Abstract
Metabolomics plays a pivotal role in systems biology, and NMR is a central tool with high precision and exceptional resolution of chemical information. Most NMR metabolomic studies are based on 1H 1D spectroscopy, severely limited by peak overlap. 13C NMR benefits from a larger signal dispersion but is barely used in metabolomics due to ca. 6000-fold lower sensitivity. We introduce a new approach, based on hyperpolarized 13C NMR at natural abundance, that circumvents this limitation. A new untargeted NMR-based metabolomic workflow based on dissolution dynamic nuclear polarization (d-DNP) for the first time enabled hyperpolarized natural abundance 13C metabolomics. Statistical analysis of resulting hyperpolarized 13C data distinguishes two groups of plant (tomato) extracts and highlights biomarkers, in full agreement with previous results on the same biological model. We also optimize parameters of the semiautomated d-DNP system suitable for high-throughput studies.
Metabolomic studies in systems biology elucidate the impacts of genetic alteration, disease, and environmental factors in broad fields such as precision medicine,1 food,2 plant,3,4 or environmental sciences.5 Untargeted metabolomics is of particular importance since it identifies crucial biomarkers without any a priori knowledge of the corresponding biological condition.6 Mass spectrometry (MS) is widely used for untargeted analysis since it benefits from a much better sensitivity, typically 3 orders of magnitude, than NMR.7 However, NMR can outperform MS in terms of reproducibility and repeatability needed to identify, with the highest achievable precision, the relative variations between metabolite concentrations from subtly distinct sample groups.8 NMR metabolomics is therefore widely used for metabolomics applications across clinical and preclinical studies,9,10 microbiology,11 forensics,12 and food2 and plant sciences.3,4 The vast majority of NMR metabolomics studies rely on one-dimensional (1D) 1H spectra, as it is by far the most sensitive NMR approach.3 However, 1H NMR suffers from limited spectral resolution, often obscuring otherwise valuable data. 13C NMR offers an appealing alternative thanks to broad 13C chemical-shift dispersion (∼200 vs ∼10 ppm for 1H) combined with relatively narrow spectral lines.11 Moreover, 13C is present in all metabolites (1.1% nat. ab.) with corresponding spectral diversity to identify, distinguish, and structurally characterize biomarkers. These advantages are long recognized13,14 but thus far limited in application due to a roughly 5880-fold sensitivity disadvantage of 13C (owing to its low natural abundance and gyromagnetic ratio) versus 1H. Selective improvement of sensitivity with 13C-enriched tracers15 yields ∼100 times better sensitivity but limits focus to specific metabolic pathways. Furthermore, resulting spectra can be complicated by undesired 13C–13C couplings (JCC).
Hyperpolarization methods16,17 can solve these issues by providing nonequilibrium 13C polarization at greater than 10 000 times sensitivity gain for natural abundance carbon sites. Dissolution-dynamic nuclear polarization (d-DNP)18−21 using 1H DNP combined with 1H → 13C cross-polarization (CP)22,23 is particularly promising owing to its ability to polarize 13C spins in a nonselective fashion.24 d-DNP hyperpolarizes solutes in frozen samples by transferring spin polarization from electrons of cosolvated radicals to nuclear spins. DNP occurs at cryogenic temperatures (1–4 K), followed by rapid dissolution with hot solvent and transfer of the hyperpolarized sample to an adjacent liquid-state, room-temperature NMR spectrometer. Dissolution and transfer do introduce some selectivity via variable losses from 13C spin relaxation.18−21
The greater than 10 000 times enhancement from d-DNP was first demonstrated in 200318 and has evolved into a general and widely used hyperpolarization method in bioanalytical chemistry.20 Nonetheless, as yet, there are only a limited number of applications to metabolomics.25−27 Lerche et al. introduced a d-DNP 13C approach to quantify metabolites in cancer cell extracts incubated with 13C-enriched glucose.25,26 While this took advantage of improved sensitivity from d-DNP and 13C enrichment, this focused approach used isotopically labeled feed material. Untargeted, natural abundance methods remain unexplored by d-DNP.
