Abstract
The evolutionarily conserved POT1 protein binds single‐stranded G‐rich telomeric DNA and has been implicated in contributing to telomeric DNA maintenance and the suppression of DNA damage checkpoint signaling. Here, we explore human POT1 function through genetics and proteomics, discovering that a complete absence of POT1 leads to severe telomere maintenance defects that had not been anticipated from previous depletion studies in human cells. Conditional deletion of POT1 in HEK293E cells gives rise to rapid telomere elongation and length heterogeneity, branched telomeric DNA structures, telomeric R‐loops, and telomere fragility. We determine the telomeric proteome upon POT1‐loss, implementing an improved telomeric chromatin isolation protocol. We identify a large set of proteins involved in nucleic acid metabolism that engage with telomeres upon POT1‐loss. Inactivation of the homology‐directed repair machinery suppresses POT1‐loss‐mediated telomeric DNA defects. Our results unravel as major function of human POT1 the suppression of telomere instability induced by homology‐directed repair.
Keywords: DNA damage response, homologous recombination, POT1, R‐loops, telomeric proteome
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination
Complete loss of the telomeric ssDNA‐binding protein in human cells induces telomeric phenotypes not anticipated from previous knockdown studies, including telomere elongation, R‐loop formation, and chromatin proteome alterations.
Introduction
Telomeres, the nucleoprotein structures at chromosome ends, are distinct from chromosome internal breaks in remarkable ways. Telomeric proteins protect chromosome ends from degradation and suppress DNA damage signaling. In addition, they prevent DNA end‐to‐end fusions by non‐homologous end joining and they suppress inter‐ and intra‐chromosomal homologous recombination between telomere repeats, which can lead to rampant telomere elongation and loss events.
Many of the telomere functions are mediated by the shelterin proteins comprising TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 which cover large parts of telomeric DNA (Lazzerini‐Denchi & Sfeir, 2016; de Lange, 2018). POT1 is a highly conserved telomere‐binding protein which binds the telomeric single‐stranded G‐rich DNA present as a 3′overhang at the end of the chromosomes (Baumann & Cech, 2001). Alternatively, when telomeres adopt a t‐loop structure in which the single‐stranded 3′ overhang is tucked into double‐stranded part of the telomere (Doksani et al, 2013), POT1 is thought to bind to the single‐stranded displaced G‐rich strand. Though POT1 binds directly to the single‐stranded telomeric DNA, it is recruited to telomeres through its physical interaction with the shelterin component TPP1 (Takai et al, 2011), which in turn interacts with TIN2 that is associated with the double strand telomere‐binding proteins TRF1 and TRF2 (de Lange, 2018). This type of molecular architecture is similar from mammals to fission yeast suggesting widespread conservation in evolution (Miyoshi et al, 2008).
Studies in a variety of eukaryotes unraveled important functions of POT1 in chromosome end protection and telomere length control. In fission yeast, Pot1‐loss leads to rapid loss of telomeric DNA and chromosome circularization suggesting roles in protecting chromosome ends from nucleolytic degradation (Baumann & Cech, 2001; Pitt & Cooper, 2010). In mammals, such dramatic telomere loss events are not seen upon depletion of POT1. In contrast to the human genome, the mouse genome contains two POT1 paralogs, Pot1a and Pot1b, and studies in this organism have contributed to the understanding of POT1 function (Hockemeyer et al, 2006; Wu et al, 2006). Deletion of conditional alleles in mouse embryonic fibroblasts (MEFs) revealed that POT1a represses ATR signaling which is triggered through RPA binding to single‐stranded telomeric DNA (Hockemeyer et al, 2006; Wu et al, 2006) whereas POT1b regulates the telomeric DNA overhang structure (Hockemeyer et al, 2006). Pot1a deletion in MEFs per se was also reported to increase homologous recombination (HR) in one study (Wu et al, 2006) but not the other (Hockemeyer et al, 2006). However, concomitant deletion of Pot1a and Pot1b induced higher incidences of telomere sister chromatid exchanges in Ku70 −/− cells suggesting contributions of mouse POT1a/b to the suppression of homology‐directed repair (HDR) in Ku70 −/− cells (Palm et al, 2009). The functions of POT1 in human cells had so far been mostly assessed by overexpression or RNAi‐mediated depletion but not knockout studies. In addition to the telomeric DNA damage response, POT1 reduction induced incorrect 5′ end processing in which telomeres lost the preferred ending with ATC‐5′ to random positions within the telomeric repeat sequence (Hockemeyer et al, 2005).
Several studies also suggest direct or indirect roles of POT1 in semiconservative DNA replication and in telomere length control by telomerase. Overexpression of POT1 mutants which are prevalent in T‐cell lymphoma in human fibrosarcoma HT1080 cells increased telomere fragility which is indicative of DNA replication defects (Pinzaru et al, 2016). This effect was suspected to be due to defects of the POT1 mutants in POT1‐assisted assembly of the CST complex (Pinzaru et al, 2016) which promotes lagging strand synthesis at telomeres. In addition, POT1 can regulate telomerase. Overexpression of a mutant form of human POT1 that lacks the DNA‐binding OB‐fold induces rapid telomere elongation (Loayza & De Lange, 2003; Zhong et al, 2012) and POT1 also inhibits telomerase in vitro (Kelleher et al, 2005). On the other hand, in conjunction with TPP1, POT1 can stimulate telomerase processivity in vitro (Latrick & Cech, 2010).
In this paper, we assess human POT1 function by analyzing the consequence of POT1 loss upon conditional deletion in HEK293E cells. We demonstrate that the complete absence of POT1 triggers telomeric DNA branching, telomere elongation, accumulation of G‐rich telomeric DNA, telomeric R‐loops, and telomere fragility. We elucidate changes in the telomeric proteome establishing an improved telomeric chromatin isolation protocol, identifying among others HDR factors that accumulate at telomeres in the absence of POT1. Significantly, the severe telomere maintenance defects but not the DNA damage response in the absence of POT1 are all suppressed upon inactivation of the HDR machinery. Thus, a major function of human POT1 is the repression of telomeric homologous recombination.
Results
POT1‐loss triggers rapid telomere elongation and a DNA damage response throughout the cell cycle
In order to study the functions of human POT1 for the maintenance of telomeric DNA structure and telomeric protein composition, we genetically modified HEK293E cells. HEK293E cells were originally derived from a human embryonic kidney preparation but are presumably of adrenal origin (Lin et al, 2014). They contain the E1A adenovirus gene and EBNA1 from Epstein–Barr virus. We first tagged using CRIPSR/Cas9 gene editing the TRF1 and TRF2 genes at their N‐termini with triple FLAG‐tags to facilitate isolation of telomere chromatin as explained below. Cell clones were isolated and genotyped by PCR. Addition of these tags in chosen clone 293E cl75 did not interfere with telomere maintenance as assessed by telomere restriction fragment length (TRF) analysis nor did it induce a telomeric DNA damage response. In a second step, we sought to develop a conditional allele of human POT1 in 293E cl75, inserting by CRISPR/Cas9 gene editing LoxP sites in the introns flanking exon 6 (Fig 1A). We screened clonal isolates by PCR and sequencing for the appropriate genotype (Fig 1B, and Appendix Fig S1A and B). 293E cl75p100 carried two POT1 alleles with the desired LoxP sites flanking exon 6 and a third allele with a frameshift mutation in exon 6 giving rise to a premature stop codon. Of note, HEK293E cells are triploid for chromosome 7, which harbors POT1. 293E cl75p100 maintains constant telomere length over several weeks of growth as assessed by TRF analysis (Appendix Fig S1C). We then transduced clone 293E cl75p100 with CreERT2 lentivirus expressing tamoxifen‐dependent Cre recombinase to allow excision of the DNA between the LoxP sites. Cells were again cloned and screened for efficient editing of the conditional POT1 alleles upon addition of 4‐hydroxytamoxifen (4OHT) scoring for loss of POT1 protein expression by Western blot analysis (Appendix Fig S1D). Two clones (293E 75p100 Cre35 and 293E 75p100 Cre45) showed efficient loss of POT1 protein and induction of the DNA damage marker γ‐H2AX as expected (Appendix Fig S1D). Phosphorylated forms of ATM and CHK1 also accumulated upon induced deletion of POT1 (Fig 1C).
Figure 1. POT1‐loss triggers rapid telomere elongation and a DNA damage response throughout the cell cycle.
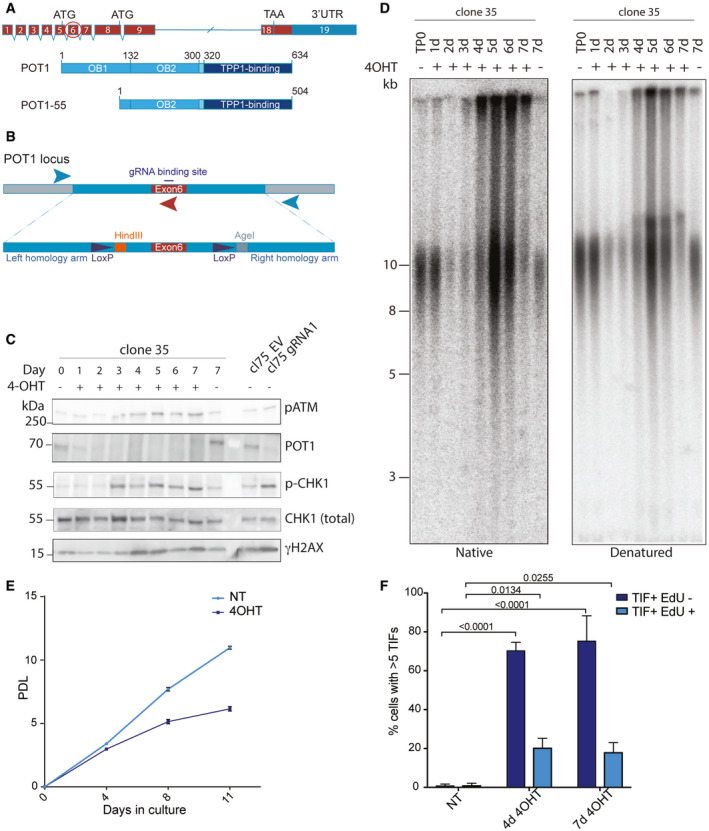
- Schematic drawing of the POT1 gene structure and two derived polypeptides. Exon 6 that has been chosen for gene editing is encircled.
- Schematic drawing of the repair template used for CRISPR/Cas9-mediated gene editing in HEK293E cells. Positions of primers for genotyping PCR are marked with arrows.
- Western blot for POT1 and DDR markers upon POT1 deletion induced with 0.5 μM 4‐OHT. Cl75—parental cell line. gRNA1—POT1 knockout in the population by transient transfection with pSpCas9(BB)‐2A-puro containing gRNA1 sequence. EV—corresponding empty vector.
- Time course of telomere length changes upon POT1 removal in clone 35. d (days) TP0—time point 0, no 4OHT.
- Growth curve for POT1 WT and POT1 knockout cells. Population doublings (PDL) are represented as mean ± SD (three biological replicates).
- Quantification for TIFs in EdU‐positive and EdU‐negative cells upon POT1 removal in clone 35. The bars show percentage of cells containing more than 5 TIFs ± SD. Experiments were performed in triplicate. At least 120 cells were analyzed per condition per replicate. Significance was determined using two‐way ANOVA. P‐values are indicated on the graph.
We assessed the effects of POT1‐loss on telomeric DNA by TRF analysis of non‐denatured DNA to detect single‐stranded G‐rich telomeric DNA and of denatured DNA to measure telomere length distribution (Fig 1D and Appendix Fig S1E). Very strikingly, loss of POT1 for more than 3–4 days caused dramatic telomere elongation, an increase of single‐stranded G‐rich telomeric DNA, and an accumulation of telomeric DNA molecules that could not enter the gel, suggestive of branched DNA structures. The viability and the proliferation of the cells upon POT1‐loss were strongly affected after 4 days of culture in presence of 4OHT (Fig 1E). Analysis of cellular DNA content by fluorocytometry indicated that during the time course, the fraction of cells in G1 was reduced and a subset of cells with a sub‐G1 DNA content accumulated indicative of cells undergoing cell death by apoptosis. The number of polyploid cells also increased indicating abnormalities in mitosis (Appendix Fig S2A). Immunofluorescence analysis of γ‐H2AX foci showed that they colocalized with telomeric DNA, in line with the crucial roles of POT1 in suppressing DNA damage specifically at chromosome ends (Appendix Fig S2B). We pulse‐labeled cells with EdU to identify cells in S phase. The DNA damage foci were detected in both pulse‐labeled EdU‐positive and EdU‐negative cells (Fig 1F), which lead us to conclude that POT1 suppresses the DNA damage response throughout the cell cycle.
