Abstract
The maleimide group is a widely used reagent for bioconjugation of peptides, proteins, and oligonucleotides employing Michael addition and Diels–Alder cycloaddition reactions. However, the utility of this functionality in chemical synthesis of peptides and proteins remains unexplored. We report, for the first time that PdII complexes can mediate the efficient removal of various succinimide derivatives in aqueous conditions. Succinimide removal by PdII was applied for the synthesis of two ubiquitin activity-based probes (Ub-ABPs) employing solid phase chemical ligation (SPCL). SPCL was achieved through a sequential three segment ligation on a polymer support via a maleimide anchor. The obtained probes successfully formed the expected covalent complexes with deubiquitinating enzymes (DUBs) USP2 and USP7, highlighting the use of our new method for efficient preparation of unique synthetic proteins. Importantly, we demonstrate the advantages of our newly developed method for the protection and deprotection of native cysteine with a succinimide group in a peptide fragment derived from thioredoxin-1 (Trx-1) obtained via intein based expression to enable ligation/desulfurization and subsequent disulfide bond formation in a one-pot process.
Introduction
Maleimide is a ubiquitous functionality often used for bioconjugation of peptides, proteins, and oligonucleotides due to its selectivity and electrophilic properties.1 Specifically, maleimide is employed as a Michael acceptor for conjugating thiols or dienophiles for Diels–Alder cycloadditions. Maleimide chemistry allows for the production of fluorescently labeled proteins,2 antibody-drug conjugates,3 PEGylated proteins,4 protein–DNA hybrids,5 and cyclic peptides among other complex conjugates.6 Despite its wide application, there are still some limitations regarding the stability of maleimide based linkers. Thiosuccinimides are susceptible to the retro-Michael reaction, which cleaves the thioether linkage under a reducing environment, leading to the loss of cargo.7 Therefore, most of the efforts have been focused on preventing the retro-Michael reaction in order to stabilize the thiosuccinimide conjugates.8 Another disadvantage is that some antibody-drug conjugates connected through cysteine-maleimide chemistry require additional linkers that can be cleaved either enzymatically9 or chemically10 to release the payload in tumor sites. Developing on demand, cleavage of the thiosuccinimide linkage has potential for expanding the utility of succinimide based linkers for antibody-drug conjugates and for chemical proteomics.11 In addition, such an approach might find useful applications in peptide and protein syntheses employing the succinimide as a reversible protecting group and as a cleavable linker.
Chemical protein synthesis permits unlimited site specific modifications of the protein target such as the introduction of posttranslational modifications (PTMs), D-amino acids, affinity and fluorophore tags.12 However, often the selection of protecting groups (PGs) is highly crucial for achieving successful synthesis of the target protein.13 In recent years our group has made significant efforts to expand this chemical toolbox, predominantly by developing palladium mediated efficient cleavage of several PGs to facilitate chemical peptide and protein synthesis.14 These PGs have enabled sequential ligation,15 selective desulfurization, and one-pot disulfide bond formation.16 This chemistry was also extended for introducing solubilizing tags17 and new linkers18 for in vitro and cellular applications. Despite their importance, the requirement for preinstallation of these PGs during synthesis of peptide segments (using SPPS), limits their applicability in particular when dealing with recombinant peptide fragments. Therefore, the development of reagents which enable on demand attachment/detachment of free Cys residues under aqueous conditions is of great interest. Despite some progress in this direction the current methods have limitations. For example, the phenacyl group was used as a Cys PG for expressed fragments to enable protein semisynthesis.19 Similarly, the trityl group was used as a Cys PG to facilitate disulfide bond formations.20 However, the instability of the phenacyl group when using methoxy amine during Thz opening19b and the trityl PG during desulfurization makes these methods less attractive.19c In addition, both methods require organic solvents for either the protection or deprotection step, which have the potential to affect peptide solubility and makes such an approach incompatible with one-pot synthesis. Therefore, the development of more efficient reagents, specifically those that are stable under native chemical ligation (NCL)/desulfurization conditions and compatible with one-pot deprotection/disulfide bond formation for protein (semi)synthesis would have useful applications.
The structural resemblances between the acetamidomethyl (Acm) and succinimide PGs inspired us to investigate the cleavage reaction of the latter PG using palladium chemistry. Herein, we demonstrate for the first time that PdII complexes mediate the efficient cleavage of succinimide PG on Cys under aqueous conditions. We further show the use of succinimide as a stable linker during solid phase chemical ligation (SPCL) for successful synthesis of ubiquitin activity-based probes (Ub-ABPs) for selective labeling of two different deubiquitinating enzymes (DUBs). In addition, we applied our strategy for the protection and deprotection of native Cys side chains in a peptide fragment obtained via intein based expression to enable the semisynthesis of the thioredoxin-1 (Trx-1) protein by means of ligation/desulfurization and disulfide bond formation in one-pot reaction.
Results and Discussion
To examine the potential of succinimide as a PG, we prepared a model peptide 1 (LYRAGC(N-Me.Suc)LYRAG), where Cys was masked with the N-methyl succinimide group (N-Me.Suc; Scheme 1). This peptide was initially treated with 10 equiv of PdCl2 in 6 M guanidine hydrochloride (Gn·HCl)/0.2 M phosphate buffer, pH 7.3 at 37 °C. Gratifyingly, the reaction furnished the unprotected peptide with a complete conversion in about 4 h (Figure S2B). Encouraged by this result, we examined the influence of other metal complexes on the succinimide decaging reaction. However, none of the examined metal complexes (AuCl, K2PtCl6, NiCl2, CuCl2, and FeCl3) were suitable, highlighting the specific role of the Pd complex in this reaction (Figure S3B). We then turned our attention to find a better Pd complex, testing different complexes such as [Pd(allyl)Cl]2, Pd2(dba)3, Pd(PPh3)4, Pd(OAc)2, Cl4Na2Pd, and PdCl2 which among them gave the best results (Figure S4B). Aiming to improve the reaction efficiency, we investigated the effect of additives for improving the kinetics of the reaction. When 50 equiv of MgCl2 was added along with 10 equiv of PdCl2 in 6 M Gn·HCl/0.2 M phosphate buffer, pH 7.3 at 37 °C, the reaction was drastically improved and produced quantitatively the decaged peptide within 45 min (Figure S5B). The addition of excess MgCl2 could play various roles including preventing the nonproductive chelation of the palladium to the side chain reactive functional groups21,15d as well affecting the chemistry of Pd by possibly forming in situ [PdCl4]2–.22 Using additional additives such as 4-mercaptophenylacetic acid (MPAA), tris(2-carboxyethyl)phosphine (TCEP), GSH, NaHSO3, Na2S2O5, MgSO4, and NaCl did not improve the reaction (Figure S5B). Optimization of the reaction buffer indicated that 6 M Gn·HCl/0.2 M phosphate buffer facilitated the fastest reaction (Figure S6B), an observation we have also made in previous systems.15d Next, different loading of PdCl2 indicated that 10 equiv was optimum for this decaging reaction (Figure S7B). We therefore concluded that the use of 10 equiv of PdCl2, 50 equiv of MgCl2 in 6 M Gn·HCl/0.2 M phosphate buffer, pH 5.5 (pH reduction after addition of the MgCl2) at 37 °C affords the best results for this reaction.
Scheme 1. Schematic Representation of Installation and Removal of Succinimide PGs on Cys in Peptides and Proteins.
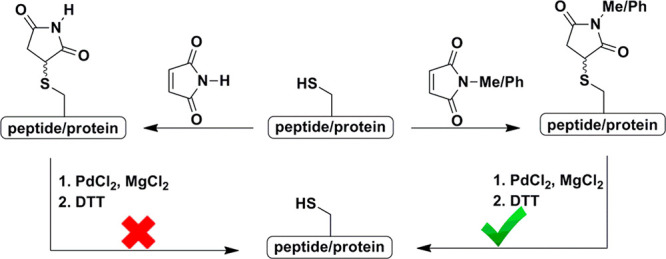
Under our established conditions, we examined the feasibility for the cleavage of additional succinimide derivatives such as the N-phenyl substituted succinimide. We therefore prepared the model peptide 2 (LYRAGC(N-Phe.Suc)LYRAG) and examined its behavior under our optimized conditions (Figure S9B). The decaged peptide was obtained with a complete conversion within 45 min, similarly to peptide 1. In contrast, the unsubstituted succinimide in model peptide 3 (LYRAGC(Suc)LYRAG) was completely stable under Pd cleavage conditions (Figure S9C), which clearly emphasizes the requirement of the phenyl or methyl substitution on the imide group to facilitate cleavage by Pd. Next, to check the generality of this method we have prepared different model peptides (4–6) and globin derivatives (7 and 8), which were exposed to the optimized decaging conditions (Figure S10). The decaging was achieved within 45 min for the model peptides (4–6), 70 min for α-globin (7), and within 2.5 h for β-globin (8). These results suggest that the decaging reaction is applicable for various peptides and proteins. As the succinimide group is easily prone to undergo hydrolysis to give succinamid acid, we examined whether the decaging is happening via succinamid acid formation. For this, model peptide 9 attached to succinamid acid was exposed to our optimized cleavage conditions; however, we did not observe any cleavage reaction (Figure S11B).
