Abstract
Nucleotide sugar transporters, encoded by the SLC35 gene family, deliver nucleotide sugars throughout the cell for various glycosyltransferase-catalyzed glycosylation reactions. GlcNAc, in the form of UDP-GlcNAc, and galactose, as UDP-Gal, are delivered into the Golgi apparatus by SLC35A3 and SLC35A2 transporters, respectively. However, although the UDP-Gal transporting activity of SLC35A2 has been clearly demonstrated, UDP-GlcNAc delivery by SLC35A3 is not fully understood. Therefore, we analyzed a panel of CHO, HEK293T, and HepG2 cell lines including WT cells, SLC35A2 knockouts, SLC35A3 knockouts, and double-knockout cells. Cells lacking SLC35A2 displayed significant changes in N- and O-glycan synthesis. However, in SLC35A3-knockout CHO cells, only limited changes were observed; GlcNAc was still incorporated into N-glycans, but complex type N-glycan branching was impaired, although UDP-GlcNAc transport into Golgi vesicles was not decreased. In SLC35A3-knockout HEK293T cells, UDP-GlcNAc transport was significantly decreased but not completely abolished. However, N-glycan branching was not impaired in these cells. In CHO and HEK293T cells, the effect of SLC35A3 deficiency on N-glycan branching was potentiated in the absence of SLC35A2. Moreover, in SLC35A3-knockout HEK293T and HepG2 cells, GlcNAc was still incorporated into O-glycans. However, in the case of HepG2 cells, no qualitative changes in N-glycans between WT and SLC35A3 knockout cells nor between SLC35A2 knockout and double-knockout cells were observed. These findings suggest that SLC35A3 may not be the primary UDP-GlcNAc transporter and/or different mechanisms of UDP-GlcNAc transport into the Golgi apparatus may exist.
Keywords: glycosylation, nucleotide sugar transporter, N-glycan, O-glycan, UDP–N-acetylglucosamine, Golgi apparatus, glycobiology, glycoconjugate, glycoprotein biosynthesis, Golgi, transporter, CRISPR/Cas, membrane transport, post-translational modification (PTM)
Glycosylation belongs to the one of the most important post-translational modifications of macromolecules, comprising N- and O-glycosylation of proteins, thus playing an essential role in the growth and development of eukaryotes, as well as in host–pathogen interactions. This modification increases the solubility and structural stability of proteins, protects against proteolysis, assists in protein folding, and plays a crucial role in immune response, cell–cell and cell–extracellular matrix recognition, and selective protein targeting (1).
The glycan moiety is formed by glycosyltransferases, the majority of whose catalytic centers face the lumen of the endoplasmic reticulum (ER) and Golgi apparatus. Nucleotide sugar transporters (NSTs) encoded by the SLC35 (solute carrier 35) gene family provide glycosyltransferases with activated sugars (nucleotide sugars), which serve as substrates for glycosylation reactions (2). NSTs also transport the nucleotide monophosphate byproduct of the glycosyltransferase reaction back to the cytoplasm. The ability of NSTs to form dimers or higher order oligomers (2) allows for multiple and simultaneous interactions between several NSTs, as well as between NSTs and glycosyltransferases (3–7). Initially, NSTs were demonstrated to be specific for the translocation of a single nucleotide sugar (8), but later multisubstrate transporters were reported (9–11). Mutations in genes encoding mammalian NSTs are associated with severe defects in development and physiology, demonstrating their requirement for proper functioning of multicellular organisms (12–16).
To date, the crystal structures of Saccharomyces cerevisiae GDP-Man transporter (Vrg4) in substrate-free, GDP-Man–bound, and GMP-bound forms (17, 18), mouse CMP–sialic acid (CMP-Sia) transporter in complex with CMP and CMP-Sia (19), and also the Zea mays analog of CMP-Sia transporter in complex with CMP (20) have been determined. The overall structure of NSTs revealed dimeric organization, the presence of 10 transmembrane α-helices connected by short loops, exposition of both N and C termini at the cytoplasmic side of the Golgi membrane, separate binding sites for the nucleotide and sugar, and a requirement for short-chain lipids.
GlcNAc is the most common among several monosaccharides used for synthesis of glycoconjugates, including all types of glycans, not only for extension of the synthesized oligosaccharide structures but importantly for generation of oligosaccharide branches. Golgi UDP-GlcNAc transporter (SLC35A3 and its homologs) has been characterized, i.e. in yeasts (21, 22), parasites (23), plants (24), and mammals (25–27). Established data suggested that SLC35A3 is the key, if not the only NST responsible for UDP-GlcNAc transport into the Golgi lumen. This assumption was made based mostly on (i) phenotypic correction of Kluyveromyces lactis mutant deficient in Golgi transport of UDP-GlcNAc using cDNA encoding SLC35A3 from Madin–Darby canine kidney cells (25), (ii) phenotypic complementation of K. lactis mutant with mouse SLC35A3 transporter (28), and (iii) UDP-GlcNAc transport using mouse SLC35A3 reconstituted in S. cerevisiae mutant and in artificial proteoliposomes (28). It is worth noting, however, that the above-mentioned approaches were based on a heterologous yeast expression system producing different types of N-glycans as compared with mammalian cells and on proteoliposomes using the protein overexpressed in S. cerevisiae cells. In contrast, our gene silencing analysis of the SLC35A3 gene did not result in significant changes in N-glycans (4). Chinese hamster ovary (CHO), Lec8 (CHO mutant cells lacking functional SLC35A2 gene), and HeLa cells with significantly decreased SLC35A3 activity exhibited only a decreased amount of highly branched tri- and tetra-antennary complex type N-glycans, whereas monoantennary and diantennary ones remained unchanged or even were accumulated.
Another important monosaccharide, galactose (Gal), is transported into the Golgi apparatus by the UDP-Gal transporter (SLC35A2) (29–36). Importantly, we demonstrated that biological function of UDP-GlcNAc and UDP-Gal transporters in glycosylation of proteins may be coupled. This hypothesis was based on the observations that (i) overexpression of UDP-GlcNAc transporter in mutant cells defective in UDP-Gal transporter partially restores galactosylation of N-glycans (37), (ii) chimeric proteins composed of regions derived from both transporters restore galactosylation in cells defective in UDP-Gal transporter (38, 39), and (iii) both overexpressed and native transporters form complexes detected not only by fluorescence lifetime imaging microscopy–FRET or in situ proximity ligation assay analyses but importantly also by coimmunoprecipitation assay, suggesting not only their close localization but also strong interaction (3, 7, 40).
This study presents novel data that broaden our knowledge of glycosylation process occurring in the Golgi apparatus. Taking into consideration that siRNA knockdown did not cause a complete inactivation of mRNA encoding SLC35A3 (4), as well as cell-specific glycosylation pattern, in this study we performed knockout of the SLC35A3 gene in three mammalian cell lines using CRISPR-Cas9 technology. Again, no significant changes in N-glycan synthesis were observed. Moreover, the knockout cells also continued to produce GlcNAc-containing O-glycans. Interestingly, we observed not only cell-specific glycosylation pattern but also differences in UDP-GlcNAc transporting activity. These findings suggest that SLC35A3 may not be the key transporter of UDP-GlcNAc and/or that different mechanisms of UDP-GlcNAc transport into the Golgi apparatus should be taken into consideration. We constructed knockout cells lacking not only the SLC35A3 transporter but also both SLC35A3 and SLC35A2 proteins. This attempt allowed us to follow changes occurring in N- and O-glycans in different cell lines resulting not only from a single deletion of the respective transporter but also from a combined deficiency of the two highly homologous and functionally related SLC35A subfamily members, which is important for broadening our knowledge on the functional glycosylation complexes and potential involvement of their members in the glycosylation process.
Results
Inactivation of the SLC35A2 and SLC35A3 genes
Taking into account the phenomenon of cell-specific glycosylation, a panel of CHO, HEK293T, and HepG2 cell lines consisting of (i) WT cells, (ii) single-knockout cells lacking SLC35A2, (iii) single-knockout cells lacking SLC35A3, and (iv) double-knockout cells lacking both SLC35A2 (A2KO) and SLC35A3 (A3KO) were analyzed. To obtain cell lines deficient in particular proteins, we employed CRISPR-Cas9 or CRISPR-Cas9 double nickase technology. The absence of SLC35A2 protein was confirmed by Western blotting analysis using highly specific anti-SLC35A2 antibody (Fig. 1). A similar approach was impossible in the case of SLC35A3 because of the lack of specific antibody. Therefore, to confirm that inactivation of the corresponding genes was effective, genomic DNA and RNA were isolated from WT cells and from several clones of the knockout cells. PCR and RT-PCR with subsequent DNA sequencing showed that the changes we detected do not allow for production of functional full-length or truncated protein, confirming that the CRISPR-Cas9 strategy successfully inactivated SLC35A3 gene in HepG2, HepG2 A2KO, HEK293T, HEK293T A2KO, CHO, and Lec8 cells (Figs. 2 and 3 and Table 1).
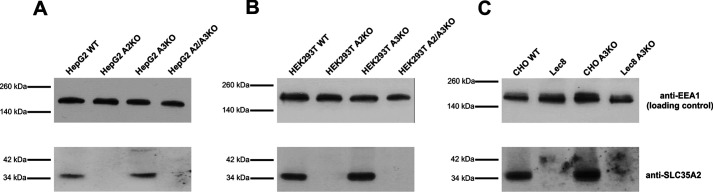
Figure 1.
Generation of SLC35A2 gene knockout in HepG2 and HEK293T cells. Proteins present in total cell lysates derived from WT or CRISPR-Cas9-generated knockout clones of HepG2 (A) and HEK293T (B) cells were separated by SDS-PAGE and analyzed by Western blotting. SLC35A2 (A2) protein was detected using rabbit anti-SLC35A2 antibody, followed by horseradish peroxidase–conjugated anti-rabbit IgG antibody and chemiluminescent staining. Protein loading was examined by immunostaining of EEA1. A2KO, SLC35A2 gene knockout; A3KO, SLC35A3 gene knockout; A2/A3KO, SLC35A2 and SLC35A3 genes knockouts. CHO (WT cells), Lec8 (SLC35A2 mutant cells), CHO A3KO (SLC35A3 gene knockout generated in CHO cells), and Lec8 A3KO (SLC35A3 gene knockout generated in Lec8 cells) cells were used to visualize specificity of anti-SLC35A2 antibody (C).
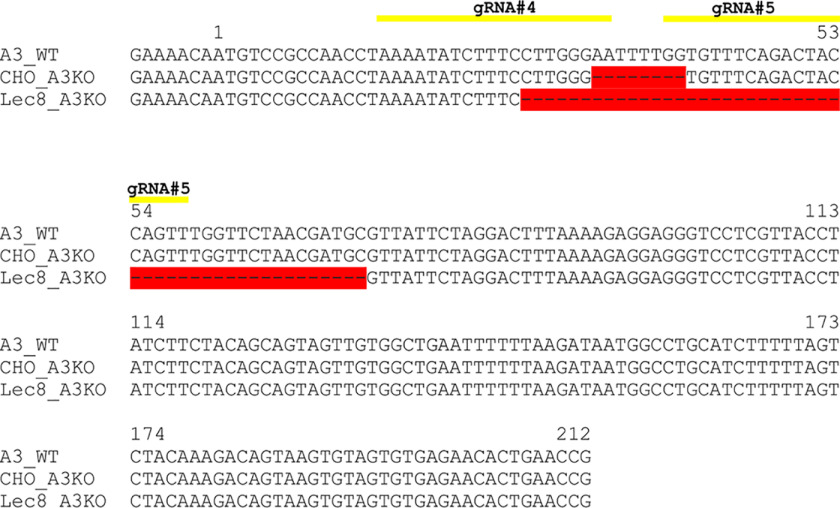
Figure 2.