POT1‐loss leads to accumulation of single‐stranded G‐rich DNA, branched DNA structures, and telomere fragility
To test whether the single‐stranded G‐rich DNA at telomeres was terminal, we treated isolated genomic DNA with commercial 3′–5′ exonuclease I in vitro and repeated the native and denatured TRF analysis upon short‐term gel electrophoresis which facilitates the quantification of the telomeric signals (Fig 2A). As expected, the G‐strand DNA signal on native gels disappeared upon exonuclease I treatment in the uninduced cells containing wild‐type POT1 because the signal stems from the telomeric 3′ overhang only (Fig 2A). Upon deletion of POT1 (lanes labeled with + 4OHT), the native signal was only partially reduced with exonuclease I, indicating that the telomeric DNA accumulated terminal as well as internal single‐stranded G‐rich DNA in the absence of POT1.
Figure 2. POT1‐loss leads to accumulation of single‐stranded G‐rich DNA, branched DNA structures, and telomere fragility.
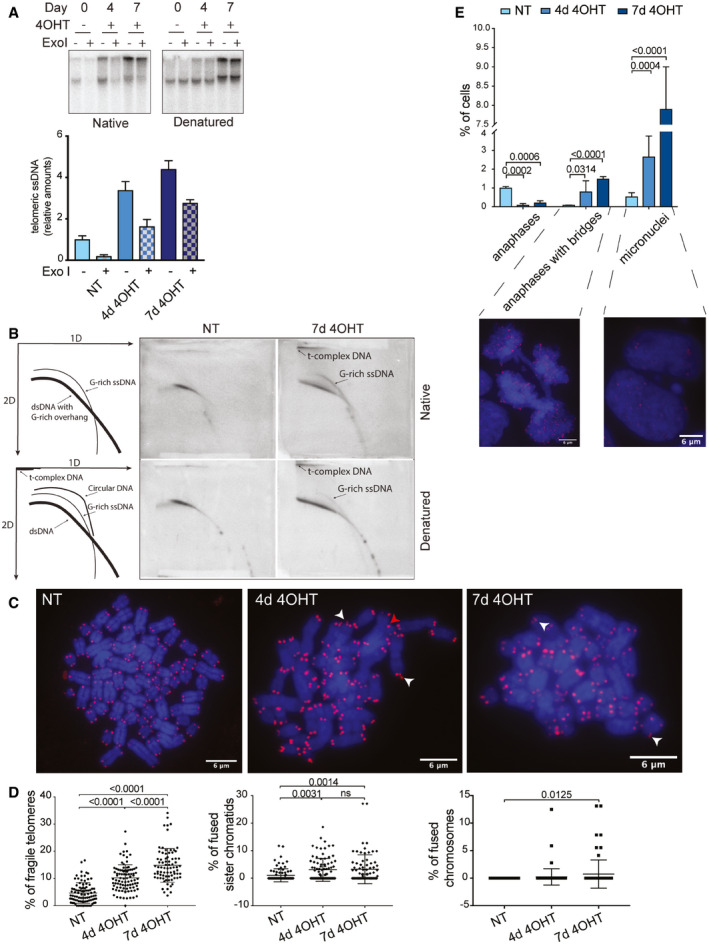
- G‐overhang assay for clone 35 upon POT1 removal. The bars show relative amounts of single‐stranded telomeric DNA ± SD. Experiments were performed in triplicate.
- 2D gels for telomeric DNA from clone 35 cells with POT1 (NT) and upon POT1 removal for 7 days (7d 4‐OHT).
- Examples of metaphase spreads for non‐treated (NT) clone 35 cells and clone 35 treated with 4‐OHT for 4 and 7 days. White arrowheads indicate fragile telomeres, the red arrowhead indicates a sister chromatid fusion. Scale bar equals 6 μm.
- Quantification of fragile telomeres (left graph), sister chromatid fusions (middle graph), and chromosome fusions (right graph) as a percentage of events per metaphase. Experiments were performed in triplicate. At least 25 metaphases were examined per condition per replicate. The mean is displayed and error bars represent ± SD. Significance was determined using one‐way ANOVA. P‐values are indicated on the graph.
- Quantification and representative images of anaphases with bridges and micronuclei for non‐treated (NT) clone 35 cells and clone 35 treated with 4‐OHT for 4 and 7 days. Scale bar equals 6 μm. Experiments were performed in triplicate. The bars show percentage of cells containing anaphases, anaphases with bridges, or micronuclei + SD. At least 650 cells were examined per condition per replicate. Significance was determined using one‐way ANOVA. P‐values are indicated on the graph. In all experiments, POT1 deletion was induced with 0.5 μM 4‐OHT.
To further characterize the telomeric DNA abnormalities, we used two‐dimensional gel electrophoresis (Wang et al, 2004). DNA was digested with HinfI and RsaI and electrophoresis in the first and second dimension was performed in the absence and presence of ethidium bromide to estimate DNA size and conformation, respectively (Fig 2B). Telomeric DNA was detected before and after denaturation. In wild‐type cells, a single arc was detected consistent with linear double‐stranded telomeric DNA with a 3′ overhang (Wang et al, 2004). POT1 deletion led to the appearance of the slow migrating so‐called t‐complex (Nabetani & Ishikawa, 2009) which was particularly retarded in the presence of ethidium bromide and which therefore is considered to represent branched DNA with double‐stranded and single‐stranded portions (Nabetani & Ishikawa, 2009). In addition, single‐stranded G‐rich DNA appeared as an arc, which may stem from unbranched molecules (Fig 2B).
Telomere replication defects lead to accumulation of so‐called fragile telomeres in which the telomeric DNA signal detected on metaphase chromosomes is smeared or double (Sfeir et al, 2009). We quantified telomere fragility as well as other telomere abnormalities in POT1 wild‐type and knockout cells (Fig 2C and D). We observed a strong increase in telomere fragility from 5 to 10% after 4 days and to 15% after 7 days of POT1‐deletion. Telomere sister chromatid fusions and chromosome end‐to‐end fusions also increased but this concerned a much smaller fraction of telomeres (Fig 2D).
Consistent with the analysis by Southern blotting, the metaphase chromosomes of POT1 knockout cells showed telomere lengthening and a striking increase in telomere length heterogeneity. It is worth noting that telomere fragility was strongly increased already on the day 4 of POT1 knockout, when telomere length in the knockout cells is very close to the wild type (Appendix Fig S3A). To make sure that increase in telomere fragility was not just a function of telomere length, we used another HEK293E clone, clone 38, that has longer telomeres than clone 35. In this clone, the background fragility was lower than in clone 35, but on day 7 of treatment with 4‐OHT the fragility increased twofold, recapitulating the phenotype we describe in clone 35 (Appendix Fig S3B and C).
When observing interphase nuclei, we noticed increased amounts of micronuclei, which is an indication for genomic instability upon POT1‐loss (Fig 2E). Finally, while anaphase cells were rare to detect in POT1 knockout cells, they displayed increased frequency of anaphase bridges, which were likely caused by the chromatid and chromosome fusions (Fig 2E).
The POT1‐loss phenotype is caused by deprotection of the G‐rich DNA overhang
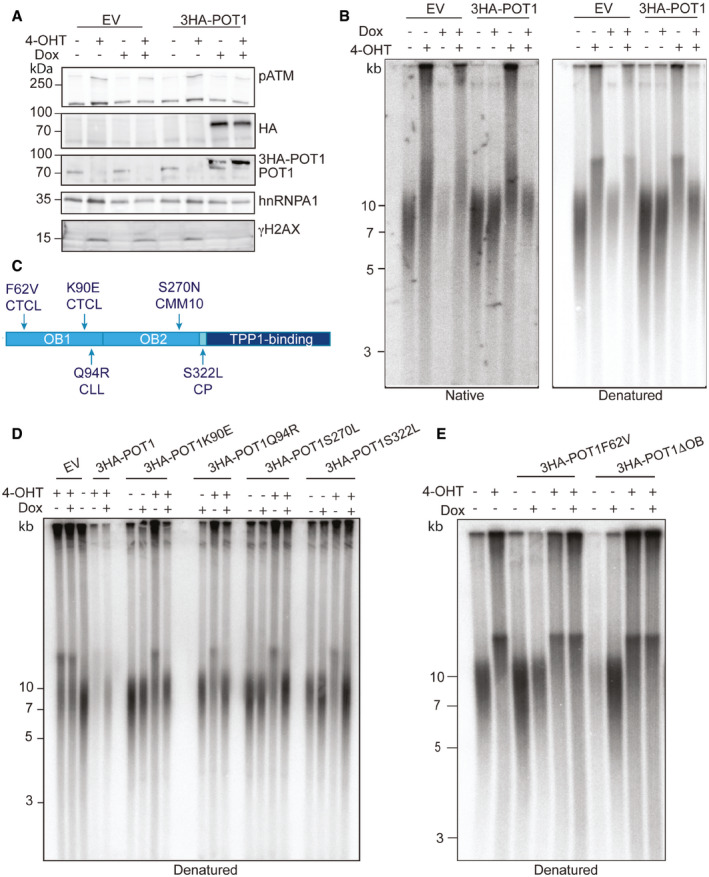
The POT1 protein contains two OB‐folds in its N‐terminal half with which it binds single‐stranded telomeric DNA (Lei et al, 2004). The C‐terminal half contains a third split OB‐fold which is bound by TPP1 via multiple interactions (Chen et al, 2017; Rice et al, 2017). For telomere recruitment, the interaction of POT1 with TPP1 is necessary and sufficient (Liu et al, 2004; Ye et al, 2004; Pinzaru et al, 2016). In order to test whether DNA binding by POT1 is required for averting the described telomere abnormalities, we developed a complementation system in which triple HA‐tagged POT1 was expressed in the POT1 knockout cells from a lentiviral construct containing a doxycycline‐inducible promoter (Fig 3A). Expression of exogenous POT1 rescued the knockout cells from the DNA damage response and the structural abnormalities in telomeric DNA (Fig 3A and B). Expression of mutant POT1 versions, some of which are associated with cancer development and which in previous analyses showed reduced but not abolished single‐stranded DNA binding, could also avert the severe telomere abnormalities when expressed from the plasmid (Fig 3C and D, and Appendix Fig S4A). However, two POT1 mutants were unable to rescue the telomere defects. The POT1F62V point mutation which diminishes single‐strand telomeric DNA binding affinity by a factor of 65 (Pinzaru et al, 2016), abolished POT1 function with regard to telomeric DNA structure (Fig 3E) as well as suppression of a DNA damage response (Appendix Fig S4B). The same was true for POT1∆OB lacking the N‐terminal OB‐fold. Therefore, we conclude that the direct binding of single‐stranded telomeric DNA by POT1 is required to prevent telomeric DNA abnormalities. The telomere recruitment via TPP1 is not sufficient for POT1 function.
Figure 3. The POT1‐loss phenotype is caused by deprotection of the G‐rich DNA overhang.
-
AWestern blot for POT1 and DDR markers upon POT1 removal in clone 35 and overexpression of ectopic WT POT1.
-
BTelomere length and G‐overhang length analysis for clone 35 upon POT1 removal and overexpression of ectopic WT POT1 (constant‐field gel electrophoresis).
-
CSchematic of disease‐associated mutations in POT1. CTCL—cutaneous T‐cell lymphoma, CMM—cutaneous malignant melanoma, CLL—chronic lymphatic leukemia, CP—Coats plus syndrome.
-
D, ETelomere length and G‐overhang length analysis for clone 35 upon POT1 removal and overexpression of ectopic POT1 variants carrying disease‐associated mutations (constant‐field gel electrophoresis).
As our knockout system did not affect a shorter polypeptide derived from the POT1 locus referred to as POT1‐55 (Hockemeyer et al, 2005; Fig 1A), we used lentiviral‐mediated delivery of shRNA that targets both POT1 forms (Hockemeyer et al, 2005) to confirm that the phenotype we observed was POT1‐55 independent (Appendix Fig S4C). Indeed, the telomere elongation and DNA damage response phenotypes upon POT1 knockout induced with 4OHT and POT1 knockdown with shRNA were identical (Appendix Fig S4D).