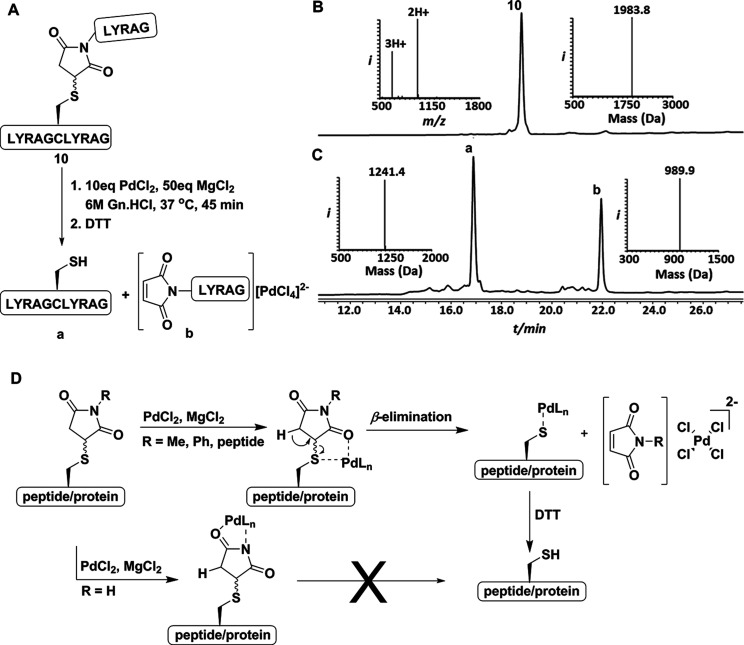
In order to gain insight into the reaction mechanism, we synthesized model peptide 10 (LYRAGC(Suc.LYRAG)LYRAG), where the succinimide group was connected with a short peptide (Figure 1A). Peptide 10 was subjected to our cleavage conditions to give two peptide segments: LYRAGCLYRAG and Mal.LYRAG-[PdCl4]2– complex. Previously it has been shown that the abstraction of the acidic hydrogen from the sulfhydryl side chain of Cys (StBu), Cys(Acm), and Cys (Trt) leads to β-elimination to cleave the thiol bond.23 In a different study, it has been shown that Pd is able to coordinate the free imide in the succinimide based derivative and generate a palladium catalyst for a cross coupling reaction.24 Based on this information and our results, we propose a plausible mechanism for the cleavage of the succinimide group, as depicted in Figure 1D. In the case of N-substituted succinimide, the chelation of the Pd complex between the thiol and carbonyl groups trigger β-elimination to cleave the C–S bond and form the thiol-Pd and maleimide-Pd complexes, respectively. The thiol-Pd complex, upon treatment with DTT liberates the free thiol. In the case of the unsubstituted succinimide, the free imide group is susceptible to chelation with the Pd to form an unproductive complex.
Figure 1.
(A) Cleavage of succinimide PG from peptide 10 under optimized conditions. (B) HPLC-MS analysis of the purified model peptide 10 with an observed mass 1983.8 ± 0.1 Da (calcd 1984.3 Da, average isotopes). (C) Cleavage of model peptide 10 under optimized conditions. Peak a corresponds to the decaged fragment (LYRAGCLYRAG) with an observed mass 1241.4 ± 0.1 Da (calcd 1242.5 Da, average isotopes). Peak b corresponds to the complex of [PdCl4]2– with (Mal.LYRAG) fragment with an observed mass 989.9 ± 0.1 Da (calcd 990.1 Da, average isotopes). (D) A proposed mechanism for the cleavage of the succinimide group.
Palladium Mediated Orthogonal Cleavage of Thiazolidine and Succinimide
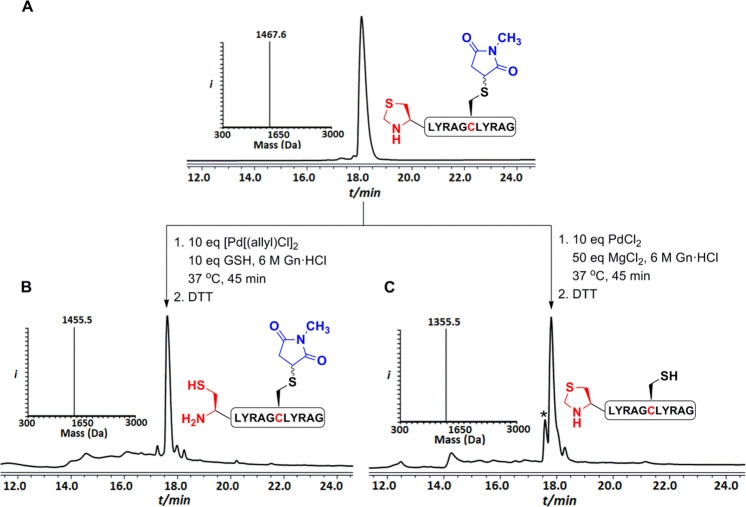
In sequential NCL, masking of the N-terminal Cys residue of a peptide fragment by thiazolidine (Thz) is highly desirable to avoid peptide cyclization or polymerization. The cleavage of Thz serving as a PG or linker was reported by our group under [Pd(allyl)Cl]2 and GSH conditions.18 To examine the orthogonality between the Thz and the succinimide PGs, we synthesized model peptide 11 (Thz-LYRAGC(N-Me.Suc)LYRAG), bearing a N-terminal Thz and methyl succinimide PGs (Figure 2A). We then treated this peptide with [Pd(allyl)Cl]2 and GSH in 6 M Gn·HCl/0.2 M phosphate buffer, pH 7.3 at 37 °C. After 45 min, we observed the full conversion of the Thz to the free N-terminal Cys, while keeping the succinimide PG completely intact (Figure 2B). However, when model peptide 11 was treated under the optimized conditions, we observed unmasking of the succinimide where the Thz remained stable (88%) along with traces of product having both the Thz and succinimide unmasking (12%; Figure 2C). This example demonstrates the influence of the specific Pd complex and additive in achieving selective unmasking of two different PGs under aqueous conditions. Notably, a model peptide bearing both the Acm and the succinimide groups did not exhibit any orthogonality due to structural resemblances.
Figure 2.
Pd-mediated orthogonal cleavage between the Thz and the succinimide PGs. HPLC-MS analysis: (A) Purified model peptide 11 with an observed mass 1467.6 ± 0.1 Da (calcd 1467.7 Da, average isotopes). (B) Crude product after Thz deprotection with an observed mass 1455.5 ± 0.1 Da (calcd 1455.6 Da, average isotopes). (C) Crude product after the succinimide deprotection with an observed mass 1355.5 ± 0.1 Da (calcd 1356.6 Da, average isotopes). * corresponds to the product with the deprotection of both Thz and succinimide PGs.
Succinimide Linker Facilitates SPCL
To date, the majority of ligation schemes in chemical protein synthesis have been conducted in aqueous solutions.12a,25 However, the shortcomings like handling losses, multiple HPLC purifications, and lyophilization steps prolong synthesis time and limit the construction of large proteins. To address these issues, one-pot multisegment condensation methods, aimed at a single purification step of the final product, have been developed.26 Another important approach is to develop different linkers to enable NCL/desulfurization on a solid support.27 The success of SPCL relies on the choice of the polymer support, the linker used, NCL/desulfurization, and cleavage conditions.
