent this regulatory mechanism occurs in intact cells. Here, we report that CaMKK2 is inactivated in cells by agonists that stimulate cAMP-PKA signaling, including the appetite suppressant liraglutide. PKA inactivates CaMKK2 by a direct mechanism that impairs Ca2+-CaM activation and an indirect mechanism involving recruitment of 14-3-3 adaptor proteins. Our data reveal how cAMP-PKA signaling negatively regulates CaMKK2 and provides a molecular rationale for using PKA-activating drugs for human diseases associated with inappropriate activation of the CaMKK2 pathway.
Results
cAMP-PKA signaling inactivates CaMKK2 by impairing Ca2+-CaM activation
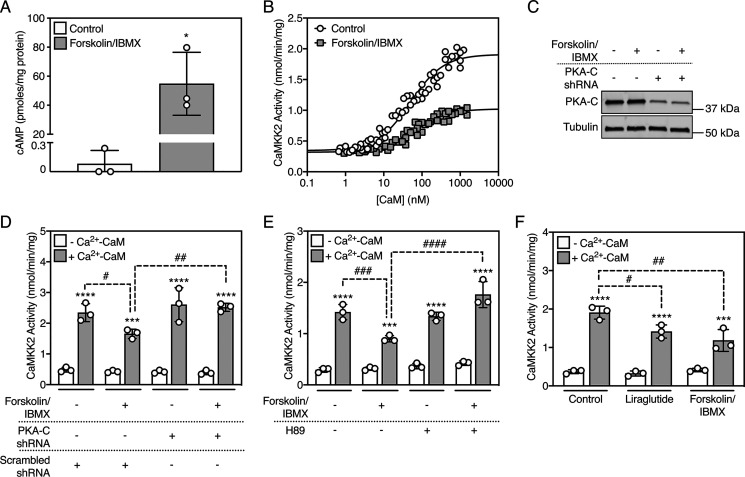
COS7 cells expressing recombinant human CaMKK2 were treated with forskolin and 3-isobutyl-1-methylxanthine (IBMX) to increase intracellular cAMP, after which we immunoprecipitated CaMKK2 and measured kinase activity over a range of CaM concentrations. Forskolin/IBMX treatment increased intracellular cAMP (680-fold over control) and impaired CaMKK2 activation by Ca2+-CaM (Fig. 1, A and B), whereas the concentration of CaM required for half-maximal activation and kinase activity in the absence of Ca2+-CaM were not significantly affected (Table 1). Co-expression with a small-hairpin RNA that decreased expression of the PKA α catalytic subunit (Fig. 1, C and D) or pretreating cells with the PKA inhibitor H89 (Fig. 1E) completely prevented forskolin/IBMX-induced inactivation of CaMKK2.
Figure 1.
cAMP-PKA–dependent signaling inactivates CaMKK2 in cells. A, cAMP levels in COS7 cells treated with vehicle (DMSO) or 50 µm forskolin/500 µm IBMX for 20 min. Data are mean ± S.D. (error bars), n = 3 independent experiments, and statistically appraised using the two-tailed, unpaired t test. *p = 0.012. B, Ca2+-CaM activation of CaMKK2 from transfected COS7 cells treated with vehicle (DMSO) or 50 µm forskolin/500 µm IBMX for 20 min. CaMKK2 was immunoprecipitated and activity was measured over a range of CaM concentrations (0–1000 nm) in the presence of 100 µm Ca2+. The data were fitted to the equation Activity = Basal + [CaM] × ((Maximum-Basal)/(A0.5 + [CaM])), where A0.5 is the concentration of CaM that gives half-maximal stimulation. n = 6 independent experiments. C, expression of endogenous PKA catalytic subunit in COS7 cells transfected with either scrambled or a PKA-Cα subunit transcript-targeted shRNA plasmid, then treated with vehicle (DMSO) or 50 µm forskolin/500 µm IBMX for 20 min. PKA-Cα and α-tubulin were detected using rabbit anti-PKA-Cα and mouse anti-α-tubulin antibodies, respectively. A representative immunoblot is shown. D, Ca2+-CaM activation of CaMKK2 from transfected COS7 cells cotransfected with PKA-Cα subunit transcript-targeted shRNA plasmid (or scrambled control plasmid), then treated with vehicle (DMSO) or 50 µm forskolin/500 µm IBMX for 20 min. CaMKK2 was immunoprecipitated and activity was measured in the presence or absence of 100 µm Ca2+ and 1 µm CaM. Data are mean ± S.D. (error bars), n = 3 independent experiments, and were statistically appraised by two-way ANOVA (treatment: F = 4.935, p = 0.0130; Ca2+-CaM: F = 369.2, p < 0.0001; interaction: F = 5.168, p = 0.0109) using Tukey's post-hoc multiple comparisons test. ****p < 0.0001, ***p = 0.0002, ##p = 0.0069, #p = 0.0399. E, Ca2+-CaM activation of CaMKK2 from transfected COS7 cells pretreated with the 20 µm H89 for 60 min, then treated with 50 µm forskolin/500 µm IBMX for 20 min. CaMKK2 was immunoprecipitated and activity was measured in the presence or absence of 100 µm Ca2+ and 1 µm CaM. Data are mean ± S.D. (error bars), n = 3 independent experiments, and were statistically appraised by two-way ANOVA (treatment: F = 17.89, p < 0.0001; Ca2+-CaM: F = 462.9, p < 0.0001; interaction: F = 11.68, p = 0.0003) using Tukey's post-hoc multiple comparisons test. ****p < 0.0001, ***p = 0.0002, ####p < 0.0001, ###p = 0.0009. F, Ca2+-CaM activation of CaMKK2 from transfected SH-SY5Y cells treated with either vehicle (DMSO), 100 nm liraglutide, or 50 µm forskolin/500 µm IBMX for 20 min. CaMKK2 was immunoprecipitated and activity was measured in the presence or absence of 100 µm Ca2+ and 1 µm CaM. Data are mean ± S.D. (error bars), n = 3 independent experiments, and were statistically appraised by two-way ANOVA (treatment: F = 7.854, p < 0.0066; Ca2+-CaM: F = 232.7, p < 0.0001; interaction: F = 9.022, p = 0.0041) using Tukey's post-hoc multiple comparisons test. ****p < 0.0001, ***p = 0.0007, ##p = 0.0012, #p = 0.0226.
Table 1.
cAMP-PKA–dependent signaling impairs CaMKK2 activation by Ca2+-CaM
Ca2+-CaM activation of CaMKK2 from transfected COS7 cells treated with vehicle (DMSO) or 50 µm forskolin/500 µm IBMX for 20 mins. CaMKK2 was immunoprecipitated and activity was measured over a range of CaM concentrations (0–1000 nM) in the presence of 100 µm Ca2+. The data were fitted to the equation Activity = Basal + [CaM] × ((Maximum-Basal)/(A0.5 + [CaM])), where A0.5 is the concentration of CaM that gives half-maximal activation. Data are n = 6 independent experiments. Best-fit values calculated for each kinetic parameter was statistically appraised using extra sum-of-squares F test. ****p < 0.0001 versus maximal activity of CaMKK2 from control cells.
| Treatment | Basal activity (nmol/min/mg) | Maximal activity (nmol/min/mg) | A0.5 (nm) |
|---|---|---|---|
| Control | 0.344 | 1.909 | 49.68 |
| Forskolin/IBMX | 0.318 | 1.022**** | 64.52 |
Activation of the glucagon-like peptide-1 (GLP-1) receptor decreases food intake and promotes weight loss via stimulation of PKA activity (18). Therefore, we examined the effect of the GLP-1 receptor agonist liraglutide on CaMKK2 activity in SH-SY5Y neuroblastoma cells that endogenously express the GLP-1 receptor (19). Similar to the effects of forskolin/IBMX, liraglutide diminished Ca2+-CaM activation but had no effect on CaMKK2 activity in the absence of Ca2+-CaM (Fig. 1F).
