Abstract
Punctin/MADD-4, a member of the ADAMTSL extracellular matrix protein family, was identified as an anterograde synaptic organizer in the nematode Caenorhabditis elegans. At GABAergic neuromuscular junctions, the short isoform MADD-4B binds the ectodomain of neuroligin NLG-1, itself a postsynaptic organizer of inhibitory synapses. To identify the molecular bases of their partnership, we generated recombinant forms of the two proteins and carried out a comprehensive biochemical and biophysical study of their interaction, complemented by an in vivo localization study. We show that spontaneous proteolysis of MADD-4B first generates a shorter N-MADD-4B form, which comprises four thrombospondin (TSP) domains and one Ig-like domain and binds NLG-1. A second processing event eliminates the C-terminal Ig-like domain along with the ability of N-MADD-4B to bind NLG-1. These data identify the Ig-like domain as the primary determinant for N-MADD-4B interaction with NLG-1 in vitro. We further demonstrate in vivo that this Ig-like domain is essential, albeit not sufficient per se, for efficient recruitment of GABAA receptors at GABAergic synapses in C. elegans. The interaction of N-MADD-4B with NLG-1 is also disrupted by heparin, used as a surrogate for the extracellular matrix component, heparan sulfate. High-affinity binding of heparin/heparan sulfate to the Ig-like domain may proceed from surface charge complementarity, as suggested by homology three-dimensional modeling. These data point to N-MADD-4B processing and cell-surface proteoglycan binding as two possible mechanisms to regulate the interaction between MADD-4B and NLG-1 at GABAergic synapses.
Keywords: ADAMTS-like, GABA receptor, Punctin/MADD-4, muscle, neuroligin, neuromuscular junction, nicotinic acetylcholine receptor, postsynaptic membrane, protein-protein interaction, synapse, ADAMTS, nicotinic acetylcholine receptors (nAChR)
Synapse formation is fundamental for communication between neurons and their target cells. The life cycle of a synapse involves attraction, recognition and interaction between cells, differentiation of the cell membrane into pre- and postsynaptic terminals, and eventually maturation and elimination (1). Proteins involved in synaptic organization fall into two categories: cell adhesion molecules acting upon contact and secreted factors acting either locally in the extracellular matrix (ECM) as scaffolding proteins or at longer range distances by diffusion (2). A cardinal pair of cell-adhesion molecules essential for excitatory (glutamatergic and cholinergic) and inhibitory (GABAergic and glycinergic) synapse function involves the membrane-tethered neuroligin (NLG) and α- or β-neurexin (NRXα/β) proteins (3–5), whose ectodomains interact trans-synaptically to bridge the pre- and postsynaptic terminals (3, 6–8). Besides the NRXs, the extracellular interactome of the NLGs also includes secreted partners, such as thrombospondin, whose interaction with NLG accelerates synapse formation (9); hevin, which is secreted by astrocytes and bridges isoforms NLG1B and NRXα1 at excitatory synapses (10); and the MAM domain-containing GPI-anchored (MDGA) proteins, which act as negative regulators of synaptic activity by challenging NRX binding to the NLGs (11). The importance of correct synapse formation for cognition is highlighted by the link between mutations in genes encoding synaptic proteins and the occurrence of neurodegenerative conditions, justifying the emerging concept of “synaptopathies” (12, 13). For instance, mutations in the NLG and/or NRX genes have been consistently detected in individuals with autism spectrum disorder (ASD), and mutations in the NRX genes have been associated with schizophrenia (14). Knockdown of the MDGA2 gene in mice resulted in increased excitatory neurotransmission and ASD-associated behavior (15), while polymorphism in the MDGA1 gene has been linked to schizophrenia (16, 17).
Genetic screens for Caenorhabditis elegans mutants with abnormal synaptic organization identified gene Punctin/madd-4, whose protein products specify cholinergic versus GABAergic identity of neuromuscular junctions (NMJs) (reviewed in Ref. 18). In C. elegans, body wall muscles are innervated by both cholinergic and GABAergic motoneurons that express and secrete distinct Punctin/MADD-4 isoforms (19). The long MADD-4L (isoforms a and c, which differ by the presence/absence of two neighboring residues) is exclusively present at cholinergic synapses, where it recruits ionotropic nicotinic acetylcholine receptors. The short MADD-4B (isoform b), which lacks the N-terminal third of MADD-4L, is present at both cholinergic and GABAergic synapses. (19). At GABAergic synapses, MADD-4B interacts with the ectodomain of NLG-1, the sole NLG in C. elegans (20, 21), to cluster GABA type A receptors (GABAARs) (19, 22). MADD-4B might also interact with NRX at GABAergic NMJs (23). Surprisingly, in the MADD-4B knockout mutant, NLG-1 and GABAAR are distributed in both the cholinergic and GABAergic synapses, indicating that MADD-4B is essential for retaining NLG-1 at GABAergic synapses, while preventing its incorrect recruitment at cholinergic synapses (19). Altogether, these data imply that specific domains of MADD-4L and MADD-4B instruct the proper recruitment of receptors at cholinergic versus GABAergic synapses. In this light, it was proposed that MADD-4L and MADD-4B undergo intermolecular association and that this event inactivates the ability of MADD-4L to recruit NLG-1 and prevents incorrect GABAAR clustering at cholinergic NMJs (19, 22).
Punctin/MADD-4 belongs to the poorly understood ADAMTSL (a disintegrin and metalloproteinase with thrombospondin repeats–like) family of secreted multidomain glycoproteins. Members in the family comprise, from the N to the C terminus, a Cys-rich domain, a “spacer” domain, a varying number of thrombospondin type I repeats (TSPs, elsewhere also referred to as “TSRs”), and unique combinations of several Ig-like domains with most often one example of protease and lacunin (PLAC) or netrin- or Kunitz-type domains (see Ref. 24 and references therein) (Fig. 1A). As such, ADAMTSL proteins resemble, in their domain composition, the C-terminal ancillary part of ADAMTS proteins, yet they are devoid of the N-terminal pro-peptide, catalytic and disintegrin domains and protease activity that characterize ADAMTS proteins (24). The importance of ADAMTS(L) proteins is highlighted by their role in binding and remodeling the ECM, an active component in synapse formation. The ECM, which is rich in heparan sulfate (HS) and chondroitin sulfate proteoglycans (25, 26), promotes the structural consolidation of formed synapses. Conversely, ECM degradation and remodeling allows synapse plasticity and promotes neurite growth (27–30). ADAMTS(L) proteins actively remodel the ECM by degrading HS and chondroitin sulfate proteoglycans (26, 31), binding fibrillins, and participating in microfibril assembly/turnover (24, 32, 33) and procollagen processing (34). In schizophrenic individuals, overexpression of chondroitin sulfate proteoglycans leads to abnormal architecture of ECM peri-neuronal nets (27, 30). ADAMTS(L) dysregulation is linked to ECM-related pathologies including arthritis (31), melanoma growth (35), bronchial epithelial dysplasia (36) among others. Interestingly, gene ADAMTSL3, one of the two human madd-4 homologs, is expressed in the central nervous system and was identified as a susceptibility gene for schizophrenia (27).
Figure 1.
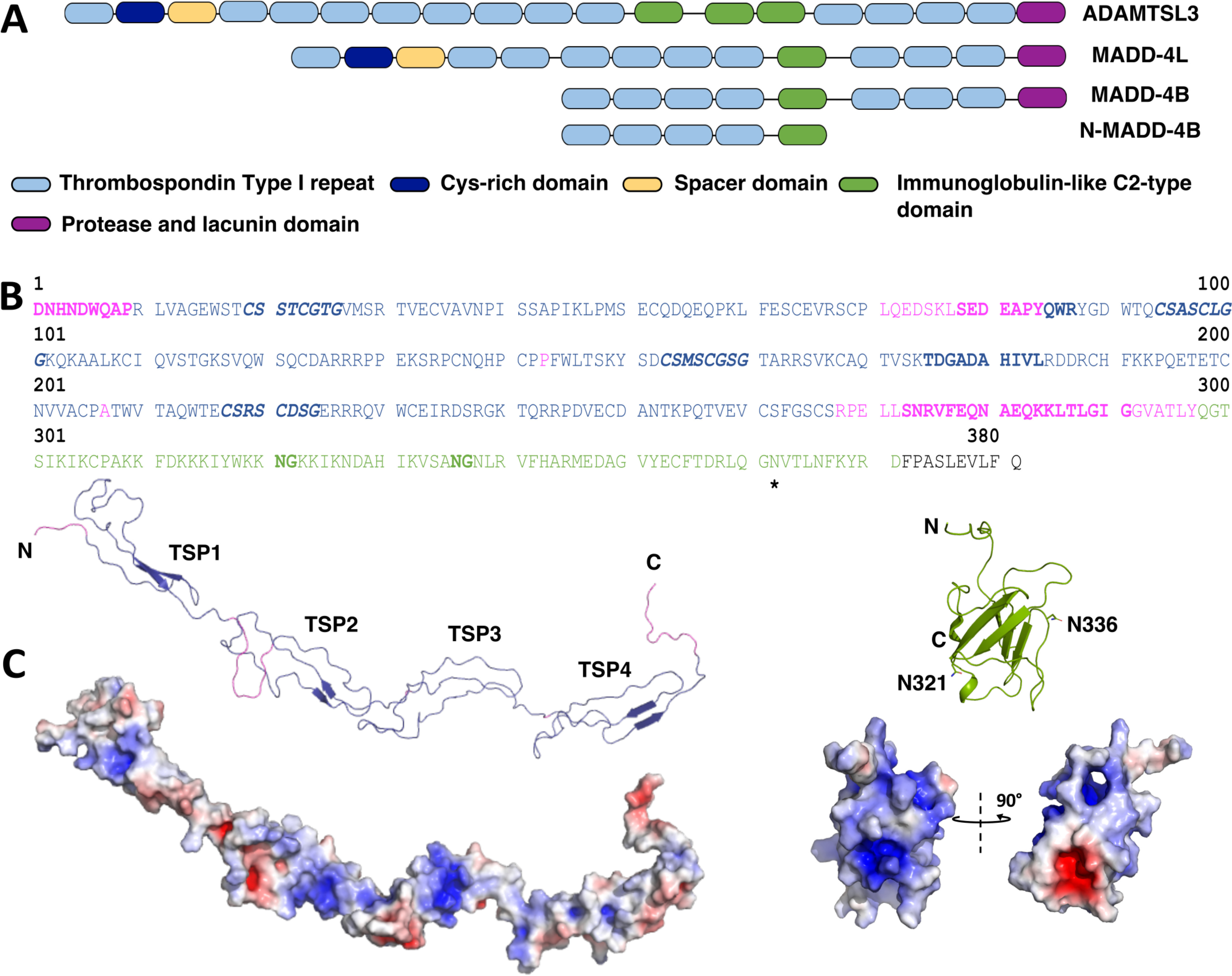
N-MADD-4B domain organization, sequence, and structural model. A, domain organization of human ADAMTSL3, C. elegans MADD-4L and MADD-4B, and the recombinant N-MADD-4B in their secreted form. B, sequence of N-MADD-4B with the TSP1–4 domains colored in blue, linker regions in magenta, and the Ig-like domain in green. CXX(S/T)CXXG consensus motifs for O-fucosylation are displayed in boldface blue italic type, and the potentially N-glycosylated Asn362 is indicated by a star. The sequences identified by Edman degradation are in boldface type. The two sites for potential deamidation (321NG322, 336NG337) in the Ig-like domain are shown in boldface green type. The C-terminal LEVLFQ hexapeptide is a leftover from the 3C-Protease cleavage site of the expressed fusion protein. C, structural models of the TSP1–4 and Ig-like domains generated by homology and displayed to scale. The ribbon representations use the same color code as in A. The molecular surface representations show the distribution of electrostatic potentials from −5 kbT/ec (red) to +5 kbT/ec (blue). Two orientations, identical to that of the ribbon model (left) and 90° apart (right), of the overall positively charged Ig-like domain (theoretical pI, 9.8) evidence two pockets, strongly positively and negatively charged, on opposite faces of the domain.