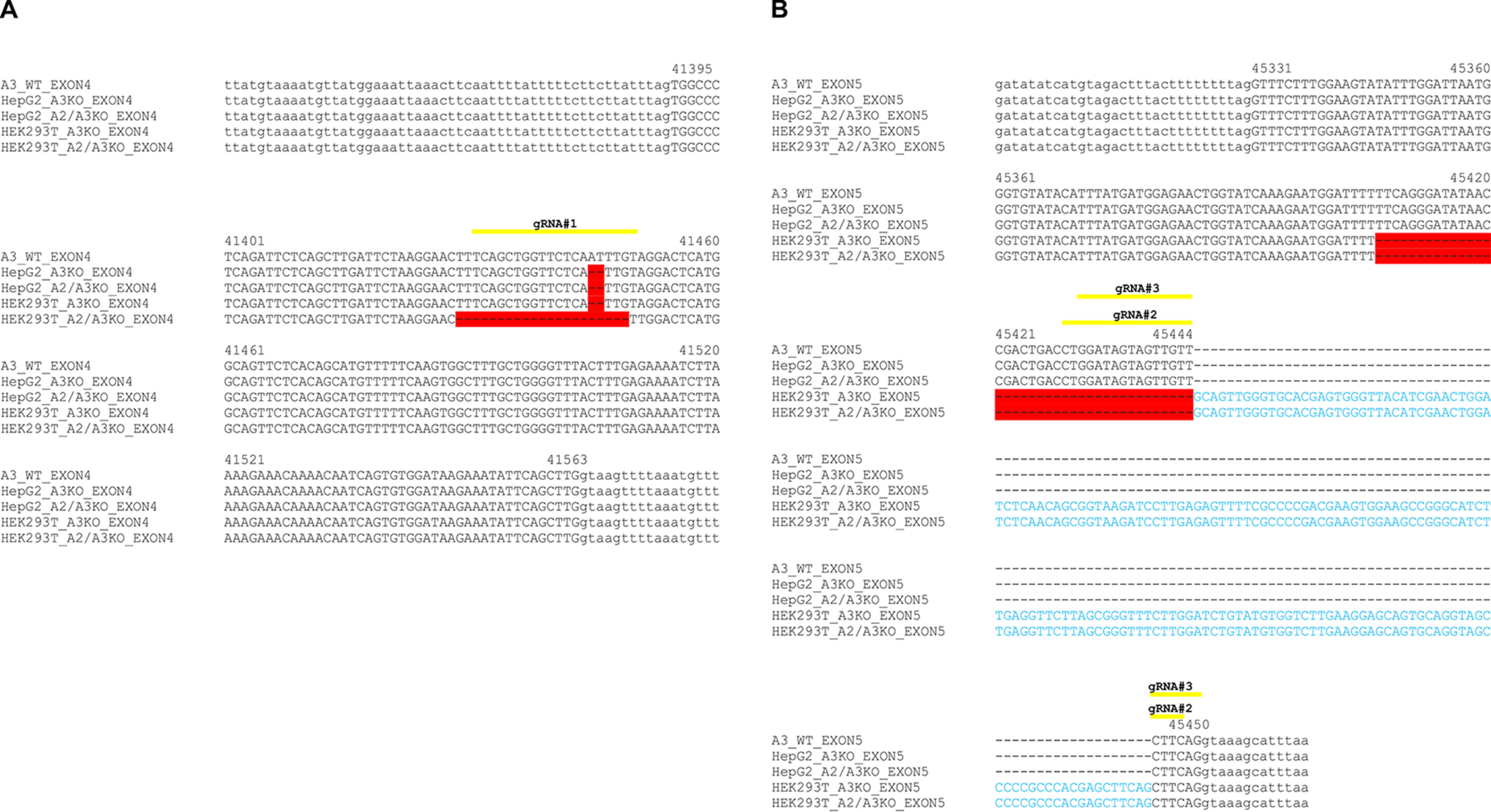
Generation of SLC35A3 gene knockout in HepG2 and HEK293T cells. A–D, DNA sequence alignment of SLC35A3 (A3) exon 4 (A) and exon 5 (B) of genomic DNA derived from WT, SLC35A3 gene knockout (A3KO), and SLC35A2 and SLC35A3 gene knockout (A2/A3KO) cells. DNA fragments were PCR-amplified and sequenced. Yellow lines indicate sequences complementary to gRNAs, red color indicates gene deletions, and blue letters indicate insertions caused by CRISPR-Cas9 approach.
Figure 3.
Evaluation of SLC35A3 gene knockout in CHO and Lec8 cells. Sequence alignment of SLC35A3 (A3) mRNA derived from CHO WT, SLC35A3 gene knockout CHO cells (CHO A3KO), and SLC35A3 gene knockout Lec8 (Lec8 A3KO) cells. RNA was isolated, RT-PCR was performed using SLC35A3 gene-specific primers, and DNA was sequenced. Yellow lines indicate sequences complementary to gRNAs, and red color indicates gene deletions caused by CRISPR-Cas9 approach.
Table 1.
List of changes caused by CRISPR-Cas9 strategy in the SLC35A3 gene
| Cell line | Nucleotide changes compared with SLC35A3 WT gene | Amino acid changes compared with SLC35A3 WT protein |
|---|---|---|
| HepG2 A3KO | Deletion of 2 nucleotides | Frameshift, premature stop codon |
| HepG2 A2/A3KO | Deletion of 2 nucleotides | Frameshift, premature stop codon |
| HEK293T A3KO | Deletion of 38 nucleotides, insertion of 178 nucleotides | Frameshift, premature stop codon |
| HEK293T A2/A3KO | Deletion of 57 nucleotides, insertion of 178 nucleotides | Changes in amino acid sequence, premature stop codon |
| CHO A3KO | Deletion of 8 nucleotides | Frameshift |
| LEC8 A3KO | Deletion of 47 nucleotides | Frameshift |
Analysis of N-glycans
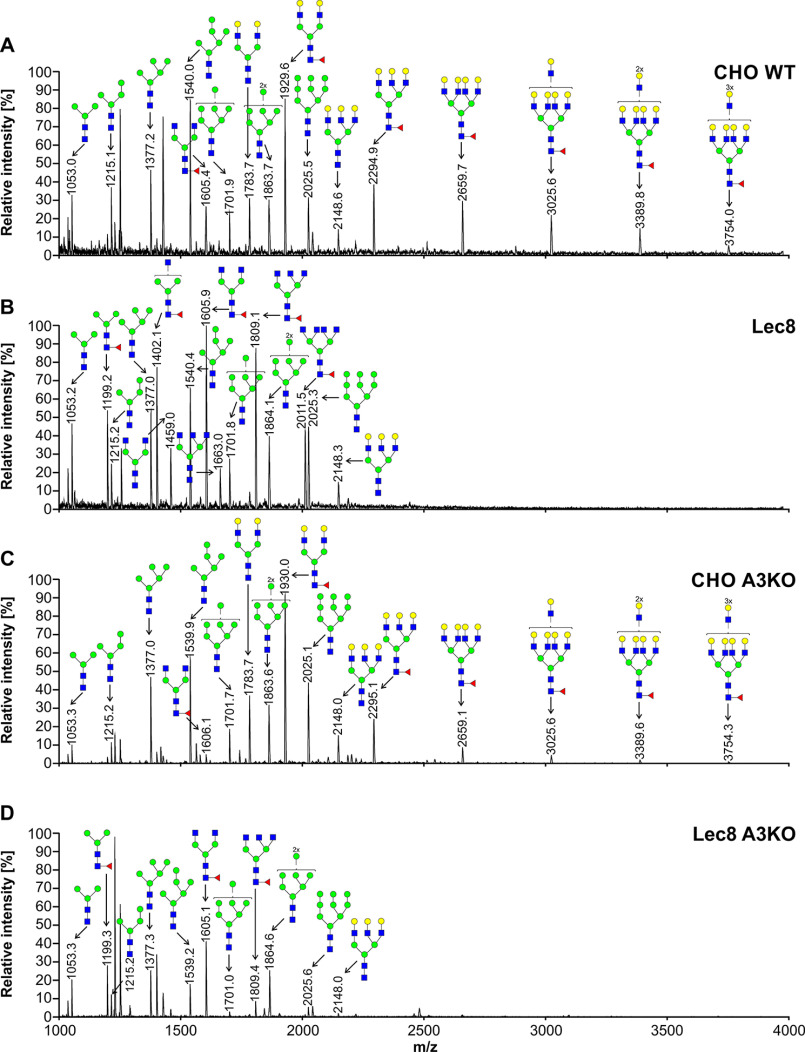
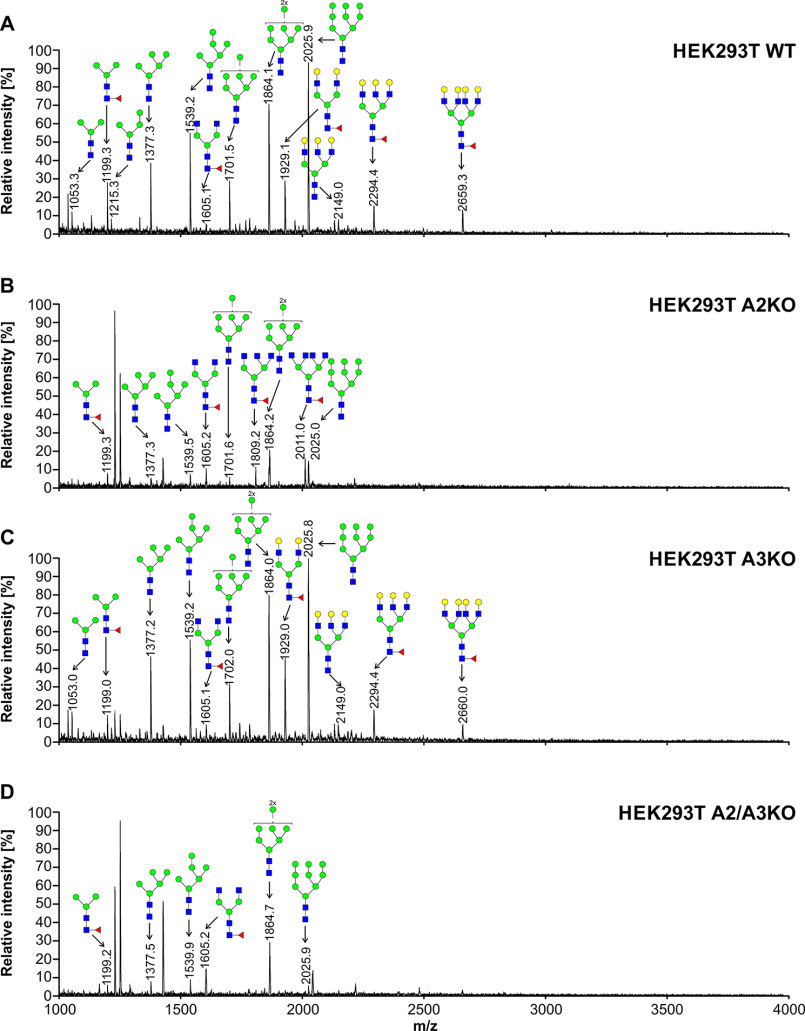
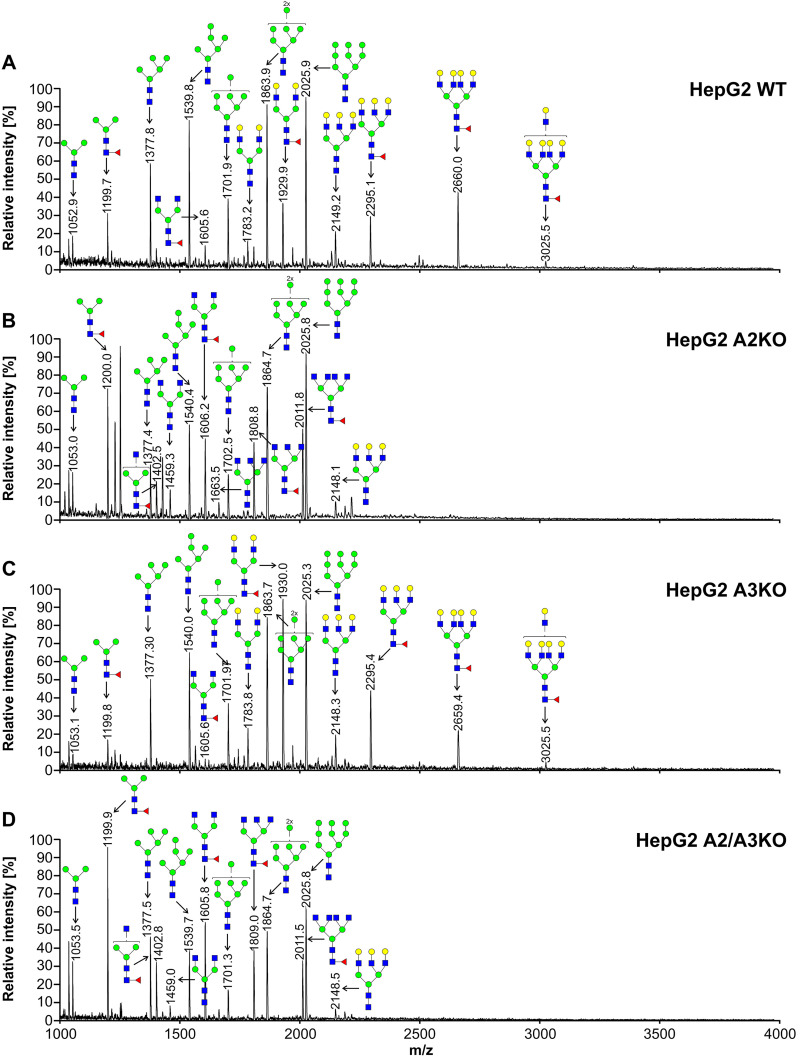
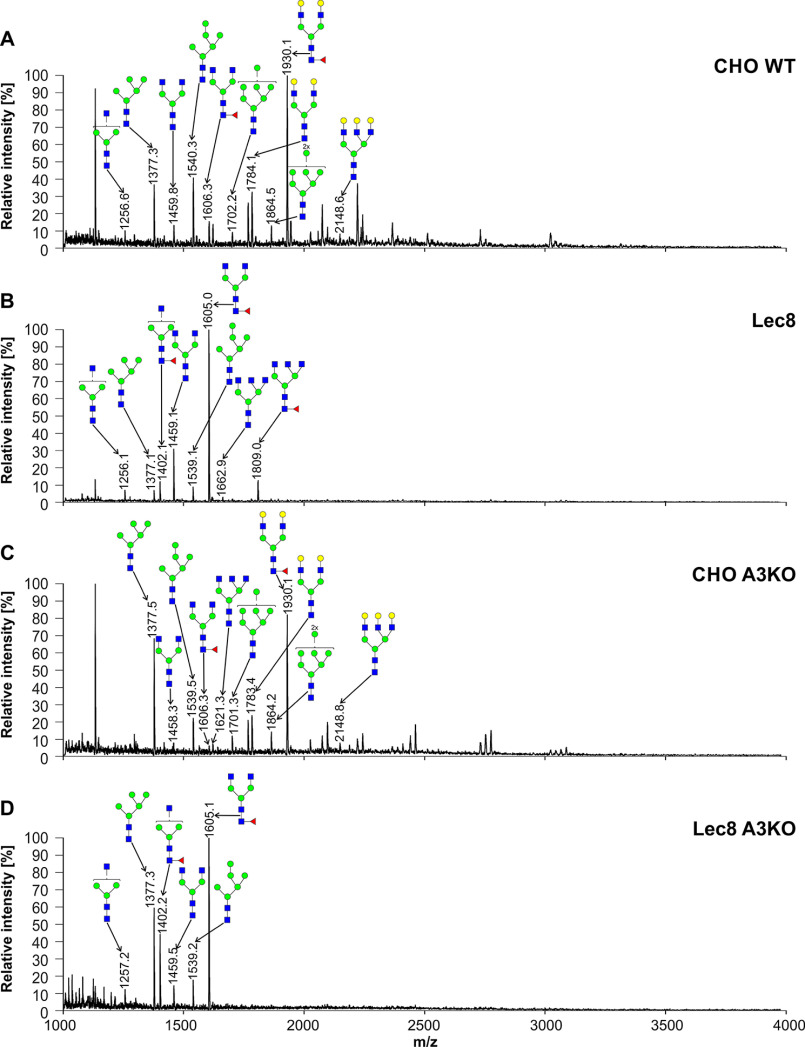
To characterize oligosaccharides produced by selected cell lines, first fluorescently labeled N-glycans were separated, and their N-glycan profiles compared. In contrast to differences clearly visible in the case of SLC35A2-deficient cells, no significant changes in N-glycan profiles between the WT and SLC35A3-deficient cells were observed (Fig. S1). Then 2-aminobenzamide (2-AB) derivatives were analyzed in detail using MALDI-TOF MS. If SLC35A3 was the main Golgi UDP-GlcNAc transporter, no complex type structures could be formed in cells lacking its activity. However, in the case of all single-knockout SLC35A3 cells, every complex type N-glycan typical of the parental cells could be detected (Figs. 4–6). However, subtle quantitative differences could be observed. In the case of CHO cells, four tetra-antennary structures were considerably decreased (Fig. 4, m/z of ∼2660, 3026, 3390, and 3754). This was less evident in the case of HEK293T cells (Fig. 5). Interestingly, the cells lacking both SLC35A2 and SLC35A3 displayed much more profound defects compared with the single-knockout cells. First of all, in the case of the double CHO and HEK293T knockout cells, a complete disappearance of a tetra-antennary, fucosylated structure could be observed (Figs. 4 and 5, m/z of ∼2011). In the case of CHO cells lacking both SLC35A2 and SLC35A3, a triantennary, nonfucosylated structure (m/z of ∼1663) was also absent from the N-glycan profile. In addition, a triantennary, fucosylated structure (m/z of ∼1802) was completely lost in the case