Development of 2‐Step QTIP uncovers substantial remodeling of the telomeric proteome upon POT1‐loss
We expected that the above‐described telomere abnormalities in the absence of POT1 would trigger substantial changes in the telomeric protein composition. In addition, we considered the possibility that the telomeric DNA abnormalities in POT1 knockout cells might be caused by inappropriate association of DNA processing enzymes with telomeric DNA in the first place. In order to address these questions, we embarked on isolating and analyzing the telomeric proteome in the presence and absence of POT1. Our laboratory previously developed a quantitative telomeric chromatin isolation protocol (QTIP) in which crosslinked telomeric chromatin is purified with antibodies against the abundant telomere‐binding proteins TRF1 and TRF2 and analyzed by mass spectrometry (Grolimund et al, 2013; Majerská et al, 2017). Different telomeric states were compared by differential SILAC labeling. Here, we wanted to compare telomeric protein composition in a larger set of different samples and therefore opted for the use of tandem mass tag reagents (TMT) (Rauniyar & Yates, 2014; Hogrebe et al, 2018; O'Connell et al, 2018). In addition, we added to the QTIP method an additional affinity purification step by first purifying the crosslinked and sonicated telomeric chromatin with anti‐FLAG sepharose beads containing monoclonal antibodies against the FLAG epitope (Fig 4A). This step became possible as our cell line contained FLAG‐tagged TRF1 and TRF2. Immunoprecipitated chromatin was eluted with excess of FLAG peptide and then re‐precipitated with polyclonal affinity‐purified anti‐TRF1 and anti‐TRF2 antibodies as in the classical QTIP method. We refer to this method as 2‐step QTIP. The two consecutive affinity purification steps yielded a recovery of telomeric DNA of 38–47% and an up to more than 1,000× fold enrichment of telomeric DNA over Alu repeat DNA (Fig 4B).
Figure 4. Development of 2‐step QTIP uncovers substantial remodeling of the telomeric proteome upon POT1‐loss.
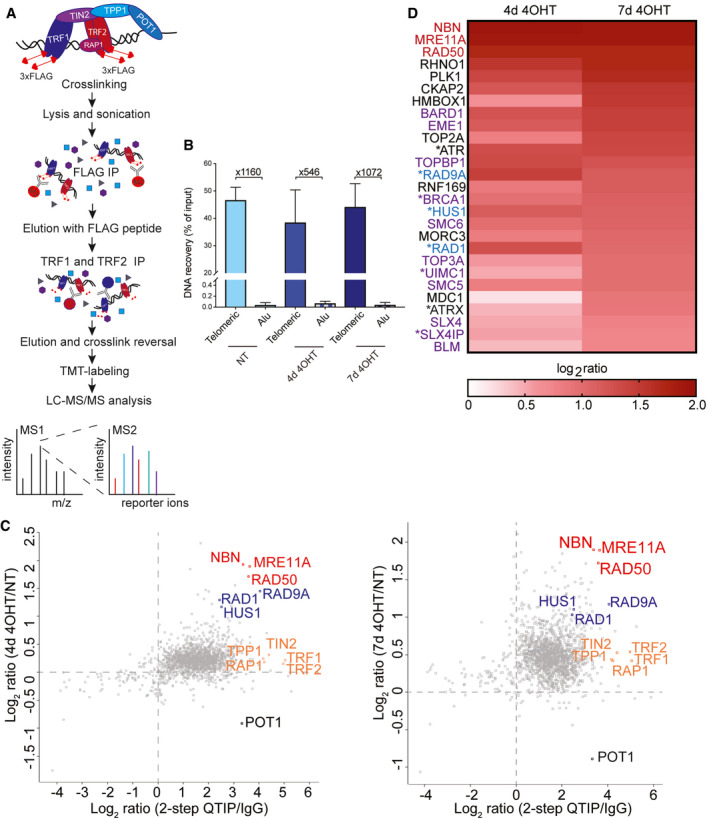
- Schematic drawing of 2‐step QTIP (improved version of the quantitative telomeric chromatin isolation protocol (QTIP)).
- Quantification of precipitated telomeric DNA in 2‐step QTIP (% of input) and fold enrichment of precipitated telomeric DNA compared with precipitated Alu repeat DNA (based on dot blot analyses). Bars represent data from three independent experiments ± SD.
- Scatter plots representing immunoprecipitation specificity (2‐step QTIP/IgG ratio) vs. difference between non‐treated cells (NT) and cells at day 4 upon 4‐OHT induction (4d 4OHT/NT ratio) or difference between non‐treated cells (NT) and cells at day 7 upon 4‐OHT induction (7d 4OHT/NT ratio), respectively (data from Table EV1). MRN complex in red, 9‐1-1 complex in blue, shelterin complex in orange except for POT1 in black. Values are the average of the 3 biological replicates.
- Heat map representing changes in levels of selected proteins involved in DNA damage (MRN complex in red, 9‐1-1 complex in blue) and HDR proteins (purple) after POT1 knockout induction. Proteins marked with * pass t‐test, but not t‐test with Benjamini–Hochberg correction (P < 0.05, q < 0.09).
We performed three biological replicates in uninduced clone 35 cells and cells from which POT1 had been deleted for 4 or 7 days (Fig 4C). To increase throughput and reduce the analytical variability and the incidence of missing data, we applied a 6‐plex TMT isobaric labeling workflow to analyze 2‐step QTIP preparations of untreated (NT) and POT1 knockout cells 4 and 7 days after induced deletion. In addition, we included IgG‐negative controls. Each of the three experimental replicates thus constituted a distinct TMT mix. After digestion, labeling, and pooling, peptides in each mix were extensively fractionated by peptide isoelectric focusing to maximize separation and increase depth of protein identification. Spike‐in proteins were used to monitor and correct signal levels across replicates and assess the dynamic range of TMT quantitation, which is known to display ratio compression phenomena (Savitski et al, 2013; Appendix Fig S5A and B). To maximize sensitivity, we chose not to apply synchronous precursor selection scan methods (McAlister et al, 2014). Analysis by mass spectrometry and comparison of 2‐step QTIP‐purified chromatin to IgG control‐purified chromatin indicated that the 2‐step QTIP approach yielded telomeric chromatin of unprecedented purity. Indeed, the vast majority of proteins detected were clearly enriched in the QTIP samples compared with the IgG controls (Fig 4C, Table EV1). The 2‐step QTIP procedure with fractionation allowed to detect, in addition to the most abundant shelterin components, more than 1,400 proteins. When comparing the telomeric chromatin in wild‐type to POT1 knockout cells, we observed a slight increase in all shelterin components except POT1, which as expected was strongly diminished (Fig 4C). The slight increase of five of the six shelterin proteins can be explained by the telomere elongation that occurred during the experiment (Figs 1D and 2C). 150 proteins were significantly enriched at telomeres that lacked POT1 at day 7 after knockout induction (paired T‐test with Benjamini–Hochberg FDR correction and threshold at 0.05 on the adjusted P‐value). Among them, a large set of proteins involved in the DNA damage response and homology‐directed DNA repair was identified (Fig 4D).
POT1 prevents telomeric accumulation of homology‐directed and other repair factors, nucleases, and cell cycle regulators
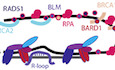
We grouped 87 proteins that were significantly enriched at telomeres in POT1 knockout cells more than 1.5‐fold (log2 > 0.6) into functional classes using the STRING database (Szklarczyk et al, 2019). The cutoff chosen takes into consideration TMT ratio compression effects (Appendix Fig S5). Apart from known telomere maintenance components, we identified checkpoint response factors, nucleases, DNA repair proteins, and proteins involved in cell cycle regulation, chromosome organization, centromere function, and RNA metabolism (Fig 5). Interestingly, the largest cluster (15 proteins) contained cell cycle and mitosis‐associated proteins (Fig 5A). We identified multiple proteins playing roles in spindle formation (AURKA, INCENP, KIFC1, TPX2, AURKB, KIF20A, PLK1). In the same cluster fall two condensin subunits (NCAPD2 and NCAPG) as well as cyclin B1 (CCNB1) and origin recognition complex subunit 1 (ORC1). In the second largest cluster (11 proteins, Fig 5B), we found DNA damage response and repair proteins as well as HR proteins: all components of MRE11/RAD50/NBS1 (MRN) complex, TOPBP1, RHNO1, BARD1, BLM, TOP3A, and the structure‐specific endonucleases ERCC1, ERCC4, and EME1. SLX4 and SLX1A, which function as Holliday junction resolvases, are also linked to this cluster. Another cluster included proteins involved in RNA metabolism (nine proteins, Fig 5C), which may relate to the increased abundance of R‐loops at POT1‐depleted telomeres (see below). We also identified a cluster of proteins that contained subunits of the SMC5‐SMC6 complex, which is involved in HR as well as TOP1, SUMO1, and SUMO2 (Fig 5D). The latter might stem from sumoylated proteins present at damaged telomeres. Also among proteins enriched at telomeres lacking POT1, we identified SMCHD1 and LRIF1 that have been previously implicated in the telomeric DNA damage response (Vančevska et al, 2020), parts of the protein phosphatase 1 complex and lamin A that had been identified at telomeres previously (Kim et al, 2009).
Figure 5. POT1 prevents telomeric accumulation of HDR and other repair factors, nucleases, and cell cycle regulators.
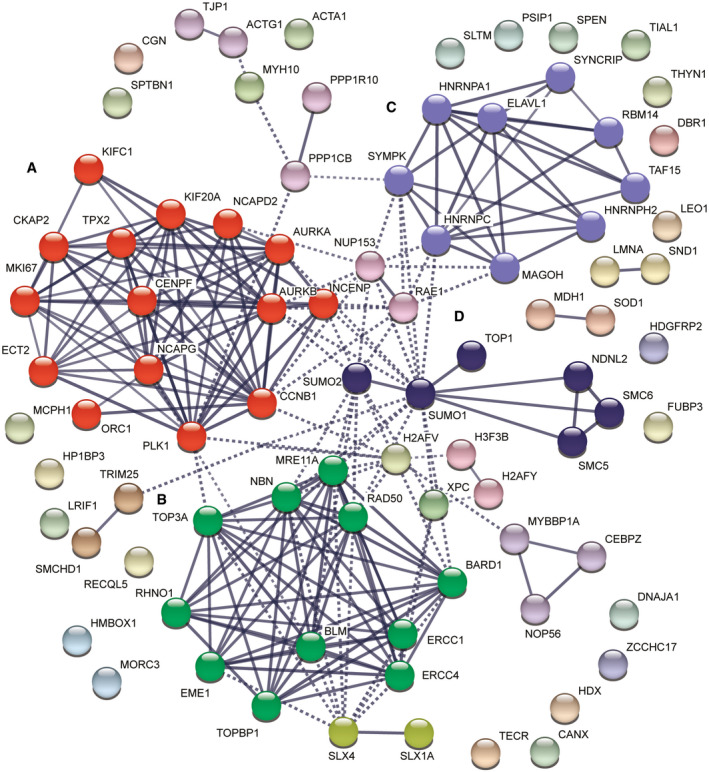
-
A–DCluster analysis of proteins enriched at telomeres lacking POT1 using STRING. The largest identified clusters are colored in red (mitosis‐associated proteins, (A), green (recombination and DNA damage response proteins, (B), purple (RNA‐binding proteins, (C), and dark blue (chromatin and DNA remodelers, (D). Line thickness indicates the strength of data support.
Telomere elongation and branching upon POT1‐loss is mediated by the HDR machinery
We next embarked on testing whether the identified proteins played roles in establishing or responding to the POT1‐loss‐mediated phenotype. Since POT1‐loss caused telomere elongation, we were particularly intrigued by the enrichment of the HDR factors MRE11, BARD1, BRCA1, and BLM at telomeres. Very strikingly, when we depleted these and other HR factors (RAD51, BRCA1, BRCA2, BARD1, BLM, POLD3) by siRNA in POT1 knockout cells, telomere elongation, telomere branching, and G‐rich single‐strand DNA accumulation were all suppressed (Figs 6A, EV1 and EV2, Appendix Fig S6A–D). Incomplete suppression of the phenotype by siRNA knockdown of BLM and POLD3 most probably is due to the incomplete depletion of these proteins. These data therefore indicate that in the absence of POT1 the HDR machinery becomes overly active at telomeres causing severe telomere instability. As POT1 was previously implicated in the regulation of telomerase (Loayza & De Lange, 2003; Kelleher et al, 2005; Latrick & Cech, 2010; Zhong et al, 2012), we also tested whether telomerase may contribute to the POT1‐loss‐mediated phenotype adding the telomerase inhibitor GRN163L to cells at the same time when POT1 deletion was induced (Fig 6B). However, although GRN163L strongly reduced telomerase activity (Appendix Fig S6E), the POT1‐loss‐mediated phenotype remained unaltered. Thus, telomerase showed no contribution to the phenotype in this experiment. Finally, we tested whether HDR inhibition influenced the DDR observed upon POT1‐loss by detecting pCHK1 or γ‐H2AX on Western blots (Appendix Fig S6A, Fig EV1B) and by detecting γ‐H2AX foci at telomeres by IF‐FISH (Appendix Fig S6B). However, in POT1 knockout cells the DDR still occurred upon suppression of HDR.