MADD-4 is the first member of the ADAMTSL family to be identified as a synaptic regulator (19). To better understand the function of MADD-4B at GABAergic synapses, we generated recombinant forms of MADD-4B and of the ectodomain of NLG-1 and carried out a comprehensive biochemical and biophysical analysis of their interaction, buttressed by a complementary in vivo analysis. We show that MADD-4B inevitable proteolysis at a well-defined site first generates a shorter entity encompassing the N-terminal four TSP domains and sole Ig-like domain (herein referred to as “N-MADD-4B”), while further processing eliminates the Ig-like domain to generate an even shorter entity comprising the TSP domains only. Quantitative characterization of the interaction of N-MADD-4B and of its maturation products with NLG-1 points to the Ig-like domain as the primary determinant responsible for NLG-1 binding in vitro and for efficient GABAAR recruitment at GABAergic synapses in vivo. Distinct relative affinities of these proteins for heparin, used as a surrogate for heparan sulfate, suggest a role of cell-surface proteoglycans as regulators of the N-MADD-4B interaction with NLG-1. We propose that N-MADD-4B maturation, resulting in elimination of the Ig-like domain, is a plausible regulation mechanism at C. elegans GABAergic synapses.
Results and discussion
The ADAMTSL protein MADD-4B comprises, from the N to the C terminus, four TSP domains, one Ig-like C2-type module, three additional TSP domains, and a PLAC domain (Fig. 1A). Initial analysis of a recombinant form of MADD-4B secreted from mammalian (HEK) cells revealed critical cleavage of most of the protein population in two pieces during the earliest steps of production. Western blotting tracking of the C-terminal Fc-tag coupled with N-terminal sequencing of the pieces led to location of the cleavage site upstream to sequence VQVSKED in the linker region that follows the Ig-like domain (data not shown). Because previous analysis of MADD-4B as a guidance cue in vivo demonstrated that the C-terminal TSP5-7 and PLAC domains were dispensable for protein functionality (37), we generated a new construct comprising only the TSP1–4 and Ig-like domains, herein referred to as “N-MADD-4B” (Fig. 1A). N-MADD-4B contains a CXX(S/T)CXXG consensus motif for O-fucosylation (38, 39) at the beginning of each TSP domain, along with an NX(T/S) consensus motif for N-glycosylation at Asn362 in the Ig-like domain (Fig. 1B and Fig. S1). Structural models of the TSP1–4 and Ig-like domains point to an elongated, flexible TSP1–4 domain arrangement, connected to the globular Ig-like domain by an equally flexible hinge (Fig. 1B and C). The net positive charge of N-MADD-4B (theoretical pI: 8.8) is reflected in the even distribution of electropositive potentials at the surface of the molecule, apart from the presence of two pockets, respectively more electropositive than the overall surface and highly electronegative, on opposite faces of the Ig domain (Fig. 1C).
N-MADD-4B processing in vitro eliminates the Ig-like domain
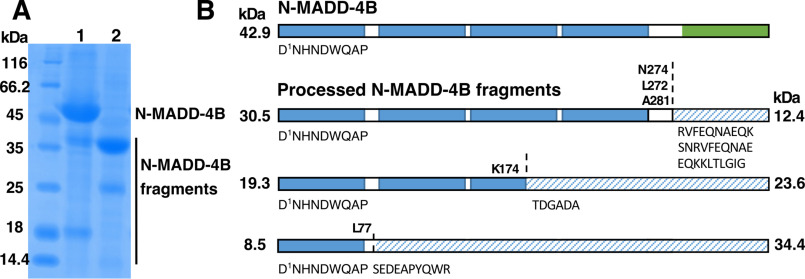
N-MADD-4B was produced, purified, and analyzed using standard procedures (see “Experimental procedures”) (Fig. 2). Electrophoretic analysis of the affinity-purified protein revealed a major entity migrating slightly above the 45 kDa marker, along with two to three smaller entities migrating at ∼35, 25, and 15 kDa, respectively (Figs. 2A and 3A). Peptide mass fingerprinting analysis confirmed that all three entities belonged to N-MADD-4B, albeit with distinct levels of C-terminal coverage (Fig. S1). Intact N-MADD-4B and the 35-kDa entity (45.781 and 31.345 kDa, respectively, by MALDI-TOF-TOF MS; Fig. S2) could be separated from each other by cation-exchange (Fig. 2B) but not size-exclusion chromatography (SEC) (Fig. 2C). Consistently, analytical SEC coupled to multiangle light scattering (SEC-MALS) pointed to two overlapping peaks of 45.7 and 33.7 kDa, respectively (Fig. S3). The ∼2.9-kDa difference between the theoretical mass of the naked N-MADD-4B polypeptide (42.9 kDa) and the experimental values obtained by electrophoresis, MS, and SEC-MALS may reflect, for example, the presence of an N-glycan linked to Asn-362 and/or O-glycans at consensus motifs (see Fig. 1 and Fig. S1). Over time, the purified 35-kDa entity further led to a ∼25-kDa piece (Fig. 3A). Thorough analysis of the fragmentation process pointed to a time- and temperature-dependent process not prevented by protease inhibitors (Fig. S4).
Figure 2.
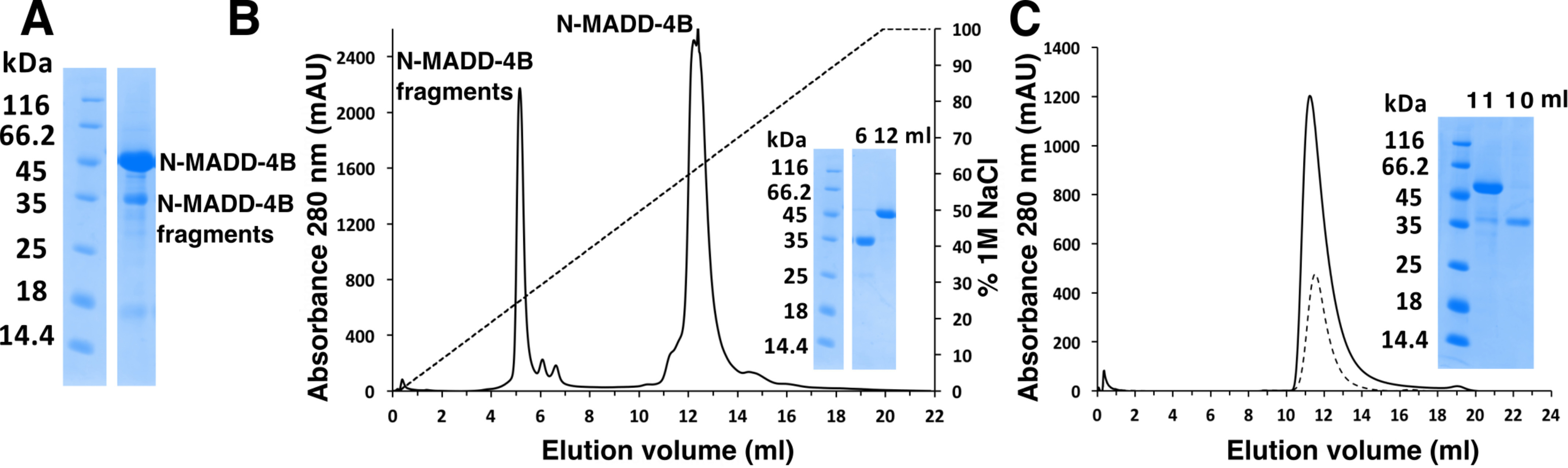
N-MADD-4B undergoes processing. A, SDS-PAGE analysis of peak elution fraction from affinity chromatography (Protein A, data not shown) evidenced intact N-MADD-4B (apparent mass, ∼45 kDa) and N-MADD-4B fragments (≤35 kDa). The protein identity in each band was confirmed by MS (see Fig. S1). B, cation-exchange chromatography (MonoS, NaCl gradient) resolves the N-MADD-4B fragments (elution volume, 5–6 ml) from intact N-MADD-4B (12–13 ml). Inset, SDS-PAGE analysis of the peak elution fractions. C, SEC (Superdex-75 10/300) does not resolve intact N-MADD-4B (solid line) from the fragments (dashed line). Inset, SDS-PAGE analysis of the peak elution fractions.
Figure 3.
C-terminal processing of N-MADD-4B eliminates the Ig-like domain. A, N-MADD-4B fragments are found as a minority in the freshly purified sample (lane 1) and as a majority in the processed sample (lane 2). The protein identity in each band was confirmed by MS (see Fig. S1). B, representation of the N-MADD-4B domains (blue, TSP; green, Ig-like) and the protein fragments identified by N-terminal sequencing (Table 1). The identified sequences account predominantly for the N-MADD-4B N terminus (∼90% abundance) and marginally for its C terminus (<10% abundance, shaded) denoting degradation within the C-terminal part of the protein. The theoretical mass of each detected fragment is indicated on either side of the scheme.