PKA-mediated inactivation of CaMKK2 depends on Ser495 phosphorylation
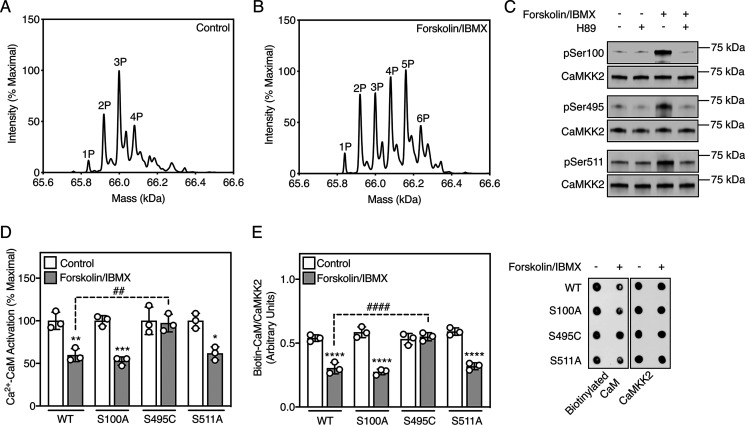
To investigate the mechanism by which cAMP-PKA signaling inactivates CaMKK2, we determined the phosphorylation profile of CaMKK2 in cells under control and PKA-activated conditions. Cells expressing human CaMKK2 were treated with or without forskolin/IBMX, after which CaMKK2 was purified and analyzed by whole-protein TOF MS. Under control conditions, the primary mass peak corresponded to a triply phosphorylated CaMKK2 species (Fig. 2A) as previously reported (14, 20). In contrast, CaMKK2 purified from forskolin/IBMX-treated cells showed a higher range of mass peaks, consistent with increased phosphorylation on at least three additional sites (Fig. 2B). Using tandem MS, we compared the spectra of tryptic peptides from CaMKK2 purified from control and forskolin/IBMX-treated cells and found that Ser100, Ser495 (which resides within the CaM-binding sequence), and Ser511 were phosphorylated in CaMKK2 from the forskolin/IBMX-treated cells but not from the control cells (Fig. S1). All three sites match the PKA phosphorylation consensus motif (Arg/Lys-Arg/Lys-X-Ser/Thr, where X is any amino acid) (21) and are conserved across a diverse range of species (Fig. S2). Using validated phospho-specific antibodies (Fig. S3), we confirmed PKA-dependent phosphorylation of all three serine residues by immunoblot (Fig. 2C).
Figure 2.
PKA signaling inactivates CaMKK2 by phosphorylating Ser495 in the CaM-binding sequence. A, representative whole-protein TOF spectra of CaMKK2 purified from transfected COS7 cells treated with DMSO vehicle control for 20 min. B, representative whole-protein TOF spectra of CaMKK2 purified from transfected COS7 cells treated with 50 µm forskolin/500 µm IBMX for 20 min. C, phosphorylation of CaMKK2 on Ser100, Ser495, and Ser511 purified from transfected COS7 cells pretreated with 20 µm H89 for 60 min, then treated with 50 µm forskolin/500 µm IBMX for 20 min. Phosphorylation and total CaMKK2 were detected using rabbit phospho-specific and mouse anti-FLAG antibodies, respectively. A representative immunoblot is shown. D, Ca2+-CaM activation of WT CaMKK2 and phosphorylation site mutants from transfected COS7 cells treated with DMSO vehicle control or 50 µm forskolin/500 µm IBMX for 20 min. CaMKK2 was immunoprecipitated and activity was measured in the presence or absence of 100 µm Ca2+ and 1 µm CaM. Data are mean ± S.D. (error bars), n = 3 independent experiments, and were statistically appraised by two-way ANOVA (treatment: F = 68.56, p < 0.0001; mutation: F = 6.624, p = 0.0041; interaction: F = 6.627, p = 0.0041) using Tukey's post-hoc multiple comparisons test. ***p = 0.0004, **p = 0.0018, *p = 0.0029, ##p = 0.0035. E, CaM overlay assay measuring binding of biotinylated CaM to WT CaMKK2 and phosphorylation site mutants purified from transfected COS7 cells treated with vehicle control (DMSO) or 50 µm forskolin/500 µm IBMX for 20 min. CaM binding and total CaMKK2 were visualized using fluorescent-labeled streptavidin and rabbit anti-FLAG antibody, respectively. Data are mean ± S.D. (error bars), n = 3 independent experiments, and were statistically appraised by two-way ANOVA (treatment: F = 200.3, p < 0.0001; mutation: F = 14.96, p < 0.0001; interaction: F = 27.86, p < 0.0001) using Tukey's post-hoc multiple comparisons test. ****p < 0.0001, ####p < 0.0001.
The function of each site was determined by treating cells expressing nonphosphorylatable mutants (S100A, S495C, and S511A) with forskolin/IBMX, after which we measured kinase activity in the presence and absence of Ca2+-CaM. We substituted Ser495 for cysteine rather than alanine because the S495A mutation impaired binding of Ca2+-CaM (Fig. S4). The S495C mutant was activated by Ca2+-CaM with similar kinetic parameters to WT CaMKK2 (Fig. S5). WT CaMKK2 and the S100A and S511A mutants from forskolin/IBMX-treated cells displayed decreased Ca2+-CaM activation; however, the S495C mutant was unaffected (Fig. 2D). This indicates that phosphorylation of Ser495 is entirely responsible for suppressing Ca2+-CaM activation in response to PKA signaling. Mirroring the impaired Ca2+-CaM activation, WT CaMKK2 and the S100A and S511A mutants from forskolin/IBMX-treated cells displayed decreased Ca2+-CaM binding, but the S495C mutant did not (Fig. 2E).
PKA-dependent phosphorylation of CaMKK2 promotes binding to 14-3-3 adaptor proteins
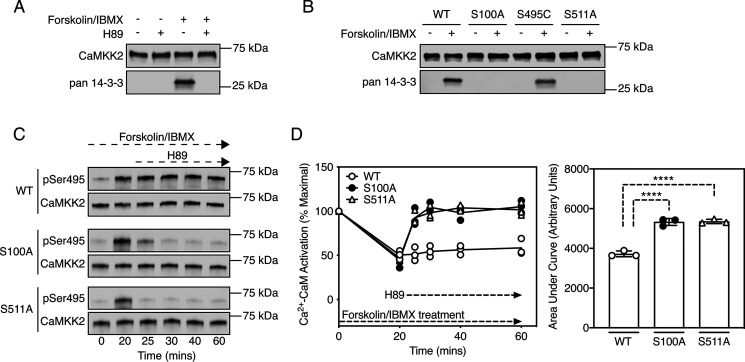
Analysis of the phosphorylation sites indicated that the sequence surrounding Ser511 conforms to a canonical 14-3-3 protein–binding motif (mode-1 motif RSXpSXP, where X is any amino acid) (22). We therefore immunoprecipitated CaMKK2 from cells treated with forskolin/IBMX and tested for copurification of endogenous 14-3-3 proteins by immunoblot. Fig. 3A shows that forskolin/IBMX treatment induced binding of 14-3-3 proteins to CaMKK2 but not in cells pretreated with H89. We detected binding of all seven isoforms of 14-3-3 by tandem MS (Table S1). 14-3-3 binding depends upon phosphorylation of both Ser100 and Ser511 (but not Ser495), because the S100A and S511A mutations were individually sufficient to abolish copurification of 14-3-3 proteins with CaMKK2 (Fig. 3B).
Figure 3.
PKA-dependent phosphorylation of Ser100 and Ser511 mediates binding of CaMKK2 to 14-3-3 adaptor proteins. A, binding of endogenous 14-3-3 proteins to WT CaMKK2 purified from transfected COS7 cells pretreated with 20 µm H89 for 60 min, followed by treatment with DMSO (vehicle control) or 50 µm forskolin/500 µm IBMX for 20 min. 14-3-3 protein and total CaMKK2 were detected using rabbit anti-pan-14-3-3 and mouse anti-FLAG antibodies, respectively. A representative immunoblot is shown. B, binding of endogenous 14-3-3 proteins to WT CaMKK2 and phosphorylation site mutants purified from transfected COS7 cells treated with DMSO (vehicle control) or 50 µm forskolin/500 µm IBMX for 20 min. 14-3-3 proteins and total CaMKK2 were detected using rabbit anti-pan-14-3-3 and mouse anti-FLAG antibodies, respectively. A representative immunoblot is shown. C, Ser495 phosphorylation of WT CaMKK2 and the S100A and S511A mutants purified from transfected COS7 cells treated with 50 µm forskolin/500 µm IBMX over a 60-min time course, with addition of 20 µm H89 to the cells at the 20-min time point. Ser495 phosphorylation and total CaMKK2 were detected using rabbit phospho-specific and mouse anti-FLAG antibodies, respectively. A representative immunoblot is shown. D, Ca2+-CaM activation of WT CaMKK2 and the S100A and S511A mutants from transfected COS7 cells treated with 50 µm forskolin/500 µm IBMX over a 60-min time course, with addition of 20 µm H89 to the cells at the 20-min time point. CaMKK2 was immunoprecipitated and activity was measured in the presence of 100 µm Ca2+ and 1 µm CaM. n = 3 independent experiments. The area under the curve is displayed as mean ± S.D. (error bars) and was statistically appraised by one-way ANOVA (F = 122.7, p < 0.0001) using Tukey's post-hoc multiple comparisons test. ****p < 0.0001.
