Abstract
New approach methodologies (NAMs) in chemical safety evaluation are being explored to address the current public health implications of human environmental exposures to chemicals with limited or no data for assessment. For over a decade since a push toward “Toxicity in the 21st Century,” the field has focused on massive data generation efforts to inform computational approaches for preliminary hazard identification, adverse outcome pathways that link molecular initiating events and key events to apical outcomes, and high-throughput approaches to risk-based ratios of bioactivity and exposure to inform relative priority and safety assessment. Projects like the interagency Tox21 and the US EPA’s ToxCast program have generated dose-response information on thousands of chemicals, identified and aggregated information from legacy systems, and created tools for access and analysis. The resulting information has been used to develop computational models as viable options for regulatory applications. This progress has introduced challenges in data management that are new, but not unique, to toxicology. Some of the key questions require critical thinking and solutions to promote semantic interoperability, including: (1) identification of bioactivity information from NAMs that might be related to a biological process; (2) identification of legacy hazard information that might be related to a key event or apical outcomes of interest; and, (3) integration of these NAM and traditional data for computational modeling and prediction of complex apical outcomes such as carcinogenesis. This work reviews a number of toxicology-related efforts specifically related to bioactivity and toxicological data interoperability based on the goals established by Findable, Accessible, Interoperable, and Reusable (FAIR) Data Principles. These efforts are essential to enable better integration of NAM and traditional toxicology information to support data-driven toxicology applications.
Keywords: Data Interoperability, Computational Toxicology, Bioinformatics, Databases, Applications
I. Introduction
Toxicology is in a period of rapid change and growth to meet the challenge of safety assessment for tens of thousands of chemicals that have potential human exposures yet lack sufficient data for hazard identification.1–4 After over a decade since the publication of the seminal National Research Council report, Toxicity Testing in the 21st Century: A Vision and a Strategy5 that called for advancements in the field of toxicology using new approach methodologies (NAMs) for hazard,6,7 substantial progress has been made. Important contributions to this reformation have initially been driven by projects including, among others, the interagency collaboration for Toxicity Testing in the 21st Century (Tox21)8,9 and the US Environmental Protection Agency (EPA) Toxicity Forecaster (ToxCast) program.10,11 These data generation efforts have produced sizeable amounts of dose-response data for chemical interactions with biological targets,2,9,12 and further motivated development of aggregated digital resources of legacy toxicity information13,14 and software for access and analysis.15–20
A state has been reached in which volumes of data can be generated, but full utilization of this information to find innovative scientific solutions absolutely necessitates taking the time to adopt improved data management practices to connect the appropriate data to a biological target and to understand the methodology employed. Data interoperability is a salient and critical need to address if computational toxicology is to succeed in supporting modern chemical safety evaluation and research in public health and toxicology. Indeed, in alignment with the amended Toxic Substances Control Act (TSCA),4 the EPA is required to develop a risk-based method for chemical prioritization, and in doing so, to use NAMs in lieu of traditional methods when practicable. Thus, data interoperability for toxicology will help meet regulatory assessment needs for diverse data from both NAM and traditional approaches. Herein we review the state of data interoperability for toxicology applications.
Many fit-for-purpose applications have been developed to understand how to use NAM data. One issue with development of these applications and data models is that information is siloed, which prevents easy integration and exchange of data (i.e. interoperability) creating problems like inconsistent versioning, lack of provenance, and unnecessary duplication. Ultimately the consequence of the lack of data interoperability is that progress in understanding biological and toxicological effects of chemical exposures is hampered despite an abundance of information. Indeed, data interoperability is an issue across all kinds of data that inform chemical safety evaluation; herein we consider toxicology or hazard information, recognizing that the issue of data interoperability to connect all of the information needed for chemical safety evaluation is immense. To fully leverage the resulting information from NAMs for toxicology and public health questions, efforts must be applied to enable connections between data sources.6 Overcoming the high opportunity cost of enabling interoperability of various toxicology-relevant data streams will help realize the following goals: rapid and reproducible associations between NAM-based information and outcomes of interest for development of computational models,21–29 hypothesis generation aimed at increased mechanistic knowledge,30–35 and systematic literature reviews,36 all of which support efficient and state-of-the-art screening level chemical safety evaluations.37
Interoperability refers to “the ability of data or tools from non-cooperating resources to integrate or work together with minimal effort.”38 Data interoperability can be accomplished through numerous means like development and adherence to controlled vocabularies (CVs), standardized chemical nomenclature, and compliance with formatting standards for exchange of data. Computational efforts in toxicology to generate and analyze large amounts of data are relatively new, so CVs and formatting standards are not widely used and accepted. Of course, data interoperability challenges are not unique to toxicology and, in fact, are one of the key challenges facing each industry from finance to social media to public health and biomedicine.38–44
As an example of how interoperability promotes greater consumption of data for biological learning, platforms from companies like Affymetrix were developed to rapidly and affordably capture and analyze transcriptomic data. The application of Affymetrix platforms and other microarray technologies in a clinical setting was aided by standardization efforts (i.e. to support interoperability) for mass distribution of kits as well as standard reporting of results, which subsequently led to development of tool suites that could consume and analyze the information.45 The adherence to data formatting standards allowed for aggregation into a single resource called the Gene Expression Omnibus (GEO), which allows data access for the research community.46,47
For toxicology, the lack of consensus on how the vast amount of concentration-response data collected from myriad heterogeneous in vitro platforms can be applied to regulatory toxicology applications has clarified the need for implementation of data management strategies that maximize interoperability. For instance, “big data” is being generated via whole-genome sequencing,48 high-content imaging,49,50 and high-throughput bioactivity screening,9,11 and how these data are formatted, processed, analyzed, stored, and accessed are dissimilar, between data types and data generators. These dissimilarities create an additional obstacle for data integration to answer applied questions. Building consensus on reporting standards, both for assay design principles and observed effects, would contribute to progress in the use of these data for regulatory applications.
Good data management practices are embodied by the FAIR data principles, defined as Findability, Accessibility, Interoperability, and Reusability.38 These principles were defined to guide existing and future endeavors in scientific research as technology advances and data are generated to support knowledge discovery. Without proper data interoperability, progress in other areas will remain limited. Strategies to promote greater data sharing and interoperability in support of public health research goals are the subject of ongoing development. In 2013, an Executive Order, “Making Open and Machine Readable the New Default for Government Information,” (M13–13) was signed to institutionalize an open data policy including data management practices that make information accessible and readable by both machines and humans. The U.S. government is developing a Federal Data Strategy (https://strategy.data.gov/) that sets forth the mission principles and practices that reflect consideration of the Fair Information Practice and Principles.51 Federal agencies are also required to make federally-funded data publicly available via the Enterprise Data Inventory,52 but the formatting and interoperability of these datasets are not currently controlled. The FAIR data principles are further echoed in the National Institutes of Health (NIH) Strategic Plan for Data Science, with emphasis on infrastructure development and support for good data management practices as a crucial effort for continued success.53 One of the first steps outlined in NIH’s approach is to update the current NIH infrastructure by connecting related systems for increased data interoperability. The answer to the overall challenge of achieving interoperability is simple to describe but difficult to implement, not only due to mountains of legacy data trapped in antiquated or difficult to process formats, but also due to ongoing rapid data generation efforts with lack of standardization, creating data “silos.” Clearly the NIH has identified data interoperability as a key measure needed to achieve near and long-term goals for health-related research, but the broader field of toxicology needs more examination of how it could be achieved in order to build and resource effective strategies.54
The objective of this review is to provide the needed introduction to the current data landscape in toxicology, including some specific examples that demonstrate a need for increased data interoperability for computational toxicology. As part of this review, research needs and key questions relevant to data interoperability in the public health and toxicology fields are highlighted.
II. Current data landscape in toxicology
Toxicology is a diverse and applied field where health-related information from models of animal and/or human toxicity inform chemical safety assessment for human and ecological health. Decisions made based on toxicity data can not only dramatically affect human health and the environment, but also have major economic implications. Thus, any changes to the existing paradigms for data collection, evaluation, and analysis due to advances in technology and science often come under intense scrutiny. However, NAM-based data generation is proceeding, and numerous ongoing efforts intend to demonstrate how this information may be used to answer regulatory toxicology questions.21–23,55–60 In this section, examples of data interoperability needs for both traditional and NAM-based toxicity information are reviewed. Table 1 provides a list of all applications, tools, and resources mentioned in this review, but is not necessarily an exhaustive listing of all resources available for computational toxicology.
