Abstract
Alfalfa (Medicago sativa L.) is one of the most important and widely cultivated forage crops. It is commonly used as a vegetable and medicinal herb because of its excellent nutritional quality and significant economic value. Based on Illumina, Nanopore and Hi-C data, we assembled a chromosome-scale assembly of Medicago sativa spp. caerulea (voucher PI464715), the direct diploid progenitor of autotetraploid alfalfa. The assembled genome comprises 793.2 Mb of genomic sequence and 47,202 annotated protein-coding genes. The contig N50 length is 3.86 Mb. This genome is almost twofold larger and contains more annotated protein-coding genes than that of its close relative, Medicago truncatula (420 Mb and 44,623 genes). The more expanded gene families compared with those in M. truncatula and the expansion of repetitive elements rather than whole-genome duplication (i.e., the two species share the ancestral Papilionoideae whole-genome duplication event) may have contributed to the large genome size of M. sativa spp. caerulea. Comparative and evolutionary analyses revealed that M. sativa spp. caerulea diverged from M. truncatula ~5.2 million years ago, and the chromosomal fissions and fusions detected between the two genomes occurred during the divergence of the two species. In addition, we identified 489 resistance (R) genes and 82 and 85 candidate genes involved in the lignin and cellulose biosynthesis pathways, respectively. The near-complete and accurate diploid alfalfa reference genome obtained herein serves as an important complement to the recently assembled autotetraploid alfalfa genome and will provide valuable genomic resources for investigating the genomic architecture of autotetraploid alfalfa as well as for improving breeding strategies in alfalfa.
Subject terms: Genome, Evolution
Introduction
Alfalfa (Medicago sativa ssp. sativa L.) is a perennial legume forage that is widely cultivated for hay, pasture and silage production (e.g., Fig. 1a). As one of the most economically valuable crops in the world1–3, alfalfa has total estimated annual sales ranging from 7.8 to 10.8 billion dollars in the USA4. Alfalfa is known as “the queen of forage crops” not only because of its high-protein content and nutritive value as an animal feed but also because of its atmospheric nitrogen (N) fixation capacity. It is used as a rotation crop to increase soil fertility and serves as an important habitat for wildlife5. In addition, alfalfa is well known for its superior contents of vitamins (A, C, E, and K), protein and minerals, such as calcium, potassium, phosphorus, and iron6. Its seed sprouts and tender tips contain all these nutrients but few calories and are often used as edible vegetables (e.g., Fig. 1b). Furthermore, alfalfa has long been used as a medicinal herb. Its seeds or dried leaves can be used as a nutritional supplement and are sold as a bulk powdered herb, capsules, and tablets in health food stores7. The extracts from alfalfa seeds and leaves have hypocholesterolemic, neuroprotective, antioxidant, hypolipidemic, and antimicrobial effects and are used in the treatment of diabetes, stroke, cancer and menopausal symptoms6,8–12 (e.g., Fig. 1c). Alfalfa also exhibits a relatively high level of disease resistance potential compared to that of other food crops13. Therefore, it provides disease prevention between planting stages and increases the stock carrying capacity. In China, the cultivated area of alfalfa reached 3.6 million hectares in 2017; however, China still imports more than 1.3 million tons per year, accounting for ~85% of the total imported hay. An increasing industrial demand, low production and a lack of multiple improved varieties with strong resistance and quality may be some of the factors accounting for such a large supply gap in the alfalfa industry14.
Fig. 1. Photographs of alfalfa uses and products.
a Photograph of alfalfa used as a widely cultivated forage crop. b Photograph of alfalfa as food. c Photograph of alfalfa as a health product. Picture from Dr. Willmar Schwabe, India Pvt., Ltd.
On the basis of advanced sequencing technologies, breeders can use DNA markers combined with genome sequences to facilitate gene discovery, trait dissection and predictive molecular breeding technology15,16. Despite the high economic value of and increasing industrial demand for alfalfa, improvements through breeding are very limited, partly due to a lack of information on the whole genome. Alfalfa is suggested to be an autotetraploid (2n = 4x = 32) subspecies in the M. sativa complex17,18. The recently published genome assembly of an autotetraploid alfalfa19 is expected to greatly facilitate the future improvement of molecular breeding strategies. However, assembling a complete autotetraploid genome is still challenging due to essential features of tetrasomic inheritance, as more than 400 Mb of contigs were not placed onto the chromosomes in the above genome assembly19. In this case, assembling the genome of the diploid progenitor could be an alternative way to obtain full genomic information for alfalfa. Indeed, genomic information for diploid progenitors has provided substantial insights into selection for several key agronomic traits and the evolutionary history of multiple polyploid food crops, such as cotton20, wheat21, and strawberry22.
Previous studies have demonstrated that M. sativa spp. caerulea (2n = 2x = 16), a perennial self-incompatible herb, is the diploid progenitor of autotetraploid alfalfa23. In this study, we assembled a chromosome-scale draft genome of M. sativa spp. caerulea voucher PI464715 (hereafter PI464715) using a combination of Illumina, Hi-C and Nanopore sequencing technologies. Using this high-quality genome, we further performed genome annotation, evolutionary analysis, and comparative genomics and identified resistance genes and genes involved in the lignin and cellulose biosynthesis pathways. Our PI464715 genome assembly provides a diploid reference for analyzing the alfalfa genome and is a valuable resource for future molecular breeding of alfalfa. This genome is also beneficial for investigating genome evolution in the genus Medicago and related taxa.