A recent study reported that 14-3-3 binding to CaMKK2 impaired dephosphorylation of phospho-Ser495 (pSer495) by protein phosphatase-1 in a cell-free assay (17), indicating that a potential biological function of 14-3-3 binding is to maintain CaMKK2 in the inactivated state by protecting pSer495 from dephosphorylation by protein phosphatases. To test whether this mechanism occurs in a cellular context, cells expressing WT CaMKK2 or the S100A and S511A mutants were treated with forskolin/IBMX, after which we measured Ser495 phosphorylation and Ca2+-CaM activation at various time points following incubation with H89. Forskolin/IBMX stimulated Ser495 phosphorylation and suppressed Ca2+-CaM activation of WT CaMKK2, which was sustained over the treatment period even after the addition of H89. In contrast, pSer495 was dephosphorylated and maximal activation was restored within 5 min for the S100A and S511A mutants in response to H89 treatment (Fig. 3, C and D). The simplest interpretation of these data is that 14-3-3 binding protects against pSer495 dephosphorylation in cells.
Dual phosphorylation of CaMKK2 increases binding affinity for 14-3-3 proteins
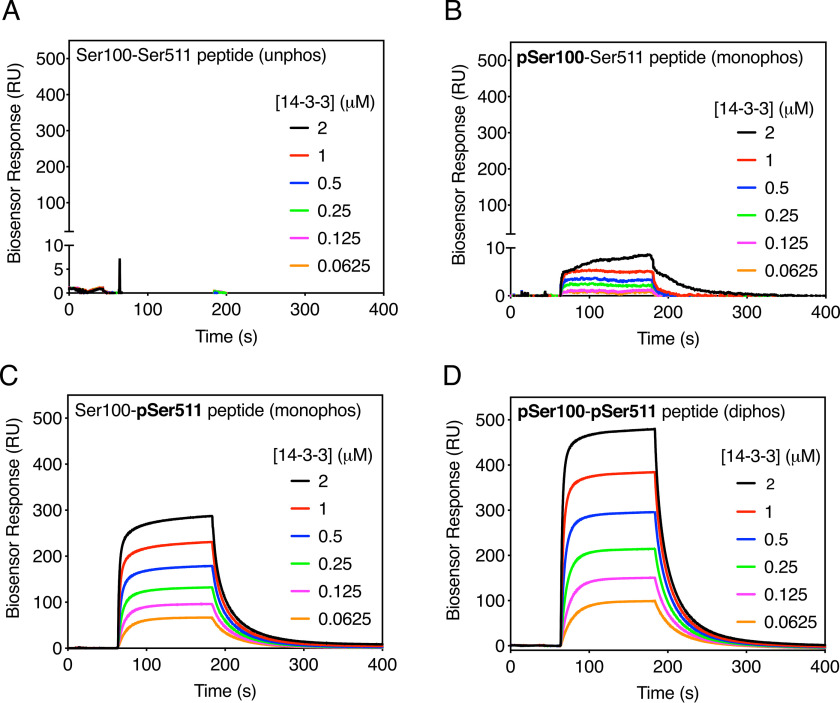
14-3-3 proteins exist functionally as dimers and therefore contain two phosphopeptide-binding pockets that can interact with two sites simultaneously, often on the same target protein (23). For 14-3-3 target proteins containing two phosphorylation sites, there is generally a primary high-affinity site that initiates 14-3-3 recruitment by binding to one monomer in the 14-3-3 dimer, which then permits binding of the weaker secondary site to the adjoining monomer to stabilize the overall interaction (24). To simulate the interaction between CaMKK2 and 14-3-3, we used surface plasmon resonance to measure 14-3-3 binding to a series of synthetic phosphopeptides corresponding to the unphosphorylated (Ser100-Ser511), monophosphorylated (pSer100-Ser511 and Ser100-pSer511), and diphosphorylated (pSer100-pSer511) species of CaMKK2 (Fig. S6). As expected, we did not observe any 14-3-3 binding to the unphosphorylated Ser100-Ser511 peptide (Fig. 4A). The pSer100-Ser511 peptide (Fig. 4B) showed modest binding to 14-3-3 (KD 1.91 µm) relative to the Ser100-pSer511 peptide, which displayed a 5-fold increase in binding affinity (KD 0.35 µm) consistent with pSer511 conforming to the canonical 14-3-3 binding motif (Fig. 4C). Phosphorylation of both sites further strengthened binding to 14-3-3 (KD 0.09 µm), displaying a 4-fold increase in affinity over the pSer511 site alone (Fig. 4D).
Figure 4.
Dual phosphorylation of Ser100 and Ser511 increases 14-3-3 binding affinity. A, surface plasmon resonance (SPR) sensorgram of 14-3-3 binding to the unphosphorylated Ser100-Ser511 peptide over a range of 14-3-3 concentrations. Data are the mean of three independent experiments. B, SPR sensorgram of 14-3-3 binding to the monophosphorylated pSer100-Ser511 peptide over a range of 14-3-3 concentrations. Data are the mean of three independent experiments. C, SPR sensorgram of 14-3-3 binding to the monophosphorylated Ser100-pSer511 peptide over a range of 14-3-3 concentrations. Data are the mean of three independent experiments. D, SPR sensorgram of 14-3-3 binding to the diphosphorylated pSer100-pSer511 peptide over a range of 14-3-3 concentrations. Data are the mean of three independent experiments.