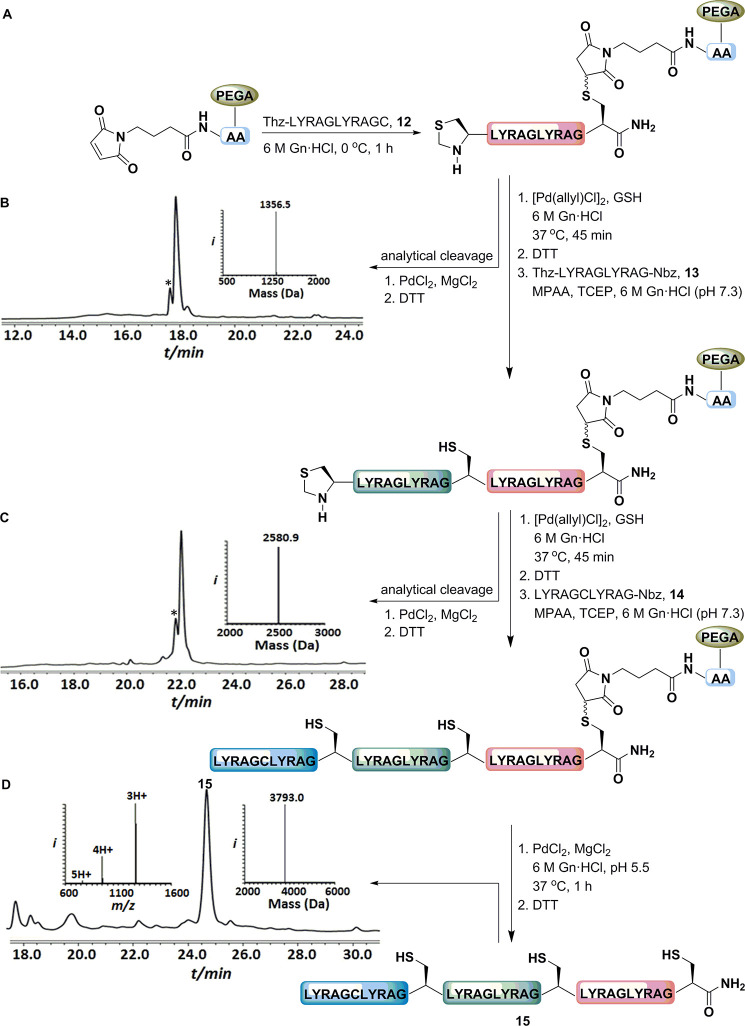
Having established conditions for the selective conversion of the Thz to Cys against the succinimide PG in model peptide 11, we wondered whether the succinimide could be employed to generate a suitable linker for SPCL (Figure 3). We chose the known PEGA resin as a solid support due to its high swelling properties and compatibility in aqueous buffers.28 Initially, the maleimide linker was attached to the PEGA polymer support through a spacer of two alanine residues. To enable SPCL for a model system, three peptide segments 12 (Thz-LYRAGLYRAG-Cys), 13 (Thz-LYRAGLYRAG-Nbz (Nbz is N-acylbenzimidazolinone),29 and 14 (LYRAGCLYRAG-Nbz) were prepared by standard Fmoc-SPPS (Figure S14). In the first step, peptide segment 12 was immobilized to the maleimide bearing solid support via a covalent bond within minutes and subsequently treated with Pd[(allyl)Cl]2 and GSH to convert the Thz to the N-terminal free Cys. After quenching the polymer support with DTT, the first ligation was performed with peptide 13 in the presence of MPAA and TCEP for 5 h. To check the ligation efficiency, small amount of polymer support was treated under succinimide cleavage conditions. As shown in Figure 3C, the succinimide linker was cleaved completely to liberate the ligated product along with traces of loss of the Thz group. Again, the solid support was treated with [Pd(allyl)Cl]2 and GSH to give the N-terminal free Cys for the subsequent ligation with peptide segment 14. In the final step, the solid support was subjected to succinimide cleavage to liberate polypeptide 15. The entire process took ∼13 h for the 6 steps and resulted in 46% isolated yield of product.
Figure 3.
Overview of our SPCL strategy employing succinimide linker. (A) The ligation strategy employing immobilization, elongation and release of the peptide from the solid support. HPLC-MS analysis: (B) Crude product after immobilization with an observed mass of 1356.5 ± 0.1 Da (calcd 1356.6 Da, average isotopes). * corresponds to product of Thz deprotection with an observed mass of 1344.5 ± 0.1 Da (calcd 1344.6 Da, average isotopes). (C) Crude product after first ligation with an observed mass of 2580.9 ± 0.1 Da (calcd 2581.1 Da, average isotopes). * corresponds to product of Thz deprotection with the observed mass of 2568.7 ± 0.1 Da (calcd 2569.1 Da, average isotopes). (D) Crude product after second ligation with an observed mass of 3793.0 ± 0.1 Da (calcd 3794.5 Da, average isotopes).
SPCL Employing Succinimide Linker for the Synthesis of Ub-ABPs
DUBs are known to cleave the isopeptide bond between the C-terminal glycine residue of Ub and the ubiquitinated proteins. DUBs play crucial roles in the ubiquitination machinery and are emerging as attractive drug targets.30 Therefore, various Ub-ABPs have been developed to study the role of DUBs in health and disease.31 Our lab developed a new strategy for the synthesis of Lys-48 and Lys-63 linked di-Ub probes to label different DUBs.32 Similarly, we also reported the synthesis of ubiquitinated α-globin probe33 and ubiquitinated H2A probe15d for labeling USP15 and Calypso/ASX, respectively. Recently, Champak and co-workers reported the semisynthesis of Ub-dehydroalanine (DHA) probe by using selenocysteine as a latent bio-orthogonal electrophile to capture the TRIM-25-associated DUB, ubiquitin-specific protease 15 (USP15).34 The Ovaa group demonstrated that Ub-DHA can be used to capture E1-E2-E3 Ub enzymes.35
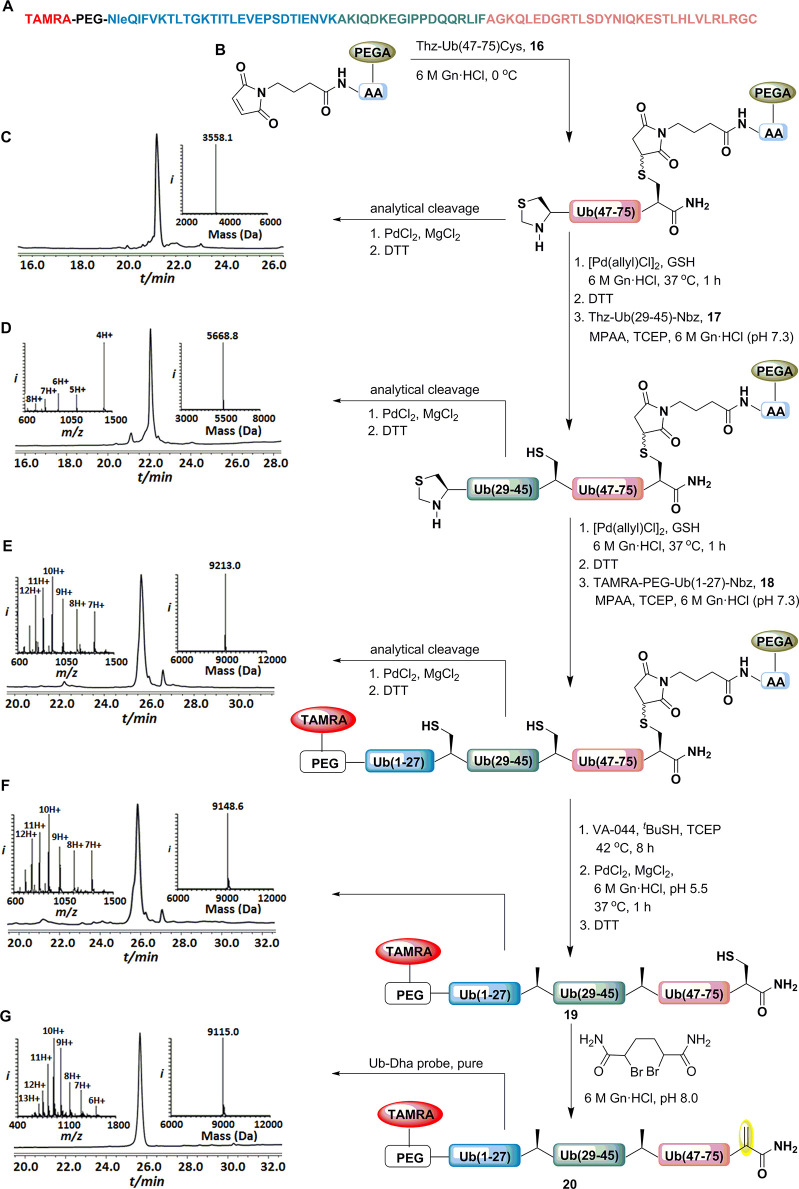
Encouraged by the success of the SPCL in producing model system 15, we sought to apply these conditions for a more complex system such as Ub-DHA, 20 (Figure 4). Our design for the construction of 20 by SPCL is shown in Figure 4. The sequence of Ub was divided into three segments, including 16 (Thz-Ub(47–75)-Cys), 17 (Thz-Ub(29–45)-Nbz), and 18 (TAMRA-PEG-Ub(1–27)-Nbz), where a fluorophore 5-carboxytetramethylrhodamine (TAMRA) was installed at the N-terminal to monitor the DUB capture by using a fluorescent gel (Figure S15). Ala 46 and 28 were temporally replaced with the Thz linkage to enable sequential SPCL. Gly 76 was temporarily mutated to Cys to facilitate the immobilization and late stage DHA formation. With the required segments in hand, the assembly of the polypeptide began with immobilization of peptide segment 16 followed by Thz conversion to the N-terminal free Cys. The first ligation was performed with peptide 17 in the presence of MPAA and TCEP for 8 h, and subsequently the Thz linkage was opened. Similarly, the second ligation was repeated with peptide 18 for 8 h and subsequently subjected to desulfurization conditions. After the desulfurization step, the solid support was treated under succinimide deprotection conditions to liberate polypeptide 19, TAMRA-PEG-UbG76C, which was isolated in 37% overall yield from the 7 steps in under 28 h. Later, the purified polypeptide 19 was treated with bisamide36 to facilitate the formation of Ub-DHA, 20 in 49% isolated yield (Figure S16). Following the same protocol, the assembly of Ubv2.3-DHA probe 23 was obtained in 51% yield (Figure S18). Ubv2.3 is an engineered Ub variant, known to selectively inhibit USP2, which has C-terminal in close proximity to the catalytic active site.37 The obtained Ub-DHA’s were subjected to inductively coupled plasma (ICP) experiment to determine the Pd content, and the amount of Pd detected was less than 0.06%.17
Figure 4.