Results
Genome sequence and assembly
Medicago sativa spp. caerulea (voucher PI464715; 2n = 2x = 16) was chosen for genome sequencing and assembly. A genome survey was first performed to assess the genome size based on 81.5 Gb of Illumina data. Using K-mer analysis, we evaluated the genome size to be ~802 Mb, with a high level of heterozygosity of 1.9% (Supplementary Table S1 and Supplementary Fig. 1). To accurately assemble this highly heterozygous genome, Illumina, Nanopore and Hi-C technologies were adopted for sequencing, and a series of methods were performed for assembly. Based on 116.5 Gb of Nanopore long reads corresponding to ~145× coverage of the estimated ~802 Mb genome, we preliminarily obtained a raw assembled genome of 1,345.8 Mb and contig N50 of 2.8 Mb by the NextGraph module. After polishing by NextPolish24 and performing deredundancy by purge_haplotigs25, we obtained the final genome assembly with a length of 793.2 Mb and a contig number of 355 and contig N50 of 3.86 Mb, constituting 98.9% of the predicted genome size (Table 1; Supplementary Table S1). We used the Benchmarking Universal Single-Copy Orthologs (BUSCO) evaluation score26 to assess the quality of the assembly, which resulted in 97.7% gene set completeness (Supplementary Table S2), indicating a very complete and high-quality genome assembly. We further connected 338 (95.2%) out of 355 contigs into eight pseudochromosomes based on ~224 Gb of Hi-C data (~279×coverage) using the hierarchical clustering strategy27 (Supplementary Fig. 2; Supplementary Tables S1 and S3). In total, 98.5% (781 Mb) of the assembly was anchored and oriented on eight pseudochromosomes, which ranged from 83.24 to 118.42 Mb in length (Supplementary Table S3), and 98.3% of transcriptomic reads and 96.4% of Illumina short reads could be properly mapped to the final genome assembly (Supplementary Tables S4 and S5).
Table 1.
Summary of the PI464715 genome assembly and annotation.
| Categories | Type | Length (bp) | No. | % of genome |
|---|---|---|---|---|
| Assembly | Contigs | 793,191,298 | 355 | - |
| Contig N50 | 3,857,628 | 65 | - | |
| Contig N90 | 1,207,163 | 205 | - | |
| Longest | 14,548,009 | 1 | - | |
| Noncoding RNAs | miRNA | 117,689 | 1023 | 0.015 |
| snRNA | 269,590 | 2438 | 0.034 | |
| rRNA | 331,884 | 1978 | 0.042 | |
| tRNA | 63,912 | 857 | 0.008 | |
| Transposable elements | DNA | 57,410,696 | - | 7.24 |
| LINE | 34,454,045 | - | 4.34 | |
| SINE | 2,527,147 | - | 0.32 | |
| LTR | 251,835,738 | - | 31.75 | |
| RC | 15,102,186 | - | 1.9 | |
| Satellite | 342,023 | - | 0.043 | |
| Simple_Repeat | 10,038,485 | - | 1.26 | |
| Unknown | 80,412,452 | - | 10.14 | |
| Low_Complexity | 1,803,306 | - | 0.23 | |
| Total | 440,637,371 | - | 55.55 | |
| Gene | Gene loci | - | 47,202 | - |
| Average gene length (bp) | 3151 | - | - | |
| Average CDS length (bp) | 1085 | - | - | |
| Average exon length (bp) | 231 | - | - | |
| Average exons per gene | - | 4.7 | - | |
| Average intron length | 615 | - | - |
Our PI464715 assembly provided significant improvement (with larger contig sizes and a higher BUSCO score) than the alfalfa genome19. Our genome has a contig N50 of 3.86 Mb, which is ~8.4-fold greater than that of the alfalfa genome (459 kb). Moreover, we placed 781 Mb of the assembly onto eight chromosomes with the aid of Hi-C data, while in the alfalfa genome, only 685 Mb (on average across the four assembled monoploid genomes) was anchored on the eight chromosomes. Our assembled genome also obtained a higher BUSCO evaluation score (97.7%) than the four monoploid genomes of alfalfa (88.5%, 88.3%, 87.5%, and 87.2%). All these comparisons indicated that our genome has better contiguity and higher quality.
Gene prediction and annotation
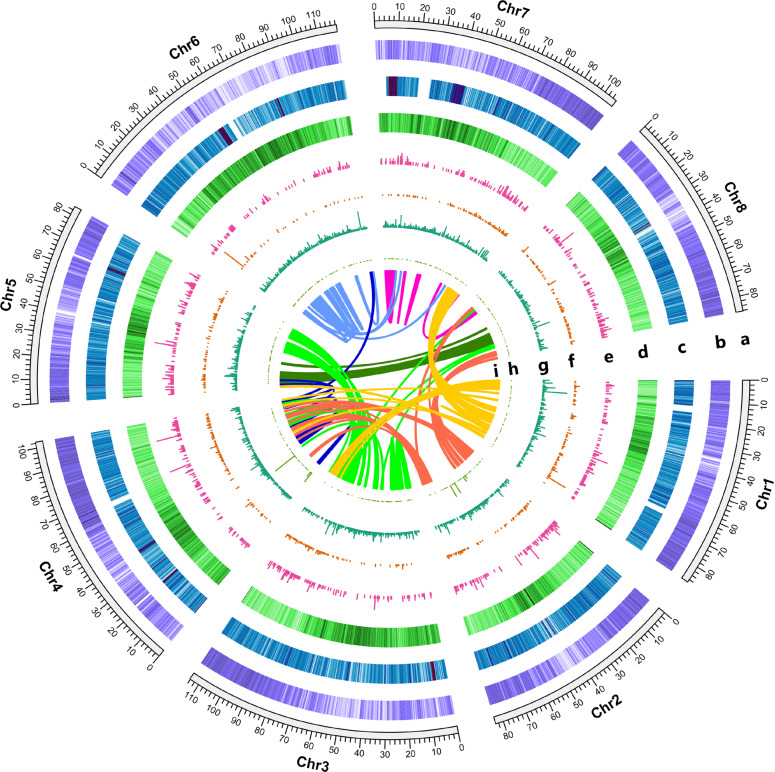
In total, we identified 47,202 protein-coding genes, with an average gene length of 3151 bp (Table 1 and Supplementary Fig. 3), based on a combined strategy using de novo, transcriptome-based and homology-based methods. The total GC content of the PI464715 genome assembly was 34.21% (Supplementary Table S3). BUSCO evaluation further showed that the annotated PI464715 genome contained 97% BUSCOs (Supplementary Table S6). Then, five protein databases, namely, InterPro, Kyoto Encyclopedia of Genes and Genomes (KEGG), SwissProt, KOG and NR, were used to compare our protein models. Overall, we assigned potential functions to 92.51% (43,669) of the protein-coding genes in the PI464715 genome (Supplementary Table S7). The gene distribution and GC content along each chromosome were calculated, and their distributions were uneven (Fig. 2b, d), as also found in many other plant species (e.g., M. truncatula). In addition, we identified 857 transfer RNAs (tRNAs), 1023 microRNAs (miRNAs), 1978 ribosomal RNAs (rRNAs), and 2438 small nuclear RNAs (snRNAs) in the PI464715 genome (Table 1 and Fig. 2e–h).