of HEK293T double-knockout cells, resulting in a unique profile completely devoid of complex type N-glycans with more than two antennae, whereas in the case of CHO cells, the relative amount of this structure was substantially reduced. In the case of HepG2 knockout cells, however, we did not observe major changes in N-glycan profiles of SLC35A3-deficient cells compared with the WT cells (Fig. 6). Importantly, in contrast to CHO and HEK293T cell lines, no significant changes could be observed in the case of SLC35A3 knockout constructed in SLC35A2-deficient cells.
Figure 4.
Structural analysis of N-glycans isolated from CHO cells. 2-AB–labeled N-glycans derived from CHO WT (A), Lec8 (B), CHO SLC35A3-deficient (CHO A3KO) (C), and Lec8 SLC35A3-deficient (Lec8 A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
Figure 5.
Structural analysis of N-glycans isolated from HEK293T cells. 2-AB–labeled N-glycans derived from HEK293T WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
Figure 6.
Structural analysis of N-glycans isolated from HepG2 cells. 2-AB–labeled N-glycans derived from HepG2 WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
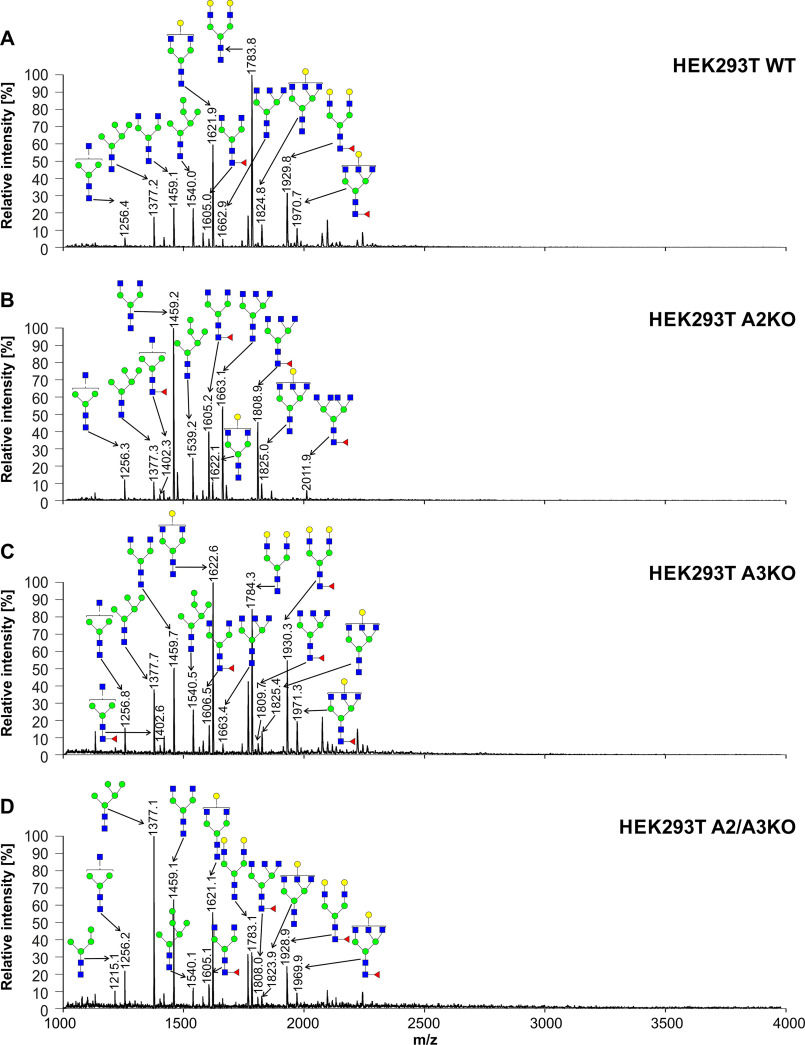
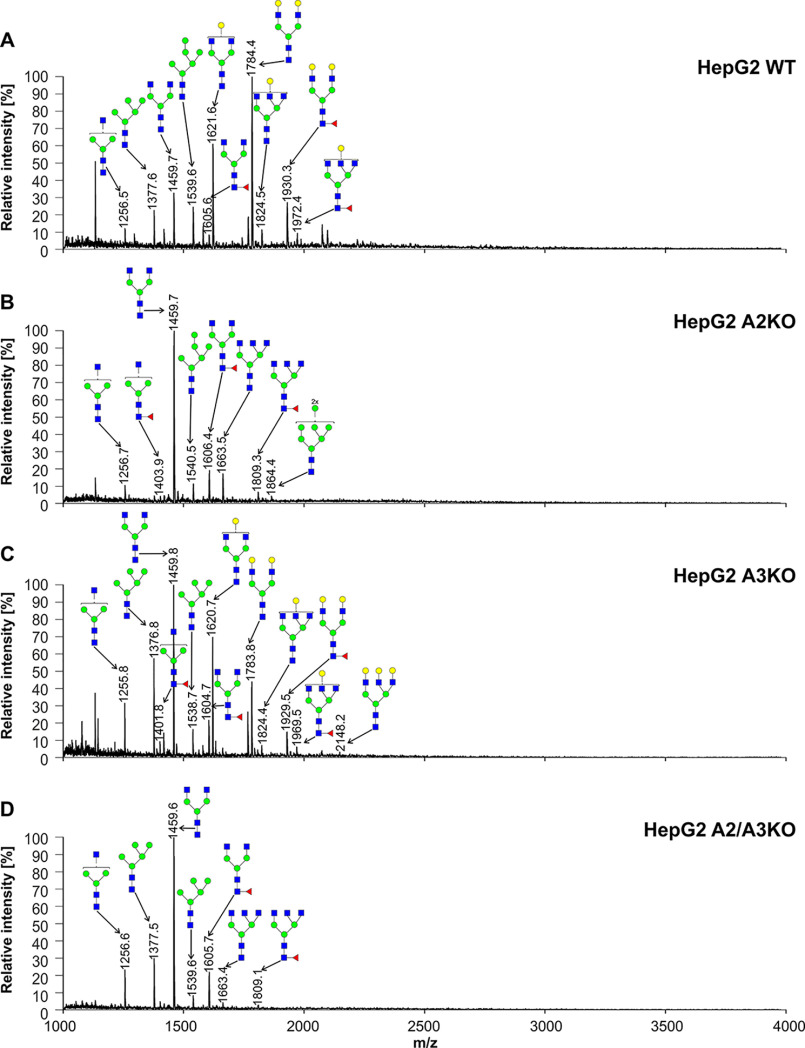
To analyze mature N-glycans in secreted proteins, we employed a reporter glycoprotein assay. For this purpose, we engineered a modified secreted alkaline phosphatase (SEAP) and established an efficient overexpression and purification protocol.1 PNGase-released and 2-AB–labeled N-glycans were separated, and N-glycan profiles were qualitatively compared in all cell lines examined in this study. No significant changes were observed between the WT and SLC35A3 knockout cells (Fig. S2). This procedure allowed us also to confirm no significant changes in sialylated N-glycans. Next, we carried out a more detailed analysis using MALDI-TOF MS in positive ion mode. N-Glycans released from SEAP secreted by SLC35A3 knockout cells were mainly of complex type, and no significant changes in GlcNAc-containing structures were observed when compared with the parental cells (Figs. 7–9). In the case of double CHO knockout cells, a complete disappearance of triantennary structures (m/z of ∼1663 and ∼1809) could be observed when compared with Lec8 cells. N-Glycans released from SEAP produced by double HepG2 knockout cells were comparable with structures derived from HepG2 SLC35A2 knockout cells. In the case of single-knockout cells deficient in SLC35A3 a triantennary, a fully galactosylated structure could be observed (m/z of ∼2148). This glycan was completely absent from the corresponding profile obtained for the WT cells, which contained only a monogalactosylated triantennary structure (m/z of ∼1824). Surprisingly, in HEK293T SLC35A2 and double-knockout cells, we observed a substantial degree of galactosylation of SEAP-derived N-glycans. Double-knockout cells contained more galactosylated glycans than single-knockout SLC35A2 cells (five versus two glycans). All N-glycan structures detected by MALDI-TOF MS are listed in Table S2.
Figure 7.
Structural analysis of N-glycans isolated from reporter glycoprotein overexpressed in CHO cells. 2-AB–labeled N-glycans derived from the SEAP purified from CHO WT (A), Lec8 (B), CHO SLC35A3-deficient (CHO A3KO) (C), and Lec8 SLC35A3-deficient (Lec8 A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
Figure 8.