Table 1: Applications, tools, and resources relevant to computational toxicology.
This table is intended to collect the applications, tools, and/or resources available for hazard information, but is not necessarily an exhaustive list.
First, in vivo models for toxicology continue to inform chemical safety assessment, whether it be directly for previously studied chemicals, or indirectly as source information for read-across, predictive modeling, or benchmarking the success of NAM approaches for prediction of specific effects. Much of the available traditional toxicology data for human health safety evaluations has been collected through animal experimentation to identify doses that do not cause adverse health effects, e.g. a no observable adverse effect level (NOAEL) in in vivo studies. This information is captured in both physical and digital text documents. Many of these documents are used for regulatory purposes and are not computationally accessible, e.g. the data are available for capture in text or PDF or in database formats that are not easily integrated. Additionally, many regulatory study reports are not public due to confidential business information (CBI) issues, and only summary reports are made available. There is always loss of detail in creating these summaries through a manual procedure and the possibility of loss of fidelity.
In addition to regulatory documents, the existing information can be found in various formats scattered across different digital systems (Table 1) such as the Integrated Risk Information System (IRIS),61 PubMed,62 Regulations.gov (https://www.regulations.gov), Chemical Effects of Biological Systems (CEBS),63 eChemPortal,64 Provisional Peer-Reviewed Toxicity Value (PPRTV),65 Carcinogenic Potency Database (CPDB),66 Toxicity Reference Database (ToxRefDB),13 ChemView,67 eChemPortal,68 the European Chemicals Agency (ECHA),69 ECOTOX,70 and the Hazard Evaluation Support System Integrated Platform (HESS).71 The multitude of database resources for in vivo study information, with different designs and applications, for existing in vivo toxicity data exemplifies the lack of interoperability that promotes duplication of information and challenges in data provenance. A more specific obstacle is identification of duplicate studies; for instance, it is difficult to identify identical National Toxicology Program (NTP) reports in the multiple databases that collect this information: CEBS,63 ToxRefDB,13 CPDB,66 HESS71 and literature compilations.72 These resources are databases that have extracted data from animal toxicity studies, including those conducted by NTP; however, the source documents are available as either full reports from various online locations or broken up as separate publications that can be found across different scientific journals.
Because of source document management that was initiated without understanding of the future database needs (i.e. lack of versioning and unique identifiers), the age of some of the studies, as well as differences in how entities like a “study” and a “record” are defined across resources, it is extremely difficult to identify the overlap among the resources. These issues primarily encompass the legacy or historical data problems the field faces, but extensive efforts are underway to increase data interoperability to mitigate such issues. Addressing these challenges is critical as the field is rapidly changing because the success of NAMs often depends on the use of legacy information as a reference or benchmark to evaluate NAM success. Further, increases in data interoperability for legacy data may improve the capacity for data sharing and use across regulatory agencies.
An example of an early implementation of a public repository that integrated information on a by-chemical basis from both in vitro bioassays and in vivo toxicology data from myriad public sources was the Aggregated Computational Toxicology Resource, or ACToR14 (Figure 1A) developed within the EPA Office of Research and Development’s National Center for Computational Toxicology (NCCT). ACToR initially provided the extracted hazard information and access to two prominent projects within NCCT, ToxCast and ToxRefDB, through a single web application and subsequently began developing Representational State Transfer (RESTful) web services73 for increased availability of the resources. ToxCast and ToxRefDB, among others, progressed the field forward because of the sheer amount of information made available to explore computational modeling approaches to examine chemical hazard.13,21–23,25,74,75 With the progress made through ToxCast and Tox21, other projects grew into defined research areas or domains including exposure3,76 that spawned the development of different databases, applications, and software packages to meet these important research needs (Figure 1B). The CompTox Chemicals Dashboard surfaces information from several databases and a long list of datasets, with additional links out to other resources such as the adverse outcome pathway Wiki (AOP-wiki) among numerous others.17 The research landscape, and the means to overcome siloed data resources, continues to evolve in real-time, within the EPA Office of Research and Development, and outside of it. Other efforts to generate NAM-based data in consortium-based projects, like SEURAT77 and EUToxRisk,78 will also generate repositories of public data and information for integration and use in screening level chemical safety assessment, highlighting the need for reporting standards across consortia and geography.
Figure 1: A historical view of the evolving infrastructure to support modern chemical safety evaluation.
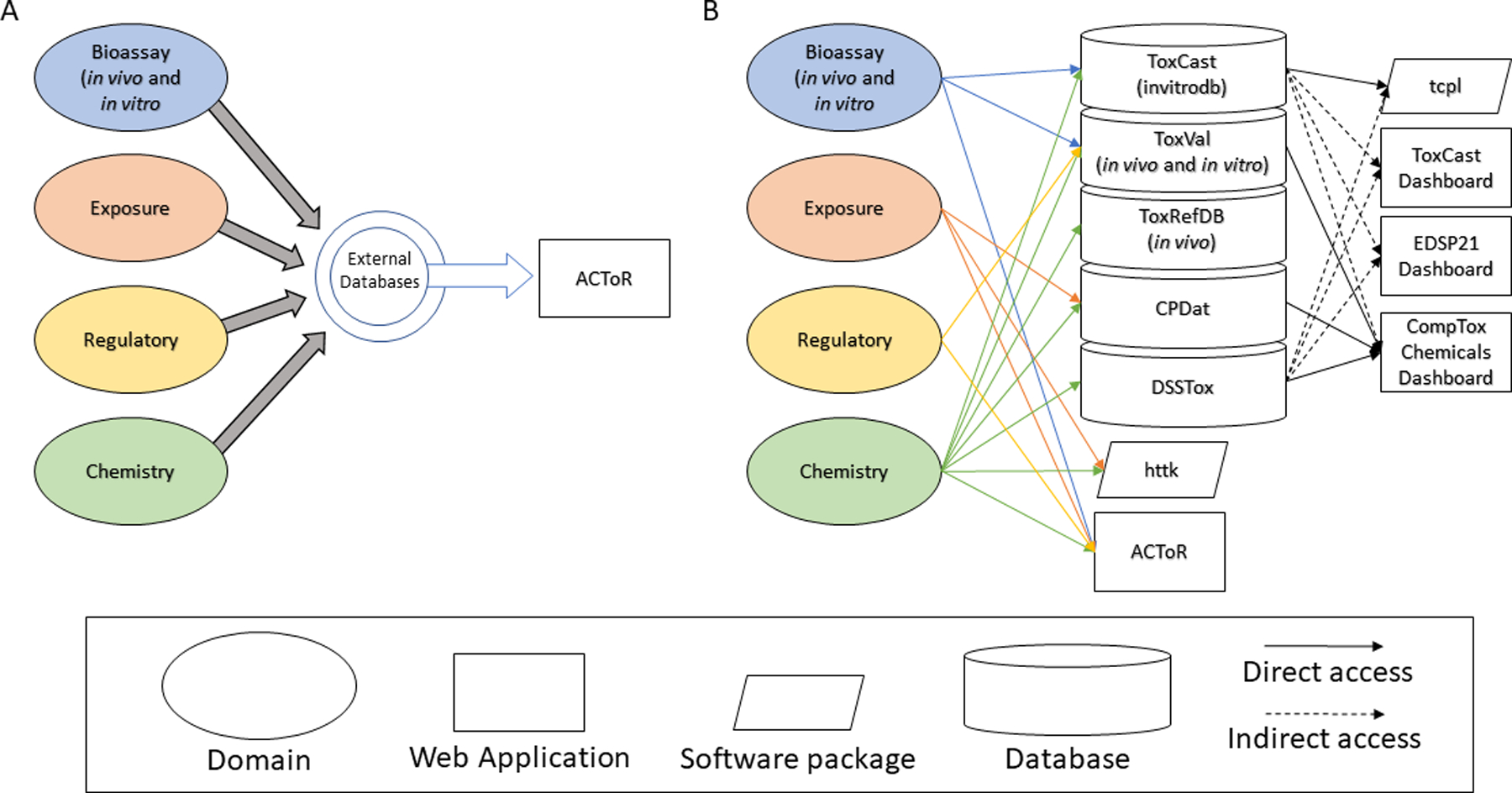
Pictured is an abstract representation of the changing infrastructure that supports USEPA’s computational toxicology efforts, presented as an example of data interoperability needs as they have evolved. (A) Initially, information across relevant domains in toxicology were aggregated from external databases to a single database accessed through a single web application called ACToR. (B) With continued success in data generation projects like ToxCast, multiple products were developed. The dashed arrows represent indirect access to the needed information. Indirect access means that the underlying information was duplicated because each web application is supported by a separate database, which is consistent with silo-ing and reinforcing data inconsistency. Note that the ToxCast Dashboard and the EDSP21 Dashboard are being sunset in 2019, with their functionality merged into the CompTox Chemicals Dashboard. CPDat = Consumer Product Database; DSSTox = distributed structure-searchable toxicity database; EDSP21 = Endocrine Disruptor Screening Program for the 21st century; httk = high-throughput toxicokinetics; tcpl= ToxCast data pipeline.