Structure of a 14-3-3-diphosphopeptide complex
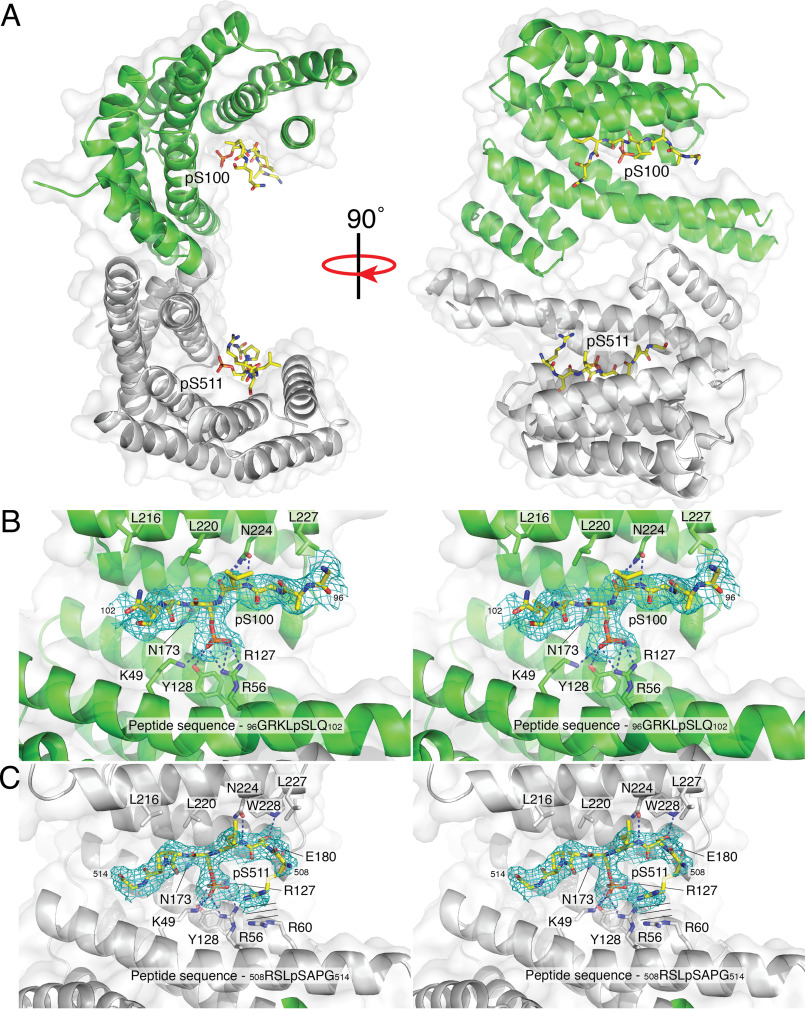
To further investigate the interaction between CaMKK2 and 14-3-3, we determined the crystal structure of 14-3-3ζ in complex with the diphosphorylated pSer100-pSer511 peptide to a resolution of 2.44 Å (Table S2). The unit cell contained four molecules of 14-3-3, arranged in a back-to-back dimer of dimers, with a 180° rotation with respect to the other dimer. Each dimer contained two phosphopeptide-binding sites, both of which showed strong electron density for the critical pSer100 and pSer511 residues (Fig. 5A). We did not observe an admixture of the pSer100-pSer511 peptide bound in both possible orientations to the 14-3-3 dimer; therefore, we were able to unambiguously place pSer100 and pSer511 into the phosphopeptide-binding sites of discrete monomers within the dimeric complex. Coordination of the phosphoserines was conserved in both binding sites (mediated by Lys-4914-3-3, Arg-5614-3-3, Arg-12714-3-3, and Tyr-12814-3-3); however, the pSer511 end of the peptide bound in site 1 makes considerably more interactions through its main chain and side chain atoms compared with the pSer100 end in site 2 (Fig. 5, B and C). The side chain of Ser509 forms hydrogen bonds with side chains of Tyr-22814-3-3 and Glu-18014-3-3, and the side chain of Arg508 makes parallel planar stacking interactions with Arg-6014-3-3. Additionally, the main chain amine from Ser509 and Leu510 form hydrogen bonds with Glu-18014-3-3 and Asn-22414-3-3, respectively. In contrast, only residues Leu99 and Leu101 on the pSer100 end make hydrogen bonds with 14-3-3 via main chain interactions with the side chains of Asn-22414-3-3 and Asn-17314-3-3, respectively.
Figure 5.
Structure of a 14-3-3 diphosphopeptide complex. A, surface representation of the 14-3-3 dimer (monomers containing the pSer100 and pSer511 binding sites are shown in green and gray, respectively) with secondary structure shown in ribbons. The pSer100 and pSer511 ends of the diphosphorylated peptide are shown as sticks in yellow. B, stereo image of difference maps of the pSer100 binding site. C, stereo image of difference maps of the pSer511 binding site. The parallel lines represent the parallel stacking interactions between Arg508 on the diphosphopeptide and Arg60 on 14-3-3.
Discussion
Herein, we report that stimulation of cAMP-PKA signaling in cells inactivates CaMKK2 by a mechanism involving tripartite phosphorylation of conserved serine residues and recruitment of 14-3-3 adaptor proteins. We found that phosphorylation of Ser495, a highly conserved site located within the CaM-binding sequence, directly impairs Ca2+-CaM binding and activation of CaMKK2. A similar mechanism of inhibitory cross-talk by the cAMP-PKA pathway, involving direct phosphorylation of CaM-binding sites, has been reported for other Ca2+-CaM–regulated proteins including CaMKK1, β-adducin, and the Ca2+-dependent K+ channel KCa3.1 (25–27). This provides a multi-level system for fine-tuning Ca2+ signaling by allowing the magnitude and duration of signal transmission to be modulated (28).
As well as impairing Ca2+-CaM activation via Ser495 phosphorylation, PKA-dependent phosphorylation of Ser100 and Ser511 mediates binding of 14-3-3 proteins, which keep CaMKK2 in the inactivated state by protecting against pSer495 dephosphorylation by cellular protein phosphatases. The absolute requirement for phosphorylation on both sites for 14-3-3 binding and phosphatase protection is consistent with the “gatekeeper” model of 14-3-3 engagement with target proteins (24). In this model, 14-3-3 target proteins that contain two phospho-binding sites generally have a primary high-affinity site that functions as the gatekeeper and a secondary lower affinity site that is not only required to stabilize the overall interaction but is also essential for the biological effect of 14-3-3 binding. Our data indicate that pSer511 is the gatekeeper site on CaMKK2 because of its greater 14-3-3 binding affinity compared with the pSer100 site, which is in accordance with the pSer511 site matching the canonical 14-3-3 consensus binding motif (23).
Our crystal structure of 14-3-3 complexed with a diphosphorylated peptide that simulates simultaneous binding of the pSer100 and pSer511 sites revealed that each site occupies the phosphopeptide-binding groove of discrete 14-3-3 monomers within the dimeric complex. Strong electron density was observed for the residues directly adjacent to the phosphoserine residues (five residues surrounding pSer100 and seven residues surrounding pSer511), whereas no electron density was visible for the flexible glycine linker region of the peptide. We also observed distinct binding of the diphosphorylated peptide to specific 14-3-3 monomers within the dimer, such that the gatekeeper pSer511 was always bound to 14-3-3 monomers B and D, whereas pSer100 was always bound to monomers A and C. This peculiarity is common to all tandem phosphopeptides/14-3-3 complex structures solved to date (29–33). Although the structures of all four monomers are highly alike (RMSD < 0.35 Å for monomers A, B, C, and D), there are perhaps subtle structural changes that allow the crystal to pack with the 14-3-3 dimer in the same orientation with respect to the phosphorylated peptide (pSer511 versus pSer100).
The 14-3-3 dimer has been described as a molecular anvil because it maintains its overall structure upon ligand binding while distorting the ligand to fit the phosphopeptide-binding groove (24). Consequently, the 14-3-3 interaction forces a new conformation on the target protein to regulate activity, localization, post-translation modifications, or protein-protein interactions. In the case of CaMKK2, 14-3-3 protein binding does not directly alter kinase activity because forskolin/IBMX treatment failed to impair Ca2+-CaM activation of the S495C mutant, despite the fact that 14-3-3 binding was unaffected. Rather, the effect of 14-3-3 binding is purely steric hindrance to protect pSer495 from dephosphorylation.
CaMKK1 (the closest human homologue to CaMKK2) is inhibited by PKA by a mechanism similar to CaMKK2 but with some notable differences. Unlike CaMKK2, there are two inhibitory PKA phosphorylation sites on CaMKK1, i.e. Ser458 located within the CaM-binding sequence and Thr108 (25, 34). Regarding the latter, the corresponding Thr145 residue in CaMKK2 has been reported to be phosphorylated in cells by AMPK rather than PKA, as part of an inhibitory feedback loop mechanism (35). This is consistent with our data, which shows that mutation of Ser495 is sufficient to prevent inactivation of CaMKK2 by PKA, reinforcing the view that Thr145 is not an inhibitory PKA phosphorylation site in cells. PKA also promotes binding of 14-3-3 proteins to CaMKK1 via phosphorylation of Ser74 and Ser475, which correspond to the Ser100 and Ser511 sites in CaMKK2, respectively. In contrast to CaMKK2, 14-3-3 protein binding directly inhibits CaMKK1 activity and blocks dephosphorylation of pThr108 (36, 37).
The physiological function of the PKA-CaMKK2 signaling axis is unclear; however, evidence in the literature hints at potential roles in metabolic control. For example, PKA RIIα regulatory subunit knockout mice that exhibit constitutive PKA catalytic subunit activation are protected against high-fat diet–induced weight gain, glucose intolerance, and insulin resistance (15, 16). This is similar to the phenotype displayed by CaMKK2 knockout mice (4), indicating that PKA-dependent regulation of whole-body energy metabolism may involve inactivation of CaMKK2. This concept is supported by our finding that CaMKK2 was inactivated in cells by the GLP-1 receptor agonist liraglutide, which is used clinically to treat obesity and type 2 diabetes. The metabolic effects of liraglutide (appetite suppression, weight loss, and increased insulin sensitivity) are likewise similar to the metabolic phenotype observed in CaMKK2 knockout mice (4, 18), raising the possibility that some of the therapeutic benefits of liraglutide may also be mediated by the PKA-CaMKK2 pathway. A recent study found that PKA negatively regulates vascular endothelial growth factor–induced AMPK activation (a pro-angiogenic signaling pathway) via CaMKK2, indicating a potential role of the PKA-CaMKK2 axis in the control of angiogenesis (38).