Structural analysis of N-glycans isolated from reporter glycoprotein overexpressed in HEK293T cells. 2-AB–labeled N-glycans derived from the SEAP purified from HEK293T WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
Figure 9.
Structural analysis of N-glycans isolated from reporter glycoprotein overexpressed in HepG2 cells. 2-AB–labeled N-glycans derived from the SEAP purified from HepG2 WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells were treated with neuraminidase and subjected to MALDI-TOF MS analysis carried out in positive ion mode with Na+ excess. N-Glycan composition was estimated using the GlycoMod tool. Identified peaks were labeled with mass information and cartoon representations of putative N-glycan chemical structures (based on biosynthetic knowledge). Representative data from two independent measurements with a similar tendency are shown. Blue squares, GlcNAc; green circles, mannose; yellow circles, galactose; red triangles, fucose.
Analysis of O-glycans
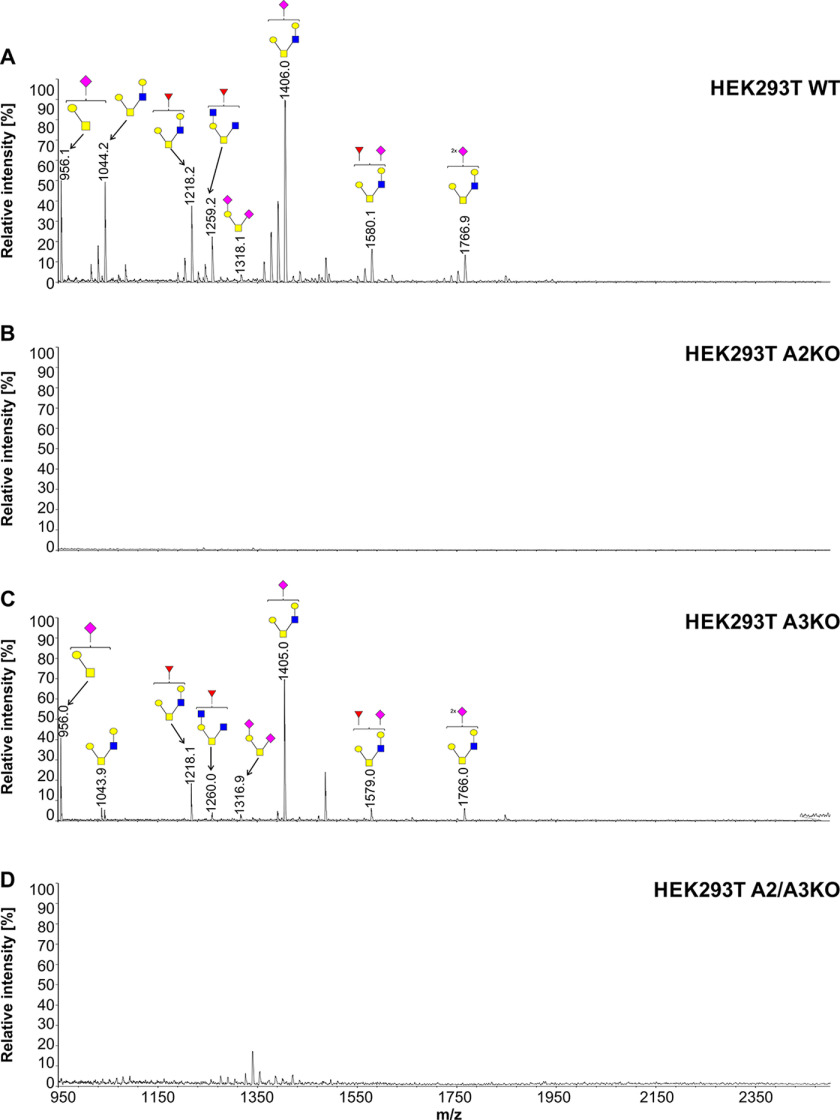
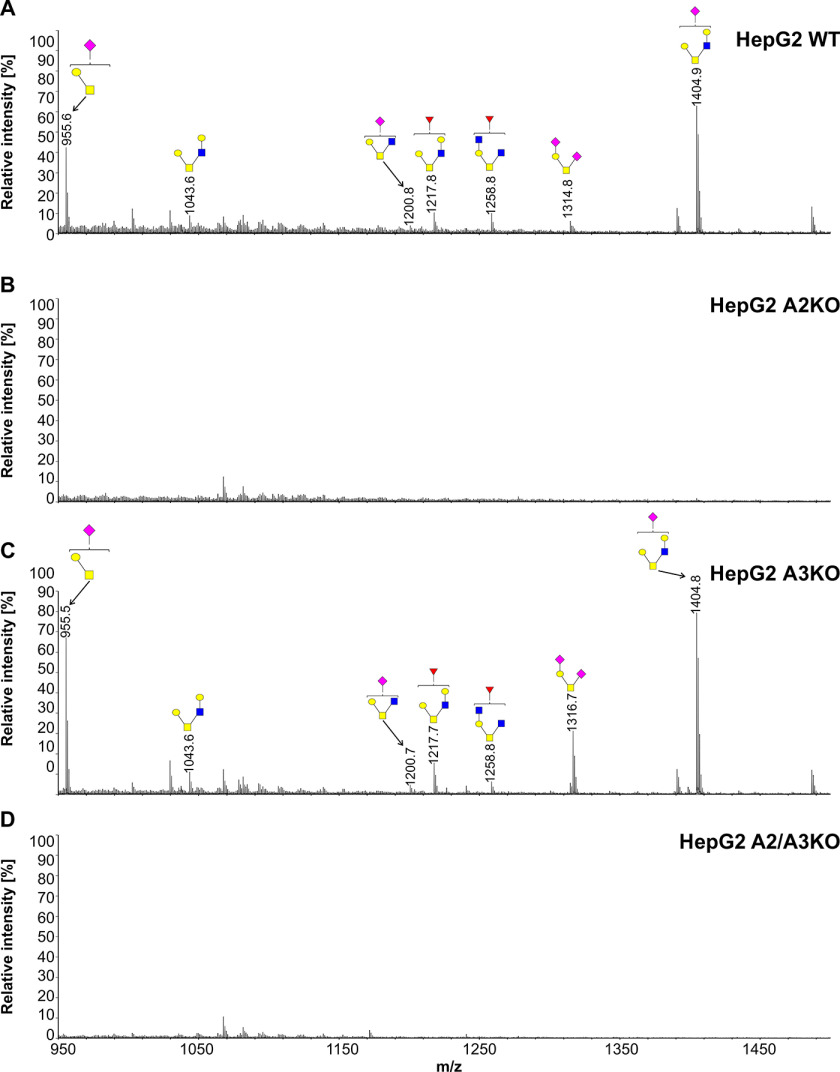
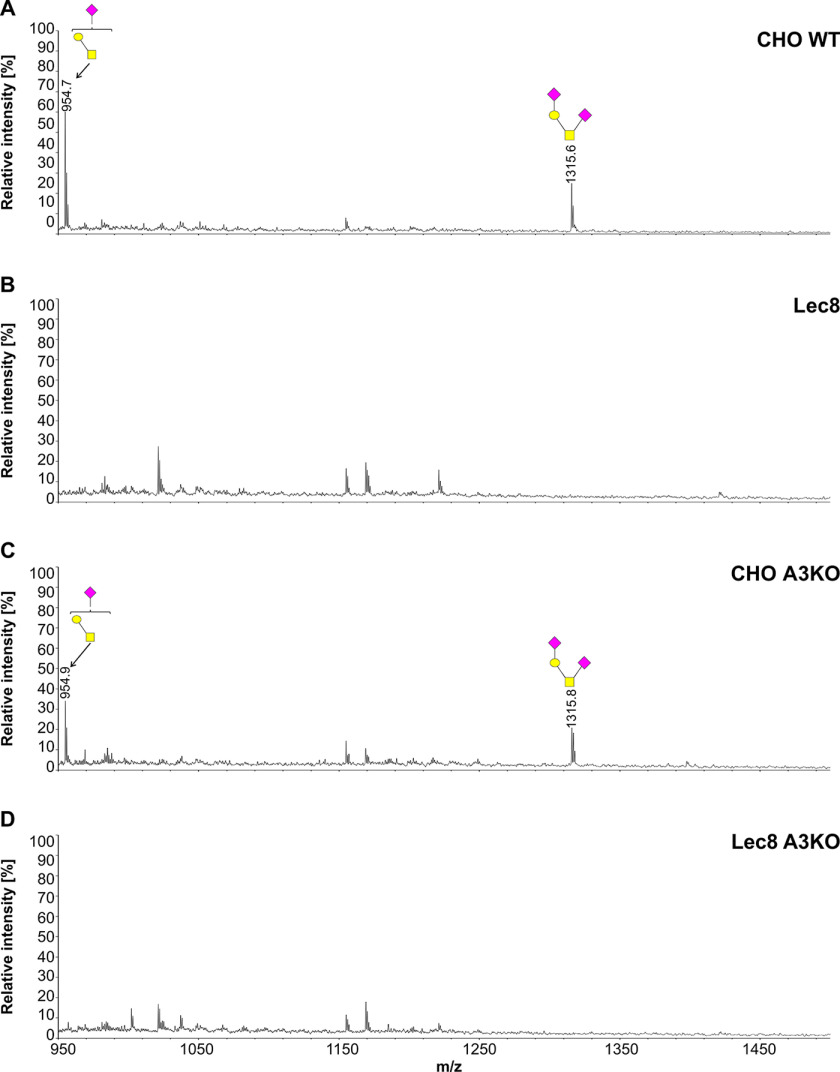
To further examine involvement of SLC35A3 in protein glycosylation, we employed the cellular O-glycome reporter/amplification strategy (41) to identify O-glycans produced by the WT and SLC35A3-deficient cells. Oligosaccharides were isolated, and their permethylated derivatives were analyzed using MALDI-TOF MS. In addition to O-glycan changes clearly visible in the case of the cells lacking SLC35A2, SLC35A3 deficiency did not prevent GlcNAc incorporation into O-glycans (Figs. 10–12). MALDI-TOF MS spectra obtained for O-glycans synthesized by the SLC35A3-deficient HEK293T and HepG2 cells showed that all GlcNAc-containing structures were still produced (Figs. 10 and 11). Even O-glycan with two incorporated GlcNAc moieties (m/z of ∼ 1260) could still be detected. In SLC35A3-deficient HEK293T cells, the relative amounts of the two nonsialylated GlcNAc-containing structures (m/z of 1044.2 and 1218.2) were substantially decreased. However, this was not visible in the case of their more complex, sialylated biosynthetic derivatives (m/z of ∼1406, ∼1580, and ∼1767). CHO cells produce only core 1–type structures (Fig. 12) and therefore are not suitable for studying this phenomenon. All O-glycan structures detected by MALDI-TOF MS are listed in Table S3.
Figure 10.
Structural analysis of O-glycans isolated from HEK293T cells. A–D, MALDI-TOF MS characterization of permethylated Bn-O-glycans secreted to culture medium by HEK293T WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells. O-Glycan composition was estimated using the GlycoWorkBench tool (2.1). Identified peaks were labeled with mass information and cartoon representations of putative O-glycan chemical structures (based on biosynthetic knowledge). Blue squares, GlcNAc; yellow squares, GalNAc; yellow circles, galactose; red triangles, fucose; pink diamonds, sialic acid.
Figure 11.
Structural analysis of O-glycans isolated from HepG2 cells. A–D, MALDI-TOF MS characterization of permethylated Bn-O-glycans secreted to culture medium by HepG2 WT (A), SLC35A2-deficient (A2KO) (B), SLC35A3-deficient (A3KO) (C), and SLC35A2- and SLC35A3-deficient (A2/A3KO) (D) cells. O-Glycan composition was estimated using the GlycoWorkBench tool (2.1). Identified peaks were labeled with mass information and cartoon representations of putative O-glycan chemical structures (based on biosynthetic knowledge). Blue squares, GlcNAc; yellow squares, GalNAc; yellow circles, galactose; red triangles, fucose; pink diamonds, sialic acid.
Figure 12.
Structural analysis of O-glycans isolated from CHO cells. A–D, MALDI-TOF MS characterization of permethylated Bn-O-glycans secreted to culture medium by CHO WT (A), Lec8 (B), CHO SLC35A3-deficient (CHO A3KO) (C), and Lec8 SLC35A3-deficient (Lec8 A3KO) (D) cells. O-Glycan composition was estimated using the GlycoWorkBench tool (2.1). Identified peaks were labeled with mass information and cartoon representations of putative O-glycan chemical structures (based on biosynthetic knowledge). Blue squares, GlcNAc; yellow squares, GalNAc; yellow circles, galactose; red triangles, fucose; pink diamonds, sialic acid.