Succinimide group facilitates the SPCL strategy for assembling Ub-DHA probe 20. (A) The sequence of Ub highlighting the ligation sites and different fragments. (B) The ligation strategy employing immobilization, NCL, desulfurization and release of the polypeptide from the solid support and Ub-DHA formation. HPLC-MS analysis: (C) Crude product after immobilization with an observed mass of 3558.1 ± 0.1 Da (calcd 3558.2 Da, average isotopes). (D) Crude product after first ligation with an observed mass of 5668.8 ± 0.1 Da (calcd 5668.6 Da, average isotopes). (E) Crude product after second ligation with an observed mass of 9213.0 ± 0.1 Da (calcd 9213.9 Da, average isotopes). (F) Crude product 19 after the desulfurization with an observed mass of 9148.6 ± 0.2 Da (calcd 9149.9 Da, average isotopes). (G) Purified Ub-DHA probe 20 with an observed mass of 9115.0 ± 0.1 Da (calcd 9115.9 Da, average isotopes).
With both the probes in hand, we then examined selectivity for labeling USP2 compared to USP7, a member of the USP family with high active site similarity to USP2 (Figure 5). We therefore treated our synthetic probes 20 and 23 with the recombinant USP2 and USP7 DUBs and compared their labeling efficiency by fluorescent gel. The labeling experiments showed that the USP2 reacted with Ubv2.3-DHA (23) three times more efficiently than with Ub-DHA (20; Figure 5B). On the other hand, USP7 almost exclusively labeled Ub-DHA (20) and showed negligible labeling of the Ubv2.3-DHA (23; Figure 5B). We then proceeded to examine this selectivity in a more complex environment by performing the labeling experiment in cellular lysate. For this study, we chose U2OS cell lines, as they are known to express substantial levels of USP7 and have minimal expression of USP2.38 Both synthetic probes 20 and 23 were incubated with the cell lysates with or without addition of recombinant USP2 and both demonstrated the appearance of a new fluorescent band after addition of USP2 corresponding to the covalent DUB-Probe formation (Figure 5C). Ubv2.3-DHA, 23 demonstrated mostly USP2 labeling and increased selectivity when compared to the Ub-DHA, 20. We also observed a side reaction corresponding to the labeling of endogenous USP7 with each probe as was confirmed by Western blotting with anti-USP7 (Figure 5C). Our strategy can substantially simplify the synthesis of existing complex probes such as Ub-like (Ubl), e.g., SUMO and ISG.
Figure 5.
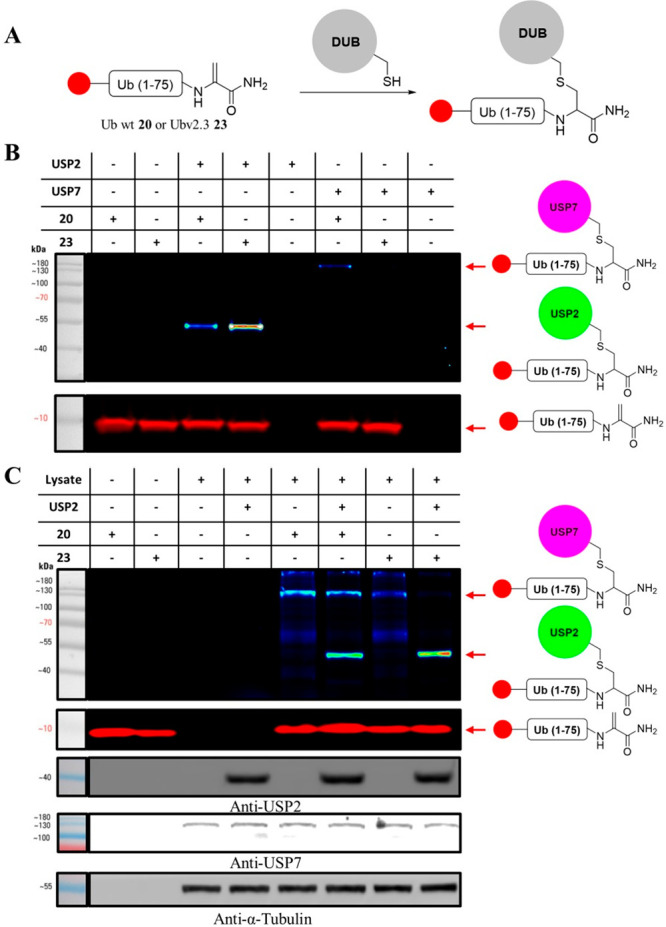
Fluorescent gel of the labeling experiment between two Ub-DHA probes and USP2/USP7. (A) Schematic reaction between Ub-DHA probes and DUBs of the USP family. (B) Fluorescent gel of the reaction between Ub-DHA probes and purified USP2 and USP7. The top part is an intensity heat map based on TAMRA fluorescence demonstrating the formation of covalent DUB-probe adducts. The bottom part is the unmodified probe from the same gel as a control for probe loading. (C) Fluorescent gel and Western blot of the reaction between Ub-DHA probes and USP2 in U2OS cell lysate with and without USP2. The top part is an intensity heat map based on TAMRA fluorescence demonstrating the formation of covalent DUB-probe adducts followed by the fluorescent image of unmodified probes. The bottom is a Western blot against USP2, USP7, and α-tubulin (loading control).