Integration efforts for these data streams that promote public accessibility have themselves been somewhat siloed endeavors that have led to duplication of information across databases and difficulty in managing this information with time and resources spent on “data cleaning,” version control, and quality assurance measures. Centralized user interfaces for accessing both NAMs and traditional data, including the CompTox Chemicals Dashboard,17 NCATS BioPlanet,18 National Library of Medicine (NLM) PubChem,79 ToxNet,80 Comparative Toxicogenomics Database,35 ECHA,69 eChemPortal,68 ChemView,67 and NTP Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) Integrated Chemical Environment (ICE),20 have all led to increased access to data. PubChem, the largest resource of downloadable bioassay information,79 contains crowdsourced bioassay data, and deposition of information is generalized in order to store heterogenous data in the single resource. PubChem allows adherence to standards, specifically the Bioassay Ontology (BAO),81,82 but this standard is not strictly enforced. Using the streamlined data model in PubChem, some information on assays and results may not be well-captured. Based on the rapid growth of these user interfaces, and their capabilities, it is clear that tools are needed to help data stakeholders better integrate and organize this information, either by chemical or by biological aspects. As the development of these tools continues, standards are essential to enable integration across data repositories and these centralized user interfaces.
III. Examples of standardization and mapping of vocabularies
As previously stated, development and adherence to formatting standards and CVs increases interoperability, especially for legacy information systems or new data streams without existing standards. The ToxRefDB has recently been updated13 and provides an example of how in vivo toxicity information can be modernized for easier integration. ToxRefDB is a large publicly available digital resource aggregating results from animal toxicity studies that was initially created for retrospective analysis and as a reference to validate both ToxCast bioactivities and computational models.28,29 The impetus for the recent update was to collect dose-response information that was not originally extracted from the studies. However, endeavors to increase interoperability were also undertaken. The studies in ToxRefDB span decades where the language for reporting adverse events is inconsistent from either subjective expert preferences or updates as knowledge about pathology has advanced. The terminology in ToxRefDB was standardized for a ToxRefDB-specific CV. The CV was mapped to concepts in the Unified Medical Language System (UMLS), which is a resource managed by the National Library of Medicine integrating over 150 biomedical vocabularies.83 By mapping to a standard that is already integrated, interoperability is achieved with any other resource that is also mapped to the same standard.
Other in vivo toxicology resources utilize CVs; CEBS also captures information from animal toxicity studies and the adverse event reporting for histopathology results adheres to a CV called the International Harmonization of Nomenclature and Diagnostic Criteria (INHAND) and is accounted for in NTP’s Nonneoplastic Lesion Atlas (NLA).84 A limitation of INHAND is that it is not currently mapped to any other resources, and thus additional mapping will be needed to further interoperability efforts. Another user of INHAND is the eTox consortium,85 which is a group of pharmaceutical companies that have compiled animal studies into a single resource. Continuing efforts of eTox include increased interoperability though CV development and mapping.86 Another resource that collects animal toxicity information is the Health Assessment Workspace Collaborative (HAWC).87 EPA has forked the HAWC project and has been actively developing a CV for data captured within HAWC for use in literature-based assessment products, with the next step to map this CV to other resources to enable interoperability.88 Finally, another resource that collects information on in vivo toxicity studies is the International Uniform Chemical Information Database (IUCLID).89 Like HAWC, IUCLID has a limited CV available as a “picklist,”90 but still lacks granularity for adverse events. IUCLID is the primary tool used by the European Chemicals Agency (ECHA) to collect and evaluate chemicals for regulatory applications. IUCLID stands out from the previously mentioned applications because it adheres to data formatting standards developed in conjunction with the Organisation for Economic Co-operation and Development (OECD) called OECD Harmonized Templates (OHTs). IUCLID can consume any data formatted according to OHTs, making it an attractive database and data reporting standard for regulatory agencies around the world. Both HAWC and IUCLID have been developed for chemical-centric regulatory applications; therefore, aggregation of information has also been primarily chemical-centric rather than adverse effect-centric. However, moving forward with research endeavors investigating NAMs and to answer questions about reproducibility in animal toxicity studies, adverse event reporting should move toward adoption of CVs and formatting standards to fully support interoperability.
A massive amount of information is readily available from each of the information systems above, yet interoperability is still lacking primarily due to lack of CVs and data formatting standards. The progress made in ToxRefDB with CV development and mapping was a manual effort; however, automatic mapping is possible. Several tools like National Center for Biomedical Ontologies (NCBO) Bioportal Annotator and UMLS MetaMap are available to map text to respective CVs using Natural Language Processing (NLP) techniques. Without definitions or full text input, these methods are limited to string comparisons, which are not always very accurate. For example, the ToxRefDB term “pathology microscopic” was manually mapped to the UMLS term “Histopathology Result.” When using the BioPortal Annotator, the UMLS terms that are mapped to “pathology microscopic” are “Pathology” and “Microscopic”, which, even together, do not represent “pathology microscopic” as well as “Histopathology Result.” In many cases, manually mapping terms may be the best option because of the accuracy, but automatic mapping pipelines, such as machine learning based methods trained to make appropriate associations, should be investigated further to facilitate rapid advancement of in vivo toxicity data interoperability.
IV. Modern chemical safety evaluation requires a framework to link relevant information
To enable modern chemical safety evaluation, a collection of information must be considered and integrated, including: (1) information on chemicals or substances; (2) information on phenotypes and toxicity associated with chemical bioactivity; and, (3) information on testing methodologies, assay principles, and intended targets. Once these linkages are made, the evidence available to associate a chemical or substance with a phenotype of interest may help inform safety evaluation.
A very basic issue with interoperability for toxicology that not only cuts across every data stream, but also is often overlooked, is chemical identity. Chemicals can be identified by their name, a chemical Abstracts Registry number (CASRN), or a chemical structure. All three of these identifiers are subject to variability and errors. It is well known that chemicals can be named in multiple ways, so name is not unique. CASRN can change over time. Chemical structures can be represented in several forms (SMILES, MOLFILE, INCHI), and may not accurately reflect the exact chemical tested (e.g. stereochemistry may be lost in SMILES representation). Further, study records may not clearly state the chemical that was tested (e.g. which salt form, what purity level). Therefore, it is important to annotate, as far as possible, the actual chemical tested in a study (in vitro or in vivo) to allow combined use of data across studies. The EPA DSSTox project has developed a standard for performing this mapping, but it is in general a manual process.17,91
Linking chemical exposure to disease or toxicity remains a challenge in part because risk assessment is chemical centric. In contrast, the development and progression of disease or toxicity in humans is due to a multitude of factors including exposure to many chemicals at different life stages all coupled with genetic predisposition. Consequently, many biological investigations are not focused on causation by one chemical, and epidemiology studies may not easily provide support for causation of a single chemical as these studies associate populations with many exposures to biological outcomes.92 Thus, biological interpretation of in vitro screening information for use in safety evaluations is dependent upon modeling the connections linking chemical activity in an assay to an effect on a biological target, and then linkage of this target to the disease or toxicity associated with perturbation of that biological target. Efforts to increase data interoperability for bioactivity, toxicity, or phenotype and testing methodology, assay principle, or intended target would enable more inferences from disease or toxicity back to chemical exposure.