Transport assay
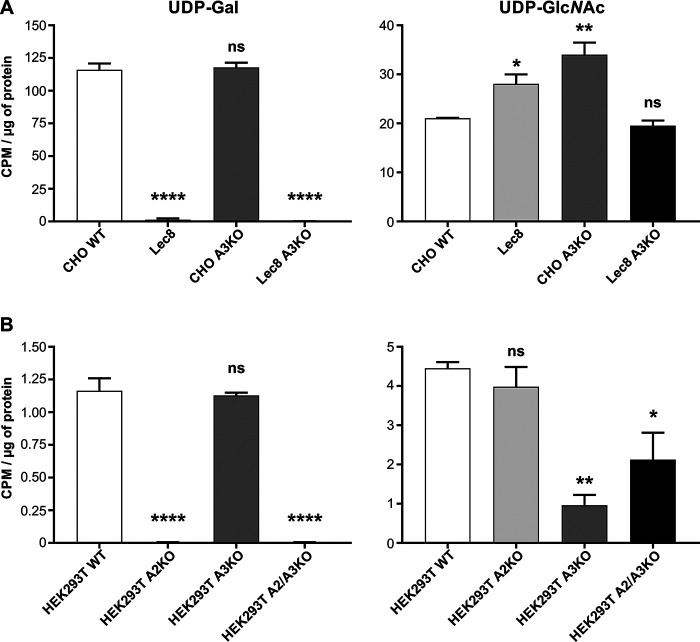
In a classical NST activity assay enriched, intact Golgi vesicles are incubated with radiolabeled NSTs and the label within the vesicles is mesured (8). However, this approach has several drawbacks. First of all, it requires enormous amounts of cells to obtain enough Golgi vesicles. Second, isolated vesicles are prone to damage during handling and storage. In addition, there is a high risk of sample contamination by the ER membranes. For all these reasons we employed an alternative approach to measure UDP-GlcNAc Golgi uptake, experimentally designed based on the procedure used for UDP-Gal uptake (42). This strategy relies on incubation of semipermeabilized cells with a radiolabeled nucleotide sugar and a membrane-permeable synthetic acceptor followed by a measurement of a cell-bound radioactivity. UDP-Gal uptake is expected to be significantly reduced in cells lacking SLC35A2 (4, 37). We indeed observed a negligible UDP-Gal uptake in single-knockout cells lacking SLC35A2, as well as in double-knockout cells lacking both SLC35A2 and SLC35A3 compared with the WT cells (Fig. 13). Next, we performed a similar assay with UDP-GlcNAc with the difference that in this case sugar incorporation into endogenous acceptors instead of a synthetic one was attempted. Surprisingly, in SLC35A3-deficient CHO cells, UDP-GlcNAc uptake did not decrease but was even higher than in the WT cells, which is in accordance with our gene silencing studies (4). In the case of HEK293T cells, however, SLC35A3 deficiency was accompanied by a statistically significant decrease in UDP-GlcNAc uptake, which was partially restored by knocking out SLC35A2 gene. Nevertheless, it must be emphasized that Golgi vesicles derived from SLC35A3-deficient HEK293T cells imported UDP-GlcNAc at a rate exceeding 25% of the WT, whereas in the case of SLC35A2-deficient HEK293T cells, UDP-Gal transport was virtually negligible. We did not examine UDP-GlcNAc transport into Golgi vesicles derived from HepG2 cells, because in their case the N-glycan profile changed upon SLC35A3 deficiency to a much lesser extent compared with CHO and HEK293T cells. A summary of all the observed phenotypic effects is presented in Table 2.
Figure 13.
Nucleotide sugar transport assay. A and B, UDP-[6-3H] galactose (UDP-Gal) and UDP-3H-N-acetyloglucosoamine (UDP-GlcNAc) transport assay in CHO WT, Lec8, SLC35A3-deficient CHO (CHO A3KO), SLC35A3-deficient Lec8 (Lec8 A3KO) cells (A) and HEK293T WT, SLC35A2-deficient (A2KO), SLC35A3-deficient (A3KO), and SLC35A2- and SLC35A3-deficient (A2/A3KO) cells (B). For each cell line transport assays were performed in three independent biological replicates. Error bars represent S.D. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, difference not statistically significant.
Table 2.
Transport and glycosylation changes in analyzed mammalian knockout cell lines
| Cell line | Inactivated gene | UDP-Gal transport | UDP-GlcNAc transport | Galactosylation of N-glycans | Antennal GlcNAc in N-glycans | Branching of N-glycans | Gal in O-glycans | GlcNAc in O-glycans |
|---|---|---|---|---|---|---|---|---|
| Lec8 | SLC35A2 | Strongly decreased | Increased | Almost completely blocked | Present | Not affected | Absent | Absent (lack of the respective enzyme) |
| CHO A3KO | SLC35A3 | Unchanged | Increased | Unchanged | Present | Subtly affected | Present | Absent (lack of the respective enzyme) |
| CHO A2/A3KO | Both SLC35A2 and SLC35A3 | Strongly decreased | Unchanged | Almost completely blocked | Present | Almost completely blocked | Absent | Absent (lack of the respective enzyme) |
| HEK293T A2KO | SLC35A2 | Strongly decreased | Unchanged | Completely blocked | Present | Not affected | Absent | Absent (lack of the respective acceptor) |
| HEK293T A3KO | SLC35A3 | Unchanged | Decreased to ∼25% | Unchanged | Present | Subtly affected | Present | Present |
| HEK293T A2/A3KO | Both SLC35A2 and SLC35A3 | Strongly decreased | Decreased to ∼50% | Completely blocked | Present | Completely blocked | Absent | Absent (lack of the respective acceptor) |
| HepG2 A2KO | SLC35A2 | NDa | NDa | Almost completely blocked | Present | Not affected | Absent | Absent (lack of the respective acceptor) |
| HepG2 A3KO | SLC35A3 | NDa | NDa | Unchanged | Present | Not affected | Present | Present |
| HepG2 A2/A3KO | Both SLC35A2 and SLC35A3 | NDa | NDa | Almost completely blocked | Present | Not affected | Absent | Absent (lack of the respective acceptor) |
aND, not determined.
Previously, we showed decreased UDP-Gal transporting activity in CHO SLC35A3 knockdown cells as compared with CHO WT cells, whereas in Lec8 SLC35A3 knockdown cells, UDP-Gal transporting activity was comparable with that determined for Lec8 cells but lower compared with CHO WT cells (4). Here, CHO cells lacking SLC35A3 exhibit transporting activity similar to CHO WT cells. These differences can be explained, at least in part, by different methods used to determine transporting activity in CHO cells. In the method used here, we can observe structures galactosylated with β1,4-Gal, whereas in the method used previously, all types of galactosylated structures were subjected to analysis. The data obtained here for HEK293T cells are very similar. However, observations made for UDP-GlcNAc transporting activity are dissimilar in both cell lines, suggesting a cell-specific effect of glycosylation.
Analysis of gene expression
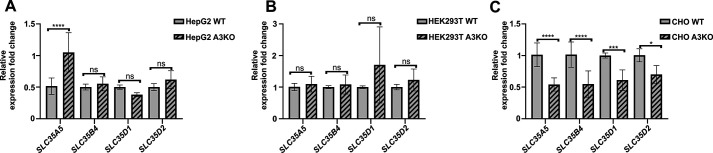
Searching for other UDP-GlcNAc transporters, we determined relative gene expression of potential candidates. In our previous study (43), we demonstrated that the first potentially strong candidate, SLC35B4, should be excluded. We showed that this protein is not involved in UDP-GlcNAc delivery for glycan synthesis because of its ER localization and the lack of glycophenotypic changes after the gene knockout. Nevertheless, this protein was further examined, together with SLC35A5, SLC35D1, and SLC35D2, in regard to transcript production. Analysis of mRNA encoding all examined potential candidates showed no dramatic changes between the WT and knockout cells (Fig. 14). Although we noticed an increase in mRNA encoding SLC35A5 in HepG2 cells lacking SLC35A3 and, at the same time, decreases in mRNAs encoding SLC35A5, SLC35B4, SLC35D1, and SLC35D2 in CHO cells lacking SLC35A3, the differences were rather subtle.
Figure 14.
Analysis of gene expression of selected nucleotide sugar transporters. Relative mRNA levels were determined in HepG2 (A), HEK293T (B), and CHO (C) WT and SLC35A3 knockout (A3KO) cells. GAPDH gene was used as a reference gene. *, p < 0.05; ***, p < 0.01; ****, p < 0.001; ns, difference not statistically significant. All samples were run in two independent experiments in triplicate for the target and reference genes.
Discussion
The goal of this study was to gain a better understanding of the role played in glycosylation by SLC35A3, which was considered for the long time the main Golgi UDP-GlcNAc transporter in mammals. GlcNAc plays a crucial role in numerous post-translational modifications of macromolecules including N- and O-glycosylation. Without the efficient delivery of UDP-GlcNAc into the Golgi lumen complex type N-glycans could not be synthesized. GlcNAc attachment is a prerequisite for galactosylation of N-glycans, which in turn enables their sialylation. Moreover, GlcNAc incorporation into N-glycans allows for their branching, i.e. formation of antennae. In addition, elongation of the α1,3-mannose arm of N-glycan with GlcNAc is considered a prerequisite for the core fucosylation. Multiantennary galactosylated, sialylated, and fucosylated structures are the fully mature forms of complex type N-glycans. These species are most capable of eliciting biological responses within the body. Our data obtained from an siRNA approach showed that the limited expression of the SLC35A3 gene causes a reduction in N-glycan branching but does not prevent GlcNAc from being incorporated into mono- and biantennary complex type structures (4). However, one cannot exclude the possibility that the residual expression of the SLC35A3 gene could have provided an intraluminal pool of UDP-GlcNAc sufficient for the biosynthesis of some complex type N-glycan species. We felt that there is a need for obtaining more convincing data on the contribution of SLC35A3 in N-glycosylation, and therefore we applied the CRISPR-Cas9 strategy to completely disable SLC35A3 expression. We also employed a larger repertoire of cell lines to avoid making conclusions from the effects that might turn out to be cell-specific. Moreover, for the first time we examined the contribution of SLC35A3 in O-glycosylation. Overall, this study has been greatly extended and refined when compared with the already published research.
Apart from the knockout cells deficient only in SLC35A3, in this work we also examined double-knockout cells in which both SLC35A2 and SLC35A3 genes were inactivated. We did it for several reasons. First of all, complex type N-glycans produced by the SLC35A2-deficient cells have simplified structures comparing with the WT cells because of the absence of galactose and sialic acid residues. In addition, because galactosylation and branching of complex type N-glycans are concurrent reactions (44), one might expect that turning off galactosylation would increase the subset of multiantennary structures and therefore provide a more suitable model for studying SLC35A3 function. Finally, we also demonstrated that SLC35A2 and SLC35A3 interact with each other and appear to be functionally related (3, 45), which raises a possibility that SLC35A2 could partially compensate for SLC35A3 deficiency in the single-knockout cells. For all the above reasons, we assumed that the putative effects of the SLC35A3 deficiency may be more pronounced and easier to demonstrate in cells that have already been deprived of SLC35A2. To obtain CHO cells lacking both NSTs, we knocked out the SLC35A3 gene in a well-established Lec8 mutant cells (34, 46). In the case of HepG2 and HEK293T cell lines, we sequentially inactivated both genes, because their SLC35A2-deficient variants were not available.
The specific decrease in the amount of tetra-antennary species in CHO and HEK293T cell lines suggests that in the absence of SLC35A3, the most compromised enzyme is Mgat5. Interestingly, we found that both SLC35A2 and SLC35A3 interact with mannoside N-acetylglucosaminyltransferases (Mgats) including Mgat5 as demonstrated by the fluorescence lifetime imaging microscopy–FRET and in situ proximity ligation assay analyses (5). Recently, several ternary complexes formed with the involvement of some Mgats, and the selected SLC35A subfamily members have been demonstrated (7). According to this study, Mgat5 was the only Mgat not being capable of interacting with other Mgats. We therefore hypothesize that SLC35A2 and SLC35A3 may play a crucial role in preventing displacement of Mgat5 from the putative multiprotein complex that possibly regulates N-glycosylation.
The relatively mild effect of SLC35A3 knockout on N-glycans produced by CHO and HEK293T cells was remarkably potentiated by the lack of SLC35A2. This appears rather surprising given the fact that GlcNAc addition is a prerequisite for Gal attachment. To our knowledge, it is not a common phenomenon that an early step of glycosylation is affected by a step occurring later within the pathway. This may suggest that in addition to transporting activity, SLC35A2 could play also a modulatory role in the multiprotein complex, because its deficiency impairs both galactosylation and GlcNAc attachment. Importantly, SLC35A2 and SLC35A3 are known to tightly interact with each other (3), and SLC35A3 overexpression in SLC35A2-deficient cells was shown to partially restore galactosylation of N-glycans (37). The current work supports the idea that these two proteins are functionally related and suggests their collaboration in the multiprotein complex performing the biosynthesis of complex type N-glycans. However, it has to be emphasized that in the case of the HepG2 cell line, knocking out SLC35A2 did not potentiate the outcome of SLC35A3 deficiency, so this phenomenon also appears to be cell-specific.