Temporal Protection of Native Cysteines to Enable Selective Desulfurization in Protein Semisynthesis
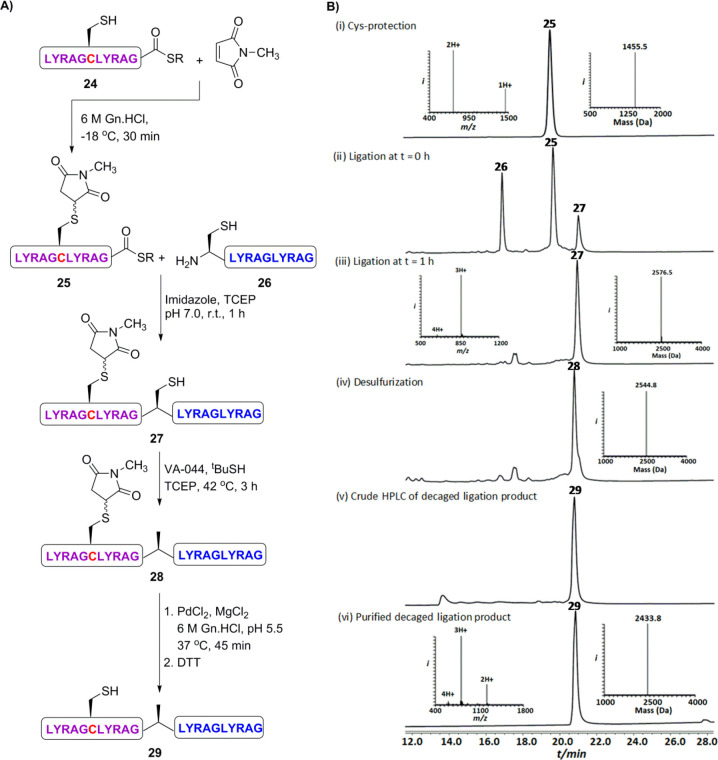
Our next goal was to employ succinimide in protein semisynthesis by using it as a temporal PG for a native Cys residue in a peptide thioester following one-pot NCL/desulfurization and final succinimide removal (Figure 6).39 However, the protection of free Cys in the presence of a thioester could be challenging as it might lead to thioester hydrolysis. To test this, we synthesized two model peptides 25, LYRAGC(N-Me.Mal)LYRAG-MMP (MMP = methyl 3-mercaptopropionate), bearing C-terminal thioester and 26, Cys-LYRAGLYRAG, bearing N-terminal free Cys. Initially, we performed Cys protection in the thioester fragment, 24, at different conditions and found that performing the reaction at −18 °C gave the desired succinimide protected thioester, 25, without any detectable hydrolysis (Figure S19A). These two peptide segments were then ligated to give 27 and desulfurized to give 28 in the presence of imidazole40 as a nonthiol catalyst and VA-044 for radical desulfurization, respectively.41 Similar to the SPCL results, the succinimide group was completely stable under imidazole NCL/desulfurization in solution. In the final step, the succinimide group on the Cys residue of 28 was removed quantitatively under our cleavage conditions to afford polypeptide 29 in 55% isolated yield.
Figure 6.
Examining the use of succinimide protection in solution and its stability during NCL/desulfurization. (A) The ligation strategy employing succinimide protection, one-pot ligation/desulfurization and cleavage the succinimide PG. HPLC-MS analysis: (B) One-pot NCL, desulfurization, and succinimide removal. (i) Purified model peptide 25 with an observed mass of 1455.5 ± 0.1 Da (calcd 1455.5 Da, average isotopes). (ii) Ligation at t = 0 h, (iii) ligation at t = 3 h with an observed mass of 2576.5 ± 0.1 Da (calcd 2576.9 Da, average isotopes). (iv) Desulfurization product 28 with an observed mass of 2544.8 ± 0.1 Da (calcd 2544.9 Da, average isotopes). (v) Crude succinimide cleavage reaction. (vi) Pure polypeptide 29 with an observed mass of 2433.8 ± 0.1 Da (calcd 2433.9 Da, average isotopes).
Next, to apply these conditions for a more complex system such as peptides or proteins containing native Cys residues involved in intramolecular disulfide bonds and to check one-pot succinimide deprotection and disulfide bond formation, we used the oxytocin peptide as a model system (Figure S21). The succinimide protected oxytocin was treated under palladium cleavage conditions and subsequently quenched with DTC for 1 h.16 The reaction afforded the desired disulfide bond formation in a one-pot reaction, highlighting the potential use of the succinimide PG for such an approach.
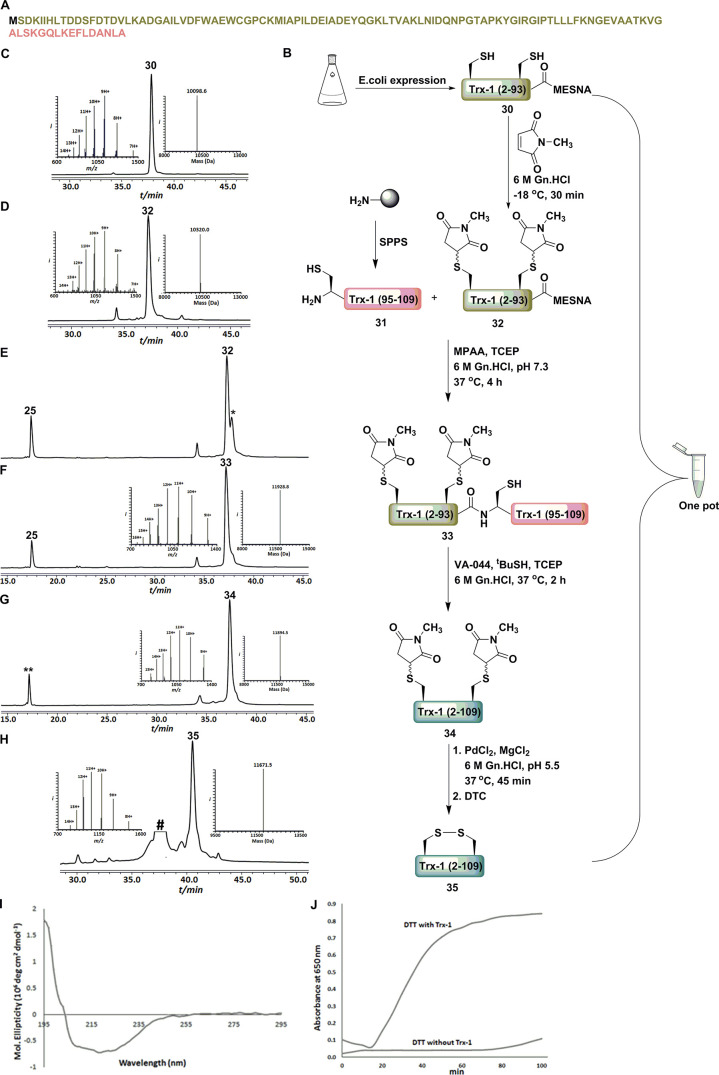
To apply our approach for production of a full semisynthetic protein, we chose E. coli Trx-1 enzyme (Figure 7). Trx-1, a thiol disulfide oxidoreductase which catalyzes the reduction of disulfide bonds in various proteins.42 Our design for the semisynthesis of Trx-1 includes a recombinant fragment 30, Trx-1 (2–93)-MESNA43 (Figure S22) and a synthetic fragment 31, Cys-Trx-1(95–109; Figure S23). The recombinant peptide containing α-thioester can also be prepared by sortase A via an enzymatic approach.44 The recombinant expressed fragment 30 bearing Cys33 and 36 was protected with the succinimide PG to form 32 and consecutively ligated45 with fragment 31. The ligation was completed after 4 h, and product 33 was dialyzed against 6 M Gn·HCl/0.2 M phosphate buffer to facilitate one-pot desulfurization.39 A free radical desulfurization step and dialysis against 6 M Gn·HCl/0.2 M phosphate buffer gave 34. Finally, succinimide was removed from the desulfurized product 34 by Pd treatment and subsequently quenched with DTC for 1 h to form the oxidized Trx-1, 35 in 34% isolated yield. Notably, the entire process of the 5 steps was achieved in one-pot. The semisynthetic Trx-1 exhibited the expected secondary structure46 (Figure 7H) as shown by circular dichroism (CD) and the expected native enzymatic activity using the reported Trx-1 activity assay47 (Figure 7J). The ICP experiment to determine the Pd content of semisynthetic Trx-1 showed ∼0.07% of Pd.
Figure 7.
Examining succinimide protection for a recombinant fragment in aqueous solution and expressed protein ligation (EPL). (A) Trx-1 sequence. (B) The ligation strategy employing one-pot succinimide protection, EPL/desulfurization, succinimide deprotection and disulfide bond formation. HPLC-MS analysis: (C) Trx-1 (2–93) recombinant fragment with an observed mass of 10098.6 ± 0.1 Da (calcd 10100.5 Da, average isotopes). (D) Succinimide protected Trx-1 (2–93) recombinant fragment with an observed mass of 10320.0 ± 0.2 Da (calcd 10322.5 Da, average isotopes). (E) Ligation at t = 0 h. * is a MPAA-peptide related to thioester fragment. (F) Ligation at t = 4 h with an observed mass of 11928.8 ± 0.2 Da (calcd 11929.5 Da, average isotopes). (G) Crude desulfurization product with an observed mass of 11894.5 ± 0.8 Da (calcd 11897.5 Da, average isotopes). ** is an unidentified product related to the Cys fragment. (H) HPLC-MS analysis of the crude disulfide bond formation with the observed mass of 11671.5 ± 0.2 Da (calcd 11673.5 Da, average isotopes). # is a nonpeptidic unidentified product related to DTC. (I) CD spectrum of semisynthesized Trx-1. (J) Semisynthesized Trx-1 activity assay for reduction of bovine insulin with and without addition of DTT. The change in turbidity of the reaction mixture was analyzed by plotting the absorbance at 650 nm versus time.