Fig. 2. Overview of the PI464715 genome assembly.
a Genomic positions (in Mb) of the eight chromosomes of PI464715. b Gene density. c Transposable element (TE) content. d Guanine-cytosine (GC) content. e miRNA density. f tRNA density. g snRNA density. h rRNA density. i Interconnections of paralogs in the genome are represented with colored lines.
We annotated repetitive sequences of the genome using both de novo and homology-based approaches. We annotated ~440 Mb (55.55%) of the PI464715 genome assembly that comprised transposable elements (TEs), of which long terminal repeat (LTR) retrotransposons were the most abundant, accounting for 57.2% of TEs and 31.75% of the assembled genome (Fig. 2c and Table 1). DNA transposons, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) accounted for 7.24%, 4.34%, and 0.32% of the genome assembly, respectively (Table 1).
Gene-family analysis
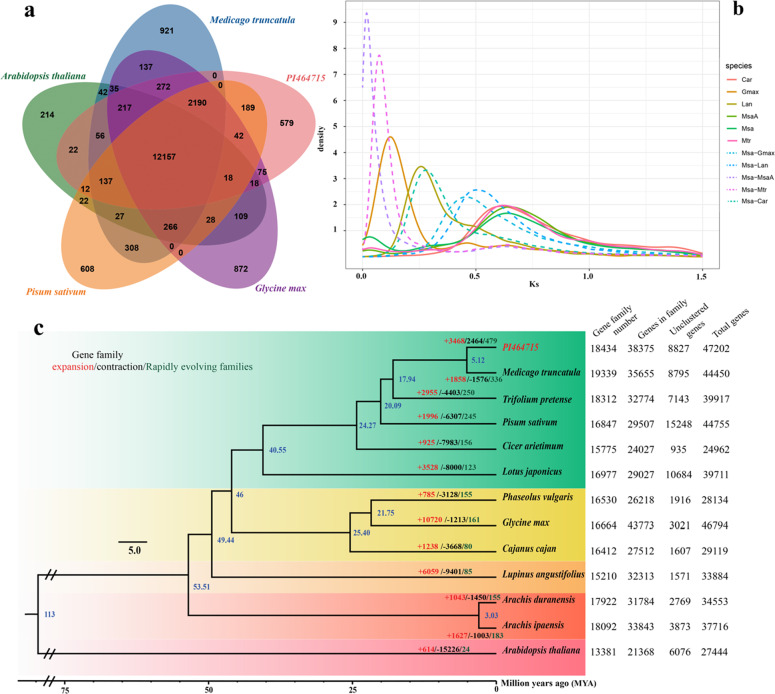
To investigate the genome evolution of PI464715, annotated genes from 11 species of the Leguminosae family (i.e., M. truncatula, Trifolium pertense, Pisum sativum, Cicer arietinum, Lotus japonicus, Phaseolus vulgaris, Glycine max, Cajanus cajan, Lupinus angustifolius, Arachis duranensis, and Arachis ipaensis) and one rosid species (Arabidopsis thaliana) were clustered into gene families. In total, 38,375 PI464715 genes (81.3%) were clustered into 18,434 gene families (Fig. 3c). PI464715 shared a total of 12,157 (65.9%) gene families with the 12 other species and contained 579 (3.1%) unique gene families (Fig. 3a,c). We determined and selected 553 single-copy orthologous genes from these 13 species for subsequent phylogenetic analysis. As expected, PI464715 displayed a close relationship with M. truncatula and phylogenetically diverged from its common ancestor ~5.12 million years ago (Fig. 3c). The phylogenetic relationships among these 13 species were the same as those recovered from a previous study28.
Fig. 3. Phylogenetic and evolutionary analyses of the PI464715 genome.
a Venn diagram of shared orthologous gene families in PI464715, A. thaliana, M. truncatula, P. sativum, and G. max. The number of gene families is listed for each component. b Distribution of synonymous substitution levels (Ks) of syntenic orthologous (dashed curves) and paralogous genes (solid curves) after evolutionary rate correction. Mtr: M. truncatula; Car: C. arietinum; Gmax: G. max; Lan: L. angustifolius; Msa: PI464715. MsaA: M. sativa subgenome 1. c Phylogenetic tree of 13 plant species and the evolution of gene families. The blue numerical value beside each node shows the estimated divergence time of each node (MYA, million years ago). The numbers of gene-family expansions, contractions and significant (p-value ≤ 0.01) gene-family expansion and contraction events are indicated by red, black and green numbers, respectively.
Among the 18,434 gene families identified in PI464715, 3468 expanded and 2464 contracted gene families were detected. Compared with its close relative M. truncatula (another important species in the genus used as a legume model species), which exhibited 1858 expanded and 1576 contracted gene families, PI464715 had more gene families (Fig. 3c). Furthermore, a higher number of gene families in PI464715 compared with M. truncatula (i.e., 479 vs. 336) exhibited significant rapid evolution (family-wide p-value ≤ 0.01) (Fig. 2c). The GO enrichment analysis suggested that reproductive processes, such as recognition of pollen (GO:0048544), pollen-pistil interaction (GO:0009875) and pollination (GO:0009856), were enriched in both the contracted and expanded gene families (Supplementary Tables S8 and S9), and these genes may be involved in the transition between self-compatibility in M. truncatula and self-incompatibility in PI464715. The GO enrichment analysis of the expanded gene families also suggested multiple response pathways (e.g., response to chemical, response to hormone, response to auxin and response to stimulus), all of which may be related to the adaptation of this species to diverse environments.