To identify the cleavage sites in N-MADD-4B, samples of intact and processed N-MADD-4B were subjected to N-terminal sequencing (Fig. 3A and Table 1). The most abundant sequence detected (∼90%) corresponded to the N-MADD-4B N terminus (Fig. 3B). Minor abundance sequences (≤10%) indicated cleavage between the TSP1 and TSP2 domains (sequence Ser78–Arg87), within the TSP3 domain (sequence Thr175–Leu184), and between the TSP4 and Ig-like domains (sequences Ser273–Glu282, Arg275–Lys284, and Glu282–Gly291). The entity cleaved between TSP1 and TSP2 corresponds to the most abundant fragment of ∼25 kDa (Fig. 3A, lane 2). A similar fragment was generated upon spontaneous processing of a recombinant “TSP1–4” protein (see below; Fig. S4). In the absence of further processing, cleavage before the Thr175–Leu184 sequence would generate N- and C-terminal moieties of ∼19 and 23 kDa, respectively, while cleavage before or within the Ser273–Glu282 sequence would generate N- and C-terminal moieties of ∼31 and 10 kDa, respectively. The two N-terminal moieties were occasionally detected by electrophoresis (Figs. 2A, 3A, and 4A), while the C-terminal moieties were detected neither by N-terminal sequencing nor by electrophoresis. These data suggest that N-MADD-4B processing readily eliminates the C-terminal Ig-like domain to generate an “N-MADD-4BΔIg” moiety comprising only the TSP1–4 domains.
Table 1.
N-terminal sequences detected by Edman degradation from liquid N-MADD-4B samples
Samples “N-MADD-4B” and “N-MADD-4B fragments” correspond to lanes 1 and 2, respectively, of the SDS-polyacrylamide gel displayed in Fig. 3A. Abundance values (rounded up) are indicated.
| N-MADD-4B, abundance | N-MADD-4B fragments, abundance |
|---|---|
| 1DNHNDWQAP9, 90% | 1DNHNDWQAP9, 90% |
| 78SEDEAPYQWR87, 10% | |
| 275RVFEQNAEQK284, 5% | 275RVFEQNAEQK284, 1% |
| 273SNRVFEQNAE282, 3% | 273SNRVFEQNAE282, 1% |
| 282EQKKLTLGIG291, 2% | 282EQKKLTLGIG291, 2.5% |
| 175TDGADAHIVL184, 2% | 175TDGADAHIVL184, 0.5% |
Figure 4.
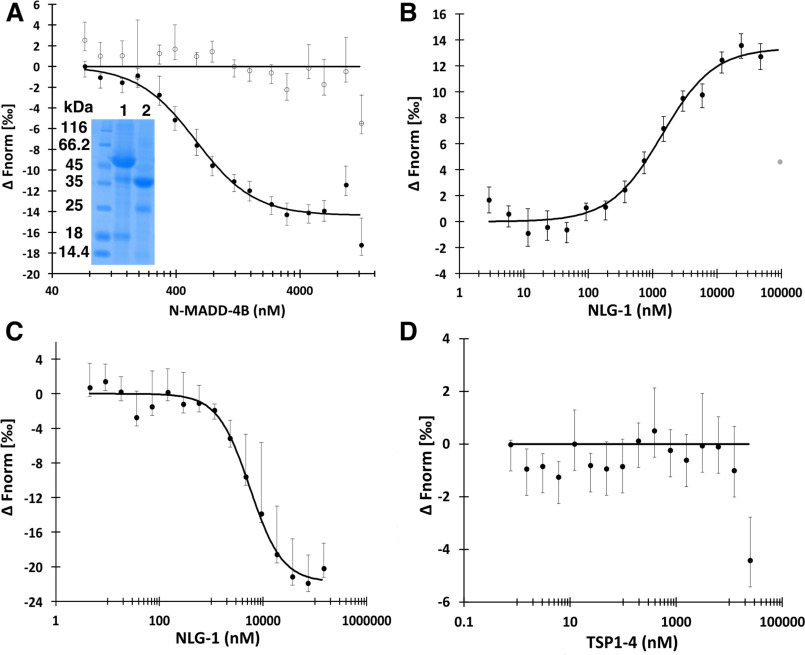
N-MADD-4B interacts with NLG-1 through the Ig-like domain. A, N-MADD-4B (73 nm to 12.5 μm) comprising mostly the intact protein (see lane 1 of inserted SDS-polyacrylamide gel (same gel as in Fig. 3A)) binds to labeled NLG-1 (35 nm) with an estimated Kd of 578 ± 77 nm (circles, n = 4) whereas N-MADD-4BΔIg (lane 2 of inserted SDS-polyacrylamide gel) does not bind (empty circles, n = 3). B, NLG-1 (38 nm to 125 μm) competes with labeled NLG-1 (35 nm) for binding to N-MADD-4B (1.25 μm) with an estimated Kd of 1396 ± 285 nm (n = 3). One outlier data point excluded from the fit is displayed in gray. C, NLG-1 (4.5 nm to 140 μm) interacts with the labeled Ig-like domain (150 nm) with an estimated Kd of 5580 ± 825 nm (n = 3). D, TSP1–4 (3.5 nm to 25 μm) does not interact with labeled NLG-1 (35 nm) (n = 3). Error bars, S.D. All fit parameters are summarized in Table 2.
Inability to prevent N-MADD-4B processing using protease inhibitors led us to suspect a maturation mechanism distinct from accidental proteolysis. Spontaneous cleavage at physiological pH was first reported for cytochrome c (40) and rabbit muscle aldolase (41) and proposed to act as a “molecular clock” i.e. a natural mechanism regulating protein activity (42). Spontaneous cleavage occurs often, but not exclusively, at Asn/Asp-Gly dipeptide sites, through nonenzymatic deamidation of Asn or dehydration of Asp, leading to intermediates prone to further hydrolysis (43). The ability of a protein to undergo a deamidation reaction depends on the conformation and/or solvent exposure of the Asn-Gly doublet (42–43). The Ig-like domain of N-MADD-4B contains two such dipeptides (321NG322 and 336NG337), likely located in surface loop regions (Fig. 1B). However, Ala substitution of these two Asn residues did not prevent the loss of the Ig-like domain (Fig. S5). Asn/Asp-X dipeptides comprising a second residue other than Gly (Ala > Ser > Leu > Val > Ile in order of reactivity) can also undergo degradation (43). Such doublets are present throughout the N-MADD-4B sequence (74DS75, 124DA125, 176DG177, 201NV202, 222DS223, 250DA251, 280NA281), of which 74DS75 and 176DG177 are near or within low-abundance N-terminal sequences (Ser78–Arg87 and Thr175–Leu184, respectively) (Fig. 3 and Table 1). The implication of deamidation in physiological responses in vivo, including signal transduction during DNA damage–induced cell apoptosis (44) and in the enhanced cell-adhesion properties of ECM proteins (45), supports the concept of deamidation regulating protein functionality. Whether this is the case for MADD-4B in vivo would be worth investigating.
The Ig-like domain of N-MADD-4B is essential for the interaction with NLG-1 in vitro
The ability of N-MADD-4B and N-MADD-4BΔIg to interact with the ectodomain of NLG-1 was assayed using microscale thermophoresis (MST), along with data fitting to binding equations considering either the law of mass action (calculation of the equilibrium dissociation constant, Kd) or the Hill model (calculation of the EC50) (Fig. 4 and Table 2). Direct binding of N-MADD-4B to labeled NLG-1 occurred with comparable Kd or EC50 values in the 0.5 μm range, whereas N-MADD-4BΔIg generated no binding signal (Fig. 4A), suggesting that the Ig-like domain is the primary determinant for N-MADD-4B interaction with NLG-1. Full competition between labeled and unlabeled NLG-1 for binding to N-MADD-4B led to Kd and EC50 values again comparable with each other and less than 3-fold higher than those obtained for direct binding (i.e. corresponding to only minimal differences in free energy requirements for binding), thereby confirming the binding specificity (Fig. 4B). If the Hill coefficient is fixed at the value of 1 (for a 1:1 binding stoichiometry), the Hill model reverts to the Kd model. However, except for binding reactions of higher-order stoichiometry, the Hill model may also apply to binding events involving multiple steps (46). Here, the multidomain organization of N-MADD-4B may trigger primary and secondary binding events accounting for the slightly better statistics obtained upon data fitting to the Hill model.
Table 2.
Fit parameters obtained for MST experiments between N-MADD-4B and labeled NLG-1 (NLG*) or NLG-1 and labeled Ig (Ig*)
The N-MADD-4B/NLG* binding data (Fig. 4A), N-MADD-4B/NLG*/NLG-1 competition data (Fig. 4B), and NLG-1/Ig* binding data (Fig. 4C) were processed using either the Kd or the Hill model (see “Experimental procedures”).
| Fit parameters | Kd model | Hill model |
|---|---|---|
| N-MADD-4B/ NLG* | ||
| Kd or EC50 (nm) | 465 | 578 |
| Kd confidence (nm) | ±130 | ±77 |
| S.E. (nm) | 1.3 | 1.2 |
| Reduced χ2 | 1.6 | 0.8 |
| Signal/noise | 14.9 | 13.1 |
| Hill coefficient | 2.3 | |
| N-MADD-4B/NLG*/NLG-1 | ||
| Kd or EC50 (nm) | 1428 | 1396 |
| Kd confidence (nm) | ±488 | ±285 |
| S.E. (nm) | 0.8 | 0.8 |
| Reduced χ2 | 1.1 | 1.1 |
| Signal/noise | 17.8 | 17.5 |
| Hill coefficient | 1.0 | |
| NLG-1/Ig* | ||
| Kd or EC50 (nm) | 6139 | 5580 |
| Kd confidence (nm) | ±1220 | ±825 |
| S.E. (nm) | 1.4 | 1.3 |
| Reduced χ2 | 0.5 | 0.2 |
| Signal/noise | 16.1 | 16.3 |
| Hill coefficient | 1.4 |
Finally, to preclude possible bias associated with the use of processed proteins, we generated recombinant forms of the TSP1–4 and Ig-like domains and explored their respective interaction with NLG-1. NLG-1 was found to bind the labeled Ig-like domain with Kd/EC50 values in the 6 μm range (Fig. 4C and Table 2) (i.e. an affinity lower, by 1 order of magnitude, than that for the intact N-MADD-4B), whereas no binding of TSP1–4 to labeled NLG-1 was observed (Fig. 4D). These data confirm the primary requirement of the Ig-like domain for N-MADD-4B interaction with NLG-1 and support secondary involvement of at least one of the TSP1–4 modules, recruited upon binding proximity when tethered to the Ig-like domain through the flexible intervening linker (see Fig. 1), albeit not able to bind NLG-1 by its/their own.
