Abstract
Introduction:
Numerous bacterial behaviors are regulated by a cell-density dependent mechanism known as Quorum Sensing (QS). QS relies on communication between bacterial cells using diffusible signaling molecules known as autoinducers. QS regulates physiological processes such as metabolism, virulence and biofilm formation. Quorum Quenching (QQ) is the inhibition of QS using chemical or enzymatic means to counteract behaviors regulated by QS.
Areas Covered:
We examine the main, diverse QS mechanisms present in bacterial species, with a special emphasis on AHL-mediated QS. We also discuss key in vitro and in vivo systems in which interference in QS was investigated. Additionally, we highlight promising developments, such as the substrate preference of the used enzymatic quencher, in the application of interference in QS to counter bacterial virulence.
Expert Opinion:
Enabled via the recent isolation of highly stable quorum quenching enzymes and / or molecular engineering efforts, the effects of the interference in QS were recently evaluated outside of the traditional model of single species culture. Signal disruption in complex microbial communities was shown to result in the disruption of complex microbial behaviors, and changes in population structures. These new findings, and future studies, may result in significant changes in the traditional views about QS.
Keywords: Quorum Sensing (QS), Quorum Quenching (QQ), signaling, lactonase, microbiome
1. Introduction
Quorum sensing (QS) was discovered in the bioluminescent marine bacteria Vibrio fischeri inhabiting the light organ of Hawaiian bobtail squid Euprymna scolopes [1]. Bioluminescence produced by the bacteria correlated with its cell density and production of small diffusible molecules called autoinducers (AI) [2]. With increasing bacterial population during growth, the AI concentration reaches a threshold and activates population-wide bioluminescence [3–5]. This phenomenon, by which AIs act as chemical signals, and how, depending on the bacterial biomass, can modulate their individual and group behaviors was termed as QS [6]. Thereafter, QS systems were found to be present in numerous bacterial species. All QS systems demonstrate a common mode of action comprising of the following three steps – (i) Intracellular production and extracellular secretion of AIs by bacteria (ii) extracellular AIs re-entry into the bacterial cells either actively (via membrane bound transporter proteins) or passively (via diffusion across the cell membrane) and bind to specific membrane or cytoplasmic receptors and (iii) signal transduction by AI receptors activates downstream signaling pathways which eventually regulate the expression of genes associated with physiological processes such as metabolism, virulence, sporulation and biofilm formation [7–9]. Regulation and disruption of QS, therefore, presents an attractive opportunity to mitigate undesirable bacterial traits controlled by signaling such as virulence and biofilm formation associated with infectious diseases [10]. Interference in QS, known as Quorum Quenching (QQ) [11], inhibits bacterial communication using chemical or enzymatic means and subsequently reduces all behaviors regulated by QS [12]. In this review, we discuss the main QS mechanisms present in bacteria and several key methods and promising developments utilizing enzymes for the disruption of QS.
2. Diversity of QS signals - it’s all about autoinducers (AIs)
Bacterial QS systems rely on AIs as specialized signaling molecules. Three main classes of AIs are shown in Fig. 1 – N-acyl Homoserine Lactones (AHLs) or Autoinducer-1, Autoinducing Peptides (AIPs) and Autoinducer-2 (AI-2) [7]. While the structural diversity of known signaling molecules is already large, it is very likely that more, new signaling molecules involved in QS or similar signaling systems are yet to discover. Known AI molecules can passively or actively diffuse across the cell membrane, bind to their cognate receptors with high specificity and trigger a cascade of signaling pathways eventually resulting in transcriptional regulation of target genes. While it is not the focus of this review, we note that signaling using the above-mentioned molecules is not limited to bacteria, but extend to eukaryotes including plants and animals [13–20].
Fig 1.
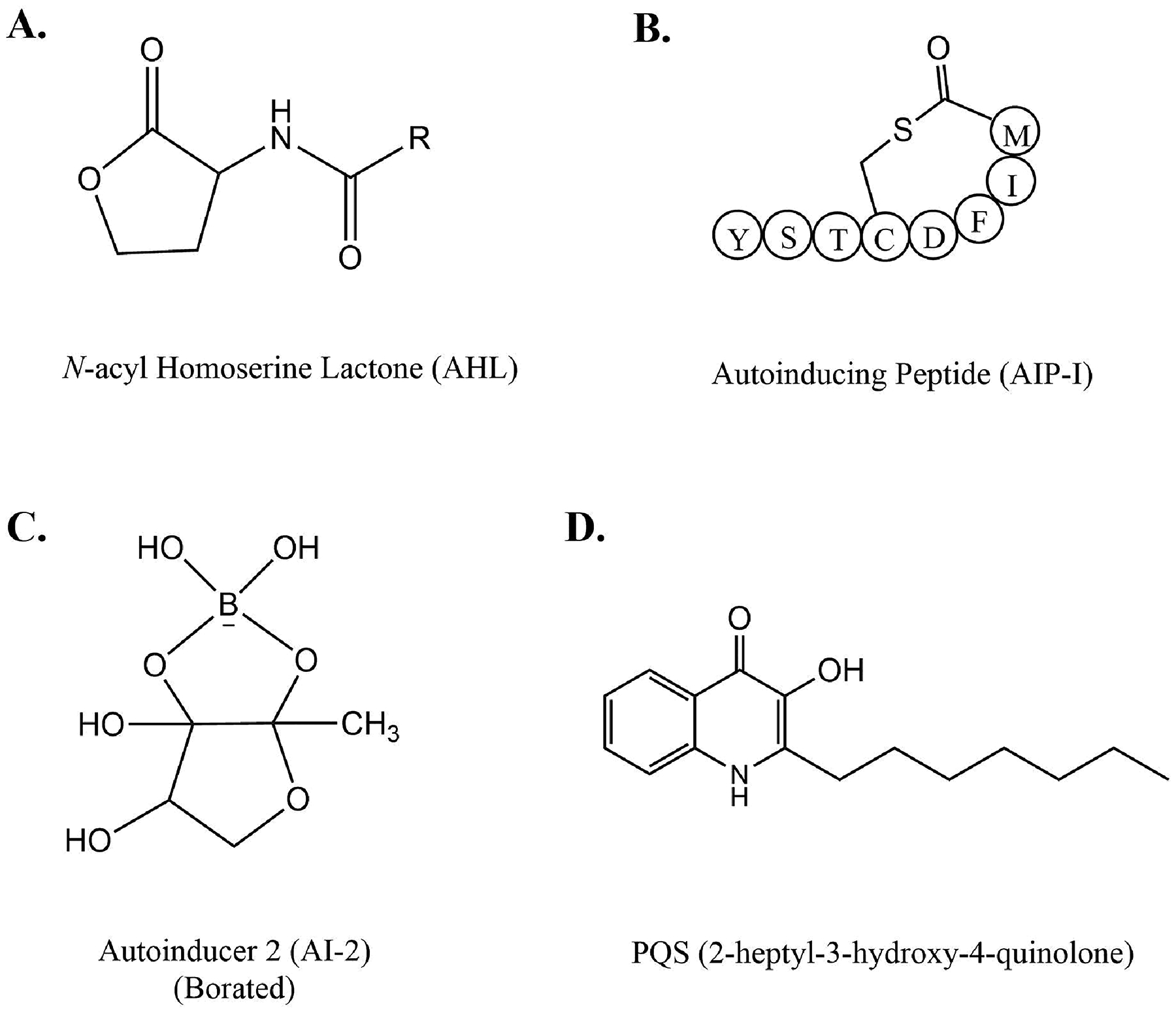
Representative chemical structures of (A) N-acyl homoserine lactone, (B) Autoinducing peptide AIP-I from S. aureus, (C) Autoinducer 2 (AI-2) from Vibrio sp., (D) PQS from P. aeruginosa.
2a. Autoinducing peptides (AIPs)
AIPs (Fig. 1B) are short cyclical peptides involved in QS found in Gram-positive bacteria [21]. AIPs are synthesized in the cytosol as pro-AIPs which are processed to form mature AIPs either in the intracellular [22] or the extracellular [23] environment depending on the species. Due to their inability to passively diffuse across the cell membrane, AIPs are chaperoned by specialized membrane transporters [24]. AIPs are detected and bound by transmembrane histidine kinases when their extracellular concentration reaches the QS threshold [25]. This binding results in phosphorylation of a downstream response regulator, which regulates the expression of target genes for pathogenic phenotypes such as competence, DNA uptake, sporulation, virulence initiation [26]. AIP based QS systems are well characterized in Staphylococcus aureus [27], Enterococcus faecalis [28], Bacillus subtilis [29], Listeria monocytogenes [30], and Clostridium perfringens [31].
2b. Autoinducer-2 (AI-2)
AI-2 (Fig. 1C) are synthesized and detected by both Gram-positive [32] and Gram-negative [33] bacteria as well as archaea [34]. AI-2 is a mixture of molecules, including borated furanone derivatives [35], derived from DPD (4,5-dihydroxy-2,3-pentanedione) synthesized by LuxS synthase in the S-adenosyl-methionine (SAM) recycling pathway [36,37]. AI-2 is imported and bound by its cognate receptor leading to a cascade of phosphorylation signaling pathways regulating phenotypes such as virulence and bioluminescence [38]. Examples of well-characterized systems are LuxPQ system in Vibrio sp. [39] and the Lsr system in Salmonella sp. and Escherichia coli [40,41].