Profiles of SEAP-derived N-glycans differed from the ones obtained for the corresponding whole cell lysates in several aspects. The most novel and striking finding was the firm presence of Gal in N-glycans derived from SEAP secreted by the double and SLC35A2 single-knockout HEK293T cells. This may mean that SLC35A2 is not the sole UDP-Gal transporter, at least in this cell line. In fact, one of our previous studies demonstrated some residual galactosylation of N-glycans synthesized by the cells deficient in SLC35A2 activity (32). Alternatively, it could be concluded that cellular and secretory proteins are not evenly glycosylated by the knockout cells.
Another interesting finding is the increase of galactosylation of N-glycans in single SLC35A3 knockout HepG2 cells compared with the WT cells as well as double-knockout HEK293T cells compared with single-knockout SLC35A2 knockout cells. It appears that in these cell lines, the lack of SLC35A3 potentiates galactosylation of complex type N-glycans.
Apart from N-glycans, in mammalian cells GlcNAc is often incorporated into O-glycans. This monosaccharide can be found in core 2–, 3–, 4–, and 6–type O-linked structures. However, the contribution of SLC35A3 in O-glycan biosynthesis has never been explored. HepG2 and HEK293T cells produce a variety of core 2–type O-glycans. These species are synthesized with the involvement of the β-1,3-galactosyl-O-glycosyl-glycoprotein β-1,6-N-acetylglucosaminyltransferase (C2GnT). Its affinity for UDP-GlcNAc has been established at 1.9 mm (46) and more recently at 0.36 mm (47). Both values significantly exceed the Km value postulated for Mgat1 (0.04 mm) (48). One therefore might expect that C2GnT activity would become severely compromised by the limited UDP-GlcNAc availability in the Golgi lumen of SLC35A3-deficient cells, and the range of the produced structures would be narrowed down to core 1. The double-knockout cells are, however, expected to produce Tn antigen only, because galactose addition to GalNAc by the glycoprotein-GalNAc 3-β-galactosyltransferase 1 (C1GalT1) is a prerequisite for C2GnT action. To our surprise, O-glycans synthesized by the SLC35A3-deficient HepG2 and HEK293T cells contained all GlcNAc-containing structures, including the one possessing two incorporated GlcNAc moieties. This finding strongly argues against the idea of SLC35A3 being a general and unfocused UDP-GlcNAc transporter in the Golgi membrane of mammalian cells.
A knockout of the gene encoding a UDP-GlcNAc transporter should typically result in a significantly decreased uptake of this nucleotide sugar into the Golgi vesicles. However, in the case of CHO cells deficient in SLC35A3, UDP-GlcNAc transport was even slightly increased, whereas in the case of HEK293T cells, it was reduced to ∼25%. This relatively high residual UDP-GlcNAc import into the Golgi apparatus in the latter cells is in line with the persistence of GlcNAc-containing N- and O-glycan structures and provides another argument against the concept of SLC35A3 being the sole UDP-GlcNAc Golgi transporter in mammals. CHO and HEK293T cell lines clearly responded differentially to SLC35A3 deficiency in terms of UDP-GlcNAc uptake. Perhaps these differences result from a generally lower transport rate in HEK293T comparing with CHO cells. Moreover, in the case of HEK239T cells, GlcNAc is incorporated both in N-and O-glycans, whereas in the case of CHO cells, GlcNAc is present only in N-glycans. In addition, it has to be emphasized that the overall uptake of both UDP-Gal and UDP-GlcNAc was 1 order of magnitude lower in HEK293T than in CHO cells. Moreover, in the WT CHO cell line, UDP-Gal transport was higher than UDP-GlcNAc transport, whereas in the case of the WT HEK293T cell line, this was completely reversed. All these discrepancies make the obtained results difficult to compare between the two cell line panels.
Although it has been reported so far that SLC35A3 is the main Golgi UDP-GlcNAc transporter in mammals, our findings do not support this idea. This is in sharp contrast to the other well-characterized, apparently monospecific NSTs related with N-glycosylation, i.e. SLC35A1, SLC35A2, and SLC35C1, for whom the postulated substrate specificity is fully supported by the phenotypes of the mutation-bearing cell lines and human individuals (Table S1). In our opinion, unlike GDP-fucose, UDP-Gal, and CMP-Sia, the main supplier of UDP-GlcNAc for N-glycan biosynthesis has yet to be identified. Obviously, SLC35A3 activity is not required for O-glycosylation. Despite a complete inactivation of the SLC35A3 gene, we were not able to obtain the expected GlcNAc-deficient N- and O-glycophenotypes. There appears to be a puzzling discrepancy between the experimentally determined UDP-GlcNAc transporting activity of SLC35A3 and its native function. However, this is not the only case in which the substrate specificity of an NST cannot be easily translated into its biological role. For instance, although SLC35A2 is considered the sole UDP-Gal transporter in mammals, its deficiency does not completely abolish galactosylation of N-glycans (Ref. 32 and this work). The recently resolved three-dimensional structure of the CMP-Sia transporter from maize clearly demonstrates a transporting capability of this protein (20). However, it is widely accepted that plants do not incorporate sialic acid into glycoconjugates (49); therefore the physiological significance of such finding is obscure. Another example is the surprising lack of glycophenotypic effects of the putative UDP-GlcNAc/UDP-xylose transporter deficiency (43). The remaining members of the SLC35A subfamily, namely SLC35A4 and SLC35A5, also appear to be members of multiprotein complex with modulatory rather than transporting properties (40, 50). For SLC35A5 a decrease in UDP-GlcNAc uptake has been demonstrated in cells deficient in this protein, although it was not reflected in the corresponding glycophenotype (50). Nevertheless, SLC35A5 may be considered as an accessory (or alternative) Golgi UDP-GlcNAc transporter. Gene expression analysis demonstrated in this study suggests that, at least at the mRNA level, no induction of SLC35A5 gene expression caused by SLC35A3 gene knockout could be observed. Another Golgi-resident NST suspected of UDP-GlcNAc delivery is SLC35D2 (51). We hypothesize that some members of the SLC35 family could have evolved toward novel biological functions while retaining transporting activity. Nevertheless, it should be realized that within cells many factors can affect transport activity and modulate substrate specificity of the putative NSTs. UDP-GlcNAc transporting activity has been assigned to SLC35A3 based on experimental approaches using conditions that poorly reflect the dynamics and complexity of the mammalian secretory pathway. We feel that the assignment of the substrate specificity to a putative NST should be made with the support of the findings resulting from a knockdown or a knockout of the corresponding gene in a cell line of interest followed by an extensive characterization of the resulting glycoconjugates.
In this work we developed and characterized single and double-knockout cell lines lacking SLC35A3 or both SLC35A2 and SLC35A3, respectively. The study involved three cell lines selected based on differences in their N- and O-glycan repertoires, e.g. the ability to produce GlcNAc-containing O-glycans, polyLacNAc extensions, and highly branched structures. The phenotypic effects of SLC35A3 depletion differed depending on the cell line used. Regardless of these differences, it can be boldly assumed that if SLC35A3 was the only UDP-GlcNAc transporter in mammals, its deficiency should disable all GlcNAc transferases including enzymes displaying high affinities toward UDP-GlcNAc. However, we clearly demonstrated that SLC35A3 deficiency does not prevent GlcNAc incorporation into N- and O-glycans. Moreover, branching of complex type N-glycans is affected to a different extent depending on the cell line used, and this phenomenon is strongly potentiated by the SLC35A2 deficiency. The lack of SLC35A3 appears to affect some specific steps of N-glycosylation, which suggests it could be a member of a catalytically more efficient multiprotein complex rather than function as a single transporter. This appears to be particularly supported by the phenotype of CHO cells lacking SLC35A3, which display perturbations in N-glycan branching despite showing no decrease in UDP-GlcNAc transport. This suggests that the role of SLC35A3 played in glycosylation is different from what has been expected so far. We believe this might be related with the complexes formed by SLC35A2, SLC35A3, and Mgats. Specific and very restricted defects in glycosylation suggest that glycoproteins produced as a result of a given step of glycosylation may not be properly committed to the next one, because consecutively performed reactions become uncoupled because of the altered composition of the corresponding multiprotein complexes. However, further studies are required to address this idea.
Our findings clearly indicate that there are some gaps in the understanding of glycosylation. First of all, the biological role of SLC35A3 has to be specified. This task might not be possible to fulfill until the architecture of glycosylation-related multiprotein complexes is fully deciphered. It may also require resolving the three-dimensional structure of SLC35A3. Second, the ways by which UDP-GlcNAc is delivered into the Golgi lumen of mammalian cells are to be clarified.
Experimental procedures
Cell maintenance, gene inactivation, and transfection
HepG2, HEK293T, CHO, and Lec8 cells were grown as described previously (3, 40, 50). For transport assay, CHO and Lec8 cells were grown in F-12K complete medium.
HepG2 and HEK293T WT cells were transfected with a pool of three plasmids (containing three different SLC35A3-specific gRNAs), according to the manufacturer's instructions (Santa Cruz Biotechnology). Preselection was performed in MEM (for HepG2 cells) or DMEM (for HEK293T cells) complete medium supplemented with 1 μg/ml puromycin (InvivoGen). After 3 weeks of preselection, clones overexpressing GFP and RFP (the second and third selection markers in the employed CRISPR-Cas9 system) were isolated. CHO and Lec8 cells were transfected with a mixture of double nickase plasmids according to the manufacturer's instructions (Santa Cruz Biotechnology) and selected according to the manufacturer recommendations. For further analyses, clones overexpressing GFP (the second selection marker in the employed CRISPR-Cas9-NIC system) were isolated.
Evaluation of the SLC35A3 and SLC35A2 knockout efficiency
To confirm the SLC35A3 gene knockout in HepG2 and HEK293T cells, genomic DNA was isolated, and PCR was performed. PCR products were separated in 1.6% (w/v) agarose gel, visualized with ethidium bromide, and sequenced as described previously (40). Confirmation of the SLC35A3 gene knockout in CHO and Lec8 cells was performed at the RNA (RT-PCR) level as reported previously (40, 43) using primers listed in Table 3. To confirm the SLC35A2 gene knockout in all cell lines studied, we used the highly specific antibody against SLC35A2 protein (Merck) and Western blotting approach (4).
Table 3.
List of primers designed and used to evaluate the SLC35A3 gene knockout efficiency
| Name | DNA sequence (5′ → 3′) |
|---|---|
| Human_Exon4_SLC35A3_Fp | GCCCTCAGATTCTCAGCTTG |
| Human_Exon4_SLC35A3_Rp | ccagatatttggatcaatagcaga |
| Human_Exon5_SLC35A3_Fp | GGAAGTATATTTGGATTAATGGGTG |
| Human_Exon5_SLC3A3_Rp | ccttgaataaactagatacgagaga |
| Hamster_SLC35A3_Fp | GAAAACAATGTCCGCCAACC |
| Hamster_SLC35A3_Rp | GCCAAGTCACAAGAGTGTG |
aLowercase letters indicate primers complementary to the sequences of introns.
Western blotting
Cell lysates were subjected to SDS-PAGE using 10% polyacrylamide gels and transferred onto nitrocellulose (Whatman) as described previously (4). Briefly, after the transfer, the membrane was cut above the 52-kDa and below the 140-kDa prestained marker bands (Thermo Fisher Scientific). The upper part of the membrane was subjected to reaction with a rabbit anti-EEA1 antibody (1:2000; Santa Cruz), and the lower part of the membrane was subjected to reaction with a rabbit anti-SLC35A2 antibody (1:1000), both followed by anti-rabbit antibody conjugated with horseradish peroxidase (1:10,000; Sigma–Aldrich). Chemiluminescence reaction products were visualized as described previously (4).
Transport assay
The cells were harvested at 70–80% confluency and permeabilized using hypotonic conditions as described by others (52). UDP-Gal transport assay was performed as reported by Ng et al. (42). UDP-GlcNAc transport assay was carried out at 37 °C using 0.75 μm UDP-3H-GlcNAc (30 Ci/mmol; GlcNAc-6-3H(N); PerkinElmer). Time and concentration of unlabeled UDP-GlcNAc were optimized for each cell line separately. Finally, for CHO cells the reaction was carried out for 10 min in the presence of 0.25 μm unlabeled UDP-GlcNAc (Sigma–Aldrich). For HEK293T the reaction was carried out for 40 min without unlabeled substrate. The reaction was terminated by addition of 10 volumes of cold transport buffer. As a negative control, the reaction was conducted in the presence of 0.1% Triton X-100, which disrupts Golgi integrity. After the transport assay, the cells were spun down (3 min, 2000 rpm), and the resulting cell pellets were sonicated in the presence of 70% ethanol. After two rounds of ethanol extraction, the pellets were dried and dissolved in 5% SDS, and the radioactivity was measured. For each cell line transport assays were performed in three independent biological replicates, using different cell preparations and performed on different days. For statistical analysis, a one-way analysis of variance test with a Bonferroni's post hoc test and ranking method was employed. All analyses were performed with GraphPad Prism (GraphPad Software, La Jolla, CA). Statistical significance was assigned to p values <0.05.