Figure 6. Telomere elongation and branching under POT1‐loss is mediated by the HDR machinery.
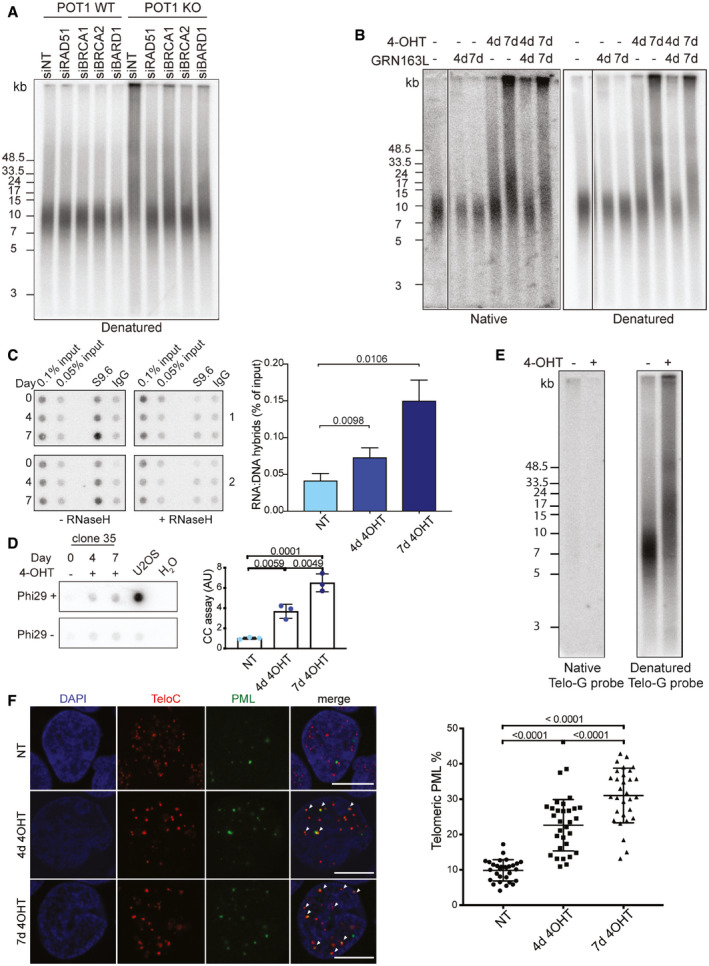
- Telomere length for clone 35 upon POT1 removal and depletion of selected recombination proteins with siRNA (pulsed‐field gel electrophoresis).
- Telomere length and G‐overhang length analysis for clone 35 upon POT1 removal and inhibition of telomerase with 1 μM GRN163L (pulsed‐field gel electrophoresis).
- DRIP assay for DNA‐RNA hybrid detection at telomeres in clone 35 upon POT1 removal with S9.6 antibody. Dot blots represent two replicates. Bars represent data from three independent experiments ± SD. The IgG signal was subtracted. Significance was determined using one‐way ANOVA, and P‐values are indicated on the graph.
- C‐circle (CC) assay for clone 35 upon POT1 removal. Bars represent data from three independent experiments ± SD. DNA from ALT positive cell line U2OS was used as positive control. Significance was determined using one‐way ANOVA, and P‐values are indicated on the graph. In all experiments, POT1 deletions were induced with 0.5 μM 4‐OHT for 4 and 7 days.
- C‐overhang length analysis for clone 35 upon POT1 removal (pulsed‐field gel electrophoresis, hybridization with Telo‐G probe).
- Quantification and representative pictures for visualization of the PML protein (IF, green) and telomeres (FISH, red) in non‐treated (NT) clone 35 cells and clone 35 treated with 4‐OHT for 4 and 7 days. Arrows indicate co‐localizations. Scale bar equals 10 μm. Bars represent data from three independent experiments ± SD. At least 150 cells were examined per condition per replicate. Significance was determined using one‐way ANOVA. P‐values are indicated on the graph. In all experiments, POT1 deletion was induced with 0.5 μM 4‐OHT.
Figure EV1. Recombination induced by absence of POT1 at telomeres depends on BLM .
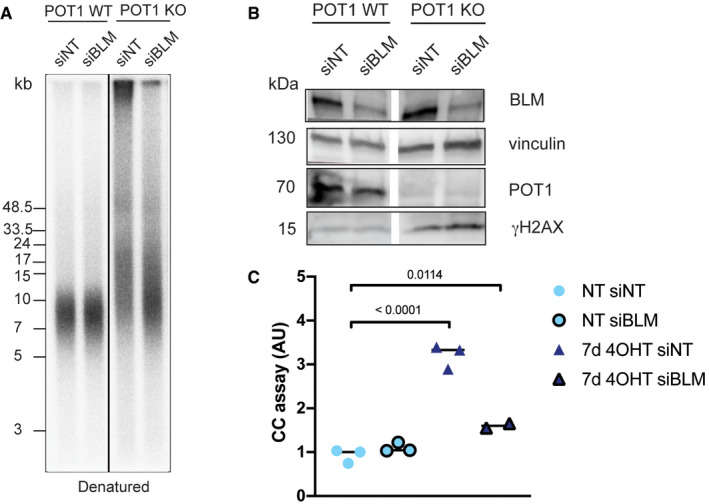
- Telomere length analysis for clone 35 upon POT1 removal and depletion of BLM with siRNA (pulsed‐field gel electrophoresis).
- Western blot for POT1, BLM, and γH2AX upon POT1 removal and BLM depletion.
- C‐circle (CC) assay for clone 35 upon POT1 removal and BLM depletion. Bars represent mean for data from three independent experiments. Significance was determined using one‐way ANOVA, and P‐values are indicated on the graph. In all experiments, POT1 deletion was induced with 0.5 μM 4‐OHT.
Figure EV2. Recombination induced by absence of POT1 at telomeres depends on POLD3.
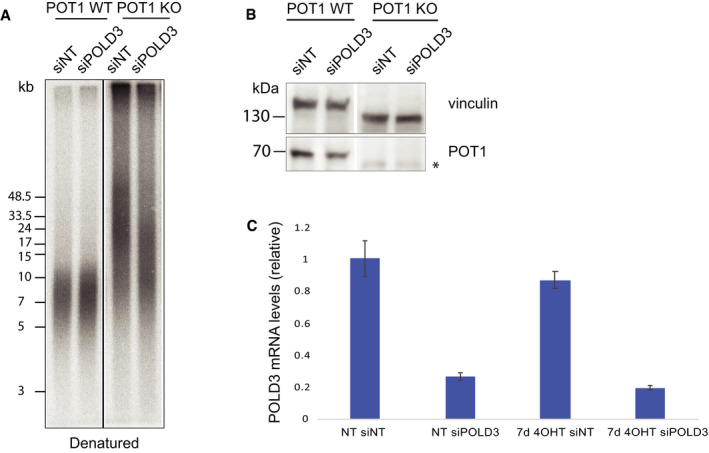
- Telomere length analysis for clone 35 upon POT1 removal and depletion of POLD3 with siRNA (pulsed‐field gel electrophoresis).
- Western blot for POT1 upon POT1 removal and POLD3 depletion. Star indicates a non‐specific band.
- qPCR to confirm depletion of POLD3 in clone 35 upon siRNA treatment. The experiment was performed in three biological and three technical replicates, error bars represent SD.
POT1‐loss leads to accumulation of telomeric R‐loops, C‐circles, and telomeric PML bodies
Next, we assessed for presence of telomeric DNA‐RNA hybrid structures which can be formed with the telomeric long noncoding RNA TERRA and are suppressed at telomeres in wild‐type cells by RNA surveillance factors (Azzalin et al, 2007; Chawla et al, 2011), RNase H (Arora et al, 2014; Graf et al, 2017), FANCM (Silva et al, 2019), and the THO‐complex (Pfeiffer et al, 2013) but can accumulate in mutant conditions interfering with telomere replication. Strikingly, telomeric R‐loops are a hallmark of ALT cells which maintain their telomeres by DNA recombination (Arora et al, 2014; Graf et al, 2017). To detect telomeric R‐loops, the structure‐specific antibody S9.6 was used to precipitate from total nucleic acid the R‐loops. Telomeric signals in precipitated nucleic acid were quantified by dot blot hybridization (Fig 6C). As a control for the specificity for R‐loop precipitation, the nucleic acid samples were treated in vitro prior to immunoprecipitation with commercial RNase H. RNase H destroys the RNA moiety in DNA/RNA hybrids. As expected, in vitro RNase H treatment abolished the telomeric signal to background levels validating the specificity of the immunoprecipitation for R‐loops (Fig 6C). POT1 deletion caused increased accumulation of telomeric R‐loops in the time course experiment. At the same time, POT1 deletion did not significantly affect TERRA expression levels as detected on Northern blots (Appendix Fig S7A) and it did not significantly affect individual TERRA molecules when the quantities stemming from six different chromosome ends were analyzed by RT–qPCR (Appendix Fig S7B).
Another hallmark of ALT cells is the formation of so‐called C‐circles which are single‐stranded circular DNA molecules containing telomeric C‐strand repeats of unknown function (Henson et al, 2017). They can be detected by rolling circle amplification followed by hybridization. POT1 deletion caused a significant increase of C‐circles, which however was lower than the amounts detected in U2OS ALT cells (Fig 6D). This increase was partially suppressed by knockdown of BLM (Fig EV1C). ALT cells also demonstrate elevated levels of C‐rich overhangs (Oganesian & Karlseder, 2011). To look at the general changes in the single‐stranded C‐rich DNA content, we performed in‐gel hybridization of non‐denatured telomeric DNA with a G‐rich telomeric probe (Fig 6E). However, we did not detect any changes of the very week signal observed in wild‐type cells.
Finally, we examined the control cells and POT1 knockout cells for presence of promyelotic leukemia (PML) bodies, which represent another marker for ALT activity (Yeager et al, 1999). The percentage of PML foci that were present at telomeres increased upon POT1 deletion significantly from 10% to more than 30% after 7 days of induced deletion (Fig 6F). Altogether, with increased telomeric R‐loops, C‐circles and PML bodies the POT1‐loss promoted the establishment of three markers that are typical for ALT cells.
POT1 knockout phenotype of HEK293E cells is reproducible in HeLa cells including increase in C‐circle formation
Finally, we recapitulated the POT1‐loss phenotype we observed in 293E cells in the HeLa cell line (Fig EV3). We disrupted the POT1 genes in HeLa cells with two guiding RNAs and CRISPR/Cas9, causing loss of POT1 protein expression (Fig EV3). POT1‐loss caused a rapid increase in telomere length and accumulation of single‐stranded branched G‐rich DNA (Fig EV3A). We performed a C‐circle assay and observed an eightfold increase in C‐circle levels, which is comparable to what we detected in 293E cells that had been deleted for POT1 (Figs 6D and EV3C). This indicates that POT1‐loss caused activation of telomeric recombination also in HeLa cells as observed in 293E cells. Interestingly, POT1 depletion with shRNA in HeLa cells was not sufficient to uncover the telomere elongation and DNA damage response phenotypes that were observed upon POT1 deletion and complete loss of the protein (Fig EV4). Therefore, very low amounts of POT1 are sufficient to prevent the pathological engagement of the recombination machinery with telomeres.
Figure EV3. The POT1 knockout phenotype is reproducible in HeLa cells and includes increase in C‐circles formation.
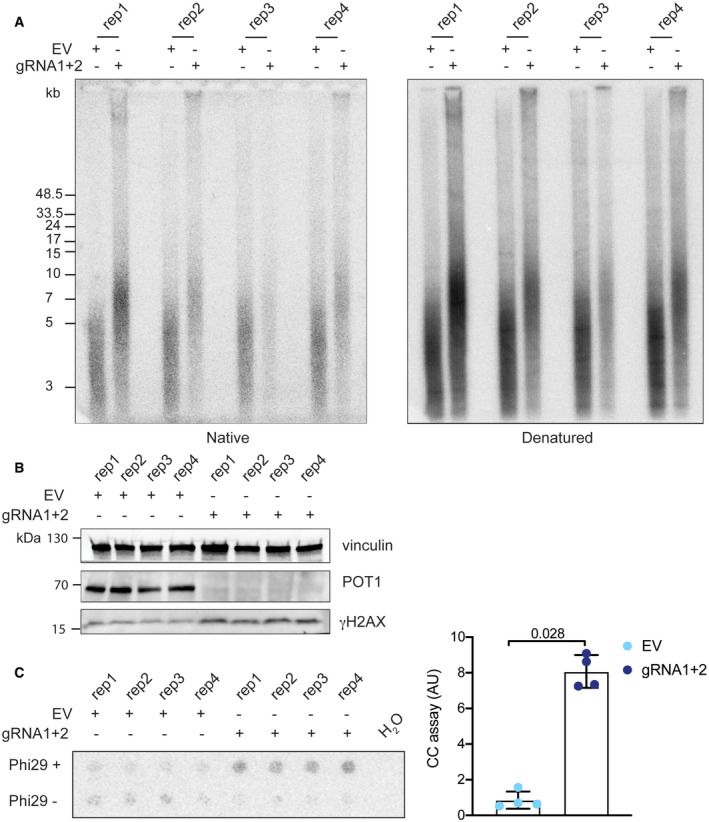
- Telomere length and G‐overhang length analysis for HeLa cells transfected with pSpCas9(BB)‐2A-puro containing gRNA1 and gRNA2 sequences or the corresponding empty vector. The experiment was done in four replicates.