2c. N-acyl homoserine lactones (AHLs)
AHLs (Fig. 1A) were previously presented to be exclusively synthesized and sensed by Gram-negative bacteria. However, in addition to Gram-negatives [9], there are some examples that in rare cases AHLs are also produced by Gram-positive bacteria [42] and archaea [43]. AHLs are small diffusible molecules composed of a lactone ring linked to an acyl chain of varying lengths (C4-C14) and C3 modifications [6,9]. AHLs are synthesized by LuxI synthases and detected by their cognate cytoplasmic LuxR receptors [44] (Fig. 2A). The specificity of AHLs binding to their receptors is determined by the structure of their acyl side chains [9]. When the AHL concentration reaches the QS threshold, typically reported to be in the nanomolar range [45–48], AHL-bound LuxR receptors act as transcriptional modulators of target genes (Fig. 2B). Many pathogenic behaviors of Gram-negative bacteria such as host adhesion, sporulation, exoenzyme production, toxin secretion, biofilm formation, siderophores and pigment production are regulated by AHL-mediated QS [9,49]. A non-exhaustive list of examples of well-characterized AHL QS systems is as follows: LuxIR in Vibrio sp. [50], TraIR in Agrobacterium tumefaciens [51], the hierarchical QS systems LasIR and RhlIR in Pseudomonas aeruginosa [52], CviIR in Chromobacterium violaceum [53], ExpIR in Erwinia carotovora [54], SmaIR in Serratia sp. [55], AhyIR and AsaIR in Aeromonas sp. [56,57]. Putative luxIR homologs have also been detected in the genomes of Gram-positive bacteria and archaea which were shown to produce AHLs [42,43].
Fig 2.
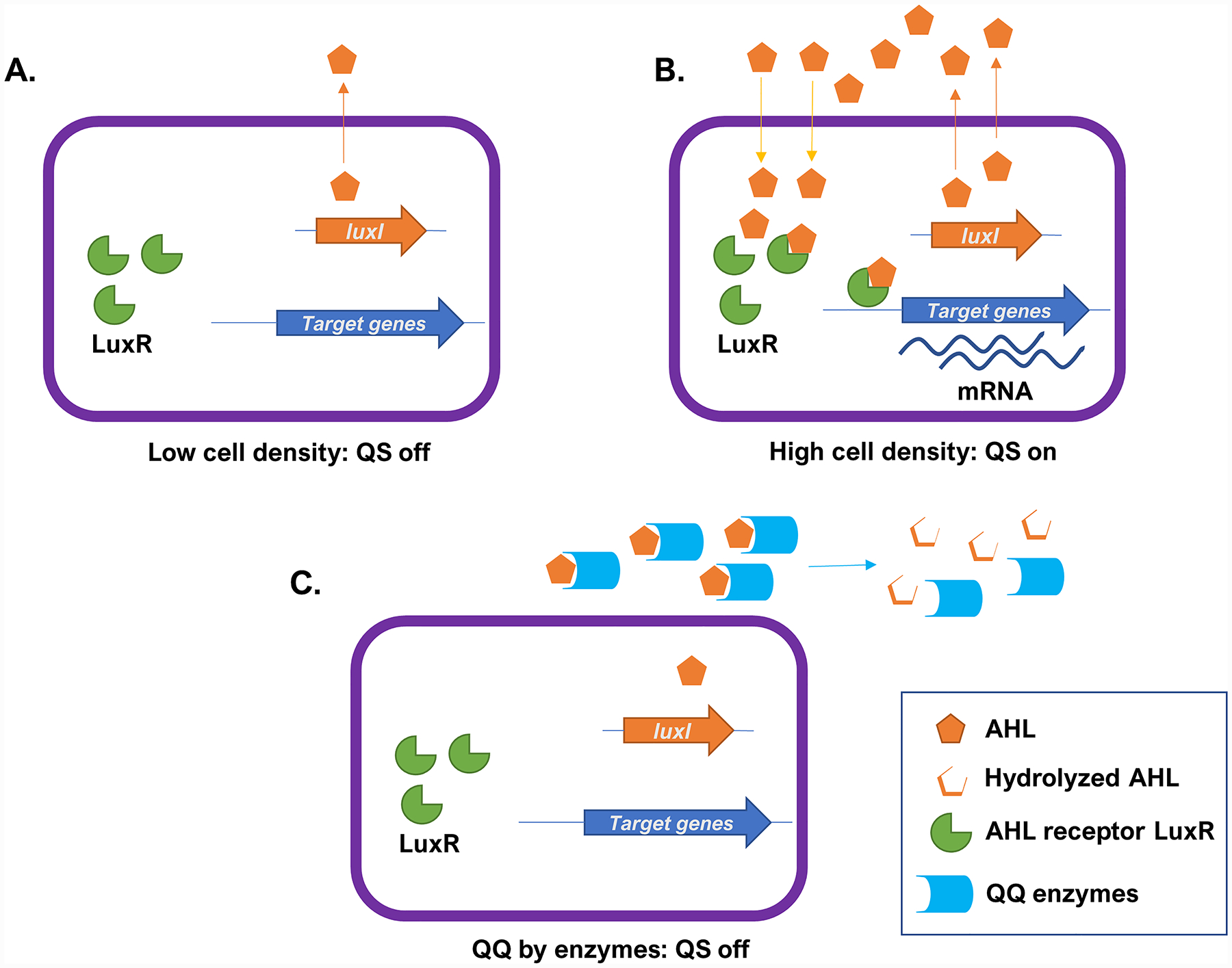
Representation of AHL mediated Quorum Sensing (QS) (A and B) and enzymatic Quorum Quenching (QQ) (C). (A) Bacteria encode a LuxI synthase producing N-acyl homoserine lactones (AHLs) in the cytoplasm, which can diffuse out of the cell. At low cell density, the extracellular concentration of AHLs remain low and QS is off. (B) When bacterial density increases, so does the extracellular AHL concentration. Upon reaching a threshold level, AHLs bind to their cognate cytoplasmic LuxR receptors. The AHL bonded LuxR acts as a transcription factor and can triggers the expression of target genes, including the production of more AHLs, and resulting in population-wide changes in gene expression profiles and bacterial behavior. (C) QQ enzymes degrade extracellular AHLs, reduce the AHL concentration, and prevents the signaling molecule to reach the concentration threshold that triggers QS.
It is interesting that LuxI homologs in certain species of α-proteobacteria can produce homoserine lactones (HSLs) with aromatic acid or branched amino acid side chains (instead of the straight fatty acyl side chains of AHLs. Notable examples include p-coumaroyl-HSL from Rhodopseudomonas palustris (RpaIR) [58], phenylacetyl-HSL from Prosthecomicrobium hirschii (HirIR) [59], isovaleryl-HSL from Bradyrhizobium japonicum (BjaIR) [60] and cinnamoyl-HSL from Bradyrhizobium strain ORS278 (BraIR) [61].
2d. Other signaling molecules
Several other molecules were found to exhibit autoinducing properties and play distinct roles in QS. For example: Alkylquinolones, underlying the Pseudomonas Quinolone Signal (PQS) QS pathway present in P.aeruginosa. Alkylquinolones (For e.g.: 2-heptyl-3-hydroxy-4-quinolone; Fig. 1D) are synthesized by the PqsABCDH system and detected by PqsR [62]. PQS responds to disparate environmental signals such as low iron conditions and response to oxidative stress [63,64] and integrates these signals into the main LasIR and RhlIR signaling systems to fine tune virulence and biofilm formation [65]. In addition to altering global transcriptional profile of genes via the PqsR dependent pathway, PQS can also function independent of PqsR by binding to hundreds of different cellular receptors [66,67], leading to modulation of host immune responses, cytotoxicity and key virulence pathways [68].
Other classes of QS signals are (a) fatty-acid derivatives – for e.g. 3-hydroxypalmitate methyl ester (3-OH-PAME) produced by Ralstonia sp. [69] and cis-11-methyl-2-dodecenoic acid (DSF, Diffusible Signal Factor) from Xanthomonas campestris [70] and (b) amino acid derivatives – for e.g. diketopiperazines produced by proteobacteria [71] and archaea [72].
3. Enzymatic quorum quenching – inactivating the messenger
The process of inhibition of QS is referred to as QQ [11]. Although QQ can be achieved by various means, in this review, we will focus on the basics, progress and potential of enzyme mediated QQ.
Disruption of AHL-mediated QS by enzymes can be achieved by degrading AHLs (Fig. 2C). As illustrated in Fig. 3, three different classes of enzymes, based on their mechanism of action, can inactivate AHLs – (i) Lactonases, that break open the lactone ring [73,74], (ii) Amidases (or acylases), that hydrolyses the amide bond of the AHLs and breaks it down into the corresponding fatty acid and homoserine lactone [75], (iii) Oxidoreductases, that either oxidize the acyl chain of AHLs or reduces 3-oxo-AHLs to their corresponding 3-hydroxy-AHL counterparts [76–78].
Fig 3.