Conclusions
We have developed for the first time a practical method for the efficient removal of succinimide derivatives by using PdII complexes to enable peptide and protein (semi)synthesis. The synthetic utility of the present method was demonstrated by assembling Ub activity-based probes by SPCL, where the succinimide group was used as a cleavable linker. PdII treatment of the polymer support liberated the polypeptide quantitatively with high yield and purity. In addition, we have shown that the succinimide group can be introduced as a protecting group in aqueous solution, especially for recombinant expressed fragments to enable EPL of complex semisynthetic targets. Further, our present protocol successfully facilitated one-pot succinimide detachment and disulfide bond formation under mild conditions. We envision the use of the succinimide group as an orthogonal protecting group and in cleavable solubilizing tags to facilitate peptide and protein synthesis. Furthermore, the succinimide group might find applications in chemical biology as a cleavable linker for antibody drug conjugates and for proteomics studies.
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant Agreement No. [831783]). A.B. holds The Jordan and Irene Tark Academic Chair. M.H.G. acknowledges NSF-BSF Grant MCB1818280.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c07663.
Discussions of materials and methods used, peptide synthesis, removal conditions for succinimide group, orthogonal removal, solid phase chemical ligation, protection of cysteine for recombinant fragments, disulfide bond formation, HPLC and MS data, CD spectrum, and enzymatic activity assay (PDF)
Author Contributions
† V.G.B. and G.S. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- a Paris C.; Brun O.; Pedroso E.; Grandas A. Exploiting Protected Maleimides to Modify Oligonucleotides, Peptides and Peptide Nucleic Acids. Molecules 2015, 20, 6389–6408. 10.3390/molecules20046389. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ravasco J. M. J. M.; Faustino H.; Trindade A.; Gois P. M. P. Bioconjugation with Maleimides: A Useful Tool for Chemical Biology. Chem. - Eur. J. 2019, 25, 43–59. 10.1002/chem.201803174. [DOI] [PubMed] [Google Scholar]
- a Renault K.; Fredy J. W.; Renard P.-Y.; Sabot C. Covalent Modification of Biomolecules through Maleimide-Based Labeling Strategies. Bioconjugate Chem. 2018, 29, 2497–2513. 10.1021/acs.bioconjchem.8b00252. [DOI] [PubMed] [Google Scholar]; b Lee T. C.; Moran C. R.; Cistrone P. A.; Dawson P. E.; Deniz A. A. Site-Specific Three-Color Labeling of α-Synuclein via Conjugation to Uniquely Reactive Cysteines during Assembly by Native Chemical Ligation. Cell Chem. Biol. 2018, 25, 797–801. 10.1016/j.chembiol.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N.; Smith S. W.; Ghone S.; Tomczuk B. Current ADC Linker Chemistry. Pharm. Res. 2015, 32, 3526–3540. 10.1007/s11095-015-1657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjula B. N.; Tsai A.; Upadhya R.; Perumalsamy K.; Smith P. K.; Malavalli A.; Vandegriff K.; Winslow R. M.; Intaglietta M.; Prabhakaran M.; Friedman J. M.; Acharya A. S. Site-Specific PEGylation of Hemoglobin at Cys-93(β): Correlation between the Colligative Properties of the PEGylated Protein and the Length of the Conjugated PEG Chain. Bioconjugate Chem. 2003, 14, 464–472. 10.1021/bc0200733. [DOI] [PubMed] [Google Scholar]
- Mukhortava A.; Schlierf M. Efficient Formation of Site-Specific Protein-DNA Hybrids Using Copper-Free Click Chemistry. Bioconjugate Chem. 2016, 27, 1559–1563. 10.1021/acs.bioconjchem.6b00120. [DOI] [PubMed] [Google Scholar]
- a Elduque X.; Pedroso E.; Grandas A. Orthogonal Protection of Peptides and Peptoids for Cyclization by the Thiol-Ene Reaction and Conjugation. J. Org. Chem. 2014, 79, 2843–2853. 10.1021/jo500427c. [DOI] [PubMed] [Google Scholar]; b Montgomery J. E.; Donnelly J. A.; Fanning S. W.; Speltz T. E.; Shangguan X.; Coukos J. S.; Greene G. L.; Moellering R. E. Versatile Peptide Macrocyclization with Diels-Alder Cycloadditions. J. Am. Chem. Soc. 2019, 141, 16374–16381. 10.1021/jacs.9b07578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Shen B.-Q.; Xu K.; Liu L.; Raab H.; Bhakta S.; Kenrick M.; Parsons-Reponte K. L; Tien J.; Yu S.-F.; Mai E.; Li D.; Tibbitts J.; Baudys J.; Saad O. M; Scales S. J; McDonald P. J; Hass P. E; Eigenbrot C.; Nguyen T.; Solis W. A; Fuji R. N; Flagella K. M; Patel D.; Spencer S. D; Khawli L. A; Ebens A.; Wong W. L.; Vandlen R.; Kaur S.; Sliwkowski M. X; Scheller R. H; Polakis P.; Junutula J. R Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat. Nat. Biotechnol. 2012, 30, 184–189. 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]; b Baldwin A. D.; Kiick K. L. Tunable degradation of maleimide-thiol adducts in reducing environments. Bioconjugate Chem. 2011, 22, 1946–1953. 10.1021/bc200148v. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Lyon R. P.; Setter J. R.; Bovee T. D.; Doronina S. O.; Hunter J. H.; Anderson M. E.; Balasubramanian C. L.; Duniho S. M.; Leiske C. I.; Li F.; Senter P. D. Self-hydrolyzing maleimides improve the stability and pharmacological properties of antibody-drug conjugates. Nat. Biotechnol. 2014, 32, 1059–1062. 10.1038/nbt.2968. [DOI] [PubMed] [Google Scholar]
- a Kalia D.; Malekar P. V.; Parthasarathy M. Exocyclic Olefinic Maleimides: Synthesis and Application for Stable and Thiol-Selective Bioconjugation. Angew. Chem., Int. Ed. 2016, 55, 1432–1435. 10.1002/anie.201508118. [DOI] [PubMed] [Google Scholar]; b Szijj P. A.; Bahou C.; Chudasama V. Minireview: Addressing the retro-Michael instability of maleimide bioconjugates. Drug Discovery Today: Technol. 2018, 30, 27–34. 10.1016/j.ddtec.2018.07.002. [DOI] [PubMed] [Google Scholar]
- a Dubowchik G. M.; Firestone R. A.; Padilla L.; Willner D.; Hofstead S. J.; Mosure K.; Knipe J. O.; Lasch S. J.; Trail P. A. Cathepsin B-Labile Dipeptide Linkers for Lysosomal Release of Doxorubicin From Internalizing Immunoconjugates: Model Studies of Enzymatic Drug Release and Antigen-Specific in vitro Anticancer Activity. Bioconjugate Chem. 2002, 13, 855–869. 10.1021/bc025536j. [DOI] [PubMed] [Google Scholar]; b Burke P. J.; Senter P. D.; Meyer D. W.; Miyamoto J. B.; Anderson M.; Toki B. E.; Manikumar G.; Wani M. C.; Kroll D. J.; Jeffrey S. C. Design, Synthesis, and Biological Evaluation of Antibody-Drug Conjugates Comprised of Potent Camptothecin Analogues. Bioconjugate Chem. 2009, 20, 1242–1250. 10.1021/bc9001097. [DOI] [PubMed] [Google Scholar]
- Rossin R.; Versteegen R. M.; Wu J.; Khasanov A.; Wessels H. J.; Steenbergen E. J.; ten Hoeve W.; Janssen H. M.; van Onzen A. H. A. M.; Hudson P. J.; Robillard M. S. Chemically triggered drug release from an antibody-drug conjugate leads to potent antitumor activity in mice. Nat. Commun. 2018, 9, 1484. 10.1038/s41467-018-03880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhang Y.; Zhou X.; Xie Y.; Greenberg M. M.; Xi Z.; Zhou C. Thiol Specific and Tracelessly Removable Bioconjugation via Michael Addition to 5-Methylene Pyrrolones. J. Am. Chem. Soc. 2017, 139, 6146–6151. 10.1021/jacs.7b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nathani R. I.; Chudasama V.; Ryan C. P.; Moody P. R.; Morgan R. E.; Fitzmaurice R. J.; Smith M. E. B.; Baker J. R.; Caddick S. Reversible protein affinity-labelling using bromomaleimide-based reagents. Org. Biomol. Chem. 2013, 11, 2408–2411. 10.1039/c3ob40239h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c McConnell E. W.; Smythers A. L.; Hicks L. M. Maleimide-Based Chemical Proteomics for Quantitative Analysis of Cysteine Reactivity. J. Am. Soc. Mass Spectrom. 2020, 31, 1697–1705. 10.1021/jasms.0c00116. [DOI] [PubMed] [Google Scholar]