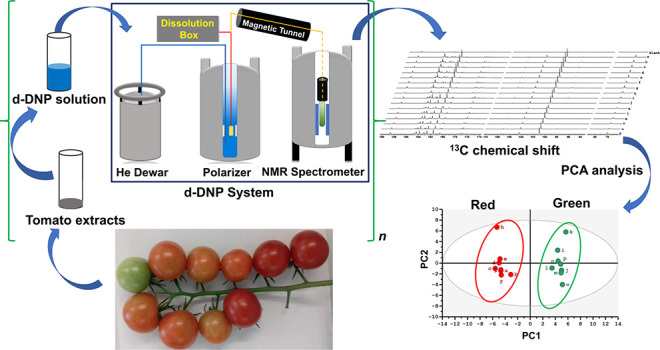
In 2015, we reported a preliminary investigation of d-DNP assisted by cross-polarization (CP) for 13C NMR of natural abundance metabolic extracts.28 This enabled single-scan hyperpolarized 13C NMR on quaternary carbons in about 30 min per sample. In contrast, classical high-field NMR without hyperpolarization required ∼12 h to produce 13C spectra with significantly sparser peaks. In a later report, repeatability better than 4% (an essential condition for metabolomics) was demonstrated on a home-built d-DNP polarizer.29 Here, we introduce a complete untargeted metabolomics approach for d-DNP NMR with real bioextracts at natural 13C abundance. It consists of sample preparation, d-DNP hyperpolarization and conversion to a solution state, data processing, and statistical analysis, ultimately highlighting the variability between sample groups and identifying putative biomarkers. The test system consists of two groups of tomato fruit extracts at ripening stages of mature–green and red–ripe. For each stage, eight extracts were prepared from a single pooled sample of fruit powder, and their pH was adjusted. Thus, concentration, pH, and spin relaxation are uniform within one of the two groups, and intragroup observational differences highlight analytical variability only.
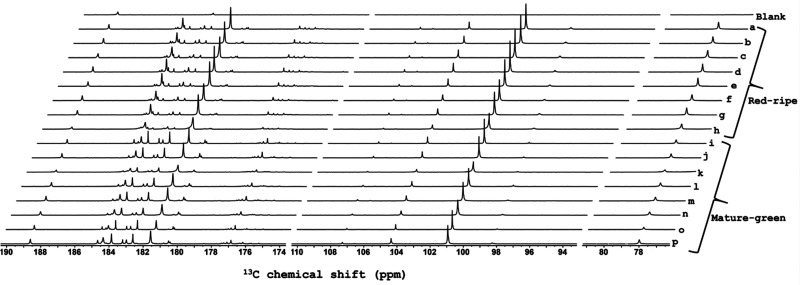
Samples were dissolved and flash-frozen in a glass-forming DNP solution with TEMPOL as a polarizing agent (see SI), then polarized for 11 min at 1.18 K with frequency-modulated30 microwave irradiation and microwave-gated31 cross-polarization (Figure S1). Hyperpolarized liquid-state spectra were acquired 12.8 s after initiation of dissolution, allowing time for fluid transfer and stabilization to a 400 MHz spectrometer equipped with a cryogenic probe. Figure S2a gives the pulse sequence for 13C acquisition in a liquid state. Figure 1 shows resulting spectra, focused on the quaternary carbon regions, the hyperpolarization of which most readily survives the sample transfer due to relatively long T1(13C). General application remains promising as most metabolites contain at least one quaternary carbon. Faster transfer times (<2 s) have been demonstrated that can enable extension to arbitrary carbon sites (e.g., methyl carbons).32,33 Nonetheless, quaternary sites are quite sufficient to define small variations among biological states. On the basis of the reasonable assumption that the individual T1 values are the same in all samples, hyperpolarized signals reflect the variations of analyte concentration between samples, but absolute concentration is inaccessible. Note that, even hyperpolarized, 13C sensitivity at natural abundance is insufficient to quantify enhancements (i.e., the thermal signals are too weak) or to estimate T1 values. Nonetheless, we typically observe 30,000 times more concentrated or 13C-enriched samples, and T1 ∼ 50–60 s for quaternary sites.