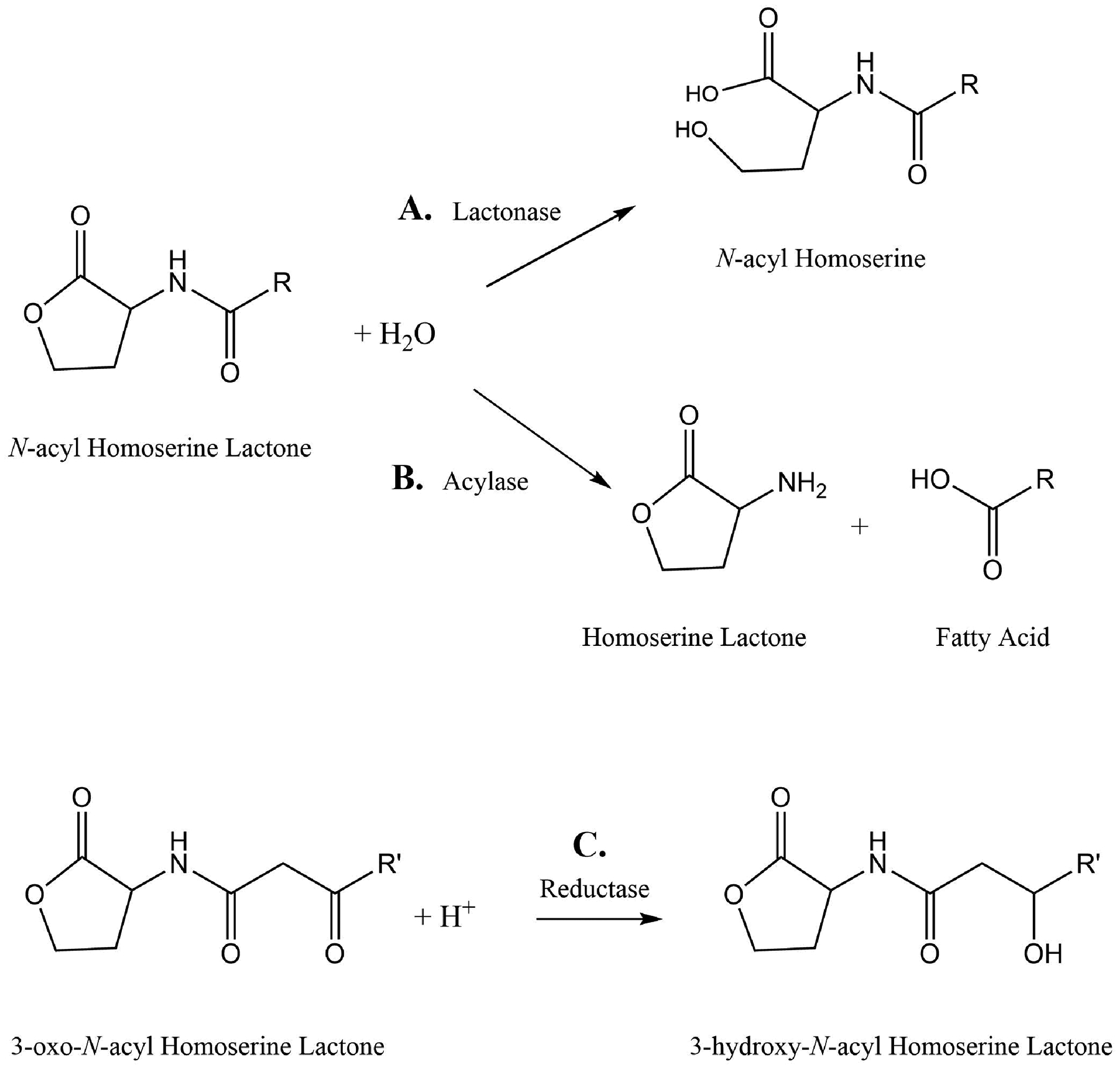
Chemical mechanisms of hydrolysis of AHLs by (A) lactonases and (B) acylases or (C) reduction of 3-oxo-AHLs by reductases into 3-hydroxy-AHLs.
AHL degrading enzymes exist naturally and are produced by bacteria [79], archaea [74] and eukaryotes [80]. Some bacteria like Pseudomonas and Agrobacterium encodes genes for AHL degrading enzymes – the corresponding enzymes, PvdQ and BlcC respectively, degrade their own AHL signals and are hypothesized to prevent their concentration from reaching toxic levels by AHL recycling [81,82].
A comprehensive list of QQ enzymes are listed in Table S1 along with their substrate specificities and description of their characteristics. Different classes of QQ enzymes demonstrate disparate substrate specificities. For e.g. while known acylases preferentially hydrolyze AHLs with long acyl chains, lactonases can demonstrate significantly higher activity against a broader (and sometimes complete) spectrum of AHL substrates. It is intriguing to note that most of the well-characterized AHL degrading enzymes fall into two categories: (i) those with very broad substrate specificities and (ii) those with marked preference for longer acyl chain AHLs.
3a. Oxidoreductases
Some oxidoreductases were characterized for their ability to modify the QS signal molecule rather than degrading it. They either oxidize the ω1, ω2 or ω3 carbon of the acyl chain or reduce C3 carboxyl group of AHLs into a hydroxyl group (Fig. 3C). This process results in alteration of the AHL signal and subsequently its capacity to bind to its cognate receptor, thereby modulating the QS response. For e.g. BpiB09 reductase (Fig. 4A) from Acidobacterium sp. reduces 3-oxo-AHLs to 3-hydroxy-AHLs and can alter the QS response of P. aeruginosa [76].
Fig 4.
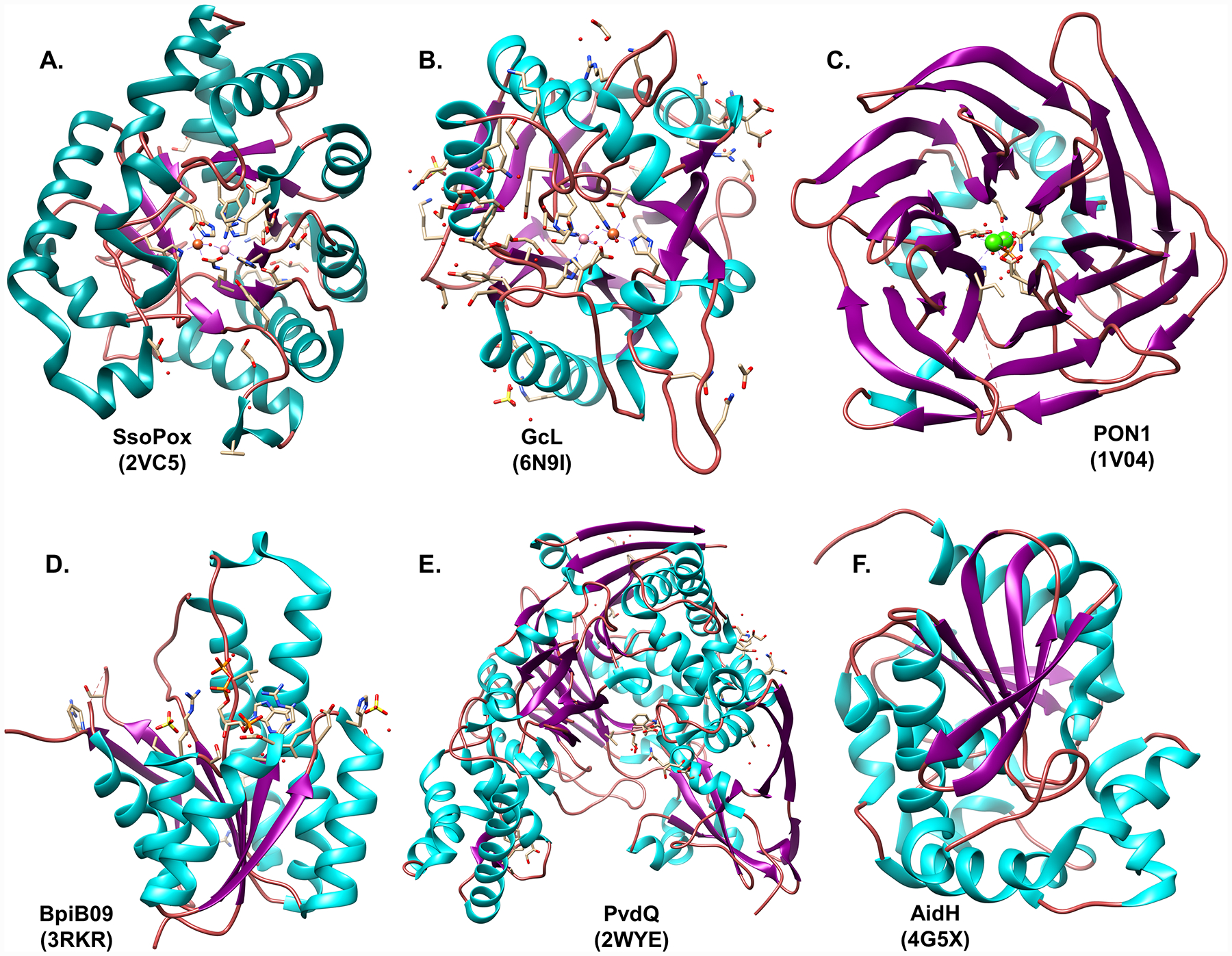
Crystal structures of representatives from different QQ enzyme families – (A) AHL reductase BpiB09 from an uncultured bacterium Bio5, (B) AHL acylase PvdQ from Pseudomonas aeruginosa, (C) phosphotriesterase-like lactonase SsoPox from Saccharolobus solfataricus, (D) metallo-β-lactamase-like lactonase GcL from Parageobacillus caldoxylosilyticus, (E) α/β hydrolase fold lactonase AidH from Ochrobactrum sp. T63. and (F) serum paraoxonase PON1 from Oryctolagus cuniculus. RCSB Protein Data Bank ID for each structure is provided in brackets.
3b. AHL Acylases
Acylases hydrolyze the amide bond of AHLs (Fig. 3B) [83]. They belong to the superfamily of N-terminal nucleophile hydrolases (Ntn-hydrolases) with a characteristic αβ/βα fold. Their active sites show a large hydrophobic binding pocket to accommodate long acyl chains of AHL substrates [84–86]. A well characterized QQ acylase is PvdQ (Fig. 4B) from P. aeruginosa [81]. PvdQ preferentially cleaves AHLs with acyl chain lengths exceeding 10 carbons [87] and plays a key role in production of pyoverdine [88].
3c. AHL Lactonases
Lactonases degrade AHLs by hydrolyzing their lactone ring (Fig. 3A) [73]. They can show preference for certain types of AHLs and other lactone substrates such as δ-, ε- and γ-lactones, and some representative from extremophiles are active under a variety of environmental conditions such as high temperature [74,89–91]. Intriguingly, different classes of lactonases were identified in a variety of protein superfamilies and folds (Table S1), including:
The Phosphotriesterase-like lactonases (PLLs) – Found in bacteria and archaea, they exhibit a (α/β)8 fold (TIM barrel). They prefer AHLs with long acyl chains. SsoPox (Fig. 4C) is a well characterized PLL representative from Saccharolobus solfataricus [91,92]. PLLs are metalloenzymes and numerous characterized PLL representatives are thermostable enzymes, a desirable property for biotechnological applications For e.g. SacPox[93], VmoLac [94], SisLac [95], and GkL [96]. PLLs are proficient lactonases that typically show a substrate preference for long acyl chain AHLs.