Inappropriate activation of CaMKK2 plays a major role in the development of prostate cancer and hepatocellular carcinoma (5, 39, 40). CaMKK2 activation has also been implicated in acquired resistance of high-grade serous ovarian cancer to chemotherapy (41). Therefore, drugs that stimulate cAMP-PKA signaling may offer potential new treatment strategies to inactivate CaMKK2 in these cancers. Indeed, the GLP-1 receptor agonist Exendin-4 has been reported to decrease proliferation of prostate cancer cells by a PKA-dependent mechanism and attenuate prostate cancer growth in a tumor xenograft mouse model (42). Exendin-4 was also shown to inhibit migration and promote apoptosis in ovarian cancer cells (43). Therefore, PKA-dependent inactivation of CaMKK2 may provide a potential mechanism for the anti-proliferative effects of Exendin-4 in both prostate and ovarian cancers.
In summary, we described a molecular mechanism by which intracellular activation of cAMP-PKA signaling inactivates CaMKK2. Further studies are required to uncover the physiological relevance of the PKA-CaMKK2 signaling node; however, our data reveal a pathway that can potentially be targeted for human diseases associated with aberrant activation of CaMKK2.
Experimental procedures
Cyclic AMP measurements
All cyclic AMP measurements were acquired using LC–MS from perchlorate extracts of forskolin- and IBMX (Sigma-Aldrich)-treated COS7 cells, using an ABSCIEX 5500 mass spectrometer operated with the turbo V ion source coupled to a Shimadzu Prominence LC-20AD UFLC pump. LC conditions were optimized for a 50-mm (length) and 2.1-mm (inner diameter) C18 column (5 µm, Vydac). The LC solvent system was (A) 100% H2O and (B) 100% acetonitrile. cAMP was eluted at a flow rate of 300 µl/min in a gradient program consisting of 100% A (5 min) and 0–70% B (10 min). Data were analyzed with Multiquant 2.0.2 utilizing the area under the LC chromatogram for the corresponding cAMP peak. Calibration curves were obtained by linear regression of the peak area ratio of a cAMP standard (Sigma-Aldrich). All data were acquired in negative mode.
MS
For TOF whole-protein MS (TOF-MS), recombinant FLAG-tagged CaMKK2 was eluted from anti-FLAG M2 agarose beads using an equal volume of 1 mg/ml FLAG-peptide in 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 10% (v/v) glycerol, then diluted in formic acid to achieve a final concentration of 1.5–2% (v/v) prior to injection onto the LC–MS. The chromatography was performed on an Agilent 6220 ESI-TOF mass spectrometer coupled to an Agilent 1260 BinPump system. CaMKK2 was resolved on an Aeris 3.6 µm WIDEPORE C4 200 Å, LC Column (150 × 2.1 mm, Phenomenex) using an elution gradient of 5–90% acetonitrile at 200 μl/min for 30 min. Buffer A was 0.1% formic acid and buffer B was 100% acetonitrile/0.1% formic acid. The mass spectrometer was set to MS1 acquisition mode and operated in positive mode with a mass range of 100–3200 m/z and scan rate of 1.10. Source gas temperature was set to 325 °C, gas flow 8 liters/min, and nebulizer 45 psi. Mass spectra were deconvoluted using Agilent MassHunter Qualitative Build 6 (v. 6.0.633.0) software.
For tandem MS, eluted CaMKK2 was pH-adjusted using 50 mm triethyl ammonium bicarbonate solution, reduced with 10 mm tris(2-carboxyethyl)phosphine for 45 min at 37 °C, and alkylated with 55 mm iodoacetamide for 30 min at room temperature in the dark. The samples were then digested with trypsin (which cleaves on the C-terminal side of lysine and arginine residues) (1:50, w/w) overnight at 37 °C. Digested tryptic peptides were cleaned up using Oasis HBL solid phase extraction cartridges (Waters Corporation) and freeze-dried overnight. Dried tryptic peptides were resuspended in 0.1% (v/v) formic acid and analyzed by LC–MS/MS using a Q-Exactive plus mass spectrometer (Thermo Fisher Scientific) fitted with nanoflow reversed-phase-HPLC (Ultimate 3000 RSLC, Dionex). The nano-LC system was equipped with an Acclaim Pepmap nano-trap column (Dionex, C18, 100 Å, 75 µm × 2 cm) and an Acclaim Pepmap RSLC analytical column (Dionex, C18, 100 Å, 75 µm × 50 cm). Typically, for each LC–MS/MS experiment, 5 μl of the peptide mix was loaded onto the enrichment (trap) column at an isocratic flow of 5 μl/min of 3% (v/v) acetonitrile containing 0.1% (v/v) formic acid for 6 min before the enrichment column was switched in-line with the analytical column. The eluents used for the LC were 0.1% (v/v) formic acid (solvent A) and 100% acetonitrile/0.1% formic acid (v/v) (solvent B). The gradient used was 3–25% B for 23 min, 25–40% B in 2 min, and 40–80% B in 2 min and maintained at 85% B for the final 2 min before equilibration for 9 min at 3% B prior to the next analysis. All spectra were acquired in positive mode with full scan MS spectra scanning from m/z 375–1400 at 70,000 resolution. Mass spectrometric raw data were converted to centroid peaklists using MSConvert (version 3.0.5047) and searched using Mascot (version 2.4) search algorithm against human SwissProt database (20,282 sequences, November 2015). Trypsin was selected as the protease with two missed cleavages allowed. MS tolerance was set to 10 ppm, MS/MS tolerance at 0.2 Da, and ion score significance threshold was p < 0.05. Cysteine carbamidomethylation was searched as a fixed modification, whereas oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were searched as variable modifications. The tandem MS proteomics raw data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD020133 (44).
Recombinant CaMKK2 expression and purification
Recombinant WT and mutant CaMKK2 was expressed in either COS7 cells or SH-SY5Y cells grown in DMEM or Eagle's Minimum Essential Medium/F-12 medium (Sigma-Aldrich), respectively, supplemented with 10% FCS at 37 °C with 5% CO2. The cells were transfected at 60% confluency using FuGene HD (Roche Applied Science) with 2 µg of pcDNA3(−) plasmid containing N-terminal FLAG-tagged human CaMKK2. For the PKA-Cα shRNA knockdown experiments, the cells were cotransfected with 2 µg of PKA-Cα targeted or scrambled shRNA plasmid (Santa Cruz Biotechnology). After 48 h, transfected cells were treated with or without forskolin and IBMX (or DMSO vehicle control) and harvested by rinsing with ice-cold PBS, followed by rapid lysis in situ using 1 ml of lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 50 mm NaF, 1 mm NaPPi, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, and 1% (v/v) Triton X-100) containing cOmplete protease inhibitor mixture (Roche Applied Science). Insoluble debris was removed by centrifugation and total protein content was quantified using the Bradford assay (Thermo Fisher Scientific). CaMKK2 was purified from 1.5 mg of total cell lysate using 100 µl of anti-FLAG M2 agarose (50% v/v) pre-equilibrated in lysis buffer, followed by successive washes in lysis buffer containing 1 m NaCl, and finally resuspended in 50 mm Hepes-NaOH, pH 7.4. CaMKK2 was eluted off the beads by incubating overnight at 4 °C with 100 µl of FLAG peptide (1 mg/ml) in 50 mm Hepes-NaOH, pH 7.4, and 10% glycerol (v/v).