Recognizing this need, the adverse outcome pathway (AOP) framework has been proposed as one method to aggregate relevant information and define how measurable perturbations in response to environmental stressors lead to an adverse outcome (AO), or an event of regulatory concern.93–96 An AOP is defined as “an analytical construct that describes a sequential chain of causally linked events at different levels of biological organization that lead to an adverse health or ecotoxicological effect”.97 An AOP is mapped as a linear progression of a series of key events (KEs) linked together by qualitative or quantitative key event relationships (KERs) across biological levels of organization, beginning with a specialized KE known as a molecular initiating event (MIE) and culminating with another specialized KE known as an adverse outcome (AO). For the purposes of understanding chemical toxicity, the linear AOPs are combined to form AOP networks to account for the complexity inherent in a biological system98–100.
As shown in Figure 2, the AOP framework connects information on phenotypes and toxicity with information on the intended target from some biological measurement of perturbation; each KE is associated with an event, event component, and biological target, detected by a biological measurement that suggests a biological explanation based on the testing methodology and assay principle employed.101,102 AOP networks provide a framework for integrating bioactivity measurements from both NAMs and traditional toxicology assays in such a way that they may be used to predict adverse outcomes.103–108
Figure 2: Integrating stressor and biological information into an Adverse Outcome Pathway workflow.
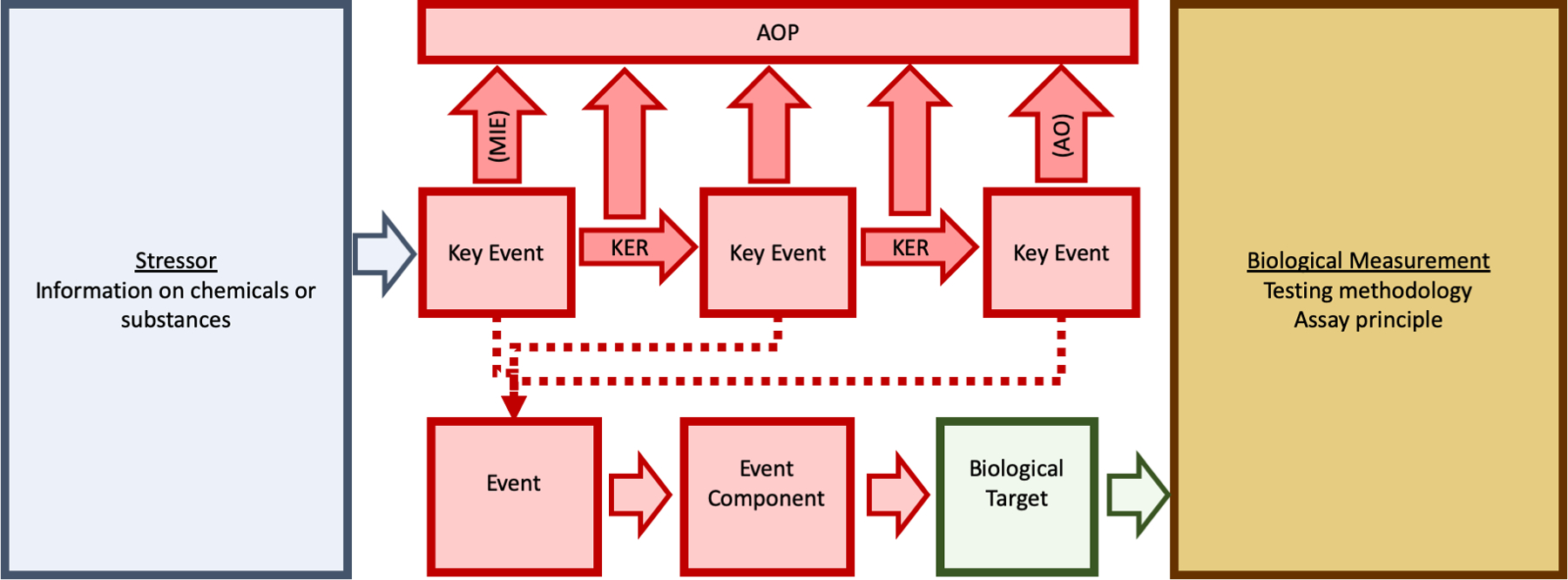
Each key event (KE) in an AOP has information about a specific biological target and subsequent biological measurement, dependent on testing methodology and the assay principle, associated with a stressor. Key event relationships (KERs) provide evidence to support linkage of KEs. Combined, all components can be organized into a series of steps that begin with a molecular initiating event (MIE) and ending with an adverse outcome (AO).
An example of an ongoing effort to promote interoperability between AOPs and NAM information is the AOP-DB.101 The AOP-DB consumes AOP descriptions from the AOP-Wiki and annotates the key events from each AOP with gene and protein identifiers based on the ontological terms assigned to each key event. These identifiers are then used to connect AOPs from the AOP-Wiki to information about chemicals, biological pathways, diseases, genetics, and toxicity assays that may be relevant for each AOP. Currently, these data are used to provide links between the EPA CompTox Chemicals Dashboard and the AOP-Wiki.109 These data have also been incorporated into workflows developed by the OpenRiskNet consortium.110,111
Current strategies for linkage of macromolecular and cellular changes with adverse outcomes to leverage NAM data for safety evaluation include extensive literature review and predictive modeling.21–23,36 One issue with using the AOP framework is that only a small fraction of the adverse effects used in regulatory toxicology decision making are the types of specific outcomes modeled in currently available AOPs. Expert evaluation of the literature is labor intensive and slow, and the lack of well-developed AOPs limits predictive modeling, which raises the following questions: (1) how can more AOPs be developed via rapid linkage of MIE or KE related information from NAM-based and traditional toxicology screening methods?; (2) how can hypotheses be generated to suggest more potential MIE to AO associations?; and, (3) how can development of putative AOPs be prioritized based on potential utility for chemical safety evaluation?
To address these persistent questions, data-driven approaches have shown promise,34,100,112 but they are restricted to toxicity information that is available through large datasets or data aggregators such as CTD. AI methods under consideration to support systematic review efforts could unlock information buried in the scientific literature113, and adaptations of literature mining tools used for chemical searches114 are being considered as well. Additional tools are needed to make the NAM and traditional information more accessible, and further, existing information from disparate resources need to be leveraged to support the discovery of novel MIE to AO relationships.
V. AOPs & Systematic Reviews
Systematic reviews may leverage existing AOPs as well as be useful for populating or support of new or existing AOPs using information mined from the literature. Systematic review methods have been successfully leveraged to increase transparency and rigor by introducing strategies that limit bias and random error115 while identifying the best available evidence relevant to a literature-based chemical assessment116,117 (Figure 3). Rigorous systematic reviews rely on the identification of all relevant information for a research question to ensure that the best available information is found, assessed, and synthesized. However, identifying all relevant information (i.e. maximizing recall) is a semantic challenge because it requires a priori knowledge of what kind of information is available and how and where it is managed in order to find it. Linguistic inconsistencies resulting from different communities using different vocabularies to describe common study characteristics, concepts, relationships, and words related to a science question must be considered when developing a search string to capture all potentially relevant studies. For literature search engines, recall is not always maximized, so “all-encompassing” search strings are built in a broad literature search step, potentially leading to the capture of non-relevant information.118 This “excess” information increases the time and resources needed to screen all retrieved literature at the title/abstract and full text levels [based on a populations, exposures, comparators, and outcomes (PECO) criteria] and can constrain scaling systematic reviews to a handful, rather than thousands, of chemicals in the exposure domain. However, depending on the need for a timely decision, rapid systematic reviews and artificial intelligence applications and tools can be deployed.113,115
Figure 3: Workflow for literature based chemical assessments based on systematic review methodology.