MADD-4 was shown to be essential in muscle arm guidance during the process of NMJ formation in C. elegans, where it acts as a molecular cue, while initial analysis of MADD-4B suggested that its TSP1- and Ig-like domains are both crucial for protein functionality (37). It was also suggested that the Ig-like domain alone is sufficient for MADD-4B interaction with NRX, while domains TSP3 and -4 may contribute some degree of synergy (23). Consistent with these reports is the radical effect of the loss of the Ig-like domain on N-MADD-4B interaction with NLG-1 in vitro. In addition, the reduced affinity of the isolated Ig-like domain for NLG-1, relative to that of N-MADD-4B, suggests that one or more of the TSP1–4 domains may reinforce the primary binding of the Ig-like domain. Intramolecular synergy has been reported within the ADAMTS proteins, where the catalytic domain alone is often not fully active and needs the disintegrin domain for substrate binding, while the ancillary domains define substrate specificity (47, 48). In turn, NLG complexes with demonstrated or proposed synergistic ligand binding are exemplified by those formed by mammalian NLGs with MDGA or NRXα, respectively. Indeed, distinct Ig-like domains of MDGA were shown to bind distinct sites at the surface of the NLG dimer, thereby clamping the two subunits together (11, 49, 50). Differently, overlaying the LNS6 domain in the L-shaped, long NRXα molecule with that forming the short NRXβ molecule as bound to NLG pointed to the LNS3 and/or LNS4 domains in NRXα as possible secondary binders reinforcing, on the same NLG subunit, the primary interaction of domain LNS6 common to the two NRX forms (51, 52).
Heparin interacts with NLG-1, N-MADD-4B, and TSP1–4 with distinct relative affinities
Proteoglycans are active components of the ECM. The negatively charged HS chains of HS proteoglycans regulate the interactions between synaptic components, such as the mammalian NRX proteins and their NLG or leucine-rich repeat transmembrane protein partners (53). To explore the potential effect of HS on the N-MADD-4B interaction with NLG-1, we used heparin as a surrogate for HS, along with complementary MST and affinity chromatography approaches (Fig. 5 and Table 3). Upon direct binding in the presence of 250 μm heparin, the affinity of intact N-MADD-4B for labeled NLG-1 was found to be reduced by ∼4-fold, indicating heparin binding to either N-MADD-4B or NLG-1, or both (Fig. 5A). Full competition between heparin binding and formation of the N-MADD-4B/labeled NLG-1 complex, with an EC50 value intermediate to those determined for formation of the N-MADD-4B/labeled NLG-1 complex in the absence/presence of heparin, confirmed heparin-driven destabilization of the N-MADD-4B interaction with NLG-1 (Fig. 5B). To preclude possible bias from molecular heterogeneity of commercial heparin, we repeated this experiment using highly purified samples of decameric heparin and HS; the same results were obtained (data not shown).
Figure 5.
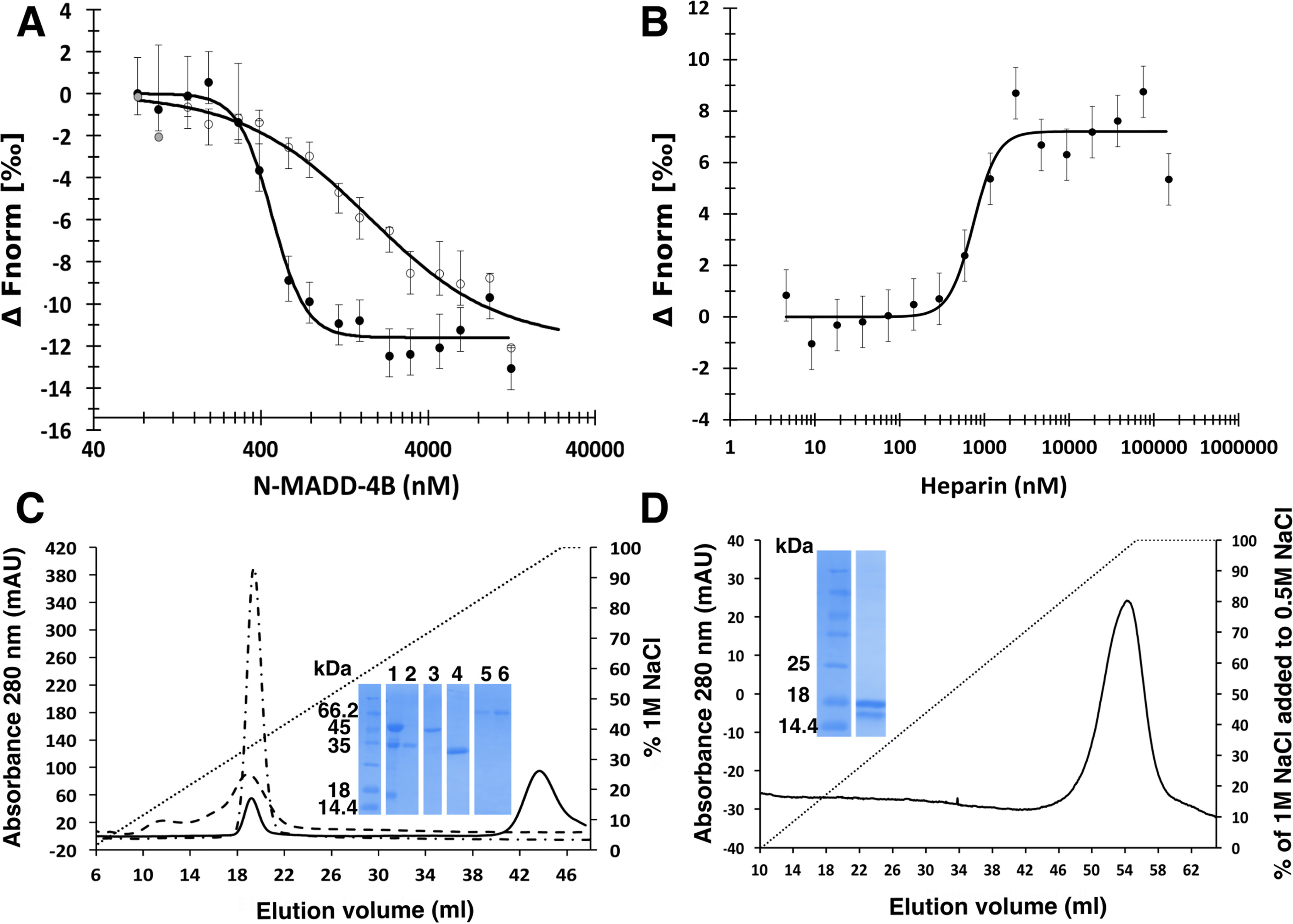
Heparin challenges the N-MADD-4B interaction with NLG-1. A, binding of N-MADD-4B (73 nm to 12.5 μm) to labeled NLG-1 (35 nm) (filled circles) is compromised in the presence of heparin (250 μm) (empty circles). Three outlier data points excluded from the fit are displayed in gray. B, heparin (4.5 nm to 150 μm) prevents binding of N-MADD-4B (1 μm) to labeled NLG-1 (35 nM) with a half-effect at ∼740 nm. C, relative affinities of N-MADD-4B fragments (solid line), TSP1–4 (dash-dotted line), and NLG-1 (dashed line) for immobilized heparin using NaCl elution (0–1 m gradient over 8 CV) and conductivity recording. Inset, SDS-PAGE of N-MADD-4B input (lane 1) and of the peak elution fractions of N-MADD-4B fragment (lane 2), intact MADD-4B (lane 3), TSP1–4 (lane 4), and NLG-1 (lanes 5 and 6). Intact N-MADD-4B is retained much more strongly (elution at 82 mS/cm; i.e. 900 mm NaCl) than N-MADD-4BΔIg (343 mm NaCl; i.e. 22.6 mS/cm), TSP1–4 (349 mm NaCl; i.e. 21.6 mS/cm), or NLG-1 (337 mm NaCl; i.e. 21.9 mS/cm). D, relative affinity of the isolated Ig-like domain for immobilized heparin (0.5–1.0 m NaCl gradient over 9 CV). The Ig-like domain is as strongly retained as intact N-MADD-4B. Inset, SDS-PAGE of the Ig-like elution fraction in which partial degradation is visible. (All proteins were loaded on the heparin column as 0.5 ml at 0.8 mg/ml.)
Table 3.
Fit parameters obtained for N-MADD-4B binding to labeled NLG-1 (NLG*) in the presence or absence of heparin
The N-MADD-4B/NLG* binding data from experiments performed in the absence or presence of heparin (Fig. 5A) and the N-MADD-4B/NLG*/heparin competition data (Fig. 5B) were processed using the Hill model (see “Experimental procedures”).
| Fit parameters | Binding without heparin | Binding with heparin | Competition experiment |
|---|---|---|---|
| Kd or EC50 (nm) | 467 | 1782 | 740 |
| Kd confidence (nm) | ±31 | ±483 | ±136 |
| S.E. (nm) | 0.9 | 0.8 | 1.0 |
| Reduced χ2 | 0.6 | 1.0 | 6.9 |
| Signal/noise | 14.5 | 17.1 | 6.7 |
| Hill coefficient | 4.3 | 1.1 | 3.0 |
The relative affinities of N-MADD-4B (processed sample containing both the intact protein and N-MADD-4BΔIg moiety), TSP14, and NLG-1 for heparin where assessed by affinity chromatography with conductivity recording and by SDS-PAGE (Fig. 5C). All proteins were retained on immobilized heparin, yet elution of N-MADD-4B required a ∼3-fold higher ionic strength (900 mm NaCl, 81.9 mS/cm) than elution of N-MADD-4BΔIg, TSP1–4, or NLG-1 (elution at ∼22 mS/cm; i.e. 340 mm NaCl). A similar analysis of the interaction of two variants of the Hedgehog morphogen (a key mediator of embryonic development) with immobilized heparin correlated elution at 37 and 52 mS/cm conductivity with Kd values of 6.8 and 0.8 μm, respectively, as measured by surface plasmon resonance (54). Linear extrapolation from these two sets of data led to estimated Kd values of ∼10 nm and 60 μm for heparin binding by N-MADD-4B and by the other three proteins, respectively. To confirm the significant (∼104-fold) difference in estimated affinities, we also loaded the recombinant Ig-like domain onto the same heparin column, albeit using a high-salt starting buffer to compensate for its lower solubility (Fig. 5D). Not only did the Ig-like domain fully bind to heparin, despite its loading at an ionic strength higher than those leading to N-MADD-4BΔIg, TSP1–4, and NLG-1 elution from the column, but also its elution required as high an ionic strength as did elution of intact N-MADD-4B. Hence, heparin-driven destabilization of the N-MADD-4B interaction with NLG-1 proceeds from specific, high-affinity binding of heparin to the Ig-like domain of N-MADD-4B.
ADAMTS proteins were shown to degrade the protein core of sulfated proteoglycans, such as aggrecan and versican (25–26). The critical contribution of the negatively charged sulfated glycan chains to this phenomenon suggested a primarily electrostatic interaction (48). Whereas the ADAMTSL proteins are devoid of the catalytic domain and activity found in their ADAMTS relatives, they share some of the ADAMTS ancillary domains (TSP, Ig, and PLAC) (Fig. 1A). Our structural model of the Ig-like domain evidences a highly electropositive surface pocket (Fig. 1C) of a suitable size for lodging heparin with high affinity. In vivo, HS or chondroitin sulfate proteoglycans are thought to generate signaling gradients guiding protein interactions and/or secreted protein release, as is the case for the HS-dependent interaction between the secreted glycoprotein, Wnt, and its partner, Frizzled (55). Binding of ADAMTS4/5 to syndecan has been proposed to trigger signaling events through activation of the mitogen-activated protein kinase pathway (28). Whether, and how, HS proteoglycans regulate the N-MADD-4B interaction with NLG-1 in C. elegans remains to be investigated.