- a Bondalapati S.; Jbara M.; Brik A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat. Chem. 2016, 8, 407–418. 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]; b Agouridas V.; El Mahdi O.; Diemer V.; Cargoët M.; Monbaliu J. -C. M.; Melnyk O. Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chem. Rev. 2019, 119, 7328–7443. 10.1021/acs.chemrev.8b00712. [DOI] [PubMed] [Google Scholar]; c Conibear A.; Watson E. E.; Payne R. J.; Becker C. F. W. Native Chemical Ligation in Protein Synthesis and Semi-Synthesis. Chem. Soc. Rev. 2018, 47, 9046–9068. 10.1039/C8CS00573G. [DOI] [PubMed] [Google Scholar]
- a Kumar K. S. A.; Spasser L.; Ohayon S.; Erlich L. A.; Brik A. Expeditious Chemical Synthesis of Ubiquitinated Peptides Employing Orthogonal Protection and Native Chemical Ligation. Bioconjugate Chem. 2011, 22, 137–143. 10.1021/bc1004735. [DOI] [PubMed] [Google Scholar]; b Siman P.; Karthikeyan S. V.; Nikolov M.; Fischle W.; Brik A. Convergent Chemical Synthesis of Histone H2B Protein Enabled Site Specific Ubiquitination at Lys34. Angew. Chem., Int. Ed. 2013, 52, 8059–8063. 10.1002/anie.201303844. [DOI] [PubMed] [Google Scholar]; c Tang S.; Si Y.-Y.; Wang Z.-P.; Mei K.-R.; Chen X.; Cheng J.-Y.; Zheng J. S.; Liu L. An Efficient One-Pot Four-Segment Condensation Method for Protein Chemical Synthesis. Angew. Chem., Int. Ed. 2015, 54, 5713–5717. 10.1002/anie.201500051. [DOI] [PubMed] [Google Scholar]
- Jbara M.; Maity S. K.; Brik A. Palladium in the Chemical Synthesis and Modification of Proteins. Angew. Chem., Int. Ed. 2017, 56, 10644–10655. 10.1002/anie.201702370. [DOI] [PubMed] [Google Scholar]
- a Jbara M.; Maity S. K.; Seenaiah M.; Brik A. Palladium Mediated Rapid Deprotection of N-Terminal Cysteine under Native Chemical Ligation Conditions for the Efficient Preparation of Synthetically Challenging Proteins. J. Am. Chem. Soc. 2016, 138, 5069–5075. 10.1021/jacs.5b13580. [DOI] [PubMed] [Google Scholar]; b Maity S. K.; Jbara M.; Laps S.; Brik A. Efficient Palladium-Assisted One-Pot Deprotection of (Acetamidomethyl) Cysteine Following Native Chemical Ligation and/or Desulfurization To Expedite Chemical Protein Synthesis. Angew. Chem., Int. Ed. 2016, 55, 8108–8112. 10.1002/anie.201603169. [DOI] [PubMed] [Google Scholar]; c Jbara M.; Eid E.; Brik A. Palladium mediated deallylation in fully aqueous conditions for native chemical ligation at aspartic and glutamic acid sites. Org. Biomol. Chem. 2018, 16, 4061–4064. 10.1039/C8OB00890F. [DOI] [PubMed] [Google Scholar]; d Jbara M.; Laps S.; Morgan M.; Kamnesky G.; Mann G.; Wolberger C.; Brik A. Palladium prompted on-demand cysteine chemistry for the synthesis of challenging and uniquely modified proteins. Nat. Commun. 2018, 9, 3154. 10.1038/s41467-018-05628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laps S.; Sun H.; Kamnesky G.; Brik A. Palladium-Mediated Direct Disulfide Bond Formation in Proteins Containing S-Acetamidomethyl-cysteine under Aqueous Conditions. Angew. Chem., Int. Ed. 2019, 58, 5729–5733. 10.1002/anie.201900988. [DOI] [PubMed] [Google Scholar]
- Maity S. K.; Mann G.; Jbara M.; Laps S.; Kamnesky G.; Brik A. Palladium-Assisted Removal of a Solubilizing Tag from a Cys Side Chain To Facilitate Peptide and Protein Synthesis. Org. Lett. 2016, 18, 3026–3029. 10.1021/acs.orglett.6b01442. [DOI] [PubMed] [Google Scholar]
- a Jbara M.; Laps S.; Maity S. K.; Brik A. Palladium-Assisted Cleavage of Peptides and Proteins Containing a Backbone with Thiazolidine Linkage. Chem. - Eur. J. 2016, 22, 14851–14855. 10.1002/chem.201603676. [DOI] [PubMed] [Google Scholar]; b Mann G.; Satish G.; Meledin R.; Vamisetti G. B.; Brik A. Palladium-Mediated Cleavage of Proteins with Thiazolidine-Modified Backbone in Live Cells. Angew. Chem., Int. Ed. 2019, 58, 13540–13549. 10.1002/anie.201906545. [DOI] [PubMed] [Google Scholar]
- a Katayama H.; Hojo H. The phenacyl group as an efficient thiol protecting group in a peptide condensation reaction by the thioester method. Org. Biomol. Chem. 2013, 11, 4405–4413. 10.1039/c3ob40644j. [DOI] [PubMed] [Google Scholar]; b Kawakami T.; Yoshikawa R.; Fujiyoshi Y.; Mishima Y.; Hojo H.; Tajima S.; Suetake I. Synthesis of histone proteins by CPE ligation using a recombinant peptide as the C-terminal building block. J. Biochem. 2015, 158, 403–411. 10.1093/jb/mvv056. [DOI] [PubMed] [Google Scholar]; c Matveenko M.; Hackl S.; Becker C. F. W. Utility of the Phenacyl Protecting Group in Traceless Protein Semisynthesis through Ligation-Desulfurization Chemistry. ChemistryOpen 2018, 7, 106–110. 10.1002/open.201700180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Mochizuki M.; Hibino H.; Nishiuchi Y. Postsynthetic Modification of Unprotected Peptides via S-Tritylation Reaction. Org. Lett. 2014, 16, 5740–5743. 10.1021/ol502773v. [DOI] [PubMed] [Google Scholar]; b Mochizuki M.; Tsuda S.; Tanimura K.; Nishiuchi Y. Regioselective Formation of Multiple Disulfide Bonds with the Aid of Postsynthetic S-Tritylation. Org. Lett. 2015, 17, 2202–2205. 10.1021/acs.orglett.5b00786. [DOI] [PubMed] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Floyd N.; Bernardes G. J. L.; Davis B. G. Allyl Sulfides Are Privileged Substrates in Aqueous Cross-Metathesis: Application to Site-Selective Protein Modification. J. Am. Chem. Soc. 2008, 130, 9642–9643. 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- Lassahn P.-G.; Lozan V.; Janiak C. Palladium (II) salts containing [PdCl4]2- and [Pd2Cl6]2- ions as pre-catalysts for the vinyl-polymerization of norbornene-evidence for the in situ formation of PdCl2 as the active species. Dalton Trans. 2003, 927–935. 10.1039/b209633a. [DOI] [Google Scholar]
- Yang Y.β-Elimination Side Reactions; Academic Press: San Diego, CA, 2016; p 33042. [Google Scholar]
- Fairlamb I. J. S.; Kapdi A. R.; Lynam J. M.; Taylor R. J. K.; Whitwood A. C. Bis(triphenylphosphine)palladium(II)succinimide as a precatalyst for Suzuki cross-coupling subtle effects exerted by the succinimide ligand. Tetrahedron 2004, 60, 5711–5718. 10.1016/j.tet.2004.05.018. [DOI] [Google Scholar]
- a Dawson P. E.; Muir T. W.; Clark-Lewis I.; Kent S. B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]; b Malins L. R.; Payne R. J. Recent extensions to native chemical ligation for the chemical synthesis of peptides and proteins. Curr. Opin. Chem. Biol. 2014, 22, 70–78. 10.1016/j.cbpa.2014.09.021. [DOI] [PubMed] [Google Scholar]
- Zuo C.; Zhang B.; Yan B.; Zheng J.-S. One-pot multi-segment condensation strategies for chemical protein synthesis. Org. Biomol. Chem. 2019, 17, 727–744. 10.1039/C8OB02610F. [DOI] [PubMed] [Google Scholar]