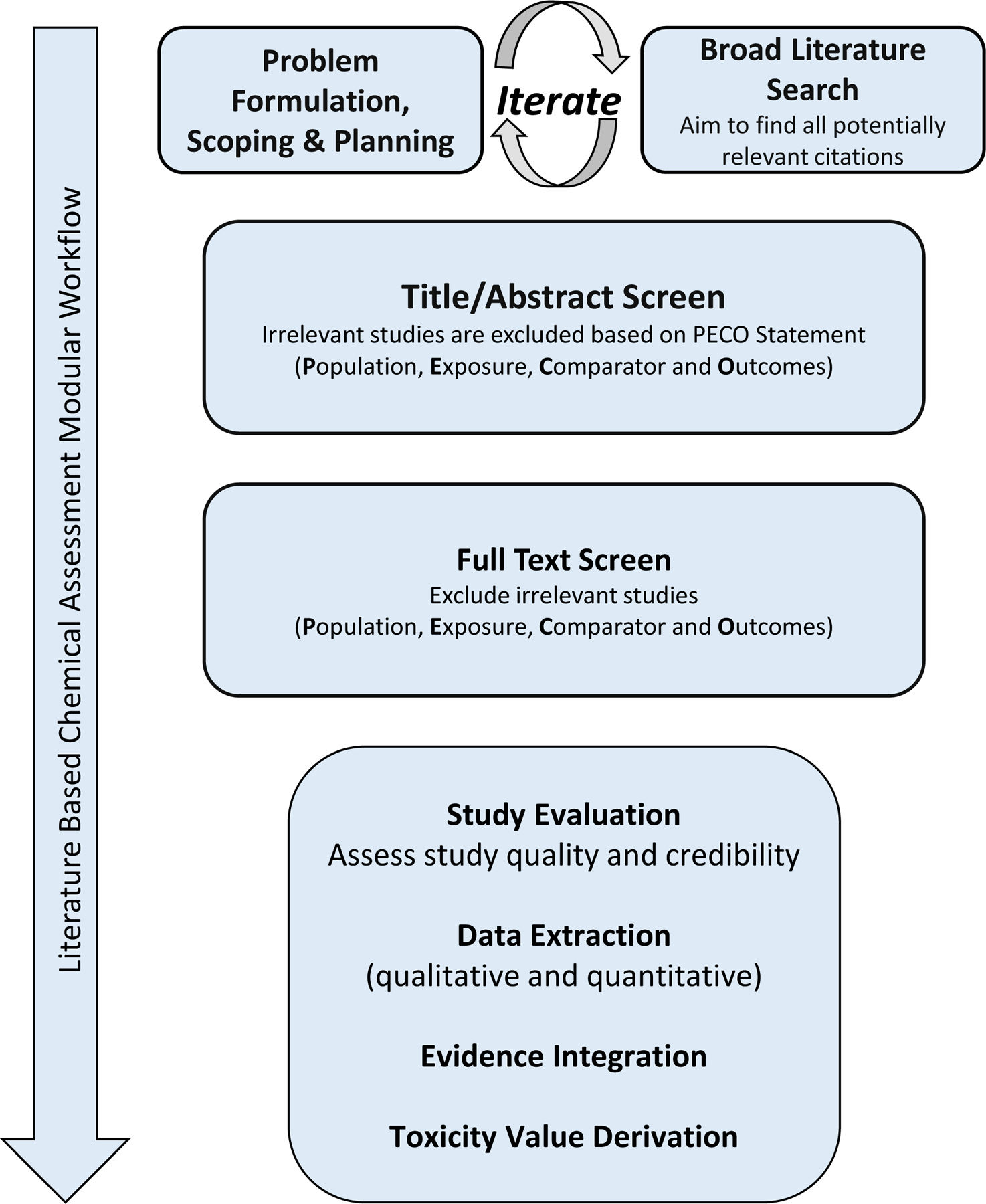
Iterations of literature-based chemical-centric systematic review methods ensure rigor and transparency while making use of the best available scientific information, requiring data interoperability to maximize unbiased data recall.
For more rapid reviews, studies can be “tagged” and inventoried (e.g., by study type, data type, chemical, outcome, population, data source) using an evidence mapping approach119 to prioritize literature for full text review, study evaluation, data extraction, and evidence integration in support of hazard identification and toxicity value derivation (Figure 3). For included studies of acceptable or higher quality these tags can be made to capture more granular study characteristics (such as summary findings, experimental methods, exposure route, and study population) that can then be used to make decisions such as where to focus research capacity, whether to broaden or narrow the scope of an assessment or focus on studies evaluating an outcome of regulatory concern.
Data interoperability via use of CVs to describe study characteristics would support integration of the information collected during a systematic review and/or evidence map into a flexible conceptual framework for aggregation and curation of literature-based information. Information gathered during a systematic review and/or evidence map could also be managed using ontology concepts adopted by the AOP community for automatic integration into a structured data format for AOPs.120 Evidence derived using systematic review methods and mapped to existing AOP content could help ground KEs and KERs, since the evidence synthesized using systematic review methods should be unbiased with respect to capturing what exists in the literature. Similarly, information extracted during a systematic review could be used in data-driven approaches to computationally predict KE and KER.34,100,112 This structured information can provide support for identifying AOPs that may be relevant to chemical safety evaluation, but that might have otherwise been missed using an expert-only guided approach.
AOP development and knowledge discovery to inform chemical safety evaluation for toxicology necessitate improvements to the FAIR data landscape, thus enabling integration of data that were previously siloed. To use NAMs (e.g. high-throughput transcriptomics, high-throughput screening, and high-content imaging data) more effectively in chemical safety evaluations, better linkages of these data to MIEs, KEs, and AOs of interest for regulatory toxicology are needed. A potential challenge in using AOPs for regulatory toxicology is that frequent determinants of adversity from in vivo testing are non-specific outcomes such as weight loss.121 Predicting these non-specific outcomes with NAMs may require multiple approaches, including mapping these endpoints to AOPs such that these non-specific indicators are put into context with more mechanistic events and/or the use of NAMs to model a threshold for systemic toxicity more generically. Identification of chemical bioactivity that may relate to AOs of regulatory interest requires NAM reporting standardization and improved computational accessibility of existing in vivo toxicity data. With this standardization, an AOP or AOP-like framework may be more useful and comprehensively populated using bioinformatic tools, including systematic review among others.
VI. Putative gene-outcome relationships for complex phenotypes
One of the most prominent challenges for adopting NAMs for chemical risk assessment is understanding how results can be applied to complex, multiple-etiology adverse outcomes like cancer. One approach from Kleinstreuer et al. (2013)25 attempted to use odds ratios between in vitro bioactivity in ToxCast assay and cancer-related phenotypes in rodents, as documented in ToxRefDB, to develop chemical cancer hazard scores. Subsequently, the biological plausibility of links between ToxCast assays and ToxRefDB cancer outcomes was manually assessed by a literature review. The limitations of this approach were made clear in Cox et al. (2016)122 stating that small changes to the dataset used in the cancer model dramatically changed the resultant associations between in vitro bioactivity and cancer-related phenotypes in rodents. This model instability could be the result of false positives i.e. the chemical bioactivity observed in ToxCast is not related to the cancer outcome observed in ToxRefDB. The approach could be improved if each ToxCast assay, which are linked to gene target(s), could be evaluated for relevance to cancer AOPs. Indeed, this type of approach was taken by the International Agency for Research on Cancer (IARC)123,124 where each ToxCast assay was reviewed and binned into the ten key characteristics of carcinogens (TKCC).125 A toxicological priority index (ToxPi) was calculated for each chemical based on the bioactivities of each assay in each TKCC. Further, Becker et al (2017)126 used the IARC binning of ToxCast assays and cancer designations by USEPA Office of Pesticides Program (OPP) Cancer Assessment Review Committee (CARC) as descriptors for machine learning models to classify chemicals as carcinogens, and ultimately concluded that ToxCast could not classify chemicals as carcinogens. Associating in vitro screening data with cancer-related outcomes continues to be an active area of research that may be transformed by generation of data that includes more of the genome, like high-throughput transcriptomic data.
Thus, a challenge remains: what are the best strategies to establish biological links between gene targets and complex AOs, based on previous understanding of the etiology and progression of these outcomes? Expert knowledge is limited due to the reliance on low-throughput manual literature review. The role that environmental chemical exposure(s) play in multiple-etiology outcomes like cancer are not well understood and may benefit from new information. Pathway analysis tools for associating gene-related data to biological processes relevant to carcinogenesis may be informative, but associations are from prior knowledge and are dependent on the specific pathway tool employed.127,128 Data-driven strategies for associating gene-target information with AO information may connect a wealth of gene information to cancer etiology or other complex phenotypes that may be difficult for an expert to identify. Novel, hypothetical associations between chemical exposure(s) and diseases may require bioinformatic tools and unsupervised approaches to putatively link chemical exposures with AOs.34,102,112 A persistent challenge with creating data driven models linking in vitro data with AOs is the relatively small data sets available, relative to the data variability.