CaMKK2 activity assay
CaMKK2 activity was determined by phosphorylation of a synthetic peptide as described previously (45). Briefly, recombinant CaMKK2 was immunoprecipitated from 10 µg of transfected cell lysate using 10 µl of anti-FLAG M2 agarose beads (50% (v/v)) (Sigma-Aldrich) and then added to a 30-µl reaction containing assay buffer (50 mm Hepes-NaOH, pH 7.4, 1 mm DTT, and 0.02% (v/v) Brij-35), 200 µm CaMKKtide (GenScript), 200 µm [γ-32P]-ATP (PerkinElmer), and 5 mm MgCl2, with or without 100 µm CaCl2 and 1 µm CaM (Sigma-Aldrich). Reactions were performed at 30 °C and terminated after 10 min by spotting 15 µl onto P81 phosphocellulose paper (GE Lifesciences), followed by extensive washing in 1% phosphoric acid (Sigma-Aldrich). Radioactivity was quantified by liquid scintillation counting. CaMKK2 activities were corrected for minor differences in expression levels by immunoblot analysis of cell lysates.
Immunoblotting
50 µg of transfected cell lysate was denatured in SDS sample buffer and resolved on a precast 4–15% Mini-Protean Gradient gel (Bio-Rad) before transferring onto Immobilon PVDF membrane (Merck Millipore). The membrane was blocked for 30 min in PBS/1% Tween-20 (PBS-T) supplemented with 2% nonfat milk and then incubated for 60 min with either mouse anti-FLAG (Cell Signaling, 8146S, Lot 3, 100 ng/ml), rabbit anti-pSer100 (Kinexus Bioinformatics, PK557, Lot 141107, 100 ng/ml), rabbit anti-pSer495 (Cell Signaling, 16737S, Lot 1, 100 ng/ml), rabbit anti-pSer511 (Cell Signaling, 12818S, Lot 1, 100 ng/ml), mouse anti-α-tubulin (Cell Signaling, 3873S, Lot 12, 100 ng/ml), rabbit anti-14-3-3 (Cell Signaling, 8312S, Lot 2, 100 ng/ml), or rabbit anti-PKA-Cα (Cell Signaling, 5842S, Lot 2, 100 ng/ml) antibodies. The membrane was then briefly washed in PBS-T, followed by incubation with goat anti-rabbit IgG IRDye 680 and goat anti-mouse IgG IRDye 800 (LI-COR) secondary antibodies for 60 min. After successive washing with PBS-T, the membranes were scanned, and the images were quantified with an Odyssey CLx IR Imager (LI-COR).
CaM overlay assay
200 ng of WT CaMKK2 and phosphorylation site mutants (S100A, S495C, and S511A) purified from DMSO vehicle control and forskolin/IBMX-treated COS7 cells were spotted onto nitrocellulose membrane (GE Lifesciences) and allowed to dry for 30 min. The membrane was then blocked in PBS-T supplemented with 2% nonfat milk for 1 h, after which it was incubated overnight at 4 °C with biotinylated CaM (500 nm, Millipore) and mouse anti-FLAG antibody (100 ng/ml) in PBS-T containing 1% nonfat milk and 10 mm CaCl2. The membranes were briefly washed in PBS-T/10 mm CaCl2 and then incubated with IRDye 680–labeled streptavidin and goat anti-mouse IgG IRDye 800 (LI-COR) for 1 h. After successive washing with PBS-T/10 mm CaCl2, the membranes were scanned, and the images were quantified with an Odyssey CLx IR Imager.
14-3-3 protein expression and purification
A cDNA sequence encoding hexahistidine-tagged (His-tag) 14-3-3ζ protein (residues 1–245) was cloned into pET15b vector for expression in BL21 Escherichia coli cells. Protein expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside for 5 h at 37 °C, after which the cells were harvested and lysed in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 10% (v/v) glycerol by sonication. Clarified lysate was applied to a nickel affinity His-trap column (GE Lifesciences) and 14-3-3 protein eluted with 500 mm imidazole. The His-tag was cleaved by overnight incubation with thrombin (Sigma-Aldrich), after which the 14-3-3 protein was further purified on a Hi-trap Q column (GE Lifesciences) and eluted using a NaCl gradient (50–500 mm) in 20 mm Tris-HCl, pH 7.5, followed by size-exclusion chromatography using a Superdex-75 gel filtration column (GE Lifesciences) pre-equilibrated with 10 mm Hepes-NaOH, pH 8.0, and 130 mm NaCl. The isolated 14-3-3ζ protein was estimated to be ≥99% pure by SDS-PAGE.
Surface plasmon resonance binding assays
The interaction between synthetic peptides corresponding to the Ser100 and Ser511 phosphorylation sites and 14-3-3ζ was measured using a BIAcore T200 instrument. N-terminal biotinylated peptides corresponding to unphosphorylated (Ser100-Ser511), monophosphorylated (pSer100-Ser511 and Ser100-pSer511), and diphosphorylated (pSer100-pSer511) species of CaMKK2 (custom synthesized by GenScript) were immobilized on streptavidin sensor chips (SAHC30M, Xantec Bioanalytics, immobilization level ∼0.8 ng/mm2). Various 14-3-3 concentrations (0–2 µm) diluted in HBS-P buffer (10 mm Hepes-NaOH, pH 7.35, 150 mm NaCl, and 0.005% Tween-20) were injected over the sensor surface and binding profiles were recorded in real time, and sensorgrams were generated by subtracting the signals from the blank streptavidin control flow cell. Following analyte injection phase completion (2 min), the dissociation was monitored in HBS-P buffer for 5 min. All flow cells were washed with 10 mm glycine, pH 3.0, to ensure all remaining analytes had been dissociated prior to the next injection cycle. All binding experiments were conducted in triplicate with consistent results. Dissociation constants were calculated by fitting the data to a single site-binding model, except for the diphosphorylated (pSer100-pSer511) peptide, which was fitted to a two-site model (BIAcore T200 evaluation software, version 2.0).
X-ray data collection and structural refinement
14-3-3ζ was crystallized in 100 mm Tris-HCl, pH 8.5, 100 mm MgCl2, 1 mm NiCl2, and 22–28% PEG3350 (w/v) using the hanging drop vapor diffusion method at 4 °C. The diphosphorylated pSer100-pSer511 peptide (custom synthesized by GenScript) (Table S3) was soaked overnight into the 14-3-3 crystals (at a molar ratio of 1:3; protein to peptide). The crystals were cryoprotected in 22% PEG400 (w/v), and X-ray diffraction data were collected on the MX2 beamline at the Australian Synchrotron (46). The crystal diffracted to 2.44 Å resolution in the P65 space group with unit-cell parameters of a = 94.66, b = 94.66, and c = 236.23 (Å) and α = 90, β = 90, and γ = 120 (°) for the 14-3-3-diphosphopeptide complex. Data were processed using XDS and scaled in Aimless (47, 48). The structure was solved by the molecular replacement method using a 14-3-3 dimer (PDB code 1QJB) as the search model (49). The asymmetric unit cell consists of a dimer of dimers, with one dimer represented by chains A and B and the second dimer represented by chains C and D. The two dimers are almost identical with an RMSD of 0.49 Å. In the final structure there are 230 residues in chains A, B, and D and 299 residues in chain C of 14-3-3. An extra N-terminal histidine residue is present in all the chains as a result of cloning of the 14-3-3 gene (denoted as residue 0). Residues 70–74 and 203–21 from chain C, residues 206–211 and 70–72 from chain A, and residues 69–72 from chain B were not modeled because of poor electron density in these regions. The structure was refined using REFMAC and Phenix (50, 51). Of the 27 residues in the diphosphopeptide, 12–14 residues were built into the 2Fo-Fc map in each dimer. The final Rwork and Rfree for the 14-3-3-diphosphopeptide complex were 0.18 and 0.23, respectively. The model had a Molprobity clash score of 5.89 (99th percentile) (52), a protein geometry score of 1.95 (95th percentile), and favored Ramachandran of 97.32% with no outliers.
Statistical analysis
The data are presented as mean values ± S.D. for at least three independent experiments. Statistical analyses were performed using GraphPad Prism (version 8.4.3). T-tests, one-way and two-way analysis of variance (ANOVA), and extra sum-of-squares F tests were used for statistical appraisal, where indicated, in the figure legends.
Data availability
Coordinates and structure factors have been deposited in the PDB with the accession code 6EF5. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (RRID:SCR_003411) with the data set identifier PXD020133.
Supplementary Material
Acknowledgments
X-ray data collection was undertaken on the MX2 beamline at the Australian Synchrotron, part of the Australian Nuclear Science and Technology Organization, using the ACRF detector. We thank the beamline staff for assistance. The 14-3-3ζ expression plasmid was a kind gift from Dr. Joanna Woodcock at the University of South Australia.