The Ig-like domain of N-MADD-4B is necessary but not sufficient for rescuing GABAAR clustering in C. elegans madd-4 null mutants
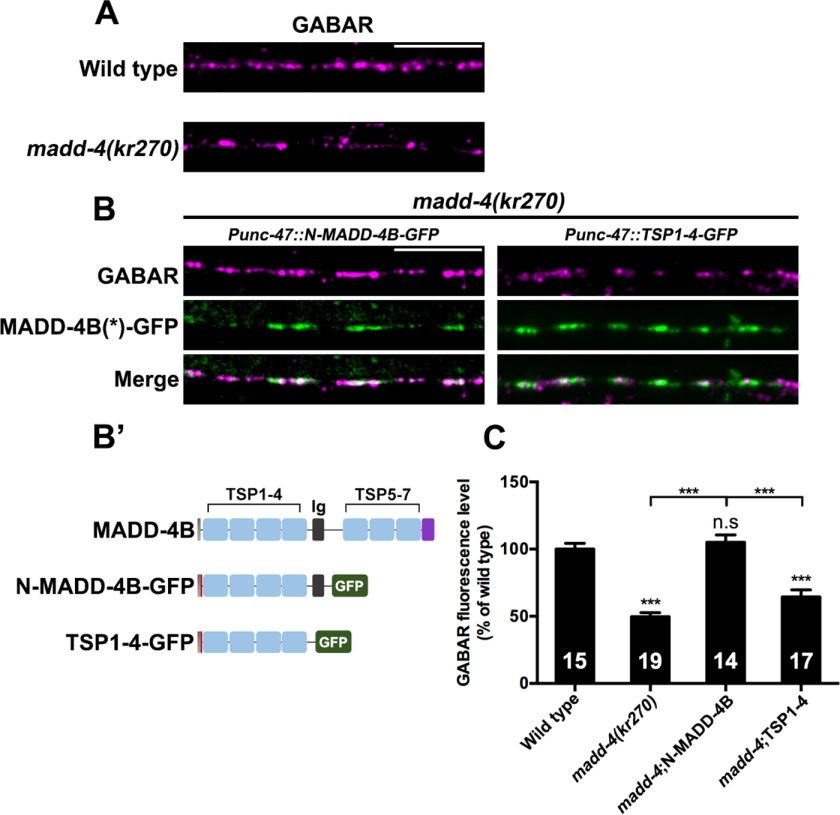
To investigate in vivo the functional relevance of the interaction between the Ig-like domain and NLG-1 observed in vitro, we expressed GFP-fused N-MADD-4B or TSP1–4 in madd-4 null mutants expressing RFP-tagged GABAAR from the CRISPR/Cas9-engineered locus unc-49::rfp(kr296) (Fig. 6, A and B′). Compared with WT animals, in madd-4 null mutants, synaptic expression of GABAAR was found to be reduced by 50% (Fig. 6, A and C). Expression of N-MADD-4B-GFP in GABAergic neurons rescued GABAAR expression to WT levels (Fig. 6, B and C). Moreover, N-MADD-4B-GFP was found to colocalize with UNC-49, a feature further supporting the hypothesis that N-MADD-4B alone can promote GABAAR clustering. In contrast, expression of TSP1–4-GFP did not rescue GABAAR clustering (Fig. 6, B and C). These data point to a critical role of the Ig-like domain for MADD-4 function at GABAergic synapses in vivo.
Figure 6.
N-MADD-4B rescues GABAAR clustering defect in the C. elegans madd-4 null mutant. A, confocal detection of RFP-GABAAR expressed from the unc-49::rfp knock-in locus kr296 in WT or madd-4(kr270) null mutant animals (magenta). B, N-MADD-4B-GFP or TSP1–4-GFP were expressed in GABAergic neurons of madd-4(kr270) mutant animals. The MADD-4B(*)-GFP row corresponds to N-MADD-4B-GFP (left panels) or TSP1–4-GFP (right panels). B′, functional domains of MADD-4B, N-MADD-4B, and TSP1–4. Gray, signal peptide; red, T7 tag; purple, PLAC domain. C, RFP-GABAAR fluorescence levels were quantified in WT, madd-4(kr270), N-MADD-4B, and TSP1–4 rescue groups and normalized to the WT group (mean ± S.E. (error bars)). One-way analysis of variance was used, followed by Tukey's multiple-comparison test. ***, p < 0.001; n.s, not significant. The numbers of animals are indicated inside the histogram boxes. Scale bar, 10 μm.
To further explore the role of this domain in vivo, we expressed a GFP-fused Ig-like domain in the madd-4 null mutants and assessed its ability to rescue the madd-4 null phenotype (Fig. S6). The Ig-GFP fusion protein did not sharply localize to GABAergic synapses, but instead, made enlarged punctae in the nerve cord region. Moreover, the Ig-GFP fusion protein did not rescue GABAAR expression defects, indicating that the Ig-like domain alone is not sufficient for GABAAR recruitment. Hence, in vivo, synergistic interaction of the Ig-like domain and one or more TSP domains with NLG-1 is likely required for ensuring N-MADD-4B synaptogenic function.
Our results, in demonstrating the essential role of the Ig-like domain of MADD-4B for NLG-1 binding in vitro and in vivo, raise the question of whether the shedding of the Ig-like domain could be a yet undescribed mechanism for regulation of GABAAR recruitment at GABAergic synapses. Autocatalytic processing of the ADAMTS2 or ADAMTS4 C terminus has been reported to generate cleavage products thought to regulate catalytic activity (34, 56). Similarity of ADAMTSL proteins to the ancillary domains of ADAMTS suggested that these domains may act as regulators of ADAMTS activity (57). Papillin, an ADAMTSL relative, was shown to act in vitro as a noncompetitive inhibitor of ADAMTS2 (58). At the C. elegans NMJ, MADD-4B processing could either be controlled by a matrix metalloprotease (potentially an ADAMTS family member) or be an autoprocessing event. Taking a step back, one may wonder whether the MADD-4B primary cleavage event identified from the conditioned culture medium reflects a regulatory mechanism in vivo, aimed at generating not only N-MADD-4B as described herein, but also a complementary “C-MADD-4B” moiety, comprising the three TSP domains and one PLAC domain C-terminal to MADD-4 (Fig. 1A). While N-MADD-4B acts at the inhibitory, GABAergic synapse, C-MADD-4B could migrate to excitatory, cholinergic NMJs, where its interaction with MADD-4L would prevent improper GABAAR recruitment, as previously proposed for intact MADD-4B (19). Whether the C-MADD-4B moiety could prevent the Ig-like domain from interacting with NLG-1 in the MADD-4L context remains to be assayed. Finally, from an evolutionary perspective, it must be pointed out that the Ig-like domain in MADD-4 resembles more, in length and sequence, the first of the three Ig-like domains in human ADAMTSL3 (36% identity, versus 23 and 26% for the other two domains (BLASTn search, data not shown)) (Fig. 1A). It would be worth investigating whether this domain plays a central role for ADAMTSL3 function, as does the Ig-like domain for MADD-4B function.
Conclusions
Previous studies identified the short MADD-4B isoform as a key molecule for the organization of GABAergic synapses (18). How MADD-4B activity is regulated remains, however, elusive. The current study evidences a critical role of the Ig-like domain contained in the central part of MADD-4B, both for in vitro binding to NLG-1 and for in vivo clustering of GABAAR, and points to two novel regulatory mechanisms of MADD-4B activity. First, we show that the Ig-like domain is the primary determinant for N-MADD-4B binding to heparin, whose competition with NLG-1 binding suggests a regulatory role by cell-surface proteoglycans at the synapse. Second, we identified an unexpected self-maturation process of N-MADD-4B in vitro, which sheds the Ig-like domain. An increasing number of studies report the shedding of synaptic protein domains by various domains in vivo. The use of self-processing also in vivo to regulate the activity of synaptic determinants is an appealing hypothesis.
Experimental procedures
Production of recombinant proteins
The C. elegans genes and construct boundaries used for recombinant protein production are as follows: (i) the ectodomain of NLG-1 (residues Tyr18–Glu607; Uniprot C40C9.5); (ii) a truncated version of MADD-4B encompassing the TSP1–4 repeats and subsequent Ig-like C2-type domain (“N-MADD-4B”) (residues Asp369–Ser743 of MADD-4 isoform a, Uniprot F53B6.2); (iii) the TSP1–4 repeats of N-MADD-4B (“TSP1–4”) (residues Asp369–Phe645 of MADD-4 isoform a); and (iv) the Ig-like C2-type domain of N-MADD-4B (“Ig-like”) (residues Glu646–Ser743 of MADD-4 isoform a).
The NLG-1–, N-MADD-4B–, and TSP1–4–encoding sequences were N-terminally fused to sequences for a signal peptide ensuring protein secretion from mammalian cells (secreted human placental alkaline phosphatase peptide for NLG-1; mouse IgK peptide for N-MADD-4B and TSP1–4), C-terminally prolonged by sequences encoding a 3C-Protease cleavage site and a human Fc domain, and inserted into a pYD7 vector (National Research Council Canada, Biotechnology Research Institute) using In-Fusion cloning (Takara-Bio). The constructs were transfected into adherent HEK293-EBNA cells (59) using polyethylenimine (Polysciences), and stable cell lines were generated using blasticidin (20–50 μg/ml) for selection pressure. The proteins were routinely produced from 500 ml of stable cell lines cultured at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and 5 μg/ml blasticidin. Proteins were then expressed over 5–8 days in Dulbecco's modified Eagle's medium supplemented with 2% (v/v) fetal bovine serum and 1.25 mm valproic acid at 37 °C for NLG-1 and at 30 °C for N-MADD-4B and TSP1–4.
The Ig-like encoding sequence was N-terminally fused to sequences for a His6 tag followed by a thrombin cleavage site and cloned into a pET28a vector (Twist Bioscience) suitable for bacterial expression. The construct was transformed into Escherichia coli (BL21_DE3), and cells were grown at 37 °C. Protein expression was induced at A600 0.2–0.6 using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and allowed to proceed overnight at 17 °C.