Comparative genomic analyses and genome expansion in PI464715
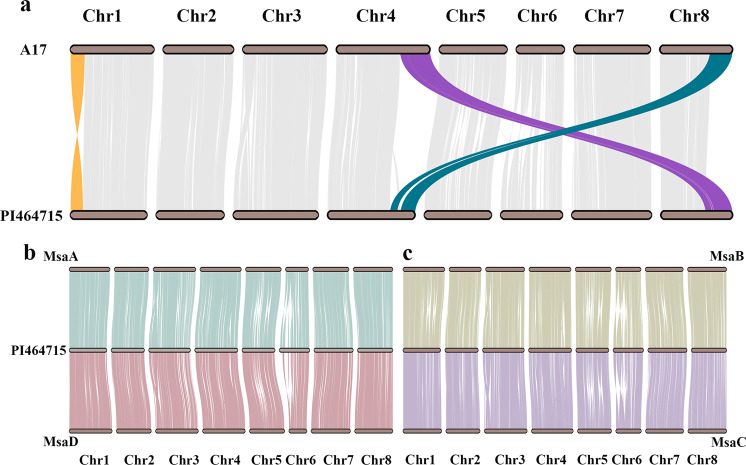
Synteny analysis was conducted between the PI464715 genome assembly, the four monoploid genomes of alfalfa19 and the M. truncatula ecotype Jemalong A17 genome v5.029 to explore their evolution. High collinearity was revealed between our genome with all four subgenomes of alfalfa and for five chromosomes between our genome and the A17 genome by visualizing syntenic blocks (Fig. 4). We further detected a pair of large interchromosomal rearrangements between chromosome 4 and chromosome 8 and a large inversion on chromosome 1, as also evident in the dot plots comparing our genome and the A17 genome (Fig. 4a and Supplementary Fig. 4). Such rearrangements and inversions were also found between the genomes of two ecotypes, A17 and R10830, but not between the PI464715 and R108 genomes (result not shown). These results indicate that the large interchromosomal rearrangements and inversion may have occurred specifically in A17 after the divergence between M. truncatula and M. sativa, but this needs further investigation.
Fig. 4. Gene synteny between the M. truncatula ecotype Jemalong A17, PI464715, and alfalfa genomes.
a Synteny comparison of homologous chromosomes between the PI464715 genome and the M. truncatula A17 genome. b Synteny comparison of homologous chromosomes between the PI464715 genome and the M. sativa genome (MsaA: M. sativa subgenome 1; MsaD: M. sativa subgenome 4). c Synteny comparison of homologous chromosomes between the PI464715 genome and the M. sativa genome (MsaB: M. sativa subgenome 2, MsaC: M. sativa subgenome 3).
Our assembled PI464715 genome is 793 Mb in size, almost two times larger than the genome of M. truncatula (420 Mb). We tested whether whole-genome duplication (WGD) events accounted for the genome expansion in PI464715. We selected the genomes of four species (i.e., M. truncatula, C. arietinum, G. max, and L. angustifolius) from the Leguminosae family and subgenome A of alfalfa and performed comparative genomic analysis with PI464715 to investigate the WGD events and divergence time between PI464715 and other species, which were evaluated by measuring the synonymous nucleotide substitution rate (Ks) of orthologous gene pairs. All six species displayed a peak Ks value of 0.62, consistent with the finding of a previous study31, and the divergence between PI464715 and the other four species occurred afterwards, suggesting a common whole-genome duplication event for all Papilionoideae species32,33. PI464715 and M. truncatula experienced no WGD events after their divergence, and the divergence between PI464715 and the diploid ancestor of alfalfa (i.e., represented by one monoploid genome, subgenome A) was the most recent (Fig. 3b).
Resistance-related (R) genes
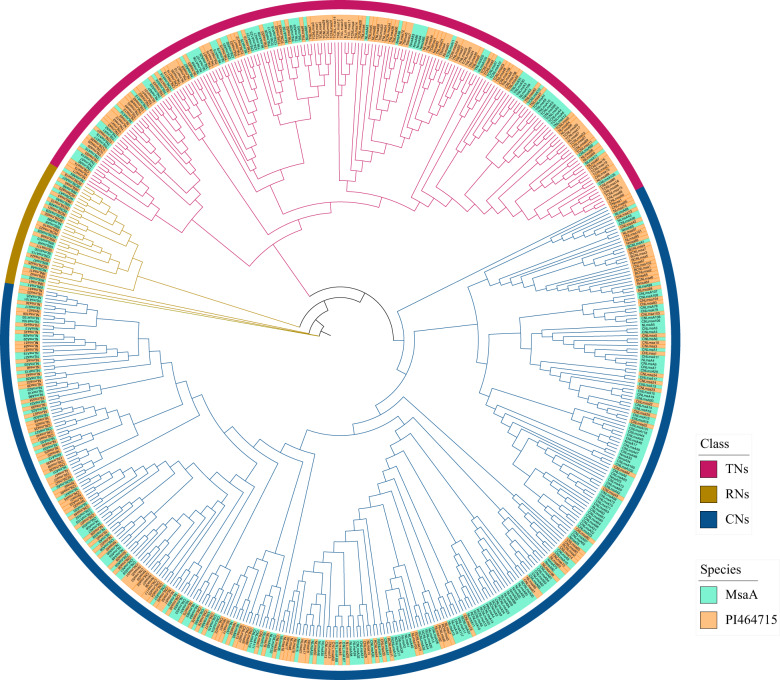
Plant resistance genes (R genes) are important gene groups that usually include an NBS (nucleotide-binding site) domain and an LRR (leucine-rich repeat) domain and play a crucial role in plant disease resistance34. Based on the types of domains in the N-terminal region, R genes belong to three groups: CNL (CC-NBS-LRR), RNL (RPW8-NBS-LRR) and TNL (TIR-NBS-LRR)35. In the PI464615 genome, 489 R genes were detected, including 117 CNL genes, 58 TNL genes and 11 RNL genes (Supplementary Table S10). The numbers of R genes detected in the four monoploid genomes of alfalfa were similar but slightly smaller than those in the PI464615 genome. In total, 1749 R genes were detected in the autotetraploid alfalfa genome. Furthermore, PI464615 had ~1.5-fold to ~2.2-fold more TNL genes but fewer CNL genes than the four monoploid genomes of alfalfa. More R genes (692) were detected in M. truncatula, including 139 CNL genes, 145 TNL genes and 15 RNL genes (Supplementary Table S10). R genes with complete domains identified from the PI464715 and M. truncatula genomes were selected to construct a phylogenetic tree. The results indicated that these R genes were clustered into the RNL, TNL and CNL groups (Fig. 5 and Supplementary Fig. 5).
Fig. 5. hylogenetic tree of the nucleotide-binding site (NBS) domain R gene-family identified in the PI464715 genome and M. sativa A subgenome.
(MsaA: M. sativa subgenome 1). Class represents the three groups of R genes. MsaA: M. sativa subgenome 1.