This article contains supporting information.
Author contributions—C. G. L., M. T. O., N. X. Y. L., T. A. D., J. S. O., B. E. K., and J. W. S. conceptualization; C. G. L., T. A. D., S. G., K. L., M. W. P., J. S. O., B. E. K., and J. W. S. supervision; C. G. L., B. E. K., and J. W. S. funding acquisition; C. G. L., M. T. O., K. R. N., L. M. M., U. D., A. H., N. X. Y. L., T. A. D., S. G., K. L., M. W. P., J. S. O., B. E. K., and J. W. S. investigation; C. G. L., M. T. O., K. R. N., L. M. M., U. D., A. H., N. X. Y. L., T. A. D., S. G., K. L., M. W. P., J. S. O., and B. E. K. methodology; C. G. L., M. T. O., B. E. K., and J. W. S. writing-original draft; C. G. L., B. E. K., and J. W. S. project administration; C. G. L., M. T. O., L. M. M., A. H., N. X. Y. L., T. A. D., S. G., K. L., M. W. P., J. S. O., B. E. K., and J. W. S. writing-review and editing; M. T. O. formal analysis.
Funding and additional information—This work was supported by National Health and Medical Research Council Grants 1138102 (to J. W. S.) and 1145265 (to J. S. O.) and the Jack Brockhoff Foundation Grant JBF-4206, 2016 (to C. G. L.). C. G. L. is a National Health and Medical Research Council Early Career Research Fellow and M. W. P. a National Health and Medical Research Council Senior Principal Research Fellow. B. E. K. was a National Health and Medical Research Council Fellow supported by the Australian Research Council (DP170101196). This project was supported in part by the Victorian Government's Operational Infrastructure Support Program. MS data were acquired at the Mass Spectrometry and Proteomics Facility, Bio21, University of Melbourne. We acknowledge the funding support of the Australian Cancer Research Foundation toward the ACRF Rational Drug Discovery Centre.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CaM
- calmodulin
- CaMK
- Ca2+-calmodulin dependent protein kinase
- PKA
- protein kinase A
- AMPK
- AMP-activated protein kinase
- IBMX
- 3-isobutyl-1-methylxanthine
- GLP
- glucagon-like peptide
- ANOVA
- analysis of variance
- SPR
- surface plasmon resonance.
References
- 1. Clapham D. E. (2007) Calcium signaling. Cell 131, 1047–1058 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 2. Hoeflich K. P., and Ikura M. (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742 10.1016/S0092-8674(02)00682-7 [DOI] [PubMed] [Google Scholar]
- 3. Marcelo K. L., Means A. R., and York B. (2016) The Ca2+/calmodulin/CaMKK2 axis: nature's metabolic CaMshaft. Trends Endocrinol. Metab. 27, 706–718 10.1016/j.tem.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson K. A., Ribar T. J., Lin F., Noeldner P. K., Green M. F., Muehlbauer M. J., Witters L. A., Kemp B. E., and Means A. R. (2008) Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 7, 377–388 10.1016/j.cmet.2008.02.011 [DOI] [PubMed] [Google Scholar]
- 5. Lin F., Marcelo K. L., Rajapakshe K., Coarfa C., Dean A., Wilganowski N., Robinson H., Sevick E., Bissig K. D., Goldie L. C., Means A. R., and York B. (2015) The CaMKK2/CaMKIV relay is an essential regulator of hepatic cancer. Hepatology 62, 505–520 10.1002/hep.27832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Penfold L., Woods A., Muckett P., Nikitin A. Y., Kent T. R., Zhang S., Graham R., Pollard A., and Carling D. (2018) CAMKK2 promotes prostate cancer independently of AMPK via increased lipogenesis. Cancer Res. 78, 6747–6761 10.1158/0008-5472.CAN-18-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Racioppi L., and Means A. R. (2012) Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J. Biol. Chem. 287, 31658–31665 10.1074/jbc.R112.356485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tokumitsu H., Wayman G. A., Muramatsu M., and Soderling T. R. (1997) Calcium/calmodulin-dependent protein kinase kinase: identification of regulatory domains. Biochemistry 36, 12823–12827 10.1021/bi971348i [DOI] [PubMed] [Google Scholar]
- 9. Scott J. W., Park E., Rodriguiz R. M., Oakhill J. S., Issa S. M., O'Brien M. T., Dite T. A., Langendorf C. G., Wetsel W. C., Means A. R., and Kemp B. E. (2015) Autophosphorylation of CaMKK2 generates autonomous activity that is disrupted by a T85S mutation linked to anxiety and bipolar disorder. Sci. Rep. 5, 14436 10.1038/srep14436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling N. X. Y., Langendorf C. G., Hoque A., Galic S., Loh K., Kemp B. E., Gundlach A. L., Oakhill J. S., and Scott J. W. (2020) Functional analysis of an R311C variant of Ca2+-calmodulin-dependent protein kinase kinase-2 (CaMKK2) found as a de novo mutation in a patient with bipolar disorder. Bipolar Disord. 1–8 10.1111/bdi.12901 [DOI] [PubMed] [Google Scholar]
- 11. Edelman A. M., Mitchelhill K. I., Selbert M. A., Anderson K. A., Hook S. S., Stapleton D., Goldstein E. G., Means A. R., and Kemp B. E. (1996) Multiple Ca2+-calmodulin-dependent protein kinase kinases from rat brain. Purification, regulation by Ca2+-calmodulin, and partial amino acid sequence. J. Biol. Chem. 271, 10806–10810 10.1074/jbc.271.18.10806 [DOI] [PubMed] [Google Scholar]
- 12. Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., and Hardie D. G. (2005) Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2, 9–19 10.1016/j.cmet.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 13. Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., and Witters L. A. (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280, 29060–29066 10.1074/jbc.M503824200 [DOI] [PubMed] [Google Scholar]
- 14. Green M. F., Scott J. W., Steel R., Oakhill J. S., Kemp B. E., and Means A. R. (2011) Ca2+/calmodulin-dependent protein kinase kinase β is regulated by multisite phosphorylation. J. Biol. Chem. 286, 28066–28079 10.1074/jbc.M111.251504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. London E., Nesterova M., Sinaii N., Szarek E., Chanturiya T., Mastroyannis S. A., Gavrilova O., and Stratakis C. A. (2014) Differentially regulated protein kinase A (PKA) activity in adipose tissue and liver is associated with resistance to diet-induced obesity and glucose intolerance in mice that lack PKA regulatory subunit type IIα. Endocrinology 155, 3397–3408 10.1210/en.2014-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schreyer S. A., Cummings D. E., McKnight G. S., and LeBoeuf R. C. (2001) Mutation of the RIIβ subunit of protein kinase A prevents diet-induced insulin resistance and dyslipidemia in mice. Diabetes 50, 2555–2562 10.2337/diabetes.50.11.2555 [DOI] [PubMed] [Google Scholar]
- 17. Psenakova K., Petrvalska O., Kylarova S., Lentini Santo D., Kalabova D., Herman P., Obsilova V., and Obsil T. (2018) 14-3-3 protein directly interacts with the kinase domain of calcium/calmodulin-dependent protein kinase kinase (CaMKK2). Biochim. Biophys. Acta Gen. Subj. 1862, 1612–1625 10.1016/j.bbagen.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 18. Hayes M. R., Leichner T. M., Zhao S., Lee G. S., Chowansky A., Zimmer D., De Jonghe B. C., Kanoski S. E., Grill H. J., and Bence K. K. (2011) Intracellular signals mediating the food intake-suppressive effects of hindbrain glucagon-like peptide-1 receptor activation. Cell Metab. 13, 320–330 10.1016/j.cmet.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y., Perry T., Kindy M. S., Harvey B. K., Tweedie D., Holloway H. W., Powers K., Shen H., Egan J. M., Sambamurti K., Brossi A., Lahiri D. K., Mattson M. P., Hoffer B. J., Wang Y., et al. (2009) GLP-1 receptor stimulation preserves primary cortical and dopaminergic neurons in cellular and rodent models of stroke and Parkinsonism. Proc. Natl. Acad. Sci. U. S. A. 106, 1285–1290 10.1073/pnas.0806720106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Brien M. T., Oakhill J. S., Ling N. X., Langendorf C. G., Hoque A., Dite T. A., Means A. R., Kemp B. E., and Scott J. W. (2017) Impact of genetic variation on human CaMKK2 regulation by Ca2+-calmodulin and multisite phosphorylation. Sci. Rep. 7, 43264 10.1038/srep43264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kemp B. E., Graves D. J., Benjamini E., and Krebs E. G. (1977) Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J. Biol. Chem. 252, 4888–4894 [PubMed] [Google Scholar]