Protein purification
All liquid chromatography was performed at room temperature using an ÄKTA Purifier apparatus (GE Healthcare). Maintaining protease inhibitors throughout all purification steps was not manageable, while their presence was reported not to prevent proteolysis of ADAMTSL proteins (32). Instead, we tested their impact on the kinetics of proteolysis of purified intact N-MADD-4B at three temperatures (see below). Conditioned culture medium (500 ml) containing the secreted, Fc-fused NLG-1, N-MADD-4B, or TSP1–4 protein was harvested, dialyzed against sodium phosphate 100 mm, pH 7.4, NaCl 200 mm (8 liters, 4 °C, overnight), and then loaded on a Protein A affinity column (5 ml; GE Healthcare). The Fc fragment was removed by in-column digestion by GST-tagged 3C-Protease (800 μl at 0.5 mg/ml, overnight, 4 °C), and the protein of interest was eluted from the Protein A column through a GSTrap column (1 ml; GE Healthcare) in the same buffer. For N-MADD-4B only, the fractions eluted from the Protein A/GST-trap columns were diluted 10-fold in Hepes 20 mm, pH 7.5, loaded on a MonoS column (1 ml; GE Healthcare), and eluted using a linear gradient of NaCl (0–1 m over 20 ml). For all three proteins, the fractions of interest were concentrated by ultrafiltration (10-kDa cut-off membrane, Amicon, Merck-Millipore) and further purified by SEC on Superdex-200 (16/60 column) or Superdex-75 (10/300 column) in Hepes 20 mm, pH 7.5, NaCl 200 mm.
Bacterial pellets containing the Ig-like domain were resuspended in 20 mm Hepes, pH 7.5, 500 mm NaCl (buffer A), supplemented with 1 mm DTT, 10 μg/ml DNase, 2 mm MgCl2 and a mixture of protease inhibitors (Roche Applied Science). The soluble fraction of the lysate was loaded on Ni Sepharose 6 Fast Flow resin (5 ml in batch; GE Healthcare), which was then washed with buffer A supplemented with NaCl (up to 1 m) and imidazole (30 mm). The protein was eluted in buffer A supplemented with 1 mm DTT and 300 mm imidazole and dialyzed against buffer A (4 °C, overnight). Finally, the protein was concentrated (3-kDa cut-off membrane, Amicon, Merck-Millipore) and further purified by SEC on Superdex-75 (10/300 column) in the dialysis buffer.
Proteins were analyzed by SDS-PAGE under denaturing and reducing conditions (loading buffer containing 4% (w/v) SDS and 500 mm DTT) using 15% (NLG-1, N-MADD-4B, and TSP1–4) or 18% gels (Ig-like) stained with Instant Blue (Expedeon). In all cases, protein identity and intact mass were verified by MS procedures (see below). Purified proteins were quantified by absorbance on a NanoDrop (Thermo Scientific) or UV-visible spectrophotometer (Carry Eclipse), using the theoretical molar extinction coefficients, ε at 280 nm, calculated using the ProtParam tool in Expasy (NLG-1, 82.65 mm−1 cm−1; N-MADD-4B, 67.06 mm−1 cm−1; TSP1–4, 55.47 mm−1 cm−1; Ig-like, 12.55 mm−1 cm−1). They were stored at 4 or −20 °C. For the time-course analysis of N-MADD-4B processing, the protein (∼0.4 mg/ml) was maintained at −20 °C, 4 °C, or room temperature in the absence or presence of an EDTA-free protease inhibitor mixture (Roche Applied Science) supplemented with 2 mm EDTA, according to the manufacturer's instructions. Aliquots were removed at selected time points, denatured, and reduced as described above and stored at 4 °C until SDS-PAGE analysis.
Heparin-affinity chromatography
Chromatography was performed using an ÄKTA Purifier apparatus equipped with an on-line conductimeter. Stock NLG-1, N-MADD-4B, and TSP1–4 proteins, diluted to 0.8 mg/ml in Hepes 20 mm, pH 7.5, to lower the NaCl concentration to less than 50 mm, were loaded on a heparin column (5 ml; GE Healthcare) pre-equilibrated in and then washed with the same buffer, and eluted using a linear gradient of NaCl (0 to 1.0 m over 40 ml). Stock Ig-like protein (0.8 mg/ml) in buffer A (see above) was loaded on the column, pre-equilibrated in and then washed with the same buffer, and eluted with a linear gradient of NaCl (500 mm to 1.0 m over 45 ml). Fractions of interest were analyzed by SDS-PAGE (see above).
SEC-MALS
The SEC-MALS-RI-UV setup consisted of an Ultimate3000 HPLC apparatus (including quaternary pump, autosampler, and UV-visible variable-wavelength (diode array) detector) (Thermo Scientific) in line with a DAWN8 multiangle laser light-scattering detector calibrated with titrated BSA, and an Optilab relative refractive interferometer (Wyatt Technology, Santa Barbara, CA). Chromatography was performed at 20 °C on Superdex-200 (10/300 column, GE Healthcare) using 20 mm Hepes, pH 7.5, 200 mm NaCl, a flow rate of 0.6 ml/min, and 30 μl of protein at 2 mg/ml. UV absorbance of the eluents was recorded at 280 nm. The Agilent software was used to control the HPLC, and the Wyatt Astra V software was used for data collection and analysis. Peak alignment and band-broadening correction between the UV, MALS, and RI detectors were performed using Astra software algorithms.
N-terminal (Edman) sequencing
Samples of intact N-MADD-4B or its processing products, either in the liquid state or transferred from SDS-polyacrylamide gels to polyvinylidene difluoride membranes, were subjected to N-terminal sequencing by automated Edman degradation. For liquid samples, 500 pmol of protein were diluted to 100 μl in 0.1% (v/v) TFA, pH 2.0, and loaded on a ProSorb membrane pretreated with 10 μl of methanol. The membrane was washed with 0.1% TFA (3 × 100 μl) to fix and desalt the protein sample and then loaded on a Shimadzu PPSQ 31B sequencer. Samples on polyvinylidene difluoride membranes were stained with Ponceau red, and then the membrane was washed three times with 90% ethanol, dried, and loaded on the sequencer as described above. 7–10 cycles were performed. N-MADD-4B was found to be preceded by an extra Asp residue arising from the cloning procedure (see Fig. S1).
MS
Protein identity in the SDS-PAGE bands was confirmed by peptide mass fingerprinting using Orbitrap LC–MS/MS, while the monoisotopic masses of intact N-MADD-4 and its processing fragments were determined by MALDI-TOF-TOF MS, as described in the supporting information.
MST
Recombinant NLG-1, N-MADD-4B, and TSP1–4 proteins were fluorescently labeled on Lys residues using Alexa Fluor 647 (Thermo Fisher Scientific) and NHS ester conjugation according to the manufacturer's instructions. Briefly, the protein at 20 μm was incubated with a 2–3-fold molar excess of dye (1 h, room temperature) and then loaded on a desalting column (NAP-5, GE Healthcare) to remove excess dye. Labeling efficiency was assessed by a protein/dye molar ratio of ∼2:1, as measured at 280 and 650 nm, respectively. In contrast, the Ig-like domain at 10 μm was labeled on the N-terminal His6 tag using the Monolith His-Tag Labeling Kit RED-tris-NTA 2nd generation (NanoTemper Technologies) and the manufacturer's instructions (5 μm, 1 h of incubation at room temperature), given for labeling half of the protein population.
Labeled NLG-1 at 35 nm was incubated with unlabeled N-MADD-4B or N-MADD-4BΔIg in the 73 nm to 12.5 μm concentration range or with unlabeled TSP1–4 in the broader 0.8 nm to 25 μm range, in the absence or presence of 250 μm heparin sodium (Euromedex) or HPLC-purified decameric heparin or HS (a gift from Hugues Lortat–Jacob, IBS, Grenoble) (1.5 h, room temperature). Samples were briefly centrifuged and loaded into standard-treated capillaries (NanoTemper Technologies). Measurements were performed at 24 °C in 20 mm Hepes, pH 7.5, 200 mm NaCl, supplemented with 2 mm CaCl2, 0.1% Tween 20 (v/v), and 0.2% BSA (w/v), using the Monolith NT.115 apparatus (NanoTemper Technologies) with 40% LED power and 80% infrared laser power. At least three independent experiments were performed, while each series of capillaries was read twice with a 20-min interval. To record the Ig-like interaction with NLG-1 with optimal thermophoretic signal amplitude, we labeled the smaller Ig-like protein and incubated it at 150 nm with unlabeled NLG-1 in the 4.5 nm to 149 μm concentration range. Here, premium capillaries, 95% LED power, and 40% infrared laser power were used for data recording. Competition experiments were performed using a mix of labeled NLG-1 (35 nm) and unlabeled N-MADD-4B (1.25 μm) along with either unlabeled NLG-1 (7.6 nm to 125 μm) or heparin (4.6 nm to 150 μm) as a competitor and the same experimental conditions as for the N-MADD-4B interaction with NLG-1.
MST data were analyzed with the NanoTemper MO.Affinity Analysis software version 2.2.4. Binding and competition curves were fitted to the data points using either the law of mass action (Kd model, with calculation of the Kd) or the Hill model (calculation of the EC50) and the equations provided by the analysis software (see supporting information).
Homology three-dimensional modeling of the TSP1–4 and Ig-like domains
Homologues of N-MADD-4B with available three-dimensional structures were identified from sequence processing by the HHpred server (60). Structures 1W0R and 1XIW were identified as suitable homologues (rank 1) for the TSP1–4 and Ig-like parts of N-MADD-4B, respectively, and used as molecular templates for homology modeling with MODELLER (61) using default parameters. Fig. 1C was generated with PyMOL (PyMOL Molecular Graphics System, version 2.2.3, Schrödinger, LLC).
Plasmids and germline transformation
The control strain used in this study was EN296 unc-49(kr296::tagRFP) (62). Plasmids were pXZ066 (Punc-47::madd-4(TSP1–4)-GFP), pXZ067 (Punc-47::madd-4(TSP1–4+Ig)-GFP), and pXZ111 (pmyo-3::madd-4(Ig)-GFP). Transgenes were created by conventional microinjection of plasmids at 15 ng/μl in the gonad of EN3218 madd-4(kr270); unc-49(kr296::tagRFP) animals and named krEx1319 and krEx1320 for Punc-47::madd-4(TSP1–4)-GFP and krEx1345 for pmyo-3::madd-4(Ig)-GFP. A single-copy insertion allele was created using the miniMos technique for pXZ067 krSi93[miniMos_Punc-47::madd-4(TSP1–4+Ig)-GFP] (63).