The Metallo-β-lactamase-like lactonases (MLLs) – Found in bacteria, archaea and eukaryotes, their main structural highlight is the presence of a conserved metal binding motif HXHXDH, exhibiting a characteristic αβ/βα fold bonded with two metal cations. These lactonases are very proficient enzymes and are found to exhibit a broad AHL substrate preference [74,89,97,98]. AiiA from Bacillus sp. is a well characterized example [99] (Fig. 4D).
The α/β hydrolase fold lactonases – Isolated from bacteria, these possess a characteristic α/β hydrolase fold but lack the conserved HXHXDH motif of MLLs [100]. These lactonases also demonstrate preference for a broad spectrum of AHL substrates [107,110–112]. One well characterized example is AidH (Fig. 4E) produced by the soil bacterium Ochrobactrum sp. [100,101].
The Paraoxonases (PONs) – They derive their name from their ability to cleave paraoxon [102], a derivative of the organophosphate insecticide parathion. PONs possess a characteristic six-bladed β-propeller fold (Fig. 4F) [103]. PONs are found in both mammals and bacteria [104]. PONs hydrolyze a wide variety of substrates which includes long acyl chain AHLs (weak activity against short chain AHLs, preference for longer acyl chains AHLs), arylesters, organophosphates, fatty acids, δ-lactones and γ-lactones [104,105].
It is interesting to note that an evolutionary convergence in the structures and catalytic mechanisms of PLLs, MLLs and PONs was observed [106]. Their active site regions and the usage of a metal cation to bind to the carbonyl oxygen of the lactone substrate leading to its subsequent hydrolysis is common to these classes of lactonases [104]. It is also intriguing to observe that most lactonases that have been identified and characterized exhibit a low variety of substrate preferences (Table S1). PLLs, specifically PLL-A [94], appear to be the most specific lactonases, with a marked preference for long chain AHLs.
3d. QQ enzymes against AI-2 and other QS signaling molecules
Proficient enzymes capable of neutralizing AI-2 remain elusive. In a known interference mechanism, AI-2 can be extracellularly phosphorylated by the kinase LsrK, which destabilizes it and prevents intracellular uptake by its cognate receptor [38,40,41,107]. Recently, oxidoreductase enzymes degrading AI-2 were discovered using metagenomic approaches [108,109] and may represent a promising way to interfere with AI-2 based QS.
The quinolone-based QS signals of P. aeruginosa (PQS) and Burkholderia sp. (AQ) can be degraded by a class of enzymes known as quinolone dioxygenases (see Table S1 for details). These enzymes open the PQS ring to form N-octanyl-anthranilic acid and carbon monoxide [119,122]. The first identified member of this class of enzymes is Hod (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxogenase) from Arthrobacter sp. Rue61a [110].Other QS signals such as 3-OH-PAME can be degraded by recently identified microbial esterases [111,112] and diketopiperazines were shown to be biologically degraded by yet unidentified enzymes in a cell-free extract of Streptomyces albulus [113].
4. Quorum quenching enzymes and biotechnology
Because QS modulates bacterial behaviors, the use of QQ enzymes is fundamentally different from the use of antimicrobials: the enzymes show no toxicity and little to no effects on growth [114–116]. Rather than entering cells or binding to a receptor, lactonases hydrolyze signaling molecules secreted in the media to affect bacteria’s behavior [117]. It is noteworthy that while QQ can also be achieved by various small molecule chemicals known as Quorum Sensing Inhibitors (QSIs) such as 5-Fluorouracil (5-FU) and halogenated furanones [120], QQ enzymes are significantly more potent than QSIs while lacking the cytotoxicity of the latter [121,122]. Resistances to these strategies has been observed. Yet, contrary to the resistance to QSIs that can be achieved by the overproduction of efflux pumps, resistance to QQ enzymes is possibly limited to cells that either lost the ability to respond to QS (e.g. social cheaters) or to cells producing a hypothetical enzyme inhibitor [115,118,119].
4a. AHL Acylases and Lactonases as anti-virulence agents
The effect of QQ enzymes has been extensively investigated with the Gram-negative pathogenic bacterium P. aeruginosa. This pathogen is associated with 8–10% of nosocomial infections in healthcare as it frequently infects immunocompromised patients and is listed among the top priority pathogens by the WHO for immediate R&D of new antimicrobials [123,124]. P. aeruginosa possesses three overlapping QS circuits – LasIR, RhlIR and the PQS systems following a strict signaling hierarchy (LasIR at the top), interwoven signaling pathways and overlapping signaling targets [65]. This sophisticated QS circuitry enables this bacterium to be a versatile and opportunistic pathogen which can adapt to a variety of environmental conditions in the host tissue and forms a robust biofilm [125]. While all these three QS systems contribute to virulence, the QS signals 3-oxo-C12 AHL (sensed by LasR), C4-AHL (sensed by RhlR) and PQS (sensed by PqsR) have been successfully targeted by QQ enzymes.
A multitude of QQ enzymes (AiiA, AiiM, SsoPox, PvdQ, MomL, BpiB09, and HodC) were shown to reduce the production of virulence factors, motility and biofilm formation of P. aeruginosa via interference in QS in vitro [76,85,98,110,121,122,126,127], as well as in vivo in various models, including Caenorhabditis elegans, Drosophila melanogaster and rodents [127–130].
As previously noted in Table S1, AHL degrading enzymes are found to exhibit two main substrate preferences: broad spectrum or preference for long acyl chain AHLs. This distinct property of QQ enzymes was recently used to specifically inhibit the different QS circuits in P. aeruginosa, i.e. systems based on C4-AHL and 3-oxo-C12-AHL. With clinical isolates from lung adapted strains, the use of two lactonases with distinct specificity resulted in different inhibitions of virulence factors, the broader spectrum enzyme being a more potent inhibitor [131] Using the same enzymes, the use of in vivo amoeba models unexpectedly showed that only the most specific enzyme could protect amoebas from infections. Analysis, including proteomics, in fact revealed large variations in protein levels involved in antibiotic resistance, biofilm formation, virulence as a function of lactonase specificity [132]. These results suggest that the specificity of the interference in signaling may be a key parameter in improving the potency of quorum quenching approaches.
In addition, numerous animal studies were performed. For e.g., the lactonase SsoPox-I was administered directly in the trachea of rats, immediately after infecting them with P. aeruginosa [129]. The treated rats showed a significantly reduced mortality rate compared to untreated rats, during a 50h observation period. A similar work was performed with the acylase PvdQ [130] that was administered to mice via the nasal route after initiating a lethal P. aeruginosa infection. Subsequently, the lung bacterial load of PvdQ treated mice was reduced 5-fold and less morbidity, reduced tissue inflammation and prolonged survival was observed compared to untreated control mice. Combinations of QQ were also tested in vivo: Combining the Rhodococcal lactonase QsdA and Mycobacterial dioxygenase AqdC resulted in enhanced QQ and reduction of virulence of P.aeruginosa in C.elegans and epithelial lung cell infection models [133]. Another study found that Deinococcus radiodurans produces a lactonase and an acylase, which demonstrated a more robust QQ of P.aeruginosa and a major reduction of virulence in a C.elegans infection model [134]. These studies demonstrate the underlying potential of QQ enzymes to inhibit infections in vivo, and that QQ strategies can putatively be improved by combining enzymes and possibly targeting multiple QS systems.
Additionally, lactonases were also shown to inhibit the virulence of numerous other pathogens in infection models, including Aeromonas hydrophila and subsequent mortality in crucian carp by AiiA [135], reduction of virulence of marine pathogenic Vibrio sp. in brine shrimp and manila clam [136] and Erwinia carotovora in plants [137]. The acylase PvdQ was also shown to be effective in an infection model. An engineered PvdQ variant hydrolyzed C8 AHL in addition to its natural long chain AHL substrates such as 3-oxo-C12 AHL, with high efficiency. Since C8 AHL is used by Burkholderia cenocepacia for QS, application of this engineered PvdQ acylase was shown to reduce virulence of B. cenocepacia and subsequently reduced mortality in a Galleria mellonella infection model [87].
It is noteworthy that QQ enzymes and QSIs can also be used in combination to achieve a more robust QQ in pathogenic bacteria. In a recent study with P. aeruginosa, it was observed that combining QQ enzyme AiiM and the QSI G1 resulted in almost complete quenching of the Las and Rhl QS systems, that was significantly better than using either AiiM or G1 separately [138].
4b. AHL Acylases and Lactonases as anti-biofilm agents
Biofilms are complex 3D structures composed of a mono- or multispecies community of bacteria attached to a surface. Bacteria in the biofilm secrete an extracellular matrix composed of exopolysaccharide (EPS), nucleic acids, proteins and lipids [139], which provide structural support, adaptability to diverse environmental conditions and resistance to a wide variety of antimicrobial compounds including antibiotics [140,141]. In most ecological niches including infected tissues [142], bacteria exist in multispecies biofilms.
The ability of QQ lactonases to inhibit biofilm formation in P. aeruginosa was reported in several reports [121,122,143]. Inhibition of biofilm formation of other pathogens was also reported, including A. baumannii. Application of QQ lactonases such as GkL [144], AaL [74] and GcL [89] resulted in reduced biofilm formation by the bacterium. In addition, engineered lactonase GkL with increased catalytic efficiency also disrupted existing biofilms of A. baumannii [144]. The effect of lactonases in inhibition or dispersal of biofilms is illustrated in Fig. 5A.
Fig 5.