- 22. Muslin A. J., Tanner J. W., Allen P. M., and Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 10.1016/S0092-8674(00)81067-3 [DOI] [PubMed] [Google Scholar]
- 23. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., and Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 10.1016/S0092-8674(00)80487-0 [DOI] [PubMed] [Google Scholar]
- 24. Yaffe M. B. (2002) How do 14-3-3 proteins work? - Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57 10.1016/S0014-5793(01)03288-4 [DOI] [PubMed] [Google Scholar]
- 25. Wayman G. A., Tokumitsu H., and Soderling T. R. (1997) Inhibitory cross-talk by cAMP kinase on the calmodulin-dependent protein kinase cascade. J. Biol. Chem. 272, 16073–16076 10.1074/jbc.272.26.16073 [DOI] [PubMed] [Google Scholar]
- 26. Wong R., and Schlichter L. C. (2014) PKA reduces the rat and human KCa3.1 current, CaM binding, and Ca2+ signaling, which requires Ser332/334 in the CaM-binding C terminus. J. Neurosci. 34, 13371–13383 10.1523/JNEUROSCI.1008-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuoka Y., Hughes C. A., and Bennett V. (1996) Adducin regulation. Definition of the calmodulin-binding domain and sites of phosphorylation by protein kinases A and C. J. Biol. Chem. 271, 25157–25166 10.1074/jbc.271.41.25157 [DOI] [PubMed] [Google Scholar]
- 28. Hofer A. M. (2012) Interactions between calcium and cAMP signaling. Curr. Med. Chem. 19, 5768–5773 10.2174/092986712804143286 [DOI] [PubMed] [Google Scholar]
- 29. Kostelecky B., Saurin A. T., Purkiss A., Parker P. J., and McDonald N. Q. (2009) Recognition of an intra-chain tandem 14-3-3 binding site within PKCepsilon. EMBO Rep. 10, 983–989 10.1038/embor.2009.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molzan M., and Ottmann C. (2012) Synergistic binding of the phosphorylated S233- and S259-binding sites of C-RAF to one 14-3-3ζ dimer. J. Mol. Biol. 423, 486–495 10.1016/j.jmb.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 31. Stevers L. M., de Vink P. J., Ottmann C., Huskens J., and Brunsveld L. (2018) A thermodynamic model for multivalency in 14-3-3 protein-protein interactions. J. Am. Chem. Soc. 140, 14498–14510 10.1021/jacs.8b09618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stevers L. M., Lam C. V., Leysen S. F., Meijer F. A., van Scheppingen D. S., de Vries R. M., Carlile G. W., Milroy L. G., Thomas D. Y., Brunsveld L., and Ottmann C. (2016) Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR. Proc. Natl. Acad. Sci. U. S. A. 113, E1152–E1161 10.1073/pnas.1516631113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalabova D., Filandr F., Alblova M., Petrvalska O., Horvath M., Man P., Obsil T., and Obsilova V. (2020) 14-3-3 protein binding blocks the dimerization interface of caspase-2. FEBS J. 287, 3494–3510 10.1111/febs.15215 [DOI] [PubMed] [Google Scholar]
- 34. Matsushita M., and Nairn A. C. (1999) Inhibition of the Ca2+/calmodulin-dependent protein kinase I cascade by cAMP-dependent protein kinase. J. Biol. Chem. 274, 10086–10093 10.1074/jbc.274.15.10086 [DOI] [PubMed] [Google Scholar]
- 35. Nakanishi A., Hatano N., Fujiwara Y., Sha'ri A., Takabatake S., Akano H., Kanayama N., Magari M., Nozaki N., and Tokumitsu H. (2017) AMP-activated protein kinase-mediated feedback phosphorylation controls the Ca2+/calmodulin (CaM) dependence of Ca2+/CaM-dependent protein kinase kinase β. J. Biol. Chem. 292, 19804–19813 10.1074/jbc.M117.805085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davare M. A., Saneyoshi T., Guire E. S., Nygaard S. C., and Soderling T. R. (2004) Inhibition of calcium/calmodulin-dependent protein kinase kinase by protein 14-3-3. J. Biol. Chem. 279, 52191–52199 10.1074/jbc.M409873200 [DOI] [PubMed] [Google Scholar]
- 37. Ichimura T., Taoka M., Hozumi Y., Goto K., and Tokumitsu H. (2008) 14-3-3 proteins directly regulate Ca2+/calmodulin-dependent protein kinase kinase α through phosphorylation-dependent multisite binding. FEBS Lett. 582, 661–665 10.1016/j.febslet.2008.01.037 [DOI] [PubMed] [Google Scholar]
- 38. Spengler K., Zibrova D., Woods A., Langendorf C. G., Scott J. W., Carling D., and Heller R. (2020) Protein kinase A negatively regulates VEGF-induced AMPK activation by phosphorylating CaMKK2 at serine 495. Biochem. J. 477, 3453–3469 10.1042/BCJ20200555 [DOI] [PubMed] [Google Scholar]
- 39. Frigo D. E., Howe M. K., Wittmann B. M., Brunner A. M., Cushman I., Wang Q., Brown M., Means A. R., and McDonnell D. P. (2011) CaM kinase kinase β-mediated activation of the growth regulatory kinase AMPK is required for androgen-dependent migration of prostate cancer cells. Cancer Res. 71, 528–537 10.1158/0008-5472.CAN-10-2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Massie C. E., Lynch A., Ramos-Montoya A., Boren J., Stark R., Fazli L., Warren A., Scott H., Madhu B., Sharma N., Bon H., Zecchini V., Smith D. M., Denicola G. M., Mathews N., et al. (2011) The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 30, 2719–2733 10.1038/emboj.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gocher A. M., Azabdaftari G., Euscher L. M., Dai S., Karacosta L. G., Franke T. F., and Edelman A. M. (2017) Akt activation by Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) in ovarian cancer cells. J. Biol. Chem. 292, 14188–14204 10.1074/jbc.M117.778464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nomiyama T., Kawanami T., Irie S., Hamaguchi Y., Terawaki Y., Murase K., Tsutsumi Y., Nagaishi R., Tanabe M., Morinaga H., Tanaka T., Mizoguchi M., Nabeshima K., Tanaka M., and Yanase T. (2014) Exendin-4, a GLP-1 receptor agonist, attenuates prostate cancer growth. Diabetes 63, 3891–3905 10.2337/db13-1169 [DOI] [PubMed] [Google Scholar]
- 43. Kosowska A., Gallego-Colon E., Garczorz W., Kłych-Ratuszny A., Aghdam M. R. F., Woz Niak M., Witek A., Wróblewska-Czech A., Cygal A., Wojnar J., and Francuz T. (2017) Exenatide modulates tumor-endothelial cell interactions in human ovarian cancer cells. Endocr. Connect. 6, 856–865 10.1530/EC-17-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Byrne S. N., Scott J. W., Pilotte J. R., Santiago A. D S., Langendorf C. G., Oakhill J. S., Eduful B. J., Couñago R. M., Wells C. I., Zuercher W. J., Willson T. M., and Drewry D. H. (2020) In depth analysis of kinase cross screening data to identify CAMKK2 inhibitory scaffolds. Molecules 25, 325 10.3390/molecules25020325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aragão D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N. P., Ericsson D. J., Gee C. L., Macedo S., Mudie N., Panjikar S., Price J. R., Riboldi-Tunnicliffe A., Rostan R., Williamson R., et al. (2018) MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 25, 885–891 10.1107/S1600577518003120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., and Yaffe M. B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153–166 10.1016/S1097-2765(00)80363-9 [DOI] [PubMed] [Google Scholar]
- 50. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. 3rd, Snoeyink J., Adams P. D., et al. (2018) MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the PDB with the accession code 6EF5. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (RRID:SCR_003411) with the data set identifier PXD020133.