- Western blot for POT1 and γH2AX upon POT1 deletion with gRNAs in HeLa cells.
- C‐circle (CC) assay for HeLa cells upon POT1 removal. Bars represent data from four independent experiments ± SD. Significance was determined using one‐way ANOVA, and P‐values are indicated on the graph.
Source data are available online for this figure.
Figure EV4. shRNA‐mediated POT1 knockdown is not enough to trigger POT1‐loss‐mediated HDR in HeLa cells.
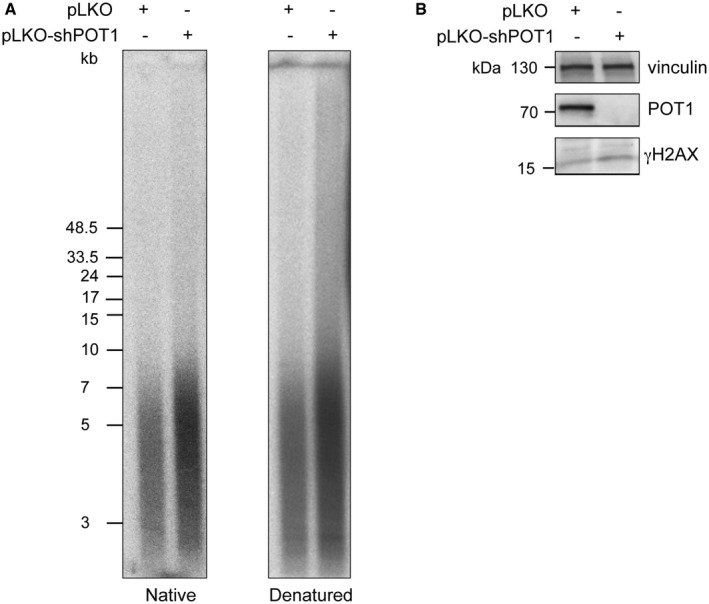
- Telomere length and G‐overhang length analysis for HeLa cells transduced with pLKO‐ lentiviral vector with or without shRNA targeting POT1. The experiment was done in four replicates.
- Western blot for POT1 and γH2AX upon POT1 depletion with shRNA in HeLa cells.
Source data are available online for this figure.
Discussion
Conditional deletion of POT1 uncovers major roles of this protein in telomere stability
In this paper, we uncover crucial roles of human POT1 in repressing HDR at telomeres which gives rise to severe telomeric DNA abnormalities including telomere elongation, telomere branching, accumulation of single‐stranded telomeric DNA, and telomeric R‐loops. The establishment of a conditional system and the application of telomere proteomics were crucial to obtain the documented insights. POT1 function had been studied before in several systems but despite its conservation in evolution and the conserved association with G‐rich single‐stranded telomeric DNA, POT1‐loss gave different phenotypes in different model systems. From the work in Schizosaccharomyces pombe, POT1 appeared to be most important for the protection of chromosome ends from nucleolytic degradation (Baumann & Cech, 2001). In mice and other rodents, the POT1 gene is duplicated and different functions have been assigned to the two paralogs indicating remarkable divergence from humans in telomere biology. With mouse cells, the data about a striking function of POT1 paralogs in the suppression of HDR were controversial (Hockemeyer et al, 2006; Wu et al, 2006), although the data seemed to support subtle contributions in the suppression of HDR at least in mutant backgrounds lacking the Ku end binding protein (Palm et al, 2009). In previous depletion studies of human POT1 (Hockemeyer et al, 2005; Kim et al, 2017), low amounts of POT1 may have been retained being sufficient to efficiently suppress telomeric HDR. Consistently, we also observed in HeLa cells that shRNA‐mediated depletion is not efficient enough to unravel the roles of POT1 in suppressing HDR (Figs EV3 and EV4). Residual amounts of POT1 in RNA interference experiments may also explain why the telomeric damage that had been detected by Hockemeyer et al (2005) was only prominent in G1, and not in S and G2 phases of the cell cycle, whereas we see DNA damage at telomeres throughout the cell cycle in our system upon POT1 deletion. From this, one may conclude that absence of POT1 can be tolerated in G1 phase of the cell cycle in immortalized cells and that low levels of POT1 are sufficient at telomeres in S and G2 cells to suppress telomeric HDR that could lead to severe telomere instability, enhanced DNA damage response and possibly, mitotic catastrophe. Also consistent with this notion, we found that POT1 mutants that had been detected in cancer were able to suppress HDR despite the fact that their binding ability of DNA is reduced.
We also observed a significant increase in telomere fragility in the cells lacking POT1 during the time course experiment (Fig 2B, Appendix Fig S3C). Telomere fragility is a hallmark of replication stress (Sfeir et al, 2009) and in ALT cells telomere fragility has been suggested to be caused by DNA recombination events among telomeric sequences (Min et al, 2017) being consistent with our findings. Interestingly, disease‐associated POT1 mutations have been shown to increase telomere fragility and cause replication fork collapse when overexpressed (Pinzaru et al, 2016). We suspect that these effects may have been due to slightly increased recombination events at telomeres due to POT1 dysfunction, which may have led to accumulation of recombination by‐products and R‐loops at telomeres that further challenged replication of telomeric sequences.
In previous work, it has been demonstrated that POT1 can regulate telomerase either negatively or positively (see Introduction). However, we observe that complete absence of human POT1 in HEK293E cells leads to rapid telomere elongation that depends on HDR and does not require telomerase (Figs 6A and B, and EV3C). Thus, our data reveal that POT1 plays crucial roles not only in regulating telomerase but also in suppressing recombination‐based mechanisms of telomere elongation. Overall, we suspect that the divergence of observed phenotypes upon POT1 depletion or POT1 knockout may not reflect fundamental differences in telomere function of POT1 in different model systems. We rather imagine that system‐specific variabilities with regard to functional redundancy and abundance of various telomeric proteins may strongly influence the phenotypic outcomes. Whereas POT1 emerges as a master in the protection and regulation of single‐strand telomeric DNA, in its absence RPA, CST, RAD51, hnRNPA1, RADX, and telomerase may all compete for binding to the G‐rich single‐stranded telomeric DNA and guide the faith of chromosome ends toward different outcomes.
2‐Step QTIP and TMT labeling unravels the telomeric proteome
We developed and implemented here a 2‐step QTIP protocol in order to obtain telomeric chromatin of high purity and TMT labeling in order to quantitatively compare the telomeric proteome changes obtained upon POT1‐loss. TMT labeling yields highest precision but it suffers from a ratio compression phenomenon and thus underestimates differences in protein abundance (Rauniyar & Yates, 2014; Hogrebe et al, 2018; O'Connell et al, 2018). Nevertheless, we could identify a large set of proteins whose association with telomeres is suppressed by POT1. However, due to variability between biological replicates, several DNA damage proteins that we used as positive controls (for example RAD9A‐HUS1‐RAD1) did not pass our stringent threshold (paired t‐test with Benjamini–Hochberg FDR correction and threshold at 0.05 on the adjusted P‐value) when considering individual subunits. Therefore, we also considered candidates with unadjusted P‐values < 0.05, especially when all subunits of multiprotein complexes behaved in a consistent manner. As expected for the ATR‐dependent DNA damage response, the 9‐1‐1 complex components and TOPBP1 were enriched at telomeres already at day 4 after induction. Their abundance decreased slightly at day 7 when recombination events became most prevalent. All the other identified DNA damage and HDR proteins were more strongly enriched at telomeres 7 days after induction of POT1‐deletion. Among the 150 proteins that were significantly enriched, 87 were enriched more than 1.5‐fold (log2 > 0.6) (Fig 5). Apart from the DNA damage proteins whose association was expected, we identified a number of functional protein groups that could not be anticipated. The increased presence of proteins involved in RNA metabolism at POT1‐depleted telomeres may be related to the increased amounts of R‐loops at telomeres lacking POT1 (Fig 6C). The observed enrichment of mitosis and spindle‐associated proteins may stem from the perturbed mitosis in cells lacking POT1 (Fig 2E). The functional relevance of HDR proteins was demonstrated in the current work while for others their exact roles remain to be defined.
Human POT1 suppresses detrimental HDR
Our work supports the following model for human POT1 function (Fig 7). In wild‐type cells, the single‐stranded telomeric DNA is to a large extent covered by POT1. Upon POT1‐loss, the telomeric C‐strand is further resected and RPA binds, triggering an ATR dependent checkpoint response. Eventually, RPA is at least partially replaced by RAD51 presumably through BRCA2‐mediated loading as during classical recombination. RAD51‐coated telomeric 3′ overhangs will invade telomeric DNA of adjacent chromosome ends, which then will be extended by DNA polymerases. The strand invasion is responsible for the observed branched DNA structures. The observed extended 3′ overhangs are due to 3′ end extension upon strand invasion and the presumed 5′ end resection, which follows POT1‐loss. In addition, the increased abundance of R‐loops between TERRA and the telomeric C‐strand should increase single‐stranded G‐rich telomeric DNA and may facilitate RAD51 binding and telomere recombination (Arora et al, 2014; Graf et al, 2017). The observed telomere fragility is due to abnormal chromatin condensation following HDR or gaps in telomeric DNA that may remain after the repair process. Overall, our data illustrate that while homology‐directed repair is highly beneficial to safeguard the genome from chromosomal breaks, it becomes detrimental when out of check at the natural ends of chromosomes, leading to severe telomere and chromosomal instability. The POT1 protein masters the repression of HDR through its sequence specific DNA binding, covering the single‐stranded telomeric DNA. POT1 therefore prevents the unwanted engagement of the HDR repair machinery at telomeres but not elsewhere in the genome.
Figure 7. Human POT1 suppresses detrimental HDR .
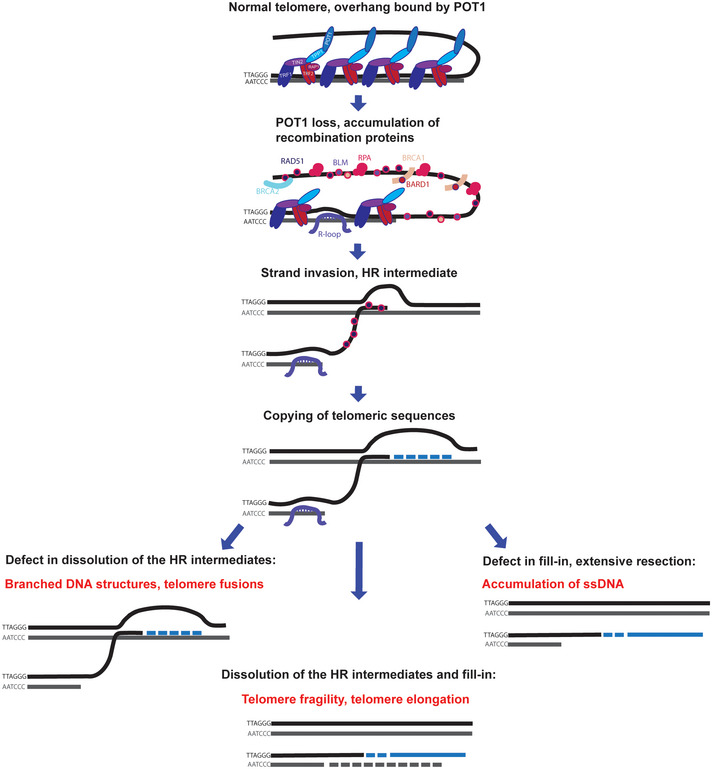
In the absence of POT1, the telomeric C‐strand is resected and RPA and recombination proteins associate with the overhang. RAD51‐coated overhangs invade telomeric DNA of adjacent chromosome ends and the invading 3′end is extended by DNA polymerases. Accumulation of R‐loops at telomeres facilitates strand invasion. In case of proper dissolution of the HR intermediates, telomeres are elongated and fragility is due to abnormal chromatin condensation after HDR. Defective dissolution of the HR intermediates leads to accumulation of branched DNA structures. Incomplete fill‐in synthesis leads to accumulation of G‐rich single‐stranded DNA.