Lignin and cellulose biosynthesis-related genes
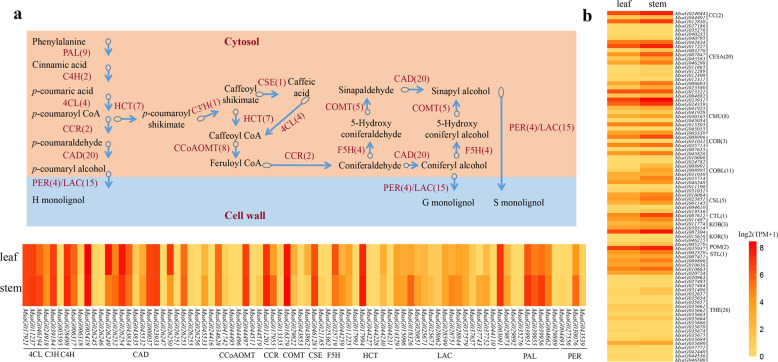
The content of lignin and cellulose is one of the important factors affecting alfalfa quality as an animal feed36, and reducing the lignin content in alfalfa can improve digestibility and, correspondingly, animal performance37. Based on a BLASTp homology search and Pfam analysis, we identified a total of 82 putative lignin biosynthesis-related genes and 85 putative cellulose biosynthesis-related genes (Fig. 6). These genes were unevenly distributed on the eight chromosomes (Supplementary Fig. 6). Hierarchical cluster analysis of transcriptomic data showed clustering of the three repeats for the leaf or stem (Supplementary Fig. 7). Transcriptomic analysis revealed that the expression patterns of these identified genes involved in the lignin and cellulose biosynthesis pathways in leaf and stem tissues were similar, but the expression levels were slightly higher in stem than in leaf tissue (Fig. 6), which is consistent with the fact that the lignin content is higher in stem than in leaf tissue. We also found that the expression levels of multiple gene copies for each gene were different. For example, among the seven gene copies that putatively encode the enzyme HCT, MsaG017994 had the highest expression level, which was 13–250 times higher than that of other gene copies (Fig. 6a). Knowing the relative expression levels of different gene copies can be useful when conducting targeted downregulation of enzymes for forage quality improvement by reducing lignin content, for example36,38,39.
Fig. 6. Expression of lignin and cellulose biosynthesis genes in leaf and stem tissues.
a Simplified diagram of lignin biosynthesis pathways in plants modified from Vanholme et al.78 and a heat map showing the expression of lignin biosynthesis-related genes in leaf and stem tissues. Values within brackets indicate the numbers of gene copies corresponding to the catalytic genes in the pathways. The enzymes involved in the lignin biosynthesis pathway are phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate/quinate 3’-hydroxylase (C3’H), caffeoyl-CoA O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), peroxidase (PER), laccase (LAC) and caffeoyl shikimate esterase (CSE). b Heat map showing the expression of cellulose-related genes in leaf and stem tissues. The enzymes involved in cellulose biosynthesis pathways are cellulose synthase (CESA), korrigan (KOR), chitinase-like (CTL), cobra (COB), cobra-like (COBL), kobito (KOB), stello (STL), cellulose synthase-microtubule uncoupling protein (CMU), companion of cellulose synthase (CC), theseus (THE), cellulose synthase interacting (POM), and tracheary element differentiation-related (TED).
Discussion
Medicago includes economically important forage crops, such as alfalfa (M. sativa) and “Jinhuacai” (M. polymorpha), in addition to a model organism (M. truncatula) in plant biology. Despite the importance of the genus, genomic resources are relatively scarce, and genome sequences are available only for M. truncatula and alfalfa, which largely slows down progress towards understanding the genome evolution and genetic code underlying molecular breeding for major crops in this genus. Here, we describe a chromosome-scale assembly of the M. sativa ssp. caerulea genome (i.e., the diploid progenitor of autotetraploid alfalfa) obtained by a combination of data from the Illumina, Nanopore and Hi-C platforms. The genome assembly was 793 Mb in length, and >98.5% of the assembled genome was placed on eight chromosomes (Table 1 and Supplementary Table S5). The BUSCO assessment revealed 97.7% complete genes in the assembled genome, which represents a more contiguous and higher-quality genome assembly than that recently published for alfalfa19. Our results further reveal that Nanopore long reads with the aid of Hi-C data can be adopted to accurately assemble a highly heterozygous and repetitive genome40.
The PI464615 genome (793 Mb) is approximately twofold larger than that of the closely related species M. truncatula (420 Mb)29. Several factors, including transposable elements (TEs) and whole-genome duplication (WGD), have been proposed to account for variation in genome size41,42. Recent analyses have shown that WGD or polyploidization seems to have occurred during the evolutionary histories of most plant species, such as the γ event43 that occurred ~140–150 Myr ago44 and is shared by all eudicots. After the γ-event, some species experienced no WGD events, such as grape and coffee, whereas other species, such as M. truncatula, kiwifruit and Asparagus setaceus, may have undergone one or two additional rounds of WGD29,45,46. All Papilionoideae within the Leguminosae family share a common WGD event, after which most species experienced no WGD events, except for G. max and L. angustifolius47–49. Our results from Ks distribution analysis reveal that both the PI464615 and M. truncatula genomes have only one peak, which precedes the divergence of the two species and is consistent with the ancestral Papilionoideae WGD event. The proliferation of TEs is another factor accounting for genome expansion. In this study, we identified ~440 Mb TEs, constituting 55.5% of the assembled PI464615 genome, which is ~234 Mb larger than the total length of TEs (~206 Mb)29 in the M. truncatula genome. Therefore, the proliferation of TEs rather than WGD and the presence of more expanded gene families than in M. truncatula resulted in genome expansion in PI464615.
In summary, we report a high-quality chromosome-level reference genome for M. sativa ssp. caerulea (voucher PI464715). We assembled a 793 Mb genome and annotated 47,202 protein-coding genes. We also identified resistance genes in the PI464715 genome and in each of the four monoploid genomes of alfalfa, which may provide a genetic basis for understanding the gain of resistance-related traits in alfalfa. We further identified 82 and 85 candidate genes that may be involved in the lignin and cellulose biosynthesis pathways, respectively, and described the expression profiles of these genes in leaf and stem tissues. Such information will be very useful for improving alfalfa quality in the future, for example, by the downregulation of targeted enzymes36,38,39 or through gene editing to decrease lignin content. The available genome sequence for the direct progenitor of autotetraploid alfalfa is an important complement to the alfalfa genome and holds great promise for further understanding fundamental aspects of genomic architecture and improving molecular breeding strategies in alfalfa. The genomic resource is also highly valuable for evolutionary studies in related species.