- a Canne L. E.; Botti P.; Simon R. J.; Chen Y.; Dennis E. A.; Kent S. B. H. Chemical Protein Synthesis by Solid Phase Ligation of Unprotected Peptide Segments. J. Am. Chem. Soc. 1999, 121, 8720–8727. 10.1021/ja9836287. [DOI] [Google Scholar]; b Ollivier N.; Desmet R.; Drobecq H.; Blanpain A.; Boll E.; Leclercq B.; Mougel A.; Vicogne J.; Melnyk O. A simple and traceless solid phase method simplifies the assembly of large peptides and the access to challenging proteins. Chem. Sci. 2017, 8, 5362–5370. 10.1039/C7SC01912B. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yu R. R.; Mahto S. K.; Justus K.; Alexander M. M.; Howard C. J.; Ottesen J. J. Hybrid phase ligation for efficient synthesis of histone proteins. Org. Biomol. Chem. 2016, 14, 2603–2607. 10.1039/C5OB02195B. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Loibl S. F.; Harpaz Z.; Zitterbart R.; Seitz O. Total chemical synthesis of proteins without HPLC purification. Chem. Sci. 2016, 7, 6753–6759. 10.1039/C6SC01883A. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Jbara M.; Seenaiah M.; Brik A. Solid phase chemical ligation employing a rink amide linker for the synthesis of histone H2B protein. Chem. Commun. 2014, 50, 12534–12537. 10.1039/C4CC06499B. [DOI] [PubMed] [Google Scholar]; f Reimann O.; Smet-Nocca C.; Hackenberger C. P. R. Traceless Purification and Desulfurization of Tau Protein Ligation Products. Angew. Chem., Int. Ed. 2015, 54, 306–310. 10.1002/anie.201408674. [DOI] [PubMed] [Google Scholar]; g Galibert M.; Piller V.; Piller F.; Aucagne V.; Delmas A. F. Combining triazole ligation and enzymatic glycosylation on solid phase simplifies the synthesis of very long glycoprotein analogues. Chem. Sci. 2015, 6, 3617–3623. 10.1039/C5SC00773A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal M. Pega: A flow stable polyethylene glycol dimethyl acrylamide copolymer for solid phase synthesis. Tetrahedron Lett. 1992, 33, 3077–3080. 10.1016/S0040-4039(00)79604-3. [DOI] [Google Scholar]
- Blanco-Canosa J. B.; Nardone B.; Albericio F.; Dawson P. E. Chemical Protein Synthesis Using a Second-Generation N-Acylurea Linker for the Preparation of Peptide-Thioester Precursors. J. Am. Chem. Soc. 2015, 137, 7197–7209. 10.1021/jacs.5b03504. [DOI] [PubMed] [Google Scholar]
- a Reyes-Turcu F. E.; Wilkinson K. D. Polyubiquitin Binding and Disassembly By Deubiquitinating Enzymes. Chem. Rev. 2009, 109, 1495–1508. 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Pfoh R.; Lacdao I. K.; Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr.-Relat. Cancer 2015, 22, T35–T54. 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ekkebus R.; Flierman D.; Geurink P. P.; Ovaa H. Catching a DUB in the act: novel ubiquitin-based active site directed probes. Curr. Opin. Chem. Biol. 2014, 23, 63–70. 10.1016/j.cbpa.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hewings D. S.; Flygare J. A.; Wertz I. E.; Bogyo M. Activity-based probes for the multicatalytic proteasome. FEBS J. 2017, 284, 1540–1554. 10.1111/febs.14016. [DOI] [PubMed] [Google Scholar]; c Gopinath P.; Ohayon S.; Nawatha M.; Brik A. Chemical and semisynthetic approaches to study and target deubiquitinases. Chem. Soc. Rev. 2016, 45, 4171–4198. 10.1039/C6CS00083E. [DOI] [PubMed] [Google Scholar]
- Haj-Yahya N.; Hemantha H. P.; Meledin R.; Bondalapati S.; Seenaiah M.; Brik A. Dehydroalanine-Based Diubiquitin Activity Probes. Org. Lett. 2014, 16, 540–543. 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- Meledin R.; Mali S. M.; Kleifeld O.; Brik A. Activity-Based Probes Developed by Applying a Sequential Dehydroalanine Formation Strategy to Expressed Proteins Reveal A Potential α-Globin-Modulating Deubiquitinase. Angew. Chem., Int. Ed. 2018, 57, 5645–5649. 10.1002/anie.201800032. [DOI] [PubMed] [Google Scholar]
- Whedon S. D.; Markandeya N.; Rana A. S. J. B.; Senger N. A.; Weller C. E.; Turecek F.; Strieter E. R.; Chatterjee C. Selenocysteine as a Latent Bioorthogonal Electrophilic Probe for Deubiquitylating Enzymes. J. Am. Chem. Soc. 2016, 138, 13774–13777. 10.1021/jacs.6b05688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M. P C; Witting K.; Berlin I.; Pruneda J. N; Wu K.-P.; Chang J.-G.; Merkx R.; Bialas J.; Groettrup M.; Vertegaal A. C O; Schulman B. A; Komander D.; Neefjes J.; El Oualid F.; Ovaa H. A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes. Nat. Chem. Biol. 2016, 12, 523–530. 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker J. M.; Gunnoo S. B.; Boutureira O.; Gerstberger S. C.; Fernandez-Gonzalez M.; Bernardes G. J. L.; Griffin L.; Hailu H.; Schofield C. J.; Davis B. G. Methods for converting cysteine to dehydroalanine on peptides and proteins. Chem. Sci. 2011, 2, 1666–1676. 10.1039/c1sc00185j. [DOI] [Google Scholar]
- Ernst A.; Avvakumov G.; Tong J.; Fan Y.; Zhao Y.; Alberts P.; Persaud A.; Walker J. R.; Neculai A.-M.; Neculai D.; Vorobyov A.; Garg P.; Beatty L.; Chan P.-K.; Juang Y.-C.; Landry M.-C.; Yeh C.; Zeqiraj E.; Karamboulas K.; Allali-Hassani A.; Vedadi M.; Tyers M.; Moffat J.; Sicheri F.; Pelletier L.; Durocher D.; Raught B.; Rotin D.; Yang J.; Moran M. F.; Dhe-Paganon S.; Sidhu S. S. A Strategy for Modulation of Enzymes in the Ubiquitin System. Science 2013, 339, 590–595. 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M.; Fagerberg L.; Hallström B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjöstedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P. H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Pontén F. Tissue-based map of the human proteome. Science 2015, 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Moyal T.; Hemantha H. P.; Siman P.; Refua M.; Brik A. Highly efficient one-pot ligation and desulfurization. Chem. Sci. 2013, 4, 2496–2501. 10.1039/c3sc50239b. [DOI] [Google Scholar]
- Sakamoto K.; Tsuda S.; Mochizuki M.; Nohara Y.; Nishio H.; Yoshiya T. Imidazole-Aided Native Chemical Ligation: Imidazole as a One-Pot Desulfurization-Amenable Non-Thiol-Type Alternative to 4-Mercaptophenylacetic Acid. Chem. - Eur. J. 2016, 22, 17940–17944. 10.1002/chem.201604320. [DOI] [PubMed] [Google Scholar]
- Wan Q.; Danishefsky S. J. Free-Radical-Based, Specific Desulfurization of Cysteine: A Powerful Advance in the Synthesis of Polypeptides and Glycopolypeptides. Angew. Chem., Int. Ed. 2007, 46, 9248–9252. 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- Ezraty B.; Gennaris A.; Barras F.; Collet J. F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385. 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- pTXB1 vector (Catalog#N6707S, New England Biolabs).
- Ling J. J.; Policarpo R. L.; Rabideau A. E.; Liao X.; Pentelute B. L. Protein Thioester Synthesis Enabled by Sortase. J. Am. Chem. Soc. 2012, 134, 10749–10752. 10.1021/ja302354v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir T. W. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003, 72, 249–289. 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Reutimann H.; Straub B.; Luisi P. L.; Holmgren A. A conformational study of thioredoxin and its tryptic fragments. J. Biol. Chem. 1981, 256, 6796–6803. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979, 254, 9627–9632. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.