Materials and Methods
Reagents and Tools table
Reagents and Tools are listed in Table EV2.
Methods and Protocols
Cell lines, CRISPR/Cas9 gene editing, transfections, and lentivirus production
HEK293E cells were grown in DMEM with 10%FCS and penicillin/streptomycin solution (1:100) at 37°C in 5% CO2. The population doubling (PDL) values were calculated using the mathematical formula PD = [(ln(N2)) − (ln(N1))]/ln(2) where N1 is the number of cells plated and N2 the number of cells harvested.
293E cl75p100 clone was generated from 293E cl75 by transfection of pSpCas9(BB)‐2A‐GFP (PX458, Ran et al, 2013) plasmid expressing gRNA targeting exon 6 of POT1 (gRNA1, 5′‐CACCGCCCCTGAATCAACTTAAGGG‐3′), a recombination reporter plasmid containing a binding site for gRNA1 [pMB1610_pRR‐Puro (Flemr & Bühler, 2015)], and pUC19 plasmid containing a repair template for exon 6 of POT1 (with a modified gRNA‐binding site) flanked with LoxP sites and homology arms of 500 bp (pUC19_RT). Cells were selected with 1 μg/ml puromycin for 5 days and clones were isolated by single cell dilution. Clones were screened using 2 genotyping PCRs (with OneTaq polymerase premix from NEB and 1.25 μM primers 1 + 2 and 1 + 3). Positive clones were kept in culture for several weeks to monitor telomere length stability and POT1 levels. The gRNA was designed using http://crispr.mit.edu/ and cloned into pSpCas9(BB)‐2A‐GFP (PX458).
Cre clones 35, 38, and 45 were generated by transducing cl75p100 cells with lentivirus containing CreERT2, selecting cells with 1 μg/ml puromycin for 5 days, and isolation of clones by single cell dilution. Isolated clones were tested for POT1 deletion by inducing Cre recombinase with 0.5 μM 4‐OHT and clones with the highest recombination efficiency were kept and tested for telomere length stability. Cells were transfected with plasmid DNA using Lipofectamine 2000 (1:200) and 2.5 μg of DNA in 2 ml of OPTIMEM per well in 6‐well plates. One day after transfection, cells were split and selected with either puromycin (1 μg/ml) or blasticidin (10 μg/ml). Cells were harvested or diluted 5 days after transfection.
The POT1 gene was disrupted in HeLa cells by two rounds of Lipofectamine 2000 transfection of pSpCas9(BB)‐2A‐GFP (PX458, Ran et al, 2013) plasmid expressing gRNA targeting exon 6 of POT1 (gRNA1, 5′‐CACCGCCCCTGAATCAACTTAAGGG‐3′) and pSpCas9(BB)‐2A‐puro (PX459, Ran et al, 2013) plasmid expressing gRNA targeting the ATG start codon of POT1 (gRNA2, 5′‐TTCTACAGAATCAATGTCTT‐3′).
Cells were transfected with siRNA pools from Dharmacon using calcium‐phosphate transfection with 10 pmol of siRNA in 1 ml of DMEM without antibiotics per well in six well plates. One day after transfection, cells were split and 72 h after transfection cells were harvested for TRF analysis, RT–qPCR, and Western blot analysis.
For lentiviral production, 293T cells were transfected in 10 cm dishes with 4 μg of the corresponding lentiviral vectors (shRNA or transgene) and packaging vectors pMD2.G (1 μg) and pCMVR8.74 (3 μg) using Lipofectamine 2000 (1:400 in OPTIMEM). The next day, the medium was changed for DMEM with 10% FCS and penicillin/streptomycin. The first harvest of viral particles was done 40 h after transfection, and fresh medium was added to the cells. Virus‐containing medium was filtered through 0.45 μm filters and 1 ml of filtrate was applied to 1 million of cells in one well of a 6‐well plate. The second harvest was done 24 h after the first harvest, and the transduction was repeated as described. The next day, transduced cells were split and selected with puromycin (1 μg/ml), blasticidin (10 μg/ml), or hygromycin (100 μg/ml) for at least 1 week. For doxycycline‐inducible expression of POT1, HA‐tagged WT POT1 or mutant POT1 cDNAs were cloned into pCW22 lentiviral vector between HpaI and PacI restriction sites. Stable cell lines were created by lentiviral transduction and blasticidin selection. Ectopic expression of POT1 constructs was induced with 1 μg/ml doxycycline. For ectopic expression of inducible Cre recombinase, ERT2CreERT2 was subcloned into pLKO1_puro (Stewart et al, 2003) digested with PpuMI.
Western blots
1 million cells were resuspended in 100 μl of Laemmli buffer (150 mM Tris–HCl pH 6.8, 4% SDS, 5% beta‐mercaptoethanol, 20% glycerol, bromophenol blue) and were boiled for 5 min at 95°C. Equivalents of 100,000 cells were loaded on Mini‐PROTEAN TGX™ 4–15% acrylamide gels. Primary antibodies were anti‐ATM pS1981 (1:1,000, ab81292; Abcam), anti‐γH2AX (1:1,000, 05‐636; Millipore), anti‐CHK1‐pS345 (1:1,000, #2348, Cell signaling), anti‐CHK1 (1:1,000, sc‐8408, Santa Cruz), anti‐RPA32 –S33 (1:1000, A300‐246A, Bethyl), anti‐vinculin (1:3,000, ab129002, Abcam), and anti‐HA (1:3,000, BLG‐901502, BioLegend). Secondary antibodies were HRP‐conjugated goat anti‐mouse (W4021, Promega) and goat anti‐rabbit (W4011, Promega). For Western blot analysis of POT1, the membrane was denatured after transfer and re‐natured with decreasing concentrations of guanidine‐HCl (Wu et al, 2007), blocked with 5% milk in TBST and incubated with anti‐POT1 (1:1,000, ab124784, Abcam) in 5% milk overnight.
Telomere restriction fragment length analysis and G‐overhang assay
Genomic DNA from 3 to 5 million cells was extracted with phenol‐chloroform‐isoamyl alcohol (25:24:1). 5–10 μg of genomic DNA were digested with 50U HinfI and 30U RsaI in CutSmart buffer (NEB) overnight at 37°C. 3 μg of digested genomic DNA per lane was resolved by pulse field gel electrophoresis in 1% agarose in 0.5× TBE. Samples were electrophoresed for 16 h at 14°C and 5.2 V with 0.5 s initial switch and 6 s final switch using a CHEF‐DRII apparatus (Bio‐Rad). The gels were dried for 3 h at 50°C, prehybridized with Church buffer (0.5 M Na2HPO4 pH 7.2, 1 mM EDTA, 7% SDS, and 1% BSA) and hybridized with a 32P‐radiolabeled telomeric probe under native conditions (for G‐overhang assay) overnight at 50°C. After hybridization, the gels were washed twice for 30 min with 2× SSC, 0.5%SDS; 1× SSC, 0.25% SDS; 0.5× SSC, 0.1% SDS and 0.5× SSC, and exposed to a phosphorimager screen. Afterward, the gel was denatured (0.5 M NaOH, 1.5 M NaCl), neutralized (0.5 M Tris pH 8.0, 1.5 M NaCl), and re‐hybridized as described above. Radioactive signals were detected with Amersham typhoon and quantified in AIDA software version 4.06.034. PRISM7 software was used to calculate relative G‐overhang amounts, averages, and P‐values.
Two‐dimensional neutral–neutral agarose gel electrophoresis
Two‐dimensional gel electrophoresis was done as described (Wang et al, 2004) with modifications. Equal amounts of HinfI+RsaI digested DNA (10–15 μg) were subjected to electrophoresis on 0.4% agarose gels at 30 V for 16 h in the dark at room temperature (first dimension). Then, the lanes were cut and overlaid on top of a second gel (second dimension) containing 1.2% agarose and 0.3 μg/ml ethidium bromide. This gel was run at 4°C and 150 V for 6 h. The gels were processed as described above for the TRF analysis.
Northern blot
10 μg of RNA per lane was loaded on a 1.2% formaldehyde agarose gel and separated by electrophoresis at 100V for 3 h. RNA was transferred to a nylon membrane (Hybond N+) overnight in 1× TBE at 15V. The membrane was UV‐crosslinked, prehybridized for 1 h in Church buffer, and hybridized with a 32P‐radiolabeled telomeric probe overnight at 55°C. The membrane was washed in 1× SSC, 0.5% SDS twice for 30 min at 60°C and exposed to a phosphorimager screen. Radioactive signals were detected with an Amersham typhoon and quantified in AIDA software version 4.06.034.
Immunofluorescence, click‐iT EdU labeling, and telomere fluorescence in situ hybridization (FISH)
Cells were pulse‐labeled with 10 μM EdU for 30 min, trypsinized, washed with PBS, and the cell concentration was adjusted to 0.5 million cells in 5% FBS in PBS. 100,000 or 200,000 cells were placed on the microscopic slides using cytospin centrifugation (Shandon cytospin 4; 5 min, 150 g) and were fixed on the slides with 4% paraformaldehyde for 15 min at room temperature. Cells were washed three times with PBS, permeabilized with 0.1% Triton X‐100, 0.02% SDS in PBS and stained with azide‐coupled fluorophore (Alexa 647) after crosslinking with Click‐iT chemistry. Cells were pre‐blocked in 2% BSA in PBS, blocked with 10% goat serum in PBS and incubated with anti‐γH2AX antibodies (1:1,000). After washing, they were incubated with secondary goat anti‐mouse antibodies conjugated with Alexa 488 (1:500). After washes with PBS, cells were incubated in 4% paraformaldehyde for 5 min at room temperature and washed with PBS again and dehydrated with increasing concentrations of ethanol. Cy3‐labeled TelC probe (PNA Bio, F1001) was diluted 1:2,000 in hybridization buffer (70% formamide, 10 mM Tris–HCl pH 7.5, 0.5% blocking reagent from Roche) applied on the slides, and slides were denatured for 3 min at 80°C. Slides were then incubated for 3 h in the dark at room temperature, washed twice in hybridization wash buffer 1 (70% formamide, 10 mM Tris–HCl pH 7.5), and washed three times in hybridization buffer 2 (0.1 M Tris–HCl 7.4, 0.15 M NaCl, 0.08% Tween‐20). The second wash contained 1 μg/ml DAPI. The slides were mounted with vectashield. Images were taken with a LSM700 upright microscope at 63× magnification. Blinded visual scoring of γH2AX foci colocalizing with telomeric FISH signal in EdU‐positive and EdU‐negative cells was done using FIJI, and statistical analysis was done using Prism7 software. Normal anaphases, anaphases with bridges, and micronuclei were counted in FIJI, and statistical analysis was done using Prism7 software.
Telomere fluorescence in situ hybridization (FISH) on metaphase spreads
Cells were treated with 0.5 μg/ml demecolcine for 2 h, harvested with trypsin, incubated with 0.056 M KCl for 7 min at 37°C, fixed with methanol:glacial acetic acid (3:1), and stored overnight at 4°C. Cells were dropped on the microscope slides, dried overnight, and fixed with 4% paraformaldehyde. Telomere FISH was conducted as described above. Images were taken with Zeiss‐Axioplan and blinded visual scoring of normal, and aberrant telomeres was done in FIJI. Telomeres were scored as fragile, when the telomeric signals from one chromosome arm was smeared or doubled instead of being a single dot. Single telomeric signal on two corresponding arms of two sister chromatids was scored as sister chromatid fusions. Single telomeric signal between two arms of different chromosomes were scored as chromosome fusions. Statistical analysis was done in Prism7.
RT–qPCR
RNA from 3 million cells was extracted using the Nucleospin RNA extraction kit (Macherey‐Nagel). 3 μg of RNA was reverse‐transcribed using 200 U super Script III reverse transcriptase (Thermo Fischer), GAPDH, and TERRA reverse primers for 1 h at 55°C, and the reaction was heat inactivated for 15 min at 70°C. 5% of the reaction was used as template for qPCR amplification with Power SYBR Green Master mix (Thermo Fisher) and 0.5 μM forward and reverse primers. The program for qPCR reaction included 10 min at 95°C, 40 cycles at 95°C for 15 s, and 60°C for 1 min in an Applied Biosystems 7900HT Fast Real‐Time System (Feretzaki & Lingner, 2017).