Spinning disk microscopy imaging and quantification
Young adult hermaphrodites (24 h post-L4 larva stage) were used for imaging. Live worms were mounted on 2% agarose dry pads with 1% polylysine beads in M9 buffer. Fluorescence images were captured using an Andor spinning disk system installed on a Nikon-IX86 microscope (Olympus, Japan) equipped with a 60/1.2 oil immersion objective and an Evolve EMCCD camera. Each animal was imaged as a stack of optical sections (0.2 μm apart) containing 30–39 slices and projected along the z axis. Images were quantified using ImageJ (version 1.48) (National Institutes of Health) with a Fiji plugin add-ons. Synaptic GABAARs were quantified as described previously (22, 64). Acquisition settings were the same for each experimental group. For fluorescence intensity measurement, 30 μm (wide) × 5 μm (high) regions along the ventral (between VD6 and VD7 neuron) or dorsal cord near the anterior mid-body were cropped and analyzed. For the MADD-4B(TSP1–4)-GFP rescue group, two independent extrachromosome arrays were quantified, and the values were pooled together.
Data availability
The raw MS data associated with Fig. S1 have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with data set identifier PXD020639.
Supplementary Material
Acknowledgments
We thank Hugues Lortat-Jacob (Institut de Biologie Structurale, Grenoble, France) for the kind gift of purified heparin and HS samples and Maria Mate (Biophysical Techniques Facility, AFMB Laboratory, Marseille, France) for SEC-MALS.
This article contains supporting information.
Author contributions—S. P., P. Mansuelle, P. F., J.-L. B. and P. Marchot data curation; S. P., X. Z., H. T., P. Mansuelle, P. F., J.-L. B. and P. Marchot formal analysis; S. P., X. Z., B. P.-L., V. D., H. T., P. Mansuelle and P. F. investigation; S. P., X. Z., P. Mansuelle, P. F. and P. Marchot visualization; S. P., B. P.-L., V. D. and P. Marchot methodology; S. P., P. Mansuelle, P. F., J.-L. B. and P. Marchot writing-original draft; S. P., B. P.-L., P. Mansuelle, P. F., Y. B., J.-L. B. and P. Marchot writing-review and editing; S. P., B. P.-L., P. Mansuelle, P. F., J.-L. B. and P. Marchot validation; B. P.-L., J.-L. B. and P. Marchot supervision; S. P., B. P.-L., Y. B., J.-L. B. and P. Marchot funding acquisition; J.-L. B. and P. Marchot project administration; B. P.-L., J.-L. B. and P. Marchot conceptualization; Y. B., J.-L. B. and P. Marchot resources.
Funding and additional information—This project was supported by the French grant ANR-15-CE11-0016 (to J.-L. B. and P. Marchot), Campus France co-financing grant PRESTIGE-2017-1-0027 (to S. P.), European Research Council grant ERC_Adg C.NAPSE 695295 (to J.-L. B.), and French Infrastructure for Integrated Structural Biology (FRISBI) grant ANR-10-INBS-0005 (to the AFMB lab). S. P. was a laureate of the Marie Sklodowska-Curie Actions Seal of Excellence Award on this project in 2017. The Marseille Proteomics (MaP) Facility is supported by “Fonds Européen de Développement Régional” (FEDER), “Infrastructures Biologie Santé et Agronomie” (IBiSA), “Plateformes Technologiques Aix-Marseille”, “Cancéropôle PACA”, “Région Sud Provence-Alpes-Côte d'Azur”, “Institut Paoli-Calmettes”, “Centre de Recherche en Cancérologie de Marseille (CRCM)”, and “Plan Cancer”.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ECM
- extracellular matrix
- ADAMTS
- a disintegrin and metalloproteinase with thrombospondin repeats
- ADAMTSL
- ADAMTS-like
- GABAAR
- GABA type A receptor
- HS
- heparan sulfate
- GPI
- glycosylphosphatidylinositol
- MDGA
- MAM domain−containing GPI-anchored
- NLG
- neuroligin
- NMJ
- neuromuscular junction
- NRX
- neurexin
- SEC
- size-exclusion chromatography
- SEC-MALS
- SEC coupled to multiangle light scattering
- TSP
- thrombospondin
- RI
- refractive index
- ASD
- autism spectrum disorder
- PLAC
- protease and lacunin
- MST
- microscale thermophoresis
- mS
- millisiemens
- CV
- column volume.
References
- 1. Biederer T., and Massimiliano S. (2008) Signalling by synaptogenic molecules. Curr. Opin. Neurobiol. 18, 261–269 10.1016/j.conb.2008.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yuzaki M. (2018) Two classes of secreted synaptic organizers in the central nervous system. Annu. Rev. Phys. 80, 243–262 10.1146/annurev-physiol-021317-121322 [DOI] [PubMed] [Google Scholar]
- 3. Graf E. R., Zhang X., Jin S.-X., Linhoff M. W., and Craig A. M. (2004) Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026 10.1016/j.cell.2004.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levinson J. N., Chéry N., Huang K., Wong T. P., Gerrow K., Kang R., Prange O., Wang Y. T., and El-Husseini E. (2005) Neuroligins mediate excitatory and inhibitory synapse formation: involvement of PSD-95 and neurexin-1 in neuroligin-induced synaptic specificity. J. Biol. Chem. 280, 17312–17319 10.1074/jbc.M413812200 [DOI] [PubMed] [Google Scholar]
- 5. Chubykin A. A., Atasoy D., Etherton M. R., Brose N., Kavalali E. T., Gibson J. R., and Südhof T. C. (2007) Activity-dependent validation of excitatory vs. inhibitory synapses by Neuroligin-1 vs. Neuroligin-2. Neuron 54, 919–931 10.1016/j.neuron.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheiffele P., Fan J., Choih J., Fetter R., and Serafini T. (2000) Neuroligin expressed in non-neuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669 10.1016/S0092-8674(00)80877-6 [DOI] [PubMed] [Google Scholar]
- 7. Fu Z., Washbourne P., Ortinski P., and Vicini S. (2003) Functional excitatory synapses in HEK293 cells expressing neuroligin and glutamate receptors. J. Neurophysiol. 90, 3950–3957 10.1152/jn.00647.2003 [DOI] [PubMed] [Google Scholar]
- 8. Ko J., Zhang C., Arac D., Boucard A. A., Brunger A. T., and Südhof T. C. (2009) Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 28, 3244–3255 10.1038/emboj.2009.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu J., Xiao N., and Xia J. (2010) Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nat. Neurosci. 13, 22–24 10.1038/nn.2459 [DOI] [PubMed] [Google Scholar]
- 10. Singh S. K., Stogsdill J. A., Pulimood N. S., Dingsdale H., Kim Y. H., Pilaz L. J., Kim I. H., Manhaes A. C., Rodrigues W. S. Jr., Pamukcu A., Enustun E., Ertuz Z., Scheiffele P., Soderling S. H., Silver D. L., et al. (2016) Astrocytes assemble thalamocortical synapses by bridging NRX1a and NL1 via hevin. Cell 164, 183–196 10.1016/j.cell.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elegheert J., Cvetkovska V., Clayton A. J., Heroven C., Vennekens K. M., Smukowski S. N., Regan M. C., Jia W., Smith A. C., Furukawa H., Savas J. N., de Wit J., Begbie J., Craig A.-M., and Aricescu A. R. (2017) Structural mechanism for modulation of synaptic neuroligin-neurexin signalling by MDGA proteins. Neuron 95, 896–913 10.1016/j.neuron.2017.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ting J. T., Peça J., and Feng G. (2012) Functional consequences of mutations in postsynaptic scaffolding proteins and relevance to psychiatric disorders. Annu. Rev. Neurosci. 35, 49–71 10.1146/annurev-neuro-062111-150442 [DOI] [PubMed] [Google Scholar]
- 13. Selkoe D. J. (2002) Alzheimer's disease is a synaptic failure. Science 298, 789–791 10.1126/science.1074069 [DOI] [PubMed] [Google Scholar]
- 14. Südhof T. C. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Connor S. A., Ammendrup-Johnsen I., Chan A. W., Kishimoto Y., Murayama C., Kurihara N., Tada A., Ge Y., Lu H., Yan R., LeDue J. M., Matsumoto H., Kiyonari H., Kirino Y., Matsuzaki F., et al. (2016) Altered cortical dynamics and cognitive function upon haploinsufficiency of the autism-linked excitatory synaptic suppressor MDGA2. Neuron 91, 1052–1068 10.1016/j.neuron.2016.08.016 [DOI] [PubMed] [Google Scholar]
- 16. Kähler A. K., Djurovic S., Kulle B., Jönsson E. G., Agartz I., Hall H., Opjordsmoen S., Jakobsen K. D., Hansen T., Melle I., Werge T., Steen V. M., and Andreassen O. A. (2008) Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1089–1100 10.1002/ajmg.b.30726 [DOI] [PubMed] [Google Scholar]
- 17. Li J., Liu J., Feng G., Li T., Zhao Q., Li Y., Hu Z., Zheng L., Zeng Z., He L., Wang T., and Shi Y. (2011) The MDGA1 gene confers risk to schizophrenia and bipolar disorder. Schizophr. Res. 125, 194–200 10.1016/j.schres.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 18. Zhou X., and Bessereau J.-L. (2019) Molecular architecture of genetically-tractable GABA synapses in C. elegans. Front. Mol. Neurosci. 12, 304 10.3389/fnmol.2019.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinan-Lucarré B., Tu H., Pierron M., Ibáñez Cruceyra P., Zhan H., Stigloher C., Richmond J. E., and Bessereau J.-L. (2014) C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature 511, 466–470 10.1038/nature13313 [DOI] [PubMed] [Google Scholar]