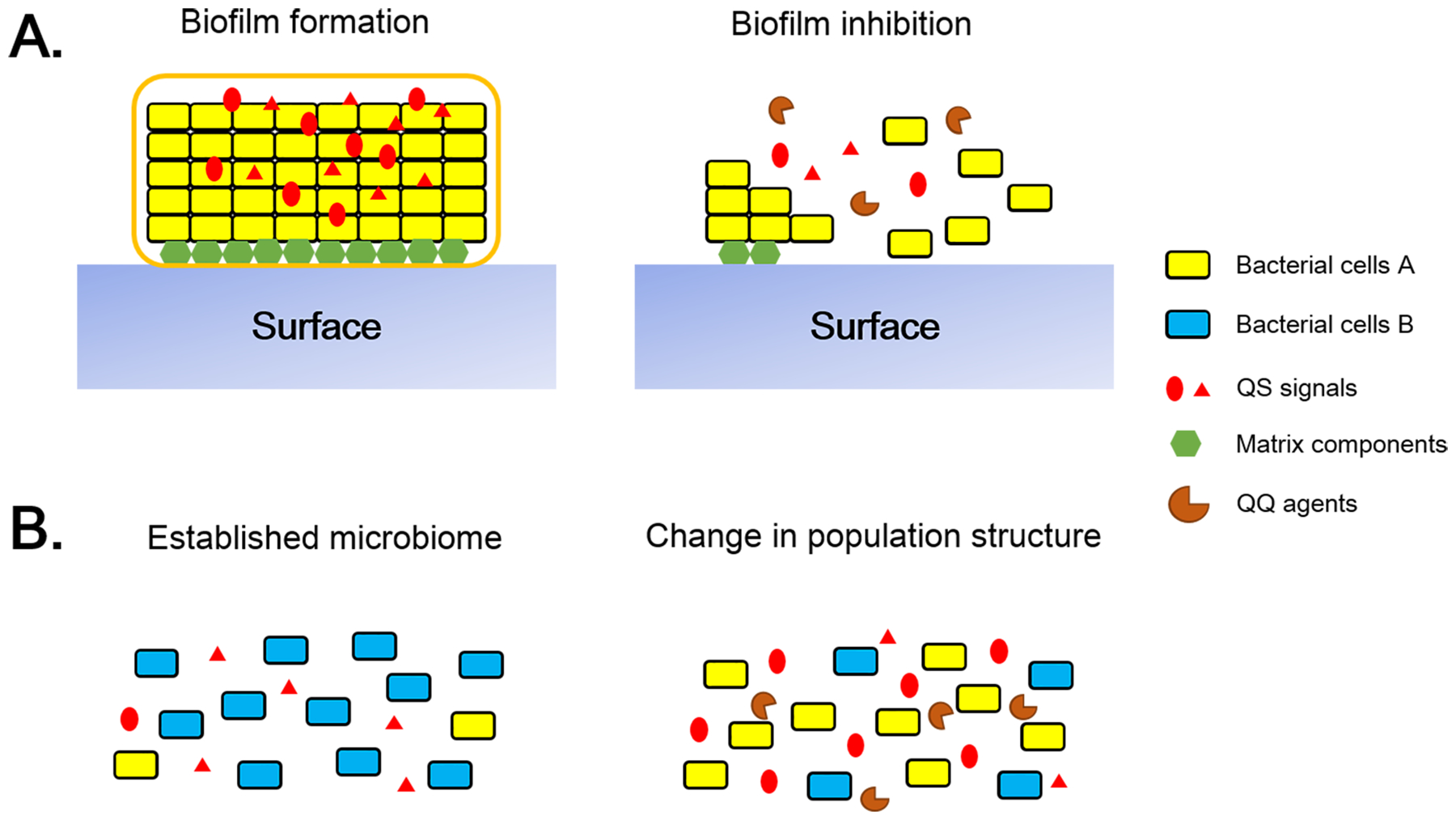
Effect of QQ on – (A) biofilm inhibition and (B) altering the species composition of the mammalian gut microbiome to prevent dysbiosis. (A) Left. The formation of stable biofilms relies extensively on QS dependent processes such as production of EPS and other components of the biofilm matrix. Right. QQ can prevent the formation of biofilms. (B) Left. Microbiome balance results from a complex interaction between several microbial species and therefore dependent on QS. Right. Because interference in QS leads to change in microbial population structure, future research will reveal interactions between microbial species and might aid in restoring healthy microbiome.
Utilization of QQ enzymes from extremophiles and/or engineering for higher stability has allowed to use these enzymes in formulations to control biofilm formation on medically and industrially relevant surfaces. Medically relevant surfaces are foreign inserts into the human body such as prosthetic valves, vascular prosthetics, catheters, endotracheal tubes, cardiac pacemakers, cerebrospinal fluid shunts, ocular prosthesis, orthopedic implants and intrauterine contraceptive devices [145]. These inserts provide a natural surface for growth of pathogenic bacteria and often results in formation of recalcitrant biofilms and subsequently difficult-to-treat infections [145,146]. The relevance of preventing biofilm formation, and thereby prophylactically preventing infection, by using surface immobilized QQ lactonases becomes more profound in the context of increasing frequency of antibiotic resistance in bacteria.
Recently, the silicone surface of a urinary catheter was coated with a QQ acylase derived from Aspergillus melleus. The immobilized enzyme imparted anti-biofilm properties to the catheter surface for a period of 7 days. This treatment caused a reduction of 75% of P. aeruginosa biofilm after 24h of incubation [147]. The same coating when combined with enzyme α-amylase, to digest the exopolysaccharide (EPS) component of biofilms, in multiple layers, demonstrated a maximal reduction of 70% of biofilm formation in a catheterized rabbit, compared to the untreated catheter after 7 days of monitoring spontaneous bacterial infection in a dynamic environment [147]. New techniques for immobilization of QQ acylases on PVC catheters by using glycidyl methacrylate, without loss of enzyme activity was recently demonstrated [148]. SsoPox was also successfully immobilized in polyurethane coatings using glutaraldehyde with no loss of activity in QQ applications with about a seven-fold reduction of biofilm formation by P. aeruginosa compared to uncoated control [121].
Conclusions
We discuss the nature and key roles of main quorum sensing (QS) circuits and signals in regulating bacterial behaviors including pathogenicity and biofilm formation. Recent efforts have demonstrated the variety of QS interference strategies, and the numerous enzymatic ways to inactivate signaling molecules. Such enzymes, through the interference in QS, can effectively behave as biofilm and virulence inhibitors. This, and their lack of cytotoxicity, are making them appealing candidates to control bacteria. Several studies have highlighted their potential to prevent bacterial infections in a variety of in vivo systems. The identification of highly stable enzymes from extremophiles, and the improvement of their properties via molecular engineering may open the path for the use of the QS interference strategy in a variety of clinical settings.
6. Expert Opinion
A significant part of the research work on QS is dedicated to the interference strategies that were discussed in this review. These strategies, using for example QQ enzymes, may allow bacterial control with little to no bactericidal activity, without the need to physically enter or contact the bacterial cells. The variety of the possible fields of applications for the anti-virulence and anti-biofilm activity of these enzymes is large, yet it will require significant improvements in the properties of these molecules to make them compatible with the constraints of industrial productions. For e.g. different enzymes demonstrate variable substrate specificities and activities, have stability issues under different application conditions such as extreme pH and temperature and dependence on the availability and nature of metal cofactors for optimal functionality. Recent efforts in these directions, such as the formulations of stable enzymatic materials [147,149,150] or preparation that can be used in the field [129,143,150], have led to promising observations. Indeed, recent work (discussed below) on microbial communities has highlighted the complex interconnection between bacteria producing and / or sensing different types of signaling molecules, and it may change traditional views about quorum sensing.
Interestingly, work using lactonases to interfere with complex biological processes revealed profound changes to the microbial population structure because of disruption of AHL signaling. This finding is surprising, since AHL interference was long studied mostly in the context of AHL producers and sensors, i.e. Gram-negative bacteria. These global changes likely pertain to the biological importance of these signaling molecules to complex microbial communities. Changes were observed numerous environments: e.g. in the context of Membrane BioReactors (MBR) treated with beads embedded with lactonases. Analysis of the microbial population structure revealed global changes to surface communities [150,151]. In waste sewage sludge system, use of the lactonase AiiM altered the microbial community, favoring Gram-positive bacteria over Gram-negative species and subsequently decreasing methane production by over 400% while increasing fermentation leading to acetic acid production [152]. This insight may be consistent with most AHL-responders being Gram negative bacteria.
Other studies, following the engineering of lactonases for higher activity and stability (e.g. Ssopox [149]), investigated the effects of signal disruption using a lactonase in the context of biocorrosion [153] and biofilm formation [154]. Both studies found that AHL signal disruption resulted in an inhibition of the tested biological processes (biofilm or biocorrosion formation) that was concomitant to significant changes in the microbial population structure (Fig. 5B). Intriguingly, observed changes in microbiomes are not easily interpretable considering the ability of bacteria to produce / sense AHLs. Indeed, some AHLs producers / sensors as well as some bacteria known not to encode genes for AHL production and sensing were either positively or negatively affected by signal disruption.
These study suggests the intricacy and intimacy of microbial relationships. Altering the AHL-based quorum sensing systems has effects that goes beyond AHL-producer and sensor microbes, and numerous, possibly indirect effects are observed. This might include changes in Gram-positive bacterial composition and abundance, most of which are not using nor sensing AHLs, but also observation of changes related to larger organisms (in the context of biofouling, e.g. mussels, algae).