RT–qPCR for mRNAs to confirm siRNA‐mediated knockdowns was performed using Luna Universal One‐Step RT‐qPCR Kit (NEB). 200 ng of RNA were used for one‐step RT–qPCR with 0.4 μM forward and reverse primers. The program for RT–qPCR included 10 min at 55°C, 1 min at 95°C, followed by 40 cycles at 95°C for 10 s and 60°C for 1 min in an Applied Biosystems 7900HT Fast Real‐Time System.
TRAP
TRAP assay was performed as described (Kim, 1997) with modifications. 1 million cells were lysed for 30 min on ice in 200 μl of CHAPS lysis buffer (10 mM Tris–HCl pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.5% (w/v) CHAPS, 10% (w/v) glycerol, 0.1 mM PMSF, 1 mM DTT) and centrifuged for 15 min at 4°C. Lysate concentrations were determined with the Bradford assay and adjusted to 1 μg/μl. Serial dilutions of extracts were used for the primer elongation reaction (1× TRAP buffer, 25 μM dNTPs, 2 ng TS primer, 2 μCi α‐32P dGTP (3,000 Ci/mmol) for 30 min at 30°C. Then, telomerase was inactivated at 94°C and 2 U of Taq polymerase (Eurobio) and 2 ng of ACX primers were added to the reaction. PCR included 27 cycles for 30 s at 94°C and 30 s at 60°C. Extracts treated with 0.01 M EDTA were used as negative controls. 20% of TRAP reactions were loaded on 15% non‐denaturing polyacrylamide gels and separated at 200 V for 4 h. Gels were dried and exposed to a phosphorimager screen. Radioactive signals were detected with Amersham Typhoon.
DRIP
20 million cells were harvested and each sample was split into two pellets of 10 million cells and lysed on ice with cold RLN buffer (50 mM Tris–HCl, pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0.5% v/v Nonidet P‐40; 1 mM DTT; and 100 U/ml RNasin Plus). After centrifugation (300 g, 2 min, 4°C), the nuclei‐enriched pellets were resuspended in 500 μl of RLT buffer with 0.13% β‐mercaptoethanol and homogenized by passing through a 0.9 × 40 mm needle. Nucleic acids were then isolated by a phenol:chloroform:isoamylalcohol (25:24:1; Biosolve BV) extraction, followed by isopropanol precipitation. The two nucleic acid pellets coming from the same sample were joined together and dissolved in 300 μl H2O. Nucleic acid solutions were sonicated using a Focused‐ultrasonicator (Covaris E220) to obtain fragments below 300 bp. 150 μl of sonicated nucleic acids were treated for 90 min at 37°C with either 10 μl of RNase H (1 U/μl; Roche) or H2O. The reaction was stopped by addition of 2 μl of 0.5 M EDTA. 50% protein G sepharose beads slurry (GE Healthcare Life Sciences) was blocked with 1 mg/ml yeast tRNA for 1 h at 4°C. The samples were then diluted with 1,200 μl of buffer 1 (10 mM Hepes‐KOH, pH 7.5; 275 mM NaCl; 0.1% SDS; 1% Triton X‐100; and 0.1% Na‐deoxycholate) and precleared for 1 h at 4°C. 650 μl of precleared extract were used per immunoprecipitation reaction together with 80 μl of blocked 50% protein G beads slurry, and 1 μg of S9.6 antibody (022715; Kerafast) or mouse IgG (sc‐2025; Santa Cruz Biotechnology). The reactions were incubated for 2 h at 4°C on a rotating wheel and then washed consecutively for 5 min each time with 1 ml of buffer 2 (50 mM Hepes‐KOH pH 7.5; 140 mM NaCl; 1% Triton X‐100; 0.1% Na‐deoxycholate; and 1 mM EDTA), buffer 3 (50 mM Hepes‐KOH pH 7.5; 500 mM NaCl; 1% Triton X‐100; 0.1% Na‐deoxycholate; and 1 mM EDTA), buffer 4 (10 mM Tris–HCl pH 8.0; 250 mM LiCl; 1% NP‐40; 1% Na‐deoxycholate; and 1 mM EDTA), and 1 × TE buffer (10 mM Tris–HCl pH 8.0, and 1 mM EDTA). Beads and inputs were incubated with 1% SDS, 0.1 M NaHCO3, 0.5 mM EDTA pH 8.0, 20 mM Tris–HCl pH 8.0, RNase‐DNase free, Roche for 1 h at 65°C and DNA was extracted using the QIAquick PCR purification kit. Samples were transferred to the Hybond N+ membrane using dot bot and UV‐crosslinked before hybridization with telomeric probe as described above.
Cell cycle
Cells were incubated with 10 μM EdU for 30 min, harvested, and washed with PBS. Cells were fixed with 70% ethanol at a concentration of 1 million cells per ml. The next day cells were washed with PBS, permeabilized with 0.5% Triton X‐100 for 20 min, stained with Alexa‐488 conjugated azide using Click‐iT chemistry for 30 min in the dark and with 1 μg/ml DAPI, transferred to FACS tubes and cell cycle profiles were acquired with Gallios (Beckman Coulter). Further analysis was done with Kaluza software.
C‐circle assay
The C‐circle assay was performed as described before (Henson et al, 2009). Briefly, 300 ng of genomic DNA was digested with 4 U/μg (each) HinfI and RsaI (NEB) in buffer 2 and 10 μg/ml RNase A for 1 h at 37°C. 30, 10, 3, and 1 ng of digested DNA was taken to perform the φ29 reaction which contained in 20 μl 0.2 mg/ml BSA, 0.1% Tween, 1 mM each of dATP, dGTP, dTTP, 1× φ29 buffer, and 7.5 U φ29 DNA polymerase. The mixture was incubated for 8 h at 30°C, and ϕ29 DNA polymerase was inactivated by incubation at 65°C for 20 min. The reaction products were dot‐blotted on a positively charged nylon membrane and UV‐crosslinked. The membrane was prehybridized at 50°C for 1 h in Church buffer and hybridized overnight at 50°C with a 32P‐radiolabeled telomeric probe. The membrane was washed three times with 4xSSC at RT and once with 4× SSC and 0.1% SDS at 50°C (30 min each wash) and exposed to phosphorimager screen. Radioactive signals were detected with Amersham typhoon and quantified in AIDA software version 4.06.034. Statistical analysis was done using Prism7.
2‐step QTIP
Telomeric chromatin precipitation was performed as described before (Majerská et al 2017) with a number of modifications. SILAC labeling was not used. One billion cells were used per IP reaction. After harvesting, cells were washed twice with PBS, fixed in 1% formaldehyde and 2 mM EGS for 15 min at 25°C at a cell concentration of 10 million cells/ml. For chromatin enrichment, cells were resuspended in the chromatin enrichment buffer (50 mM Tris–HCl pH 8.0, 1% SDS, 1 mM EDTA) at 10 million cells/ml, incubated on the wheel for 5 min at 25°C, and centrifuged at 2,057 g for 5 min. Chromatin enriched pellets were resuspended at 20 million cells/ml in four batches of 25 ml of LB3 buffer (10 mM Tris–HCl, pH 8.0; 200 mM NaCl; 1 mM EDTA–NaOH, pH 8.0; 0.5 mM EGTA–NaOH, pH 8.0; 0.1% w/v sodium deoxycholate; 0.25% w/v sodium lauryl sarcosinate; and EDTA‐free protease inhibitor complex (Roche)) and sonicated using a Branson Tip sonicator (30% power, 10 s constant pulse, and 20 s pause for a total sonication time of 9 min). Sonicated extracts were dialyzed against immunoprecipitation (IP) buffer (50 mM Tris–HCl pH 8.0, 600 mM NaCl, 10 mM EDTA pH 8, 0.75% Triton X‐100) and a first IP was performed using either ANTI‐FLAG M2 Affinity Agarose Gel (Sigma) or mouse IgG coupled to sepharose beads that had been blocked with yeast tRNA (1 mg/ml) for 1 h at 4°C. 1.6 ml of 50% beads slurry was used per IP, reactions were performed overnight at 4°C. After five rounds of washes with IP buffer for 5 min each, five rounds of elutions were performed with 100 μg/ml of FLAG peptide (Sigma). A second IP was performed overnight at 4°C with home‐made affinity‐purified anti‐TRF1 and anti‐TRF2 antibodies coupled to protein G sepharose beads that had been blocked with yeast tRNA. 0.8 ml of 50% beads slurry was used per IP. After washes with buffer 1 (20 mM Tris–HCl pH 8.0, 0.1% SDS, 1% Triton X‐100, 2 mM EDTA pH 8.0, 300 mM NaCl), buffer 2 (20 mM Tris–HCl pH 8.0, 0.1% SDS, 1% Triton, 2 mM EDTA pH 8.0, 500 mM NaCl), buffer 3 (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 1% NP‐40, 1% Na‐Deoxycholate, 1 mM EDTA pH 8.0) and buffer 4 (10 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0), beads were dried and resuspended in 2.5 bed bead volume equivalent of 0.25 M ammonium hydroxide for elution for 15 min at 37°C. Four rounds of elution were performed, and eluates were pooled, flash‐frozen, and lyophilized.
Protein digestion
Lyophilized samples were digested following a modified version of the iST method (Kulak et al, 2014). Briefly, pellets were resuspended in 100 μl miST lysis buffer (100 mM Tris–HCl pH 8.6, 1% sodium deoxycholate, 10 mM DTT) by vigorous vortexing. An internal standard protein (alcohol dehydrogenase from Saccharomyces cerevisiae, Merck‐Sigma A7011, 0.02 μg) was spiked in all samples, while 0.05 μg of bovine beta‐lactoglobulin (Merck‐Sigma L3908) were added only to the TRF1/2 QTIP immunoprecipitates in order to quantify TMT ratio compression effects. Resuspended samples were heated at 95°C for 30 min to denature proteins and reverse formaldehyde‐induced crosslinking. Samples were then diluted 1:1 (v:v) with water containing 4 mM MgCl2 and Benzonase (Merck‐Novagen 70746‐4, 1:100 of reaction volume = 51.6 Units) and incubated for 1 h at RT to digest nucleic acids crosslinked to proteins. Reduced disulfides were alkylated by adding ¼ vol (50 μl) of 160 mM chloroacetamide (final 32 mM) and incubating at RT for 45 min in the dark. Samples were adjusted to 3 mM EDTA and digested with 1 μg Trypsin/LysC mix (Promega V5073) for 1 h at 37°C, followed by a second overnight digestion with a second, identical aliquot of proteases. To remove sodium deoxycholate, digests were phase extracted by adding 600 μl of ethyl acetate containing 1% TFA, vortexing for 2 min, and centrifugation. The bottom aqueous phase containing peptides was collected, diluted 2× with 0.5% formic acid and desalted on strong cation exchange (SCX) cartridges (SOLA Cartridges (Thermo Fisher: 60209‐002)) by centrifugation. After washing the SCX cartridges with 0.5% formic acid, 20% acetonitrile, peptides were eluted in 200 μl of 80% MeCN, 19% water, and 1% (v/v) ammonia.
An aliquot of 1/15 of samples from experiment replicate 1 (2‐2) was analyzed directly by LC‐MS and label‐free quantitation to assess the quality of the digestion, the specificity of the preparation and, by comparison with known amounts of a standard HeLa digest, obtain an approximate estimate of the total amount (by mass) of peptides present before TMT labeling. The samples appeared to contain approximately 30 μg of material, with antibody chains used for purification seemingly quantitatively dominant, thus making up the bulk of protein material.
TMT labeling
Eluates after SCX desalting were dried, resuspended in 100 μl water, and dried again to remove excess ammonia. Digests were then resuspended in 40 μl of 50 mM TEAB buffer, pH 8.0 and reacted with 0.4 mg of TMT reagent for 1 h at RT, after which excess reagent was quenched with 8 μl of 50% hydroxylamine for 20 min at RT. For each replicate experiment, untreated (NT, day 0) and knockout samples (day 4, day 7) were mixed with their corresponding negative IgG controls in a six‐plex TMT experiment using, respectively, TMT channels 126, 127N, 128N, 129N, 130N, and 131. Neutron‐encoded C labels were not used). TMT multiplex samples were dried and desalted on C18 SepPak well plates. An aliquot (5%) was injected before fractionation to assess labeling completion (> 98% peptide spectrum matches) by database search with TMT as variable modification (MASCOT software).
Peptide fractionation
Dried desalted eluates were dissolved in 4M urea containing 0.1% ampholytes pH 3–10 (GE Healthcare) and fractionated by off‐gel focusing as described (Geiser et al, 2011). The 24 peptide fractions obtained were desalted on a SepPak microC18 96‐well plate (Waters Corp., Milford, MA), dried, and redissolved in 30 μl of 0.05% trifluroacetic acid, 2% (v/v) acetonitrile for LC‐MS/MS analysis.