Material and methods
Plant materials, DNA extraction, and estimation of genome size
Seeds of M. sativa spp. caerulea voucher PI464715 were obtained from the National Plant Germplasm System (NPGS) of the United States Department of Agriculture (USDA) and planted in a greenhouse. Fresh leaves of a growing plant cultivated in a greenhouse were used to extract genomic DNA using a DNA Secure Plant Kit (Tiangen Biotech, Co., Ltd., Beijing, China). Paired-end libraries with insert sizes of 270 bp were constructed, and the Illumina HiSeq X Ten platform was used to generate Illumina short reads, which were first used to estimate genome size. We generated ~81.5 Gb of reads and determined the abundance of 17-K-mers in the generated Illumina data using Kmerfreq50. K-mer curve fitting was also performed under different gradient combinations of heterozygosity to estimate the heterozygosity of the genome.
Genome sequencing and assembly
Total genomic DNA was fractionated into 10–50 kb fragments with BluePippin, which was used to construct the libraries following the Nanopore library construction protocol. The generated libraries were then submitted for sequencing at the Nextomics Biosciences Company (Wuhan, China) using the GridION X5 sequencer platform (Oxford Nanopore Technologies, UK). The quality-controlled reads were used for assembly with the software Nextdenovo v. 2.3.051 following three steps. First, the NextCorrect module was applied to correct sequencing errors. Second, a preliminary assembly was generated based on the NextGraph module, which resulted in a genome size of 1345.8 Mb, with a contig number of 1154 and contig N50 of 2.8 Mb. Then, we polished the preliminary assembly using the Nextpolish v. 1.2.424 module. At this stage, Nanopore long reads and Illumina short reads were used repetitively three times for genome correction. Finally, allelic haplotigs were removed using purge_haplotigs25 software to obtain the final genome sequence. BUSCO v. 2.026, with 1,350 genes from Embryophyta odb10, was used to evaluate the completeness and accuracy of the assembled genome.
Chromosome-scale assembly with Hi-C data
Approximately 2 g of fresh leaves collected from the same PI464715 accession was used for Hi-C sequencing. Hi-C libraries were constructed following Miele et al.52 with chromatin extraction; digestion; and DNA ligation, purification and fragmentation. Hi-C sequencing was performed using the Illumina HiSeq X Ten platform (Illumina, CA, USA). A preliminary assembly was carried out to correct errors in contigs by splitting contigs into 100 kb segments on average. BWA v. 0.7.1753 was used to map the Hi-C data to these segments. The uniquely mapped Hi-C data were retained, clustered, ordered and placed onto the eight pseudochromosomes using LACHESIS28. A heat map of the interaction matrix of all pseudochromosomes was plotted with a resolution of 100 kb.
Repetitive sequence and gene annotation
Repetitive elements in the PI464715 genome assembly were identified based on a combination of homology-based and de novo approaches at both the protein and DNA levels. First, TRF v. 4.0.754 was applied to identify the tandem repeats in the genome assembly. Then, TEs were identified using RepeatMasker v. 4.1.055 and RepeatProteinMask (http://www.repeatmasker.org/) with Repbase51 as the query library. Next, RepeatModeler v. 5.8.856 (http://www.repeatmaskerorg/) was used to construct a de novo repeat library for the identification of TEs that were not found in the Repbase library.
We predicted protein-coding genes using a combination of de novo prediction, homology-based prediction and transcriptome-based prediction. Augustus v. 3.3.257, GlimmerHMM v. 3.0.458, Geneid v. 1.4.559, and Genscan60 software were used for de novo prediction. GeMoMa v. 1.3.161 was used for homology prediction, with protein sequences from M. truncatula, C. arietinum, G. max, P. vulgaris, P. persica and A. thaliana. For transcriptome-based predictions, we first sequenced the RNA library generated from mixed stem, leaf and flower tissues, and the RNA-seq reads were assembled into transcripts using Trinity v. 2.1.162 with default parameters. In addition, we mapped all the RNA-seq reads to the final assembled genome by PASA v. 2.1.063 to assess genome assembly quality. To annotate the noncoding RNAs, tRNAscan-SE v. 1.3.164 was applied for identifying tRNA genes with eukaryotic parameters. BLAST65 was applied to search the rRNA sequences in the PI464715 genome assembly with default parameters. MiRNA and snRNA were identified using INFERNAL v. 1.166 software based on covariance models deposited in the Rfam v. 13.067 database.
Gene functions were annotated by performing BLAST65 (E-value ≤ 1e−5) searches against four protein databases, i.e., SwissProt, KOG, NR, and KEGG. The InterPro database with BLAST or InterProScan v. 4.868 was used to annotate the functions of protein-coding genes. UniProt and GO annotations were assigned for each protein based on the results of alignment.
Gene families and phylogenetic analysis
We used OrthoFinder v. 2.2.769 to identify the orthologous groups among 12 Leguminosae species (PI464715, M. truncatula, T. pretense, P. sativum, C. arietimum, L. japonicus, P. vulgaris, G. max, C. cajan, L. angustifolius, A. duranensis, A. ipaensis) and one rosid species (A. thaliana). We then extracted the single-copy orthologous genes from the orthologous clustering results. We used CAFÉ v. 2.270 software to identify the expanded and contracted gene families in the 13 species, which were further subjected to GO enrichment analysis. For phylogenetic analysis, we first used MAFFT to perform multiple alignments of protein sequences of single-copy orthologous genes with default parameters. Then, the protein sequence alignments were converted into codon alignments. Second, Gblocks v. 0.9171 was used to delete regions with poor alignment or large differences in the results of multiple sequence alignments. Finally, the codon alignment results of all single-copy orthologs were connected to form a supergene for phylogenetic analysis. RAxML v. 8.2.072 was used to construct the phylogenetic tree. We calculated the average substitution rate along each branch and estimated species divergence time using r8s v. 1.8173.
Gene collinearity and Ks analysis
Syntenic blocks between PI464715 and the four monoploid genomes of alfalfa and M. truncatula ecotype Jemalong A17 were detected using MCScanX74. The number of synonymous substitutions per synonymous site (Ks) on each branch was estimated using the codeml program in the PAML v. 4.0 package75, and the median Ks value was representative of the collinear blocks.