An example of a bioinformatic resource to connect gene-target and AO information is the recently published Entity MeSH Co-occurrence Network (EMCON);32 EMCON can be used to identify genes that are linked to complex disorders like breast cancer. EMCON was also used as a data stream in Grashow et al. (2018)129 as part of a comprehensive gene prioritization framework to identify a breast cancer gene panel. The traditional approach to analyze gene expression results is gene set enrichment analysis (GSEA), where pathways or other concepts are identified from overrepresented differentially-expressed genes in reference gene sets that are primarily manually curated.130 Relevant gene sets for understanding links between chemical exposures and complex phenotypes are not readily available. Most gene sets are available through the Molecular Signatures Database (MSigDB)130–132 or other resources for GSEA like Enrichr.133,134 A commonly used resource that links genes to disease is Online Mendelian Inheritance in Man® (OMIM),135 which links genetic variants to disease; however, variants that have been linked to complex phenotypes have primarily been identified in genome wide association studies (GWAS) and are not always easy to mechanistically characterize. A well-curated resource that houses linkages between chemical exposures and disease is the Comparative Toxicogenomics Database (CTD).35 CTD extracts chemical-gene, gene-disease, and chemical-disease interactions from literature and integrates gene-disease relationships primarily from OMIM. The chemical-disease links are inferred by looking for genes common to both chemical-gene interactions and gene-disease relationships.136 CTD is a high-quality resource, but dependent on manual curation, which is low-throughput. Manual curation efforts cannot keep pace with the rate of publication, which highlights a need for alternative methods for data extraction including text-mining tools such as named entity recognition (NER),137,138 with EMCON providing just one example of a bioinformatic tool using NER to connect gene and AO information.
To identify environmental exposures that could potentially influence susceptibility to a complex disease, more complex gene networks may be important to identify. Other manual literature curation efforts continue for gene and gene function information,139 proteins,140,141 pathways,142–145 diseases,146 and chemicals.35,87,147 Aggregating these curated target-disease associations with chemical concentration-response information from NAMs targeting different levels of biological information could lead to rapid putative AOP development and development of robust computational models for chemical hazard.
VII. Conclusion
The current regulatory framework for toxicology is adapting to keep pace with modern chemical safety evaluation needs and the development of NAMs. However, a major obstacle to streamlining chemical safety evaluation and uptake of NAMs is data interoperability. Improving data interoperability will enable researchers to more comprehensively interrogate available data to better understand the existing knowledge landscape, thereby identifying data gaps, and perhaps identifying how environmental chemical exposure may influence complex AOs. Initial efforts in toxicology to promote interoperability demonstrate immense progress and promise, yet, for continued success, more work is needed in development and adherence to CVs and data formatting standards as well as implementing modern data infrastructures to support the large amounts of data and data analyses.
From here, both immediate and more long-term actions are required to meet a standard of data interoperability. Using CVs and data formatting standards for existing legacy toxicology data are critical first steps. Development and implementation of NAM data formatting standards in parallel will enable greater uptake of this information by various stakeholders. The complete implementation of chemical identity reporting standards, e.g. the use of DSSTox identifiers that link to unique substances, will cut across any kind of toxicology data integration. The development and use of more automated, computational bioinformatic tools to both identify and populate relationships like AOPs or exposure-disease hypotheses are needed to leverage existing data. Increased support and development of research computing environments that enable easy integration of datasets, in a reproducible manner, continue to be developed. Ultimately, avoiding the generation of siloed data, e.g. unstructured data in PDF documents, would be supported by extension of the resources available to build complex data systems for sharing and extraction of data.
In this review, some of the possibilities enabled by increased efforts toward data interoperability are presented. Obstacles for further progress on data interoperability include the need for resources and the diversity of toxicology data stakeholders. Data interoperability in toxicology requires balancing the need for domain-specific details with the need to reduce complexity to enable broader use of the data. The needs of data scientists for available tools and datasets may differ from the needs of the public for data transparency and availability as well as the needs of regulatory toxicologists charged with making public health decisions. Thus, any data strategy and data interoperability solutions must provide a sufficient amount of detail for the many applications of toxicology data.
VIII. Acknowledgments
The authors wish to thank Kathie Dionisio, Maureen Gwinn, and Antony Williams (of the US EPA) and Matthew T. Martin (Pfizer, Inc.) for helpful comments on previous versions of this manuscript.
Footnotes
Publisher's Disclaimer: Disclaimer: The United States Environmental Protection Agency (U.S. EPA) through its Office of Research and Development has subjected this article to Agency administrative review and approved it for publication. Mention of trade names or commercial products does not constitute endorsement for use. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the US EPA.
References
- 1.Richard AM et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol 29, 1225–1251 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Judson R et al. The toxicity data landscape for environmental chemicals. Env. Heal. Perspect 117, 685–695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egeghy PP et al. The exposure data landscape for manufactured chemicals. Sci Total Env. 414, 159–166 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Shimkus JM Frank R Lautenberg Chemical Safety for the 21st Century Act. United States Statutes at Large 448–513 (2014). [Google Scholar]
- 5.National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. (The National Academies Press, 2007). doi:doi: 10.17226/11970 [DOI] [Google Scholar]
- 6.ECHA. New Approach Methodologies in Regulatory Science. in (2016). [Google Scholar]
- 7.OCSPP, U. & USEPA. Strategic Plan to Promote the Development and Implementation of Alternative Test Methods Within the TSCA Program. (2018). [Google Scholar]
- 8.Thomas RS et al. The US Federal Tox21 Program: A strategic and operational plan for continued leadership. ALTEX 35, 163–168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tice RR, Austin CP, Kavlock RJ & Bucher JR Improving the human hazard characterization of chemicals: a Tox21 update. Env. Heal. Perspect 121, 756–765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Using 21st Century Science to Improve Risk-Related Evaluations. (2017). doi: 10.17226/24635 [DOI] [PubMed] [Google Scholar]
- 11.Dix DJ et al. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 95, 5–12 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Judson RS et al. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Env. Heal. Perspect 118, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watford S et al. ToxRefDB version 2.0: Improved utility for predictive and retrospective toxicology analyses. Reprod. Toxicol (2019). doi: 10.1016/J.REPROTOX.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Judson RS et al. Aggregating data for computational toxicology applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) System. Int J Mol Sci 13, 1805–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce RG, Setzer RW, Strope CL, Wambaugh JF & Sipes NS httk: R Package for High-Throughput Toxicokinetics. J Stat Softw 79, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filer DL, Kothiya P, Setzer RW, Judson RS & Martin MT tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics 33, 618–620 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Williams AJ et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 9, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bioplanet. Available at: https://tripod.nih.gov/bioplanet/. (Accessed: 28th December 2018)
- 19.Tox21 Toolbox. 2018,