Interestingly, these global effects do not appear to be limited to autoinducer-1 (AHL) based systems, but were also observed with autoinducer-2. The effects of the administration of bacteria producing AI-2 in a gut microbiome model. Because AI-2 is used for QS by numerous species of Firmicutes, administration of E. coli overproducing AI-2 was used to restore the population of Firmicutes and subsequent health of the microbiota [155,156]. Similarly, Blautia obeum, a gut commensal bacterium, produces the QS molecule DPO which can prevent colonization by V. cholerae [157]. Therefore, oral route of delivery of QQ enzymes to facilitate selective QQ and allow for healthy microbial composition in the human gut holds promise. However, there remains concerns that these enzymes could lose stability and activity once exposed to the extremes of pH and proteases in the gastrointestinal (GI) tract. These concerns could be alleviated by coating the enzymes inside specialized pH-responsive capsules [158] which limit their exposure to low pH and/or by bioengineering these enzymes for better protease and pH tolerance [90,91,143].
These discoveries may represent the tip of the iceberg in the understanding of the biological role(s) of signaling molecules in complex microbiomes, and the potential utilization of these molecules to alter them. Future discoveries in the field of QS and QQ will reveal the mechanisms for the cellular responses of bacteria to signaling molecules, including the context of stress (e.g. antibiotics [159] or phage infection [160]). New, efficient ways to interfere with other microbial languages will be identified (e.g. AI-2). More studies, focusing on complex biological processes and communities will likely provide unprecedented understanding of interactions, interdependences, cooperation, and competitions at the microbiome level. This will ultimately inform us about the biological importance of bacterial signaling in complex communities.
Supplementary Material
Article highlights:
Quorum quenching strategies can interfere with microbial signaling both in vivo and in vitro.
Enzymatic quenchers show low diversity in substrate preference, yet the latter property is key in interference strategies.
Highly stable and active enzymatic quenchers allow to investigate signaling in complex communities.
Interference strategies in complex communities leads to global changes in population structures and behaviors.
Funding
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM133487. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of interest
M Elias deposited the patent WO2014167140 A1 that was licenced to Gene&GreenTK, a company that he co-founded in 2013. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970. October;104(1):313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985. September;163(3):1210–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egland KA, Greenberg EP. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol Microbiol. 1999. February;31(4):1197–204. [DOI] [PubMed] [Google Scholar]
- 4.Perez PD, Hagen SJ. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One. 2010. November 16;5(11):e15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shadel GS, Baldwin TO. Positive autoregulation of the Vibrio fischeri luxR gene. LuxR and autoinducer activate cAMP-catabolite gene activator protein complex-independent and -dependent luxR transcription. J Biol Chem. 1992. April 15;267(11):7696–702. [PubMed] [Google Scholar]
- 6.Schuster M, Sexton DJ, Diggle SP, et al. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. [DOI] [PubMed] [Google Scholar]
- 7.Hense BA, Schuster M. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev. 2015. March;79(1):153–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019. June;17(6):371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016. August 11;14(9):576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** A comprehensive review of QS systems in Gram-negative bacteria.
- 10.Hartman G, Wise R. Quorum sensing: potential means of treating gram-negative infections? Lancet. 1998. March 21;351(9106):848–9. [DOI] [PubMed] [Google Scholar]; * Summary of initial developments of using QQ approaches for reducing virulence in Gram negative pathogens.
- 11.Grandclement C, Tannieres M, Morera S, et al. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev. 2016. January;40(1):86–116. [DOI] [PubMed] [Google Scholar]; ** A comprehensive review of QQ approaches including non-enzymatic processes.
- 12.Manefield M, Rasmussen TB, Henzter M, et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology. 2002. April;148(Pt 4):1119–27. [DOI] [PubMed] [Google Scholar]
- 13.Ismail AS, Valastyan JS, Bassler BL. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe. 2016. April 13;19(4):470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harder T, Campbell AH, Egan S, et al. Chemical mediation of ternary interactions between marine holobionts and their environment as exemplified by the red alga Delisea pulchra. J Chem Ecol. 2012. May;38(5):442–50. [DOI] [PubMed] [Google Scholar]
- 15.Mathesius U, Mulders S, Gao M, et al. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci U S A. 2003. February 4;100(3):1444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenk ST, Hernandez-Reyes C, Samans B, et al. N-Acyl-Homoserine Lactone Primes Plants for Cell Wall Reinforcement and Induces Resistance to Bacterial Pathogens via the Salicylic Acid/Oxylipin Pathway. Plant Cell. 2014. June;26(6):2708–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telford G, Wheeler D, Williams P, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infection and immunity. 1998. January;66(1):36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith RS, Fedyk ER, Springer TA, et al. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001. July 1;167(1):366–74. [DOI] [PubMed] [Google Scholar]
- 19.Tateda K, Ishii Y, Horikawa M, et al. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infection and immunity. 2003. October;71(10):5785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008. February;6(2):111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Very important review on how prokaryotes can communicate with eukaryotes via QS.
- 21.Sturme MH, Kleerebezem M, Nakayama J, et al. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek. 2002. August;81(1–4):233–43. [DOI] [PubMed] [Google Scholar]
- 22.Le KY, Otto M. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol. 2015;6:1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slamti L, Lereclus D. Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group. J Bacteriol. 2005. February;187(3):1182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouillaut L, Perchat S, Arold S, et al. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008. June;36(11):3791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon GJ, Wright JS, Muir TW, et al. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry. 2002. August 6;41(31):10095–104. [DOI] [PubMed] [Google Scholar]
- 26.Monnet V, Juillard V, Gardan R. Peptide conversations in Gram-positive bacteria. Crit Rev Microbiol. 2016. May;42(3):339–51. [DOI] [PubMed] [Google Scholar]; ** A comprehensive review of AIP mediated QS in Gram-positive bacteria.
- 27.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. [DOI] [PubMed] [Google Scholar]
- 28.Ali L, Goraya MU, Arafat Y, et al. Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches. Int J Mol Sci. 2017. May 3;18(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bareia T, Pollak S, Eldar A. Self-sensing in Bacillus subtilis quorum-sensing systems. Nat Microbiol. 2018. January;3(1):83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zetzmann M, Sanchez-Kopper A, Waidmann MS, et al. Identification of the agr Peptide of Listeria monocytogenes. Front Microbiol. 2016;7:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtani K, Yuan Y, Hassan S, et al. Virulence gene regulation by the agr system in Clostridium perfringens. J Bacteriol. 2009. June;191(12):3919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Xue T, Shang F, et al. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infection and immunity. 2010. August;78(8):3506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beeston AL, Surette MG. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002. July;184(13):3450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols JD, Johnson MR, Chou CJ, et al. Temperature, not LuxS, mediates AI-2 formation in hydrothermal habitats. FEMS Microbiol Ecol. 2009. May;68(2):173–81. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Schauder S, Potier N, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002. January 31;415(6871):545–9. [DOI] [PubMed] [Google Scholar]
- 36.Guo M, Gamby S, Zheng Y, et al. Small molecule inhibitors of AI-2 signaling in bacteria: state-of-the-art and future perspectives for anti-quorum sensing agents. Int J Mol Sci. 2013. August 29;14(9):17694–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999. February 16;96(4):1639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013. March;37(2):156–81. [DOI] [PubMed] [Google Scholar]; * A review of AI-2 mediated QS in bacteria.
- 39.Neiditch MB, Federle MJ, Miller ST, et al. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005. May 27;18(5):507–18. [DOI] [PubMed] [Google Scholar]
- 40.Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003. November;50(4):1411–27. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Attila C, Wang L, et al. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J Bacteriol. 2007. August;189(16):6011–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswa P, Doble M. Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS Microbiol Lett. 2013. June;343(1):34–41. [DOI] [PubMed] [Google Scholar]; * Discovery of AHL mediated QS in Gram-positive bacteria.
- 43.Zhang G, Zhang F, Ding G, et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 2012. July;6(7):1336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Discovery of AHL-mediated QS in archaea.
- 44.Watson WT, Minogue TD, Val DL, et al. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002. March;9(3):685–94. [DOI] [PubMed] [Google Scholar]
- 45.Erickson DL, Endersby R, Kirkham A, et al. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infection and immunity. 2002. April;70(4):1783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlton TS, de Nys R, Netting A, et al. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ Microbiol. 2000. October;2(5):530–41. [DOI] [PubMed] [Google Scholar]
- 47.Leipert J, Treitz C, Leippe M, et al. Identification and Quantification of N-Acyl Homoserine Lactones Involved in Bacterial Communication by Small-Scale Synthesis of Internal Standards and Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2017. December;28(12):2538–2547. [DOI] [PubMed] [Google Scholar]
- 48.Kim YW, Sung C, Lee S, et al. MALDI-MS-based quantitative analysis for ketone containing homoserine lactones in Pseudomonas aeruginosa. Anal Chem. 2015. January 20;87(2):858–63. [DOI] [PubMed] [Google Scholar]
- 49.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012. November 1;2(11). [DOI] [PMC free article] [PubMed] [Google Scholar]; * Summary of initial developments of using QQ approaches for reducing virulence in Gram negative pathogens.
- 50.Verma SC, Miyashiro T. Quorum sensing in the squid-Vibrio symbiosis. Int J Mol Sci. 2013. August 7;14(8):16386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dessaux Y, Faure D. Quorum Sensing and Quorum Quenching in Agrobacterium: A “Go/No Go System”? Genes (Basel). 2018. April 16;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009. April;12(2):182–91. [DOI] [PubMed] [Google Scholar]
- 53.Stauff DL, Bassler BL. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol. 2011. August;193(15):3871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnard AM, Bowden SD, Burr T, et al. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2007. July 29;362(1483):1165–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Houdt R, Givskov M, Michiels CW. Quorum sensing in Serratia. FEMS Microbiol Rev. 2007. July;31(4):407–24. [DOI] [PubMed] [Google Scholar]
- 56.Garde C, Bjarnsholt T, Givskov M, et al. Quorum sensing regulation in Aeromonas hydrophila. J Mol Biol. 2010. March 5;396(4):849–57. [DOI] [PubMed] [Google Scholar]
- 57.Swift S, Karlyshev AV, Fish L, et al. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997. September;179(17):5271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaefer AL, Greenberg EP, Oliver CM, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008. July 31;454(7204):595–9. [DOI] [PubMed] [Google Scholar]; * Discovery of aryl homoserine lactones as QS signals.