Analysis of fluorescently labeled N- and O-glycans
Total cellular N-glycans were enzymatically released, isolated, fluorescently labeled with 2-AB, and separated as reported previously (37). Briefly, the cells were collected and lysed using the CelLytic M detergent solution (Sigma). Proteins from whole cell lysates were precipitated with cold acetone. Precipitates were dissolved in denaturation buffer and digested using peptide:N-glycosidase F (New England Biolabs). Released N-glycans were purified with Hypercarb Hypersep 200 mg SPE columns (Thermo Scientific) and labeled with 2-AB. Prior to HPLC analysis, the samples were purified on blotting filter paper (Whatman). Prior to MALDI-TOF MS analysis, 2-AB–labeled glycans were desialylated by using α2-3,6,8,9-neuraminidase A (New England Biolabs) and purified using Hypercarb Hypersep with 10–200-µl tips (Thermo Scientific). The last steps were also applied to purification of SEAP-derived N-glycans.
To analyze O-glycans, the procedure reported by Kudelka et al. (41) was adopted, as reported recently (50, 51). Briefly, the cells were incubated with 50 μm peracetylated O-glycan precursor (Ac3GalNAcBn) in medium containing 5% serum and grown for 72 h. Then the medium was collected and centrifuged (1000 × g), and the supernatant was subjected to glycan extraction procedure. Isolated N-glycans and permethylated O-glycans were subsequently analyzed using MALDI-TOF MS (4).
MALDI-TOF-MS experiments were performed on an Axima Performance TOF spectrometer (Shimadzu Biotech, Manchester, UK), equipped with a nitrogen laser (337 nm). The pulsed extraction ion source accelerated the ions to a kinetic energy of 20 keV. The data were obtained in a positive-ion linear mode. The calibration of the linear mode analysis was done using PEG in mass range up to 5000 Da. The accuracy of the product ion calibration is ∼1.5 Da. The mass calibration was conducted based on the average masses. The samples were dissolved in 20% acetonitrile in water. As a matrix, 2,5-dihydroxybenzoic acid (20 mg/ml) dissolved in 20 mm sodium acetate in 20% acetonitrile in water was used. The sample and matrix were combined at a 1:1 ratio. The resulting solution (1 μl) was spotted on a 384-well MALDI-TOF plate, followed by evaporation of the solvent at ambient temperature without any assistance. Each mass spectrum was accumulated from at least 200 laser shots and processed by Biotech Launchpad version 2.9.1 program (Shimadzu).
Secreted glycoprotein reporter assay
To overexpress and purify reporter glycoprotein, we constructed genetically modified vector, enabling improved expression and purification of SEAP.1 Briefly, psiTEST plasmid (Invivogen) was used as a template to add His6 at the C terminus of the SEAP. In addition, using a Q5 site-directed mutagenesis kit (New England Biolabs), a new N-glycosylation site at 278-amino acid position was introduced. The cells were transiently transfected with the modified plasmid using FuGENE 6 transfection reagent (Promega) at 3:1 transfection reagent:DNA ratio and grown in respective serum-containing medium. Media containing SEAP-His6 were collected 24 h after transfection by centrifugation (500 × g, 5 min) and examined with QUANTI-Blue phosphatase reagent according to the manufacturer's protocol (Invivogen). Recombinant SEAP-His6 was purified using nickel–nitrilotriacetic acid magnetic agarose beads (Jena Bioscience) according to the modified protocol.3 Purified SEAP-His6 obtained after elution with 2× glycoprotein denaturing buffer (New England Biolabs) was subjected to peptide:N-glycosidase F (New England Biolabs) treatment. Released N-glycans were isolated, labeled, and separated as described previously (37).
Quantitative reverse transcriptase–PCR
RNA was isolated from 1.2 × 106 cells using the ExtractMe total RNA kit (Blirt). Isolated RNA was treated with DNase I and purified using the Clean-Up RNA concentrator kit (A&A Biotechnology). RNA integrity was verified using a spectrophotometric method. Reverse transcription was carried out using 1 μg of RNA and the SensiFAST cDNA synthesis kit (Bioline). Real-time PCR was carried out using the RT HS-PCR mix SYBR A kit (A&A Biotechnology) and the LightCycler 96 System (Roche). The amplification reaction included initial denaturation at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The melting curves were analyzed to monitor the quality of PCR products. The relative quantification of expression of respective genes was determined compared with the human or hamster GAPDH gene as a reference, using the ΔΔCq method and LightCycler 96 System software (Roche). All samples were run in two independent experiments in triplicate for the target and reference genes. No-template controls were included on each reaction plate to check for contamination. All primers used in this study are listed in Table 4.
Table 4.
List of primers designed and used in quantitative RT-PCR
| Analyzed gene | DNA sequence (5′ → 3′) |
||
|---|---|---|---|
| HEK293T and HepG2 cells | CHO cells | ||
| SLC35A5 | Forward | CTGCTAGGTGCCATATTCATT | TGGTCCATTCCTGCCTTTCT |
| Reverse | AACACAGAATGACACAAGCAC | TGCGAAGTTTTGGTACTGGC | |
| SLC35B4 | Forward | ATGCGCCCGGCCTTGGCGGT | CCATGTTCTTCACCGTGAGC |
| Reverse | ACGCTCACGGTGAAGAACATG | ACTGGGAAGTCACCTGCTTT | |
| SLC35D1 | Forward | ATGGCGGAAGTTCATAGACGT | CTCTCCTGTGTGATGGGGTT |
| Reverse | TGAGCACGCTCTTATTCACCA | GTTTGCTCAGCTGCTCTTCA | |
| SLC35D2 | Forward | TCCTTCCTCATCGTGCTTGT | TGCTGACCACCTACGGTTTTC |
| Reverse | CAACGTAGAGGAGAGGCAGA | CACGGTGAACATTGGCAAGTC | |
| GAPDH | Forward | AGGTCGGAGTCAACGGATTT | GAAAGCTGTGGCGTGATGG |
| Reverse | TGACAAGCTTCCCGTTCTCA | TACTTGGCAGGTTTCTCCAG | |
Data availability
The MS data has been deposited in the GlycoPost under ID GPST000080. Some unpublished data are available upon request from the corresponding author (E-mail address: mariusz.olczak@uwr.edu.pl).
Supplementary Material
Acknowledgments
We thank Karolina Chabowska for generation of SLC35A2-deficient HEK293T cells.
This article contains supporting information.
Author contributions—B. S., D. M.-S., and M. O. conceptualization; B. S. data curation; B. S., P. S., and M. O. formal analysis; B. S., P. S., E. S., A. S., and M. O. investigation; B. S., P. S., T. O., H. H. F., and M. O. writing-review and editing; B. S. and P. S. visualization; D. M.-S. and T. O. writing-original draft; M. O. resources; M. O. supervision; M. O. project administration.
Funding and additional information—This work was supported by National Science Center (Krakow, Poland) Grants 2014/15/B/NZ3/00372 (to M. O.) and 2017/27/N/NZ3/00369 (to B. S.), funds from the Rocket Fund, and National Institutes of Health Grant R01DK099551 (to H. H. F). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
M. Olczak and B. Szulc, submitted for publication.
- UDP-Gal
- UDP-galactose
- UDP-GlcNAc
- UDP–N-acetylglucosamine
- CMP-Sia
- CMP–sialic acid
- NST
- nucleotide sugar transporter
- Mgat
- mannoside N-acetylglucosamine transferase
- ER
- endoplasmic reticulum
- CHO
- Chinese hamster ovary
- 2-AB
- 2-aminobenzamide
- SEAP
- secreted alkaline phosphatase
- C2GnT
- β-1,3-galactosyl-O-glycosyl-glycoprotein β-1,6-N-acetylglucosaminyltransferase.
References
- 1. Song Z. (2013) Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol. Aspects Med. 34, 590–600 10.1016/j.mam.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 2. Hadley B., Litfin T., Day C. J., Haselhorst T., Zhou Y., and Tiralongo J. (2019) Nucleotide sugar transporter SLC35 family structure and function. Comp. Struct. Biotechnol. J. 17, 1123–1134 10.1016/j.csbj.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maszczak-Seneczko D., Sosicka P., Majkowski M., Olczak T., and Olczak M. (2012) UDP–N-acetylglucosamine transporter and UDP-galactose transporter form heterologous complexes in the Golgi membrane. FEBS Lett. 586, 4082–4087 10.1016/j.febslet.2012.10.016 [DOI] [PubMed] [Google Scholar]
- 4. Maszczak-Seneczko D., Sosicka P., Olczak T., Jakimowicz P., Majkowski M., and Olczak M. (2013) UDP–N-acetylglucosamine transporter (SLC35A3) regulates biosynthesis of highly branched N-glycans and keratan sulfate. J. Biol. Chem. 288, 21850–21860 10.1074/jbc.M113.460543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maszczak-Seneczko D., Sosicka P., Kaczmarek B., Majkowski M., Luzarowski M., Olczak T., and Olczak M. (2015) UDP-galactose (SLC35A2) and UDP–N-acetylglucosamine (SLC35A3) transporters form glycosylation-related complexes with mannoside acetylglucosaminyltransferases (Mgats). J. Biol. Chem. 290, 15475–15486 10.1074/jbc.M115.636670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maszczak-Seneczko D., Sosicka P., Olczak T., and Olczak M. (2016) In Situ proximity ligation assay (PLA) analysis of protein complexes formed between Golgi-resident, glycosylation-related transporters and transferases in adherent mammalian cell cultures. Methods Mol. Biol. 1496, 133–143 10.1007/978-1-4939-6463-5_11 [DOI] [PubMed] [Google Scholar]
- 7. Khoder-Agha F., Sosicka P., Escriva Conde M. E., Hassinen A., Glumoff T., Olczak M., and Kellokumpu S. (2019) N-Acetylglucosaminyltransferases and nucleotide sugar transporters form multi-enzyme–multi-transporter assemblies in Golgi membranes in vivo. Cell. Mol. Life Sci. 76, 1821–1832 10.1007/s00018-019-03032-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirschberg C. B., Robbins P., and Abeijon C. (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49–69 10.1146/annurev.biochem.67.1.49 [DOI] [PubMed] [Google Scholar]
- 9. Suda T., Kamiyama S., Suzuki M., Kikuchi N., Nakayama K., Narimatsu H., Jigami Y., Aoki T., and Nishihara S. (2004) Molecular cloning and characterization of a human multisubstrate specific nucleotide-sugar transporter homologous to Drosophila fringe connection. J. Biol. Chem. 279, 26469–26474 10.1074/jbc.M311353200 [DOI] [PubMed] [Google Scholar]
- 10. Muraoka M., Kawakita M., and Ishida N. (2001) Molecular characterization of human UDP-glucuronic acid/UDP–N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 495, 87–93 10.1016/S0014-5793(01)02358-4 [DOI] [PubMed] [Google Scholar]
- 11. Caffaro C. E., and Hirschberg C. B. (2006) Nucleotide sugar transporters of the Golgi apparatus: from basic science to diseases. Acc. Chem. Res. 39, 805–812 10.1021/ar0400239 [DOI] [PubMed] [Google Scholar]
- 12. Thomsen B., Horn P., Panitz F., Bendixen E., Petersen A. H., Holm L. E., Nielsen V. H., Agerholm J. S., Arnbjerg J., and Bendixen C. (2006) A missense mutation in the bovine SLC35A3 gene, encoding a UDP–N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. 16, 97–105 10.1101/gr.3690506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edvardson S., Ashikov A., Jalas C., Sturiale L., Shaag A., Fedick A., Treff N. R., Garozzo D., Gerardy-Schahn R., and Elpeleg O. (2013) Mutations in SLC35A3 cause autism spectrum disorder, epilepsy and arthrogryposis. J. Med. Genet. 50, 733–739 10.1136/jmedgenet-2013-101753 [DOI] [PubMed] [Google Scholar]
- 14. Marini C., Hardies K., Pisano T., May P., Weckhuysen S., Cellini E., Suls A., Mei D., Balling R., Jonghe P. D., Helbig I., Garozzo D., and Guerrini R., Euro-EPINOMICS Consortium AR Working Group (2017) Recessive mutations in SLC35A3 cause early onset epileptic encephalopathy with skeletal defects. Am. J. Med. Genet. A 173, 1119–1123 10.1002/ajmg.a.38112 [DOI] [PubMed] [Google Scholar]