- 20.Bell SM et al. An Integrated Chemical Environment to Support 21st-Century Toxicology. Environ. Health Perspect 125, 054501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstreuer NC et al. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem Res Toxicol 30, 946–964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haggard DE et al. High-Throughput H295R Steroidogenesis Assay: Utility as an Alternative and a Statistical Approach to Characterize Effects on Steroidogenesis. Toxicol Sci 162, 509–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judson RS et al. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol Sci 148, 137–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J et al. Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure. Chem Res Toxicol 28, 738–751 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kleinstreuer NC et al. In vitro perturbations of targets in cancer hallmark processes predict rodent chemical carcinogenesis. Toxicol Sci 131, 40–55 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Kleinstreuer N et al. A computational model predicting disruption of blood vessel development. PLoS Comput Biol 9, e1002996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sipes NS et al. Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol Sci 124, 109–127 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Martin MT et al. Profiling the reproductive toxicity of chemicals from multigeneration studies in the toxicity reference database. Toxicol Sci 110, 181–190 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Martin MT, Judson RS, Reif DM, Kavlock RJ & Dix DJ Profiling chemicals based on chronic toxicity results from the U.S. EPA ToxRef Database. Env. Heal. Perspect 117, 392–399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pigliucci M Genotype-phenotype mapping and the end of the ‘genes as blueprint’ metaphor. Philos Trans R Soc L. B Biol Sci 365, 557–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner B Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet 14, 168–178 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Watford SM et al. Novel application of normalized pointwise mutual information (NPMI) to mine biomedical literature for gene sets associated with disease: use case in breast carcinogenesis. Comput. Toxicol (2018). doi: 10.1016/j.comtox.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452 e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oki NO & Edwards SW An integrative data mining approach to identifying adverse outcome pathway signatures. Toxicology 350–352, 49–61 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Davis AP et al. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 45, D972–D978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooney AA, Boyles AL, Wolfe MS, Bucher JR & Thayer KA Systematic review and evidence integration for literature-based environmental health science assessments. Env. Heal. Perspect 122, 711–718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavlock RJ et al. Accelerating the Pace of Chemical Risk Assessment. Chem Res Toxicol 31, 287–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M, Mao S & Liu Y Big Data: A Survey. Mob. Networks Appl 19, 171–209 (2014). [Google Scholar]
- 40.Raja K et al. A Review of Recent Advancement in Integrating Omics Data with Literature Mining towards Biomedical Discoveries. Int J Genomics 2017, 6213474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jagodnik KM et al. Developing a framework for digital objects in the Big Data to Knowledge (BD2K) commons: Report from the Commons Framework Pilots workshop. J Biomed Inf. 71, 49–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bui AAT, Van Horn JD & Consortium NBKC Envisioning the future of ‘big data’ biomedicine. J Biomed Inf. 69, 115–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sagiroglu S & Sinanc D Big data: A review. in 2013 International Conference on Collaboration Technologies and Systems (CTS) 42–47 (2013). doi: 10.1109/CTS.2013.6567202 [DOI] [Google Scholar]
- 44.Begoli E & Horey J Design Principles for Effective Knowledge Discovery from Big Data. in 2012 Joint Working IEEE/IFIP Conference on Software Architecture and European Conference on Software Architecture 215–218 (2012). doi: 10.1109/WICSA-ECSA.212.32 [DOI] [Google Scholar]
- 45.Affymetrix Standards Program. Available at: http://www.affymetrix.com/about_affymetrix/outreach/standards_program/standards-program.affx. (Accessed: 15th January 2019)
- 46.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–10 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett T et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merrick BA, Paules RS & Tice RR Intersection of toxicogenomics and high throughput screening in the Tox21 program: an NIEHS perspective. Int J Biotechnol 14, 7–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrill J Development and Use of a High Content Imaging-Based Phenotypic Profiling Assay for Chemical Bioactivity Screening. (2018). doi: 10.23645/epacomptox.7003181.v1 [DOI] [Google Scholar]
- 50.Shah I et al. Using ToxCast™ Data to Reconstruct Dynamic Cell State Trajectories and Estimate Toxicological Points of Departure. Environ. Health Perspect 124, 910–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Privacy Act of 1974 5 U.S.C. § 552a, as amended. Available at: https://www.justice.gov/opcl/privacy-act-1974. (Accessed: 19th July 2019)
- 52.Datasets - Data.gov. Available at: https://catalog.data.gov/dataset. (Accessed: 19th July 2019)
- 53.NIH STRATEGIC PLAN FOR DATA SCIENCE. doi: 10.1371/journal.pbio.1002195 [DOI] [Google Scholar]
- 54.Mahony C, Currie R, Daston G, Kleinstreuer N & van de Water B Highlight report: ‘Big data in the 3R’s: outlook and recommendations’, a roundtable summary. Arch. Toxicol 92, 1015–1020 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Judson RS et al. Estimating Toxicity-Related Biological Pathway Altering Doses for High-Throughput Chemical Risk Assessment. Chem. Res. Toxicol 24, 451–462 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Judson R et al. In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme. Basic Clin Pharmacol Toxicol 115, 69–76 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Barton-Maclaren T, Gwinn M, Thomas R, Kavlock R & Rasenberg M INSIGHT: New Approaches to Chemical Assessment—a Progress Report. (2019). Available at: https://news.bloombergenvironment.com/environment-and-energy/insight-new-approaches-to-chemical-assessmenta-progress-report. (Accessed: 24th January 2019)
- 58.Fitzpatrick JM & Patlewicz G Application of IATA – A case study in evaluating the global and local performance of a Bayesian network model for skin sensitization. SAR QSAR Environ. Res 28, 297–310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fitzpatrick JM, Roberts DW & Patlewicz G An evaluation of selected (Q)SARs/expert systems for predicting skin sensitisation potential. SAR QSAR Environ. Res 29, 439–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts DW & Patlewicz G Non-animal assessment of skin sensitization hazard: Is an integrated testing strategy needed, and if so what should be integrated? J. Appl. Toxicol 38, 41–50 (2018). [DOI] [PubMed] [Google Scholar]
- 61.US EPA, O. Integrated Risk Information System.
- 62.NCBI. PubMed. Available at: https://www.ncbi.nlm.nih.gov/pubmed/. (Accessed: 24th January 2019) [Google Scholar]
- 63.Lea IA, Gong H, Paleja A, Rashid A & Fostel J CEBS: a comprehensive annotated database of toxicological data. Nucleic Acids Res 45, D964–D971 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.eChemPortal - Substance Search. Available at: https://www.echemportal.org/echemportal/substancesearch/substancesearchlink.action. (Accessed: 6th February 2019)
- 65.US EPA, O. Provisional Peer-Reviewed Toxicity Values (PPRTVs).
- 66.The Carcinogenic Potency Project (CPDB). Available at: https://toxnet.nlm.nih.gov/cpdb/index.html. (Accessed: 6th February 2019)
- 67.Chemview | US EPA.
- 68.eChemPortal - Home. Available at: https://www.echemportal.org/echemportal/index.action. (Accessed: 7th May 2019)
- 69.Registered substances - ECHA. Available at: https://echa.europa.eu/information-on-chemicals/registered-substances. (Accessed: 7th May 2019)
- 70.ECOTOX | Home. Available at: https://cfpub.epa.gov/ecotox/. (Accessed: 7th May 2019)
- 71.Hazard Evaluation Support System Integrated Platform (HESS): | Chemical Management | National Institute of Technology and Evaluation (NITE). Available at: https://www.nite.go.jp/en/chem/qsar/hess-e.html. (Accessed: 7th May 2019)
- 72.Wignall JA et al. Standardizing benchmark dose calculations to improve science-based decisions in human health assessments. Env. Heal. Perspect 122, 499–505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ACToR Web Services. Available at: https://actorws.epa.gov/actorws/. (Accessed: 21st January 2019)
- 74.Reif DM et al. Endocrine Profiling and Prioritization of Environmental Chemicals Using ToxCast Data. Environ. Health Perspect 118, 1714–1720 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rusyn I, Sedykh A, Low Y, Guyton KZ & Tropsha A Predictive modeling of chemical hazard by integrating numerical descriptors of chemical structures and short-term toxicity assay data. Toxicol Sci 127, 1–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dionisio KL et al. The Chemical and Products Database, a resource for exposure-relevant data on chemicals in consumer products. Sci. Data 5, 180125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daston G et al. SEURAT: Safety Evaluation Ultimately Replacing Animal Testing—Recommendations for future research in the field of predictive toxicology. Arch. Toxicol 89, 15–23 (2015). [DOI] [PubMed] [Google Scholar]
- 78.EU-ToxRisk - About EU-ToxRisk. Available at: http://www.eu-toxrisk.eu/page/en/about-eu-toxrisk.php. (Accessed: 23rd April 2019) [Google Scholar]
- 79.Kim S et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.TOXNET. Available at: https://toxnet.nlm.nih.gov/. (Accessed: 7th May 2019)
- 81.Vempati UD et al. Formalization, Annotation and Analysis of Diverse Drug and Probe Screening Assay Datasets Using the BioAssay Ontology (BAO). PLoS One 7, e49198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abeyruwan S et al. Evolving BioAssay Ontology (BAO): modularization, integration and applications. J. Biomed. Semantics 5, S5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bodenreider O The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 32, D267–70 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cesta MF et al. The National Toxicology Program Web-based nonneoplastic lesion atlas: a global toxicology and pathology resource. Toxicol Pathol 42, 458–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Briggs K, Barber C, Cases M, Marc P & Steger-Hartmann T Value of shared preclinical safety studies - The eTOX database. Toxicol Rep 2, 210–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ravagli C, Pognan F & Marc P OntoBrowser: a collaborative tool for curation of ontologies by subject matter experts. Bioinformatics 33, 148–149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shapiro AJ et al. Software Tools to Facilitate Systematic Review Used for Cancer Hazard Identification. doi: 10.1289/EHP4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.EPA Health Assessment Workspace Collaborative (HAWC). Available at: https://hawcprd.epa.gov/. (Accessed: 19th July 2019)