- 59.Liao L, Schaefer AL, Coutinho BG, et al. An aryl-homoserine lactone quorum-sensing signal produced by a dimorphic prosthecate bacterium. Proc Natl Acad Sci U S A. 2018. July 17;115(29):7587–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindemann A, Pessi G, Schaefer AL, et al. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 2011. October 4;108(40):16765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahlgren NA, Harwood CS, Schaefer AL, et al. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc Natl Acad Sci U S A. 2011. April 26;108(17):7183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wade DS, Calfee MW, Rocha ER, et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005. July;187(13):4372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bredenbruch F, Geffers R, Nimtz M, et al. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol. 2006. August;8(8):1318–29. [DOI] [PubMed] [Google Scholar]
- 64.Diggle SP, Matthijs S, Wright VJ, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007. January;14(1):87–96. [DOI] [PubMed] [Google Scholar]
- 65.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015. January;6(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Discussion of the complex QS signal hierarchy in Pseudomonas aeruginosa.
- 66.Rampioni G, Falcone M, Heeb S, et al. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS pathogens. 2016. November;12(11):e1006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dandela R, Mantin D, Cravatt BF, et al. Proteome-wide mapping of PQS-interacting proteins in Pseudomonas aeruginosa. Chem Sci. 2018. February 28;9(8):2290–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J, Cheng J, Wang Y, et al. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front Cell Infect Microbiol. 2018;8:230. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Summary of the PQS system in Pseudomonas aeruginosa.
- 69.Flavier AB, Clough SJ, Schell MA, et al. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol. 1997. October;26(2):251–9. [DOI] [PubMed] [Google Scholar]; *Discovery of 3-OH-PAME as a new class of QS molecules.
- 70.Ryan RP, An SQ, Allan JH, et al. The DSF Family of Cell-Cell Signals: An Expanding Class of Bacterial Virulence Regulators. PLoS pathogens. 2015. July;11(7):e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Summary of the DSF class of QS signals.
- 71.Holden MT, Ram Chhabra S, de Nys R, et al. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol Microbiol. 1999. September;33(6):1254–66. [DOI] [PubMed] [Google Scholar]
- 72.Tommonaro G, Abbamondi GR, Iodice C, et al. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb Ecol. 2012. April;63(3):490–5. [DOI] [PubMed] [Google Scholar]
- 73.Uroz S, Oger PM, Chapelle E, et al. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Applied and environmental microbiology. 2008. March;74(5):1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergonzi C, Schwab M, Naik T, et al. Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties. Sci Rep. 2018. July 26;8(1):11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin YH, Xu JL, Hu J, et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003. February;47(3):849–60. [DOI] [PubMed] [Google Scholar]
- 76.Bijtenhoorn P, Mayerhofer H, Muller-Dieckmann J, et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS One. 2011;6(10):e26278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bijtenhoorn P, Schipper C, Hornung C, et al. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J Biotechnol. 2011. August 20;155(1):86–94. [DOI] [PubMed] [Google Scholar]
- 78.Chowdhary PK, Keshavan N, Nguyen HQ, et al. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry. 2007. December 18;46(50):14429–37. [DOI] [PubMed] [Google Scholar]
- 79.Dong YH, Xu JL, Li XZ, et al. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A. 2000. March 28;97(7):3526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mackness M, Mackness B. Human paraoxonase-1 (PON1): Gene structure and expression, promiscuous activities and multiple physiological roles. Gene. 2015. August 1;567(1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sio CF, Otten LG, Cool RH, et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infection and immunity. 2006. March;74(3):1673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chai Y, Tsai CS, Cho H, et al. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J Bacteriol. 2007. May;189(9):3674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000. December;182(24):6921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wahjudi M, Papaioannou E, Hendrawati O, et al. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiol-Sgm. 2011. July;157:2042–2055. [DOI] [PubMed] [Google Scholar]
- 85.Utari PD, Vogel J, Quax WJ. Deciphering Physiological Functions of AHL Quorum Quenching Acylases. Frontiers in Microbiology. 2017. June 19;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bokhove M, Jimenez PN, Quax WJ, et al. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. P Natl Acad Sci USA. 2010. January 12;107(2):686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koch G, Nadal-Jimenez P, Reis CR, et al. Reducing virulence of the human pathogen Burkholderia by altering the substrate specificity of the quorum-quenching acylase PvdQ. Proc Natl Acad Sci U S A. 2014. January 28;111(4):1568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Drake EJ, Gulick AM. Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis. ACS Chem Biol. 2011. November 18;6(11):1277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bergonzi C, Schwab M, Naik T, et al. The Structural Determinants Accounting for the Broad Substrate Specificity of the Quorum Quenching Lactonase GcL. Chembiochem. 2019. July 15;20(14):1848–1855. [DOI] [PubMed] [Google Scholar]
- 90.Hiblot J, Gotthard G, Elias M, et al. Differential active site loop conformations mediate promiscuous activities in the lactonase SsoPox. PLoS One. 2013;8(9):e75272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hiblot J, Gotthard G, Chabriere E, et al. Characterisation of the organophosphate hydrolase catalytic activity of SsoPox. Sci Rep. 2012;2:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elias M, Dupuy J, Merone L, et al. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J Mol Biol. 2008. June 20;379(5):1017–28. [DOI] [PubMed] [Google Scholar]
- 93.Bzdrenga J, Hiblot J, Gotthard G, et al. SacPox from the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius is a proficient lactonase. BMC Res Notes. 2014. June 3;7:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hiblot J, Bzdrenga J, Champion C, et al. Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia. Sci Rep. 2015. February 11;5:8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hiblot J, Gotthard G, Chabriere E, et al. Structural and enzymatic characterization of the lactonase SisLac from Sulfolobus islandicus. PLoS One. 2012;7(10):e47028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chow JY, Xue B, Lee KH, et al. Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily. J Biol Chem. 2010. December 24;285(52):40911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang LH, Weng LX, Dong YH, et al. Specificity and enzyme kinetics of the quorum-quenching N-Acyl homoserine lactone lactonase (AHL-lactonase). J Biol Chem. 2004. April 2;279(14):13645–51. [DOI] [PubMed] [Google Scholar]
- 98.Tang K, Su Y, Brackman G, et al. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Applied and environmental microbiology. 2015. January;81(2):774–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong YH, Wang LH, Xu JL, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001. June 14;411(6839):813–7. [DOI] [PubMed] [Google Scholar]
- 100.Gao A, Mei GY, Liu S, et al. High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase. Acta Crystallogr D Biol Crystallogr. 2013. January;69(Pt 1):82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mei GY, Yan XX, Turak A, et al. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Applied and environmental microbiology. 2010. August;76(15):4933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aldridge WN. Serum esterases. II. An enzyme hydrolysing diethyl p-nitrophenyl phosphate (E600) and its identity with the A-esterase of mammalian sera. Biochem J. 1953. January;53(1):117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harel M, Aharoni A, Gaidukov L, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004. May;11(5):412–9. [DOI] [PubMed] [Google Scholar]
- 104.Bar-Rogovsky H, Hugenmatter A, Tawfik DS. The evolutionary origins of detoxifying enzymes: the mammalian serum paraoxonases (PONs) relate to bacterial homoserine lactonases. J Biol Chem. 2013. August 16;288(33):23914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Draganov DI, Teiber JF, Speelman A, et al. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J Lipid Res. 2005. June;46(6):1239–47. [DOI] [PubMed] [Google Scholar]
- 106.Elias M, Tawfik DS. Divergence and convergence in enzyme evolution: parallel evolution of paraoxonases from quorum-quenching lactonases. J Biol Chem. 2012. January 2;287(1):11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roy V, Fernandes R, Tsao CY, et al. Cross species quorum quenching using a native AI-2 processing enzyme. ACS Chem Biol. 2010. February 19;5(2):223–32. [DOI] [PubMed] [Google Scholar]
- 108.Weiland-Brauer N, Kisch MJ, Pinnow N, et al. Highly Effective Inhibition of Biofilm Formation by the First Metagenome-Derived AI-2 Quenching Enzyme. Front Microbiol. 2016;7:1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weiland-Brauer N, Malek I, Schmitz RA. Metagenomic quorum quenching enzymes affect biofilm formation of Candida albicans and Staphylococcus epidermidis. PLoS One. 2019;14(1):e0211366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pustelny C, Albers A, Buldt-Karentzopoulos K, et al. Dioxygenase-Mediated Quenching of Quinolone-Dependent Quorum Sensing in Pseudomonas aeruginosa. Chem Biol. 2009. December 24;16(12):1259–1267. [DOI] [PubMed] [Google Scholar]
- 111.Shinohara M, Nakajima N, Uehara Y. Purification and characterization of a novel esterase (beta-hydroxypalmitate methyl ester hydrolase) and prevention of the expression of virulence by Ralstonia solanacearum. J Appl Microbiol. 2007. July;103(1):152–62. [DOI] [PubMed] [Google Scholar]