Identification of resistance (R) genes
We used both BLAST searches and the hidden Markov model (HMM) to obtain R genes in the PI464715 genome, the four monoploid genomes of alfalfa and M. truncatula genomes. All of the protein sequences annotated in these genomes were first searched by using the HMM profile of the NB-ARC domain (Pfam no. PF00931) in a hmmscan subprocess of HMMER 3.2.1 (http://hmmer.org/). We used BLASTp to search the amino acid sequences of the NB-ARC domain against all annotated protein sequences in each genome. We merged all hits obtained from both analyses and removed the redundant hits. The sequences were further subjected to Pfam analysis and coiled-coil (CC) analysis to identify TIR, LRR, RPW8, zf-BED and CC domains. The method was similar to that used in a previous study76. We used paircoil277 (the threshold value was set to 0.025) and coils software to identify CC domains.
Identification and expression of lignin and cellulose biosynthesis genes
To identify the genes involved in the lignin and cellulose biosynthesis pathways in PI464715, we used the genes listed in the diagram of lignin biosynthesis pathways in plants by Vanholme et al.78 and cellulose biosynthesis genes identified in the A. thaliana database as references79. Then, the BLASTp algorithm and Pfam analysis were used to search our genome for homologs. The locations of all identified lignin and cellulose biosynthesis genes were marked on the eight chromosomes by MapChart v. 2.32 software80.
To examine the expression of these lignin and cellulose biosynthesis-related genes, we carried out RNA sequencing of two tissues (i.e., leaf and stem, each with three replicates) through 2 × 151-bp paired-end libraries using an Illumina HiSeq 4000. Leaf and stem tissues were obtained from a voucher PI464715 plant. Raw Illumina reads of low quality (when the percentage of low-quality bases was over 50% in a read) and with unknown bases (>10%) were filtered out to obtain clean reads. Then, the clean reads were mapped to the genome assembly using HISAT2 v. 2.0.481 with default parameters. Read alignments for transcripts in each sample were extracted using StringTie v. 1.2.382. The expression level of each gene was measured by transcripts per million (TPM) values estimated in StringTie.
Supplementary information
Acknowledgements
This work was supported equally by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502) and the National Natural Science Foundation of China (31971391) and further supported by the National Natural Science Foundation of China (41901056 and 31722055).
Author contributions
G.-P.R. and Y.-Z.Y. conceived and designed the project. Q.-W.D. provided the seeds. Z.-P.L. helped plant the seeds. A. Li. and A. Liu. collected the materials, assembled the genome, and performed gene annotation, gene-family and evolutionary analyses. X.D. conducted resistance gene identification. M.Y., J.-Y.C, H.-Y.H, S.-D.W., and H.-Q.W. helped with data analyses. A.Li and G.-P.R. wrote the manuscript with help from N.S., Y.-Z.Y., and J.-Q.L. All authors read and approved the final version of the manuscript.
Data availability
The whole-genome sequence data (including Illumina short reads, Nanopore long reads and Hi-C interaction reads), the final assembled genome and the transcriptomes of different tissues used in this study have been deposited in the NCBI database under BioProject ID PRJNA657344. The genome annotation information has been uploaded to Figshare.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Ao Li, Ai Liu
Contributor Information
Yong-Zhi Yang, Email: yangyongzhi2008@gmail.com.
Guang-Peng Ren, Email: rengp@lzu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00417-7).
References
- 1.Zhou Q, et al. MYB transcription factors in alfalfa (Medicago sativa): genome-wide identification and expression analysis under abiotic stresses. PeerJ. 2019;7:e7714. doi: 10.7717/peerj.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, et al. Global transcriptome sequencing using the Illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS ONE. 2013;8:e83549. doi: 10.1371/journal.pone.0083549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Brummer EC. Applied genetics and genomics in alfalfa breeding. Agronomy. 2012;2:40–61. doi: 10.3390/agronomy2010040. [DOI] [Google Scholar]
- 4.United States Department of Agriculture-National Agriculture Statistics Service. Crop Production Historical Track Records, April 2018. https://downloads.usda.library.cornell.edu/usda-esmis/files/c534fn92g/6q182n624/v405sd06x/htrcp-04-12-2018.pdf. (2019).
- 5.Russelle MP, Birr AS. Large-Scale assessment of symbiotic dinitrogen fixation by crops. Agron. J. 2004;96:1754–1760. doi: 10.2134/agronj2004.1754. [DOI] [Google Scholar]
- 6.Bora KS, Sharma A. Phytochemical and pharmacological potential of Medicago sativa: a review. Pharm. Biol. 2011;49:211–220. doi: 10.3109/13880209.2010.504732. [DOI] [PubMed] [Google Scholar]
- 7.Brinker, F. Herb Contraindications and Drug Interactions. Eclectic Medical Publications (Eclectic Medical Publications, 2010).