- 89.Heidorn CJ et al. IUCLID: an information management tool for existing chemicals and biocides. J Chem Inf Comput Sci 43, 779–786 (2003). [DOI] [PubMed] [Google Scholar]
- 90.IUCLID format - IUCLID. Available at: https://iuclid6.echa.europa.eu/format. (Accessed: 31st January 2019)
- 91.Grulke CM, Williams AJ, Thillanadarajah I & Richard AM EPA’s DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research. Comput. Toxicol 12, 100096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gwinn MR et al. Chemical Risk Assessment: Traditional vs Public Health Perspectives. Am J Public Heal 107, 1032–1039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ankley GT et al. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Env. Toxicol Chem 29, 730–741 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Villeneuve DL et al. Adverse Outcome Pathway (AOP) Development I: Strategies and Principles. Toxicol. Sci 142, 312–320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villeneuve DL et al. Adverse Outcome Pathway Development II: Best Practices. Toxicol. Sci 142, 321–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edwards SW, Tan YM, Villeneuve DL, Meek ME & McQueen CA Adverse Outcome Pathways-Organizing Toxicological Information to Improve Decision Making. J Pharmacol Exp Ther 356, 170–181 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Adverse Outcome Pathways, Molecular Screening and Toxicogenomics - OECD. Available at: http://www.oecd.org/chemicalsafety/testing/adverse-outcome-pathways-molecular-screening-and-toxicogenomics.htm. (Accessed: 21st January 2019)
- 98.Knapen D, Vergauwen L, Villeneuve DL & Ankley GT The potential of AOP networks for reproductive and developmental toxicity assay development. Reprod. Toxicol 56, 52–55 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Knapen D et al. Adverse outcome pathway networks I: Development and applications. Environ. Toxicol. Chem 37, 1723–1733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Villeneuve DL et al. Adverse outcome pathway networks II: Network analytics. Environ. Toxicol. Chem 37, 1734–1748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pittman ME, Edwards SW, Ives C & Mortensen HM AOP-DB: A database resource for the exploration of Adverse Outcome Pathways through integrated association networks. Toxicol. Appl. Pharmacol 343, 71–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oki NO, Nelms MD, Bell SM, Mortensen HM & Edwards SW Accelerating Adverse Outcome Pathway Development Using Publicly Available Data Sources. Curr. Environ. Heal. Reports 3, 53–63 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Brockmeier EK et al. The Role of Omics in the Application of Adverse Outcome Pathways for Chemical Risk Assessment. Toxicol. Sci 158, 252–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kleinstreuer NC et al. Adverse outcome pathways: From research to regulation scientific workshop report. Regul. Toxicol. Pharmacol 76, 39–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wittwehr C et al. How Adverse Outcome Pathways Can Aid the Development and Use of Computational Prediction Models for Regulatory Toxicology. Toxicol. Sci 155, 326–336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tollefsen KE et al. Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA). Regul. Toxicol. Pharmacol 70, 629–640 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Juberg DR et al. FutureTox III: Bridges for Translation. Toxicol. Sci 155, 22–31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buesen R et al. Applying ‘omics technologies in chemicals risk assessment: Report of an ECETOC workshop. Regul. Toxicol. Pharmacol 91, S3–S13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.CompTox Chemistry Dashboard. Available at: https://comptox.epa.gov/dashboard/dsstoxdb/results?search=DTXSID8022292#exec_sum. (Accessed: 19th July 2019)
- 110.AOP-DB: The Adverse Outcome Pathway Database • OpenRiskNet. Available at: https://openrisknet.org/events/60/. (Accessed: 19th July 2019)
- 111.Connecting Adverse Outcome Pathways, knowledge and data with AOPLink workflows • OpenRiskNet. Available at: https://openrisknet.org/events/70/. (Accessed: 19th July 2019)
- 112.Bell SM, Angrish MM, Wood CE & Edwards SW Integrating Publicly Available Data to Generate Computationally Predicted Adverse Outcome Pathways for Fatty Liver. Toxicol. Sci 150, 510–520 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Jaspers S, De Troyer E & Aerts M Machine learning techniques for the automation of literature reviews and systematic reviews in EFSA. EFSA Support. Publ 15, (2018). [Google Scholar]
- 114.Baker N, Knudsen T & Williams A Abstract Sifter: a comprehensive front-end system to PubMed. F1000Research 6, 2164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schünemann HJ & Moja L Reviews: Rapid! Rapid! Rapid! …and systematic. Syst. Rev 4, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cano-Sancho G et al. Human epidemiological evidence about the associations between exposure to organochlorine chemicals and endometriosis: Systematic review and meta-analysis. Environ. Int 123, 209–223 (2019). [DOI] [PubMed] [Google Scholar]
- 117.Yost EE et al. Hazards of diisobutyl phthalate (DIBP) exposure: A systematic review of animal toxicology studies. Environ. Int 125, 579–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Relevo R Effective Search Strategies for Systematic Reviews of Medical Tests. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolffe TAM, Whaley P, Halsall C, Rooney AA & Walker VR Systematic evidence maps as a novel tool to support evidence-based decision-making in chemicals policy and risk management. Environ. Int 130, 104871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ives C, Campia I, Wang R-L, Wittwehr C & Edwards S Creating a Structured AOP Knowledgebase via Ontology-Based Annotations. Appl. Vitr. Toxicol 3, 298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown JM, Watford S & Paul-Friedman K Bioinformatic Integration of in vivo Data and Literature-based Gene Associations for Prioritization of Adverse Outcome Pathway Development. (2019). doi: 10.23645/epacomptox.7844555.v1 [DOI] [Google Scholar]
- 122.Anthony Tony Cox L, Popken DA, Kaplan AM, Plunkett LM & Becker RA How well can in vitro data predict in vivo effects of chemicals? Rodent carcinogenicity as a case study. Regul Toxicol Pharmacol 77, 54–64 (2016). [DOI] [PubMed] [Google Scholar]
- 123.Chiu WA, Guyton KZ, Martin MT, Reif DM & Rusyn I Use of high-throughput in vitro toxicity screening data in cancer hazard evaluations by IARC Monograph Working Groups. ALTEX 35, 51–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guyton KZ et al. Application of the key characteristics of carcinogens in cancer hazard identification. Carcinogenesis 39, 614–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith MT et al. Key Characteristics of Carcinogens as a Basis for Organizing Data on Mechanisms of Carcinogenesis. Env. Heal. Perspect 124, 713–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker RA et al. How well can carcinogenicity be predicted by high throughput ‘characteristics of carcinogens’ mechanistic data? Regul Toxicol Pharmacol 90, 185–196 (2017). [DOI] [PubMed] [Google Scholar]
- 127.Papatheodorou I, Oellrich A & Smedley D Linking gene expression to phenotypes via pathway information. J Biomed Semant. 6, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanoudi S, Donato M & Draghici S Identifying biologically relevant putative mechanisms in a given phenotype comparison. PLoS One 12, e0176950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grashow RG, De La Rosa VY, Watford SM, Ackerman JM & Rudel RA BCScreen: A gene panel to test for breast carcinogenesis in chemical safety screening. Comput. Toxicol 5, 16–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liberzon A et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen EY et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Online Mendelian Inheritance in Man, OMIM®. Available at: https://omim.org/. [PubMed]
- 136.King BL, Davis AP, Rosenstein MC, Wiegers TC & Mattingly CJ Ranking transitive chemical-disease inferences using local network topology in the comparative toxicogenomics database. PLoS One 7, e46524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hirschman L, Yeh A, Blaschke C & Valencia A Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics 6 Suppl 1, S1 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smith L et al. Overview of BioCreative II gene mention recognition. Genome Biol 9 Suppl 2, S2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brown GR et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res 43, D36–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pundir S, Martin MJ & O’Donovan C UniProt Protein Knowledgebase. Methods Mol Biol 1558, 41–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Suzek BE et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kanehisa M, Sato Y, Kawashima M, Furumichi M & Tanabe M KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D457–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fabregat A et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 44, D481–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD) Available at: https://www.omim.org/. (Accessed: 30th January 2019) [Google Scholar]
- 147.Judson R et al. Workflow for Defining Reference Chemicals for Assessing Performance of In Vitro Assays. ALTEX (2018). doi: 10.14573/altex.1809281 [DOI] [PMC free article] [PubMed] [Google Scholar]