MS analysis
Data‐dependent LC‐MS/MS analysis of extracted peptide mixtures after digestion was carried out on a Fusion Tribrid Orbitrap mass spectrometer (Thermo Fisher Scientific) interfaced through a nano‐electrospray ion source to a RSLC 3000 HPLC system (Dionex). Peptides were separated on a reversed‐phase custom packed 40 cm C18 column (75 μm ID, 100 Å, Reprosil Pur 1.9 μm particles, Dr. Maisch, Germany) with a 4–76% acetonitrile gradient in 0.1% formic acid (total time 140 min). Full MS survey scans were performed at 120,000 resolution. In data‐dependent acquisition controlled by Xcalibur 4.0.27 software (Thermo Fisher Scientific), a data‐dependent acquisition method was used that optimized the number of precursors selected (“topspeed”) of charge 2+ to 5+ while maintaining a fixed scan cycle of 1.5 s. The ion isolation window used was 0.7 Th.
Peptides were fragmented by higher energy collision dissociation (HCD) with a normalized energy of 35%. MS2 scans were done at a resolution of 30,000 in the Orbitrap cell, which is sufficient to resolve 6‐plex TMT reporter ions. The m/z of fragmented precursors was then dynamically excluded from selection during 60 s.
MS data analysis
Data files were analyzed with MaxQuant 1.6.3.4 (Cox & Mann, 2008; Cox et al, 2011) incorporating the Andromeda search engine (Cox et al, 2011). Cysteine carbamidomethylation and TMT labeling (peptide N‐termini and lysine side chains) were selected as fixed modification while methionine oxidation and protein N‐terminal acetylation were specified as variable modifications. The sequence database used for searching was the human Reference Proteome based on the UNIPROT database (www.uniprot.org), version of October 29th, 2017 (2017_10) containing 71,803 sequences. This was completed with custom databases containing most usual environmental contaminants (keratins, trypsin, etc.), the sequence of yeast ADH, and a collection of mouse (554) and sheep (37) sequences of immunoglobulins (also from UNIPROT). Mass tolerance was 4.5 ppm on precursors (after recalibration) and 20 ppm on HCD fragments. Both peptide and protein identifications were filtered at 1% FDR relative to hits against a decoy database built by reversing protein sequences. For unlabeled samples, iBAQ values (Schwanhäusser et al, 2011) generated by MaxQuant in label‐free quantitation were used. For TMT analysis, the raw reporter ion intensities generated by MaxQuant and summed for each protein group were used in all following steps to derive quantitation. Only identified peptide ions with a precursor intensity fraction (PIF parameter) greater than 0.75 were accepted and used for TMT quantitation.
Processing of quantitative data and statistical tests
The MaxQuant output table “proteinGroups.txt” for the three TMT replicate samples was first processed to remove proteins matched to the contaminant database as well as proteins identified only by modified peptides and reverse database hits, yielding a first unfiltered list of 2,674 identified proteins. As the composition of positive and negative samples was very different, no global normalization was applied to the data. After log‐2 transformation of all intensity values, a plot of the intensity of the spiked‐in control (yeast ADH) showed a slight decrease of signal intensity correlating with the order of MS injection (Appendix Fig S5). A global correction factor was applied per TMT sample mix to compensate for this effect, which is probably in partly due to gradual deterioration of the performance of the LC column over the > 80 h of continuous MS measurement. The largest correction factor was +0.28 (log2 scale) relative to the median of all samples. Next, the table was filtered to retain only proteins identified by a minimum of two “razor or unique” peptides and having at least 12 out of 18 total quantitative values (1,695 proteins left). Ratios (difference in log scale) were calculated for each sample relative to its IgG control.
A one‐sample t‐test (P‐value filter at 0.05) was performed on all values by comparing the 9 TRF1/2 IP's vs. 9 IgG controls to identify a set of invariant proteins that did not significantly change in intensity between IgG controls and positive IP's. 249 proteins (including many keratins and common contaminants) passed the test and were removed from the dataset, leaving 1,446 protein groups. The obtained ratio QTIP/IgG was recorded in the tables as “specificity ratio”. To determine proteins significantly changing between NT and the knockout (day 4, day 7) conditions, we applied a paired t‐test (each sample against its IgG control) with Benjamini–Hochberg FDR correction (Benjamini & Hochberg, 1995) and threshold at 0.05 on the adjusted P‐value. No protein passed the test at day 4 while 150 proteins passed the test at day 7. Analysis was performed using Perseus (Tyanova et al, 2016).
Author contributions
GG carried out all experiments except for the mass spectrometry analysis, which was done by MQ. A‐SB carried out initial experiments demonstrating that RAD51 depletion rescues from telomeric DNA abnormalities observed in POT1 knockout cells. GG and JL wrote the paper. The study was conceptualized by GG and JL.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 6
Acknowledgements
We thank members of the Lingner laboratory for fruitful discussions, the Protein Expression Core Facility at EPFL for 293E cells, Gérald Lossaint for 293E cl75, Pierre Gönzy for U2OS cells, Etienne Meylan for the pCW22 construct and Jerry Shay (UT Southwestern) for GRN163L. Research in J.L.'s laboratory was supported by the Swiss National Science Foundation (SNSF grant 310030_184718), the SNSF funded NCCR RNA and disease network (grant 182880), an Initial Training Network (ITN) grant (aDDRess) from the European Commission's Seventh Framework Programme (grant No 812829), the Swiss Cancer League (KLS‐3824‐02‐2016) and EPFL.
The EMBO Journal (2020) 39: e104500
Contributor Information
Galina Glousker, Email: galina.glousker@epfl.ch.
Joachim Lingner, Email: joachim.lingner@epfl.ch.
Data availability
All raw MS data together with MaxQuant output tables are available via the ProteomXchange data repository (https://www.ebi.ac.uk/pride/archive) with the accession number PXD016826.
References
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM (2014) RNaseH1 regulates TERRA‐telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5: 5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end‐binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol) 57: 289–300 [Google Scholar]
- Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM (2011) Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading‐strand replication. EMBO J 30: 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Gu P, Wu J, Chen X, Niu S, Sun H, Wu L, Li N, Peng J, Shi S et al (2017) Structural insights into POT1‐TPP1 interaction and POT1 C‐terminal mutations in human cancer. Nat Commun 8: 14929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Michalski A, Mann M (2011) Software lock mass by two‐dimensional minimization of peptide mass errors. J Am Soc Mass Spectrom 22: 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X (2013) Super‐resolution fluorescence imaging of telomeres reveals TRF2‐dependent T‐loop formation. Cell 155: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Lingner J (2017) A practical qPCR approach to detect TERRA, the elusive telomeric repeat‐containing RNA. Methods 114: 39–45 [DOI] [PubMed] [Google Scholar]
- Flemr M, Bühler M (2015) Single‐step generation of conditional knockout mouse embryonic stem cells. Cell Rep 12: 709–716 [DOI] [PubMed] [Google Scholar]
- Geiser L, Dayon L, Vaezzadeh AR, Hochstrasser DF (2011) Shotgun proteomics: a relative quantitative approach using Off‐Gel electrophoresis and LC‐MS/MS. Methods Mol Biol 681: 459–472 [DOI] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, Luke B (2017) Telomere length determines TERRA and R‐loop regulation through the cell cycle. Cell 170: 72–85.e14 [DOI] [PubMed] [Google Scholar]
- Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J (2013) A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun 4: 2848 [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AYM, Pickett HA, Reddel RR (2009) DNA C‐circles are specific and quantifiable markers of alternative‐lengthening‐of‐telomeres activity. Nat Biotechnol 27: 1181–1185 [DOI] [PubMed] [Google Scholar]
- Henson JD, Lau LM, Koch S, Martin La Rotta N, Dagg RA, Reddel RR (2017) The C‐Circle Assay for alternative‐lengthening‐of‐telomeres activity. Methods 114: 74–84 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T (2005) POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24: 2667–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels J‐P, Takai H, de Lange T (2006) Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hogrebe A, von Stechow L, Bekker‐Jensen DB, Weinert BT, Kelstrup CD, Olsen JV (2018) Benchmarking common quantification strategies for large‐scale phosphoproteomics. Nat Commun 9: 1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J (2005) Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro . Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N (1997) Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res 25: 2595–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee O‐H, Xin H, Chen L‐Y, Qin J, Chae HK, Lin S‐Y, Safari A, Liu D, Songyang Z (2009) TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol 16: 372–379 [DOI] [PubMed] [Google Scholar]
- Kim H, Li F, He Q, Deng T, Xu J, Jin F, Coarfa C, Putluri N, Liu D, Songyang Z (2017) Systematic analysis of human telomeric dysfunction using inducible telosome/shelterin CRISPR/Cas9 knockout cells. Cell Discov 3: 17034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic‐sample processing applied to copy‐number estimation in eukaryotic cells. Nat Methods 11: 319–324 [DOI] [PubMed] [Google Scholar]
- de Lange T (2018) Shelterin‐mediated telomere protection. Annu Rev Genet 52: 223–247 [DOI] [PubMed] [Google Scholar]
- Latrick CM, Cech TR (2010) POT1‐TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini‐Denchi E, Sfeir A (2016) Stop pulling my strings – what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol 17: 364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR (2004) Structure of human POT1 bound to telomeric single‐stranded DNA provides a model for chromosome end‐protection. Nat Struct Mol Biol 11: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Lin Y‐C, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R et al (2014) Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun 5: 4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C‐YG, Näger A‐C, Lunardi T, Vančevska A, Lossaint G, Lingner J (2020) The human telomeric proteome during telomere replication. bioRxiv 10.1101/2020.06.14.150524 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z (2004) Telosome, a mammalian telomere‐associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Majerská J, Redon S, Lingner J (2017) Quantitative telomeric chromatin isolation protocol for human cells. Methods 114: 28–38 [DOI] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86: 7150–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, Shay JW (2017) Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break‐induced replication processes. Mol Cell Biol 37: e00226‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F (2008) Fission yeast Pot1‐Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F (2009) Unusual telomeric DNAs in human telomerase‐negative immortalized cells. Mol Cell Biol 29: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JD, Paulo JA, O'Brien JJ, Gygi SP (2018) Proteome‐wide evaluation of two common protein quantification methods. J Proteome Res 17: 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Karlseder J (2011) Mammalian 5′ C‐rich telomeric overhangs are a mark of recombination‐dependent telomere maintenance. Mol Cell 42: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T (2009) Functional dissection of human and mouse POT1 proteins. Mol Cell Biol 29: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J (2013) The THO complex component Thp2 counteracts telomeric R‐loops and telomere shortening. EMBO J 32: 2861–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaru AM, Hom RA, Beal A, Phillips AF, Ni E, Cardozo T, Nair N, Choi J, Wuttke DS, Sfeir A et al (2016) Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep 15: 2170–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt CW, Cooper JP (2010) Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res 38: 6968–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauniyar N, Yates JR (2014) Isobaric labeling‐based relative quantification in shotgun proteomics. J Proteome Res 13: 5293–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C, Shastrula PK, Kossenkov AV, Hills R, Baird DM, Showe LC, Doukov T, Janicki S, Skordalakes E (2017) Structural and functional analysis of the human POT1‐TPP1 telomeric complex. Nat Commun 8: 14928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, Pachl F, Kuster B, Bantscheff M (2013) Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J Proteome Res 12: 3586–3598 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Pentz R, Figueira AM, Arora R, Lee YW, Hodson C, Wischnewski H, Deans AJ, Azzalin CM (2019) FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R‐loops. Nat Commun 10: 2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al (2003) Lentivirus‐delivered stable gene silencing by RNAi in primary cells. RNA 9: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta‐Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res 47: D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T (2011) Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Vančevska A, Pfeiffer V, Feretzaki M, Ahmed W, Lingner J (2020) SMCHD1 promotes ATM‐dependent DNA damage signaling and repair of uncapped telomeres. EMBO J 39: e102668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T‐loop‐sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme‐Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM et al (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Wu Y, Li Q, Chen X‐Z (2007) Detecting protein–protein interactions by far western blotting. Nat Protoc 2: 3278–3284 [DOI] [PubMed] [Google Scholar]
- Ye JZ‐S, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T (2004) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR (1999) Telomerase‐negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59: 4175–4179 [PubMed] [Google Scholar]
- Zhong FL, Batista LFZ, Freund A, Pech MF, Venteicher AS, Artandi SE (2012) TPP1 OB‐fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 6
Data Availability Statement
All raw MS data together with MaxQuant output tables are available via the ProteomXchange data repository (https://www.ebi.ac.uk/pride/archive) with the accession number PXD016826.