- 8.Malinow MR, McLaughlin P, Naito HK, Lewis LA, McNulty WP. Effect of alfalfa meal on shrinkage (regression) of atherosclerotic plaques during cholesterol feeding in monkeys. Atherosclerosis. 1978;30:27–43. doi: 10.1016/0021-9150(78)90150-8. [DOI] [PubMed] [Google Scholar]
- 9.Malinow MR, McLaughlin P, Stafford C. Alfalfa seeds: effects on cholesterol metabolism. Experientia. 1980;36:562–564. doi: 10.1007/BF01965801. [DOI] [PubMed] [Google Scholar]
- 10.Seida A, El-Hefnawy H, Abou-Hussein D, Mokhtar FA, Abdel-Naim A. Evaluation of Medicago sativa L. sprouts as antihyperlipidemic and antihyperglycemic agent. Pak. J. Pharm. Sci. 2015;28:2061–2074. [PubMed] [Google Scholar]
- 11.Sadeghi L, Tanwir F, Yousefi BV. Antioxidant effects of alfalfa can improve iron oxide nanoparticle damage: Invivo and invitro studies. Regul. Toxicol. Pharmacol. 2016;81:39–46. doi: 10.1016/j.yrtph.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Hong YH, Chao WW, Chen ML, Lin BF. Ethyl acetate extracts of alfalfa (Medicago sativa L.) sprouts inhibit lipopolysaccharide-induced inflammation in vitro and in vivo. J. Biomed. Sci. 2009;16:64. doi: 10.1186/1423-0127-16-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Shi S. Physiological and Proteomic Responses of Contrasting Alfalfa (Medicago sativa L.) Varieties to PEG-Induced Osmotic Stress. Front. Plant Sci. 2018;9:242. doi: 10.3389/fpls.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan, X. et al. Current Situation and Prospect of Alfalfa Industry. J. Green Sci. Technol. 4, 104–107 (2017). (in Chinese)
- 15.Rusk, Nicole Cheap third-generation sequencing. Nat. Methods. 2009;6:244–244. doi: 10.1038/nmeth0409-244a. [DOI] [PubMed] [Google Scholar]
- 16.Choi, Chul S. On the study of microbial transcriptomes using second- and third-generation sequencing technologies. J. Microbiol. 2016;54:527–536. doi: 10.1007/s12275-016-6233-2. [DOI] [PubMed] [Google Scholar]
- 17.Matheson NK, Small DM, Copeland L. β- d-mannanases in germinating lucerne (alfalfa) seeds. Carbohyd. Res. 1980;82:325–331. doi: 10.1016/S0008-6215(00)85706-7. [DOI] [PubMed] [Google Scholar]
- 18.Yu CY, Dong JG, Hu SW, Xu AX. Exposure to trace amounts of sulfonylurea herbicide tribenuron-methyl causes male sterility in 17 species or subspecies of cruciferous plants. BMC Plant Biol. 2017;17:95. doi: 10.1186/s12870-017-1019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020;11:2494. doi: 10.1038/s41467-020-16338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang G, et al. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020;52:516–524. doi: 10.1038/s41588-020-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling HQ, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- 22.Edger PP, et al. Author correction: origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019;51:765. doi: 10.1038/s41588-019-0380-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small E, Jomphe M. A synopsis of the genus Medicago (Leguminosae) Can. J. Bot. 2011;67:3260–3294. doi: 10.1139/b89-405. [DOI] [Google Scholar]
- 24.Hu J, Fan J, Sun Z, Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 25.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinforma. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simao FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 27.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreplak J, et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019;51:1411–1422. doi: 10.1038/s41588-019-0480-1. [DOI] [PubMed] [Google Scholar]
- 29.Pecrix Y, et al. Whole-genome landscape of Medicago truncatula symbiotic genes. Nat. Plants. 2018;4:1017–1025. doi: 10.1038/s41477-018-0286-7. [DOI] [PubMed] [Google Scholar]
- 30.Karen, et al. Strategies for optimizing BioNano and Dovetail explored through a second reference quality assembly for the legume model, Medicago truncatula. BMC Genomics. 2017;18:578. doi: 10.1186/s12864-017-3971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, et al. Hierarchically aligning 10 legume genomes establishes a family-level genomics platform. Plant Physiol. 2017;174:284–300. doi: 10.1104/pp.16.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young ND, Debellé F, Oldroyd GED, Geurts R, Roe BA. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannon SB, et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol. Biol. Evol. 2014;32:193–210. doi: 10.1093/molbev/msu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozano R, Hamblin MT, Prochnik S, Jannink JL. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics. 2015;16:360. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang L, et al. Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species. BMC Genomics. 2017;18:292. doi: 10.1186/s12864-017-3682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy MS, et al. Targeted down‐regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.) Proc. Natl Acad. Sci. USA. 2005;102:16573–16578. doi: 10.1073/pnas.0505749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barros J, Temple S, Dixon RA. Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotech. 2019;56:48–54. doi: 10.1016/j.copbio.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Shadle G, et al. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry. 2007;68:1521–1529. doi: 10.1016/j.phytochem.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 39.Bhattarai K, et al. Agronomic performance and lignin content of HCT down-regulated alfalfa (Medicago sativa L.) Bioenerg. Res. 2018;11:505–515. doi: 10.1007/s12155-018-9911-6. [DOI] [Google Scholar]
- 40.Kang M, et al. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine. Hortic. Res. 2020;7:18. doi: 10.1038/s41438-020-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiley GP, Burleigh JG. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evol. Biol. 2015;15:194. doi: 10.1186/s12862-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl Acad. Sci. USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S, Han B, Jiao Y. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant. 2020;13:59–71. doi: 10.1016/j.molp.2019.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Tang H, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li SF, et al. Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus. Hortic. Res. 2020;7:48. doi: 10.1038/s41438-020-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, et al. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 2019;6:117. doi: 10.1038/s41438-019-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barker DG, et al. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 1990;8:40–49. doi: 10.1007/BF02668879. [DOI] [Google Scholar]
- 48.Cook DR. Medicago truncatula-A model in the making! Curr. Opin. Plant Biol. 1999;2:301–304. doi: 10.1016/S1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- 49.Kreplak J, et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019;51:1411–1422. doi: 10.1038/s41588-019-0480-1. [DOI] [PubMed] [Google Scholar]
- 50.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin HH, Liao YC, Dutilh BE. Evaluation and validation of assembling corrected Pacbio long reads for microbial genome completion via hybrid approaches. PLoS ONE. 2015;10:e0144305. doi: 10.1371/journal.pone.0144305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miele A, Dekker J. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C) Methods Mol. Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tempel S. Using and understanding RepeatMasker. Methods Mol. Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- 56.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 57.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:435–439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 59.Blanco E, Parra G, Guigó R. Using geneid to Identify Genes. Curr. Protoc. Bioinforma. 2018;65:56. doi: 10.1002/cpbi.56. [DOI] [PubMed] [Google Scholar]
- 60.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keilwagen J, et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44:e89. doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grabherr MG, et al. Trinity: reconstructing a full-lentgh transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalvari I, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46:335–342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 71.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 72.Alexandros S. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:9. doi: 10.1093/bioinformatics/btt255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 76.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 77.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 78.Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019;56:230–239. doi: 10.1016/j.copbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 79.Lampugnani ER, et al. Cellulose synthesis-central components and their evolutionary relationships. Trends Plant Sci. 2019;24:402–412. doi: 10.1016/j.tplants.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 80.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 81.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequence data (including Illumina short reads, Nanopore long reads and Hi-C interaction reads), the final assembled genome and the transcriptomes of different tissues used in this study have been deposited in the NCBI database under BioProject ID PRJNA657344. The genome annotation information has been uploaded to Figshare.