- 15. Edmondson A. C., Bedoukian E. C., Deardorff M. A., McDonald-McGinn D. M., Li X., He M., and Zackai E. H. (2017) A human case of SLC35A3-related skeletal dysplasia. Am. J. Med. Genet. A 173, 2758–2762 10.1002/ajmg.a.38374 [DOI] [PubMed] [Google Scholar]
- 16. Bruneel A., Cholet S., Drouin-Garraud V., Jacquemont M. L., Cano A., Mégarbané A., Ruel C., Cheillan D., Dupré T., Vuillaumier-Barrot S., Seta N., and Fenaille F. (2018) Complementarity of electrophoretic, mass spectrometric, and gene sequencing techniques for the diagnosis and characterization of congenital disorders of glycosylation. Electrophoresis 39, 3123–3132 10.1002/elps.201800021 [DOI] [PubMed] [Google Scholar]
- 17. Parker J. L., Corey R. A., Stansfeld P. J., and Newstead S. (2019) Structural basis for substrate specificity and regulation of nucleotide sugar transporters in the lipid bilayer. Nat. Commun. 10, 4657 10.1038/s41467-019-12673-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parker J. L., and Newstead S. (2017) Structural basis of nucleotide sugar transport across the Golgi membrane. Nature 551, 521–524 10.1038/nature24464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahuja S., and Whorton M. R. (2019) Structural basis for mammalian nucleotide sugar transport. eLife 8, e45221 10.7554/eLife.45221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nji E., Gulati A., Qureshi A. A., Coincon M., and Drew D. (2019) Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat. Struct. Mol. Biol. 26, 415–423 10.1038/s41594-019-0225-y [DOI] [PubMed] [Google Scholar]
- 21. Abeijon C., Robbins P. W., and Hirschberg C. B. (1996) Molecular cloning of the Golgi apparatus uridine diphosphate–N-acetylglucosamine transporter from Kluyveromyces lactis. Proc. Natl. Acad. Sci. U.S.A. 93, 5963–5968 10.1073/pnas.93.12.5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guillen E., Abeijon C., and Hirschberg C. B. (1999) The genes for the Golgi apparatus N-acetylglucosaminyltransferase and the UDP–N-acetylglucosamine transporter are contiguous in Kluyveromyces lactis. J. Biol. Chem. 274, 6641–6646 10.1074/jbc.274.10.6641 [DOI] [PubMed] [Google Scholar]
- 23. Baptista C. G., Rodrigues E. C., Morking P., Klinke A., Zardo M. L., Soares M. J., de Aguiar A. M., Goldenberg S., and Ramos A. S. (2015) Identification of a Golgi-localized UDP–N-acetylglucosamine transporter in Trypanosoma cruzi. BMC Microbiol. 15, 269 10.1186/s12866-015-0601-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ebert B., Rautengarten C., McFarlane H. E., Rupasinghe T., Zeng W., Ford K., Scheller H. V., Bacic A., Roessner U., Persson S., and Heazlewood J. L. (2018) A Golgi UDP-GlcNAc transporter delivers substrates for N-linked glycans and sphingolipids. Nat. Plants 4, 792–801 10.1038/s41477-018-0235-5 [DOI] [PubMed] [Google Scholar]
- 25. Guillen E., Abeijon C., and Hirschberg C. B. (1998) Mammalian Golgi apparatus UDP–N-acetylglucosamine transporter: molecular cloning by phenotypic correction of a yeast mutant. Proc. Natl. Acad. Sci. U.S.A. 95, 7888–7892 10.1073/pnas.95.14.7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishida N., Yoshioka S., Chiba Y., Takeuchi M., and Kawakita M. (1999) Molecular cloning and functional expression of the human Golgi UDP–N-acetylglucosamine transporter. J. Biochem. 126, 68–77 10.1093/oxfordjournals.jbchem.a022437 [DOI] [PubMed] [Google Scholar]
- 27. Andersen P. K., Veng L., Juul-Madsen H. R., Vingborg R. K., Bendixen C., and Thomsen B. (2007) Gene expression profiling, chromosome assignment and mutational analysis of the porcine Golgi-resident UDP–N-acetylglucosamine transporter SLC35A3. Mol. Membr. Biol. 24, 519–530 10.1080/09687680701459877 [DOI] [PubMed] [Google Scholar]
- 28. Toscanini M. A., Favarolo M. B., Gonzalez Flecha F. L., Ebert B., Rautengarten C., and Bredeston L. M. (2019) Conserved Glu-47 and Lys-50 residues are critical for UDP–N-acetylglucosamine/UMP antiport activity of the mouse Golgi-associated transporter Slc35a3. J. Biol. Chem. 294, 10042–10054 10.1074/jbc.RA119.008827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miura N., Ishida N., Hoshino M., Yamauchi M., Hara T., Ayusawa D., and Kawakita M. (1996) Human UDP-galactose translocater: molecular cloning of a complementary DNA that complements the genetic defect of a mutant cell line deficient in UDP-galactose translocater. J. Biochem. 120, 236–241 10.1093/oxfordjournals.jbchem.a021404 [DOI] [PubMed] [Google Scholar]
- 30. Ishida N., Miura N., Yoshioka S., and Kawakita M. (1996) Molecular cloning and characterization of a novel isoform of the human UDP-galactose transporter, and of related complementary cDNAs belonging to the nucleotide-sugar transporter gene family. J. Biochem. 120, 1074–1078 10.1093/oxfordjournals.jbchem.a021523 [DOI] [PubMed] [Google Scholar]
- 31. Oelmann S., Stanley P., and Gerardy-Schahn R. (2001) Point mutations identified in Lec8 Chinese hamster ovary glycosylation mutants that inactivate both the UDP-galactose and CMP–sialic acid transporters. J. Biol. Chem. 276, 26291–26300 10.1074/jbc.M011124200 [DOI] [PubMed] [Google Scholar]
- 32. Maszczak-Seneczko D., Olczak T., Wunderlich L., and Olczak M. (2011) Comparative analysis of involvement of UGT1 and UGT2 splice variants of UDP-galactose transporter in glycosylation of macromolecules in MDCK and CHO cell lines. Glycoconj. J. 28, 481–492 10.1007/s10719-011-9348-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brändli A. W., Hansson G. C., Rodriguez-Boulan E., and Simons K. (1988) A polarized epithelial cell mutant deficient in translocation of UDP-galactose into the Golgi complex. J. Biol. Chem. 263, 16283–16290 [PubMed] [Google Scholar]
- 34. Stanley P. (1983) Lectin-resistant CHO cells: selection of new mutant phenotypes. Somatic Cell Genet. 9, 593–608 10.1007/BF01574260 [DOI] [PubMed] [Google Scholar]
- 35. Hara T., Endo T., Furukawa K., Kawakita M., and Kobata A. (1989) Elucidation of the phenotypic change on the surface of Had-1 cell, a mutant cells line of mouse FM3A carcinoma cells selected by resistance to Newcastle disease virus infection. J. Biochem. 106, 236–247 10.1093/oxfordjournals.jbchem.a122838 [DOI] [PubMed] [Google Scholar]
- 36. Yoshioka S., Sun-Wada G. H., Ishida N., and Kawakita M. (1997) Expression of the human UDP-galactose transporter in the Golgi membranes of murine Had-1 cells that lack the endogenous transporter. J. Biochem. 122, 691–695 10.1093/oxfordjournals.jbchem.a021810 [DOI] [PubMed] [Google Scholar]
- 37. Maszczak-Seneczko D., Olczak T., Jakimowicz P., and Olczak M. (2011) Overexpression of UDP-GlcNAc transporter partially corrects galactosylation defect caused by UDP-Gal transporter mutation. FEBS Lett. 585, 3090–3094 10.1016/j.febslet.2011.08.038 [DOI] [PubMed] [Google Scholar]
- 38. Olczak M., Maszczak-Seneczko D., Sosicka P., Jakimowicz P., and Olczak T. (2013) UDP-Gal/UDP-GlcNAc chimeric transporter complements mutation defect in mammalian cells deficient in UDP-Gal transporter. Biochem. Biophys. Res. Commun. 434, 473–478 10.1016/j.bbrc.2013.03.098 [DOI] [PubMed] [Google Scholar]
- 39. Sosicka P., Jakimowicz P., Olczak T., and Olczak M. (2014) Short N-terminal region of UDP-galactose transporter (SLC35A2) is crucial for galactosylation of N-glycans. Biochem. Biophys. Res. Commun. 454, 486–492 10.1016/j.bbrc.2014.10.098 [DOI] [PubMed] [Google Scholar]
- 40. Sosicka P., Maszczak-Seneczko D., Bazan B., Shauchuk Y., Kaczmarek B., and Olczak M. (2017) An insight into the orphan nucleotide sugar transporter SLC35A4. Biochim. Biophys. Acta Mol. Cell Res. 1864, 825–838 10.1016/j.bbamcr.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 41. Kudelka M. R., Antonopoulos A., Wang Y., Duong D. M., Song X., Seyfried N. T., Dell A., Haslam S. M., Cumming R. D., and Ju T. (2016) Cellular O-glycome reporter/amplification to explore O-glycans of living cells. Nat. Methods 13, 81–86 10.1038/nmeth.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng B. G., Sosicka P., Agadi S., Almannai M., Bacino C. A., Barone R., Botto L. D., Burton J. E., Carlston C., Chung B. H., Cohen J. S., Coman D., Dipple K. M., Dorrani N., Dobyns W. B., et al. (2019) SLC35A2-CDG: functional characterization, expanded molecular, clinical, and biochemical phenotypes of 30 unreported Individuals. Hum. Mutat. 40, 908–925 10.1002/humu.23731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bazan B., Wiktor M., Maszczak-Seneczko D., Olczak T., Kaczmarek B., and Olczak M. (2018) Lysine at position 329 within a C-terminal dilysine motif is crucial for the ER localization of human SLC35B4. PLoS One 13, e0207521 10.1371/journal.pone.0207521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McDonald A. G., Hayes J. M., Bezak T., Głuchowska S. A., Cosgrave E. F. J., Struwe W. B., Stroop C. J. M., Kok H., van de Laar T., Rudd P. M., Tipton K. F., and Davey G. P. (2014) Galactosyltransferase 4 is a major control point for glycan branching in N-linked glycosylation. J. Cell Sci. 127, 5014–5026 10.1242/jcs.151878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maszczak-Seneczko D., Olczak T., and Olczak M. (2011) Subcellular localization of UDP-GlcNAc, UDP-Gal and SLC35B4 transporters. Acta Biochim. Pol. 58, 413–419 [PubMed] [Google Scholar]
- 46. Deutscher S. L., and Hirschberg C. B. (1986) Mechanism of galactosylation in the Golgi apparatus: a Chinese hamster ovary cell mutant deficient in translocation of UDP-galactose across Golgi vesicle membranes. J. Biol. Chem. 261, 96–100 [PubMed] [Google Scholar]
- 47. Ropp P. A., Little M. R., and Cheng P. W. (1991) Mucin biosynthesis: purification and characterization of a mucin β 6N-acetylglucosaminyltransferase. J. Biol. Chem. 266, 23863–23871 [PubMed] [Google Scholar]
- 48. Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., and Dennis J. W. (2007) Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 10.1016/j.cell.2007.01.049 [DOI] [PubMed] [Google Scholar]
- 49. Varki A., and Schnaar R. L. (2017) Schnauer, R Sialic acids and other nonulosonic acids. In Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., Seeberger P. H., eds) 3rd edition, Chapter 15, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 50. Sosicka P., Bazan B., Maszczak-Seneczko D., Shauchuk Y., Olczak T., and Olczak M. (2019) SLC35A5 protein: a Golgi complex member with putative nucleotide sugar transport activity. Int. J. Mol. Sci. 20, 276 10.3390/ijms20020276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ishida N., Kuba T., Aoki K., Miyatake S., Kawakita M., and Sanai Y. (2005) Identification and characterization of human Golgi nucleotide sugar transporter SLC35D2, a novel member of the SLC35 nucleotide sugar transporter family. Genomics 85, 106–116 10.1016/j.ygeno.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 52. Kim S., Miura Y., Etchison J. R., and Freeze H. H. (2001) Intact Golgi synthesize complex branched O-linked chains on glycoside primers: evidence for the functional continuity of seven glycosyltransferases and three sugar nucleotide transporters. Glycoconj. J. 18, 623–633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data has been deposited in the GlycoPost under ID GPST000080. Some unpublished data are available upon request from the corresponding author (E-mail address: mariusz.olczak@uwr.edu.pl).