- 112.Lee MH, Khan R, Tao W, et al. Soil metagenome-derived 3-hydroxypalmitic acid methyl ester hydrolases suppress extracellular polysaccharide production in Ralstonia solanacearum. J Biotechnol. 2018. March 20;270:30–38. [DOI] [PubMed] [Google Scholar]
- 113.Kanzaki H, Imura D, Nitoda T, et al. Enzymatic conversion of cyclic dipeptides to dehydro derivatives that inhibit cell division. J Biosci Bioeng. 2000;90(1):86–9. [DOI] [PubMed] [Google Scholar]
- 114.Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS pathogens. 2010. July 8;6(7):e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gerdt JP, Blackwell HE. Competition studies confirm two major barriers that can preclude the spread of resistance to quorum-sensing inhibitors in bacteria. ACS Chem Biol. 2014. October 17;9(10):2291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garcia-Contreras R, Maeda T, Wood TK. Can resistance against quorum-sensing interference be selected? ISME J. 2016. January;10(1):4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dong YH, Zhang LH. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005. February;43 Spec No:101–9. [PubMed] [Google Scholar]
- 118.Garcia-Contreras R, Maeda T, Wood TK. Resistance to quorum-quenching compounds. Applied and environmental microbiology. 2013. November;79(22):6840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krzyzek P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front Microbiol. 2019;10:2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garcia-Contreras R, Perez-Eretza B, Jasso-Chavez R, et al. High variability in quorum quenching and growth inhibition by furanone C-30 in Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Pathog Dis. 2015. August;73(6):ftv040. [DOI] [PubMed] [Google Scholar]
- 121.Guendouze A, Plener L, Bzdrenga J, et al. Effect of Quorum Quenching Lactonase in Clinical Isolates of Pseudomonas aeruginosa and Comparison with Quorum Sensing Inhibitors. Front Microbiol. 2017;8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lopez-Jacome LE, Garza-Ramos G, Hernandez-Duran M, et al. AiiM Lactonase Strongly Reduces Quorum Sensing Controlled Virulence Factors in Clinical Strains of Pseudomonas aeruginosa Isolated From Burned Patients. Front Microbiol. 2019;10:2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Richards MJ, Edwards JR, Culver DH, et al. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999. May;27(5):887–92. [DOI] [PubMed] [Google Scholar]
- 124.Wagner S, Sommer R, Hinsberger S, et al. Novel Strategies for the Treatment of Pseudomonas aeruginosa Infections. J Med Chem. 2016. July 14;59(13):5929–69. [DOI] [PubMed] [Google Scholar]
- 125.Lee K, Yoon SS. Pseudomonas aeruginosa Biofilm, a Programmed Bacterial Life for Fitness. J Microbiol Biotechnol. 2017. June 28;27(6):1053–1064. [DOI] [PubMed] [Google Scholar]
- 126.Anandan K, Vittal RR. Quorum quenching activity of AiiA lactonase KMMI17 from endophytic Bacillus thuringiensis KMCL07 on AHL- mediated pathogenic phenotype in Pseudomonas aeruginosa. Microb Pathog. 2019. July;132:230–242. [DOI] [PubMed] [Google Scholar]
- 127.Papaioannou E, Wahjudi M, Nadal-Jimenez P, et al. Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob Agents Chemother. 2009. November;53(11):4891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stoltz DA, Ozer EA, Ng CJ, et al. Paraoxonase-2 deficiency enhances Pseudomonas aeruginosa quorum sensing in murine tracheal epithelia. Am J Physiol Lung Cell Mol Physiol. 2007. April;292(4):L852–60. [DOI] [PubMed] [Google Scholar]
- 129.Hraiech S, Hiblot J, Lafleur J, et al. Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat pneumonia. PLoS One. 2014;9(10):e107125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Utari PD, Setroikromo R, Melgert BN, et al. PvdQ Quorum Quenching Acylase Attenuates Pseudomonas aeruginosa Virulence in a Mouse Model of Pulmonary Infection. Front Cell Infect Microbiol. 2018;8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mahan K, Martinmaki R, Larus I, et al. Effects of Signal Disruption Depends on the Substrate Preference of the Lactonase. Front Microbiol. 2019;10:3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Remy B, Plener L, Decloquement P, et al. Lactonase Specificity Is Key to Quorum Quenching in Pseudomonas aeruginosa. Front Microbiol. 2020;11:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Birmes FS, Saring R, Hauke MC, et al. Interference with Pseudomonas aeruginosa Quorum Sensing and Virulence by the Mycobacterial Pseudomonas Quinolone Signal Dioxygenase AqdC in Combination with the N-Acylhomoserine Lactone Lactonase QsdA. Infection and immunity. 2019. October;87(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koch G, Nadal-Jimenez P, Cool RH, et al. Deinococcus radiodurans can interfere with quorum sensing by producing an AHL-acylase and an AHL-lactonase. FEMS Microbiol Lett. 2014. July;356(1):62–70. [DOI] [PubMed] [Google Scholar]
- 135.Zhang B, Zhuang X, Guo L, et al. Recombinant N-acyl homoserine lactone-Lactonase AiiAQSI-1 Attenuates Aeromonas hydrophila Virulence Factors, Biofilm Formation and Reduces Mortality in Crucian Carp. Mar Drugs. 2019. August 27;17(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Torres M, Reina JC, Fuentes-Monteverde JC, et al. AHL-lactonase expression in three marine emerging pathogenic Vibrio spp. reduces virulence and mortality in brine shrimp (Artemia salina) and Manila clam (Venerupis philippinarum). PLoS One. 2018;13(4):e0195176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garge SS, Nerurkar AS. Attenuation of Quorum Sensing Regulated Virulence of Pectobacterium carotovorum subsp. carotovorum through an AHL Lactonase Produced by Lysinibacillus sp. Gs50. PLoS One. 2016;11(12):e0167344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fong J, Zhang C, Yang R, et al. Combination Therapy Strategy of Quorum Quenching Enzyme and Quorum Sensing Inhibitor in Suppressing Multiple Quorum Sensing Pathways of P. aeruginosa. Sci Rep. 2018. January 18;8(1):1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Billings N, Millan M, Caldara M, et al. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS pathogens. 2013;9(8):e1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Solano C, Echeverz M, Lasa I. Biofilm dispersion and quorum sensing. Curr Opin Microbiol. 2014. April;18:96–104. [DOI] [PubMed] [Google Scholar]
- 141.Flemming HC, Wingender J, Szewzyk U, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016. August 11;14(9):563–75. [DOI] [PubMed] [Google Scholar]
- 142.Fux CA, Costerton JW, Stewart PS, et al. Survival strategies of infectious biofilms. Trends in microbiology. 2005. January;13(1):34–40. [DOI] [PubMed] [Google Scholar]
- 143.Bzdrenga J, Daude D, Remy B, et al. Biotechnological applications of quorum quenching enzymes. Chem Biol Interact. 2017. April 1;267:104–115. [DOI] [PubMed] [Google Scholar]
- 144.Chow JY, Yang Y, Tay SB, et al. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob Agents Chemother. 2014;58(3):1802–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bryers JD. Medical biofilms. Biotechnol Bioeng. 2008. May 1;100(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gbejuade HO, Lovering AM, Webb JC. The role of microbial biofilms in prosthetic joint infections. Acta Orthop. 2015. April;86(2):147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ivanova K, Fernandes MM, Mendoza E, et al. Enzyme multilayer coatings inhibit Pseudomonas aeruginosa biofilm formation on urinary catheters. Appl Microbiol Biotechnol. 2015. May;99(10):4373–85. [DOI] [PubMed] [Google Scholar]
- 148.Costoya A, Velazquez Becerra LE, Melendez-Ortiz HI, et al. Immobilization of antimicrobial and anti-quorum sensing enzymes onto GMA-grafted poly(vinyl chloride) catheters. Int J Pharm. 2019. March 10;558:72–81. [DOI] [PubMed] [Google Scholar]
- 149.Remy B, Plener L, Poirier L, et al. Harnessing hyperthermostable lactonase from Sulfolobus solfataricus for biotechnological applications. Sci Rep. 2016. November 23;6:37780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim SR, Lee KB, Kim JE, et al. Macroencapsulation of quorum quenching bacteria by polymeric membrane layer and its application to MBR for biofouling control. J Membrane Sci. 2015. January 1;473:109–117. [Google Scholar]
- 151.Oh HS, Yeon KM, Yang CS, et al. Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in microporous membrane. Environ Sci Technol. 2012. May 1;46(9):4877–84. [DOI] [PubMed] [Google Scholar]
- 152.Nguyen PDT, Mustapha NA, Kadokami K, et al. Quorum sensing between Gram-negative bacteria responsible for methane production in a complex waste sewage sludge consortium. Appl Microbiol Biotechnol. 2019. February;103(3):1485–1495. [DOI] [PubMed] [Google Scholar]
- 153.Huang S, Bergonzi C, Schwab M, et al. Evaluation of biological and enzymatic quorum quencher coating additives to reduce biocorrosion of steel. PLoS One. 2019;14(5):e0217059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwab M, Bergonzi C, Sakkos J, et al. Signal Disruption Leads to Changes in Bacterial Community Population. Front Microbiol. 2019;10:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xavier KB. Bacterial interspecies quorum sensing in the mammalian gut microbiota [C. R. Biologies 341 (2018) 10.1016/j.crvi.2018.03.006] C R Biol. 2018. May-Jun;341(5):300. [DOI] [PubMed] [Google Scholar]
- 156.Thompson JA, Oliveira RA, Djukovic A, et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015. March 24;10(11):1861–71. [DOI] [PubMed] [Google Scholar]
- 157.Hsiao A, Ahmed AM, Subramanian S, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014. November 20;515(7527):423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu L, Yao W, Rao Y, et al. pH-Responsive carriers for oral drug delivery: challenges and opportunities of current platforms. Drug Deliv. 2017. November;24(1):569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gupta P, Chhibber S, Harjai K. Efficacy of purified lactonase and ciprofloxacin in preventing systemic spread of Pseudomonas aeruginosa in murine burn wound model. Burns. 2015. February;41(1):153–62. [DOI] [PubMed] [Google Scholar]
- 160.Mion S, Remy B, Plener L, et al. Quorum Quenching Lactonase Strengthens Bacteriophage and Antibiotic Arsenal Against Pseudomonas aeruginosa Clinical Isolates. Front Microbiol. 2019;10:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