- 20. Hunter J. W., Mullen G. P., McManus J. R., Heatherly J. M., Duke A., and Rand J. B. (2010) Neuroligin-deficient mutants of C. elegans have sensory processing deficits and are hypersensitive to oxidative stress and mercury toxicity. Dis. Model. Mech. 3, 366–376 10.1242/dmm.003442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Z., Hom S., Kudze T., Tong X.-J., Choi S., Aramuni G., Zhang W., and Kaplan J. M. (2012) Neurexin and neuroligin mediate retrograde synaptic inhibition in C. elegans. Science 337, 980–984 10.1126/science.1224896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tu H., Pinan-Lucarré B., Ji T., Jospin M., and Bessereau J.-L. (2015) C. elegans punctin clusters GABAA receptors via neuroligin binding and UNC-40/DCC recruitment. Neuron 86, 1407–1419 10.1016/j.neuron.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 23. Maro G. S., Gao S., Olechwier A. M., Hung W. L., Liu M., Özkan E., Zhen M., and Shen K. (2015) MADD-4/Punctin and neurexin organize C. elegans GABAergic postsynapses through neuroligin. Neuron 86, 1420–1432 10.1016/j.neuron.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hubmacher D., and Apte S. S. (2015) ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 47, 34–43 10.1016/j.matbio.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferrer-Ferrer M., and Dityatev A. (2018) Shaping synapses by the neural extracellular matrix. Front. Neuroanat. 12, 40–56 10.3389/fnana.2018.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krishnaswamy V. R., Benbenishty A., Blinder P., and Sagi I. (2019) Demystifying the extracellular matrix and its proteolytic remodeling in the brain: structural and functional insights. Cell. Mol. Life Sci. 76, 3229–3248 10.1007/s00018-019-03182-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dow D. J., Huxley-Jones J., Hall J. M., Francks C., Maycox P. R., Kew J. N. C., Gloger I. S., Mehta N. A. L., Kelly F. M., Muglia P., Breen G., Jugurnauth S., Pederoso I., St.Clair D., Rujescu D., et al. (2011) ADAMTSL3 as a candidate gene for schizophrenia: gene sequencing and ultra-high density association analysis by imputation. Schizophr. Res. 127, 28–34 10.1016/j.schres.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 28. Hamel M. G., Ajmo J. M., Leonardo C. C., Zuo F., Sandy J. D., and Gottschall P. E. (2008) Multimodal signaling by the ADAMTSs (a disintegrin and metalloproteinase with thrombospondin motifs) promotes neurite extension. Exp. Neurol. 210, 428–440 10.1016/j.expneurol.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chelini G., Pantazopoulos H., Durning P., and Berretta S. (2018) The tetrapartite synapse: a key concept in the pathophysiology of schizophrenia. Eur. Psychiatry 50, 60–69 10.1016/j.eurpsy.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pantazopoulos H., Woo T.-U. W., Lim M. P., Lange N., and Berretta S. (2010) Extracellular matrix-glial abnormalities in the amygdala and entorhinal cortex of subjects diagnosed with schizophrenia. Arch. Gen. Psychiatry 67, 155–166 10.1001/archgenpsychiatry.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterso B., and Nagase H. (2007) Proteolytic activities of human ADAMTS-5 comparative studies with ADAMTS-4. J. Biol. Chem. 282, 18294–18306 10.1074/jbc.M701523200 [DOI] [PubMed] [Google Scholar]
- 32. Tsutsui K., Manabe R., Yamada T., Nakano I., Oguri Y., Keene D. R., Sengle G., Sakai L. Y., and Sekiguchi K. (2010) ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J. Biol. Chem. 285, 4870–4882 10.1074/jbc.M109.076919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bader H. L., Wang L. W., Ho J. C., Tran T., Holden P., Fitzgerald J., Atit R. P., Reinhardt D. P., and Apte S. S. (2012) A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module. Matrix Biol. 31, 398–411 10.1016/j.matbio.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colige A., Ruggiero F., Vandenberghe I., Dubail J., Kesteloot F., Van Beeumen J., Beschin A., Brys L., Lapière C. M., and Nusgens B. (2005) Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I–III and V. J. Biol. Chem. 280, 34397–34408 10.1074/jbc.M506458200 [DOI] [PubMed] [Google Scholar]
- 35. Rao N., Ke Z., Liu H., Ho C.-J., Kumar S., Xiang W., Zhu Y., and Ge R. (2013) ADAMTS4 and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice. Int. J. Cancer 133, 294–307 10.1002/ijc.28037 [DOI] [PubMed] [Google Scholar]
- 36. Hubmacher D., Wang L. W., Mecham R. P., Reinhardt D. P., and Apte S. S. (2015) Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia – a novel mouse model providing insights into geleophysic dysplasia. Disease Model. Mech. 8, 487–499 10.1242/dmm.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seetharaman A., Selman G., Puckrin R., Barbier L., Wong E., D'Souza S. A., and Roy P. J. (2011) MADD-4 is a secreted cue required for midline-oriented guidance in Caenorhabditis elegans. Dev. Cell 21, 669–680 10.1016/j.devcel.2011.07.020 [DOI] [PubMed] [Google Scholar]
- 38. Vasudevan D., and Haltiwanger R. S. (2014) Novel roles for O-linked glycans in protein folding. Glycoconj. J. 31, 417–426 10.1007/s10719-014-9556-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozma K., Keusch J. J., Hegemann B., Luther K. B., Klein D., Hess D., Haltiwanger R. S., and Hofsteenge J. (2006) Identification and characterization of a β1,3-glucosyltransferase that synthesizes the Glc-β1,3-Fuc disaccharide on thrombospondin type 1 repeats. J. Biol. Chem. 281, 36742–36751 10.1074/jbc.M605912200 [DOI] [PubMed] [Google Scholar]
- 40. Flatmark T., and Sletten K. (1968) Multiple forms of cytochrome c oxidase in the rat. J. Biol. Chem. 243, 1623–1629 [PubMed] [Google Scholar]
- 41. Midelfort C. F., and Mehler A. H. (1972) Deamidation in vivo of an asparagine residue of rabbit muscle aldolase. Proc. Natl. Acad. Sci. U. S. A. 69, 1816–1819 10.1073/pnas.69.7.1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson N. E., and Robinson A. B. (2001) Molecular clocks. Proc. Natl. Acad. Sci. U. S. A. 98, 944–949 10.1073/pnas.98.3.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stephenson R. C., and Clarke S. (1989) Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 264, 6164–6170 [PubMed] [Google Scholar]
- 44. Deverman B. E., Cook B. L., Manson S. R., Niederhoff R. A., Langer E. M., Rosová I., Kulans L. A., Fu X., Weinberg J. S., Heinecke J. W., Roth K. A., and Weintraub S. J. (2002) Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell 111, 51–62 10.1016/S0092-8674(02)00972-8 [DOI] [PubMed] [Google Scholar]
- 45. Curnis F., Longhi R., Crippa L., Cattaneo A., Dondossola E., Bachi A., and Corti A. (2006) Spontaneous formation of l-isoaspartate and gain of function in fibronectin. J. Biol. Chem. 281, 36466–36476 10.1074/jbc.M604812200 [DOI] [PubMed] [Google Scholar]
- 46. Wilkinson K. D. (2004) Quantitative analysis of protein-protein interactions. Methods Mol. Biol. 261, 15–32 10.1385/1-59259-762-9:015 [DOI] [PubMed] [Google Scholar]
- 47. Gao G., Westling J., Thompson V. P., Howell T. D., Gottschall P. E., and Sandy J. D. (2002) Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J. Biol. Chem. 277, 11034–11041 10.1074/jbc.M107443200 [DOI] [PubMed] [Google Scholar]
- 48. Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., and Nagase H. (2004) Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 279, 10109–10119 10.1074/jbc.M312123200 [DOI] [PubMed] [Google Scholar]
- 49. Kim J. A., Kim D., Won S. Y., Han K. A., Park D., Cho E., Yun N., An H. J., Um J. W., Kim E., Lee J. O., Ko J., and Kim H. M. (2017) Structural insights into modulation of neurexin-neuroligin trans-synaptic adhesion by MDGA1/neuroligin-2 complex. Neuron 94, 1121–1131 10.1016/j.neuron.2017.05.034 [DOI] [PubMed] [Google Scholar]
- 50. Gangwar S. P., Zhong X., Seshadrinathan S., Chen H., Machius M., and Rudenko G. (2017) Molecular mechanism of MDGA1: regulation of neuroligin 2: neurexin trans-synaptic bridges. Neuron 94, 1132–1141 10.1016/j.neuron.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miller M. T., Mileni M., Comoletti D., Stevens R. C., Harel M., and Taylor P. (2011) The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure 19, 767–778 10.1016/j.str.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen F., Venugopal V., Murray B., and Rudenko G. (2011) The structure of neurexin 1α reveals features promoting a role as synaptic organizer. Structure 19, 779–789 10.1016/j.str.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang P., Lu H., Peixoto R. T., Pines M. K., Ge Y., Oku S., Siddiqui T. J., Xie Y., Wu W., Archer-Hartmann S., Yoshida K., Tanaka K. F., Aricescu A. R., Azadi P., Gordon M. D., et al. (2018) Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell 174, 1450–1464 10.1016/j.cell.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Whalen D. M., Malinauskas T., Gilbert R. J. C., and Siebold C. (2013) Structural insights into proteoglycan-shaped Hedgehog signalling. Proc. Natl. Acad. Sci. U. S. A. 110, 16420–16425 10.1073/pnas.1310097110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ai X., Do A.-T., Lozynska O., Kusche-Gullberg M., Lindahl U., and Emerson C. P. Jr. (2003) QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 162, 341–351 10.1083/jcb.200212083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flannery C. R., Zeng W., Corcoran C., Collins-Racie L. A., Chockalingam P. S., Hebert T., Mackie S. A., McDonagh T., Crawford T. K., Tomkinson K. N., LaVallie E. R., and Morris E. A. (2002) Autocatalytic cleavage of ADAMTS-4 (aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J. Biol. Chem. 277, 42775–42780 10.1074/jbc.M205309200 [DOI] [PubMed] [Google Scholar]
- 57. Colige A. C. (2020) Challenges and solutions for purification of ADAMTS proteases: an overview. Methods Mol. Biol. 2043, 45–53 10.1007/978-1-4939-9698-8_4 [DOI] [PubMed] [Google Scholar]
- 58. Kramerova I. A., Kawaguchi N., Fessler L. I., Nelson R. E., Chen Y., Kramerova A., Kusche-Gullberg M., Kramer J. M., Ackley B. D., Sieron A. L., Prockop D. J., and Fessler J. (2000) Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 127, 5475–5485 [DOI] [PubMed] [Google Scholar]
- 59. Tom R., Bisson L., and Durocher Y. (2008) Culture of HEK293-EBNA1 cells for production of recombinant proteins. CSH Protoc. 3, 1–5 10.1101/pdb.prot4976 [DOI] [PubMed] [Google Scholar]
- 60. Zimmermann L., Stephens A., Nam S.-Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A. N., and Alva V. (2018) A completely re-implemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 61. Sanchez R., and Sali A. (1997) Evaluation of comparative protein structure modelling by MODELLER-3. Proteins 1, 50–58 [DOI] [PubMed] [Google Scholar]
- 62. Zhou X., Gueydan M., Jospin M., Ji T., Valfort A., Pinan-Lucarré B., and Bessereau J.-L. (2020) The netrin receptor UNC-40/DCC assembles a postsynaptic scaffold and sets the synaptic content of GABAA receptors. Nat. Commun. 11, 2674 10.1038/s41467-020-16473-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frøkjær-Jensen C., Davis M. W., Sarov M., Saroy M., Taylor J., Flibotte S., LaBella M., Pozniakivsky A., Moerman D. G., and Jorgensen E. M. (2014) Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11, 529–534 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boulin T., Rapti G., Briseño-Roa L., Stigloher C., Richmond J. E., Paoletti P., and Bessereau J.-L. (2012) Positive modulation of a Cys-loop acetylcholine receptor by an auxiliary transmembrane subunit. Nat. Neurosci. 15, 1374–1381 10.1038/nn.3197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw MS data associated with Fig. S1 have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with data set identifier PXD020639.