Figure 1.
Stacked plot of d-DNP-enhanced single-scan 13C NMR spectra of 16 tomato samples. At right, labels “a” to “h” refer to spectra of red–ripe fruit extracts, and “i” to “p” refer to mature–green fruit extracts. Selections from 76–82, 94–110, and 174–190 ppm are shown to focus on spectrally populated regions (full spectrum in Figure S7). The sample for the blank control spectrum (top) has signals only from Na-TSP-d4 (188.6 ppm) and EDTA·Na2·2H2O (182.9 ppm).
The overall experiment duration (30 min from sample loading, through hyperpolarization, dissolution, and observation) is fully compatible with high-throughput metabolomics. In addition, the 13C-hyperpolarized spectra can be processed through automated spectra and data analysis methods for conventional metabolomics. Spectra were processed as described in the SI, then divided into user-defined spectral regions (“buckets”) (Figure S3), whose integral values were normalized with respect to that of sodium 3-trimethylsilylpropionate-d4 (Na-TSP-d4) (internal reference) at 188.6 ppm and to the weight of the fruit tissue powder. Then, as in standard NMR metabolomics, principal component analysis (PCA) was applied to the normalized set of 13C integrals from each sample in order to distinguish biological variability.34,35 Relevant details of the widely accepted PCA method are given in the SI. Figure 2 shows the resulting scores plot, where each point represents a single sample, the coordinates of which are determined by the collective contributions of all corresponding bucket integrals (weighed with the PCA “loadings” values). Point proximity indicates sample similarity, whereas group separation shows clear discrimination along the first-principal component (PC1) for mature–green and red–ripe tomato fruit extracts. This reflects the robustness of the entire analytical workflow, including the complexities of d-DNP, since biological variability within each of the groups is not expected, as all red–ripe fruit samples were prepared from the same source and likewise for mature–green. The unexpected intragroup deviations (termed as “analytical” variability) of samples “h” (red) and “k” (green) along the PC2 axis results from the two times poorer line shape of spectra “h” and “k” (Figure 1). Still, the deviations on PC2, that makes a relatively modest 20.0% contribution to total variance, are not critical, as PC1 is the dominant component (explaining 60.5% of total variance).
Figure 2.
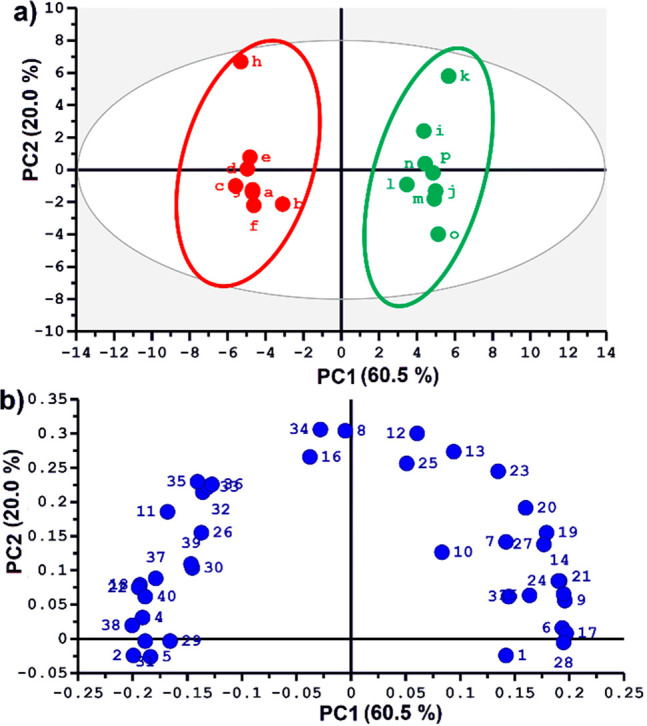
(a) Scores plot and (b) loadings plot of the principal component analysis (PCA) obtained from 40 spectral buckets from hyperpolarized 13C spectra of 16 tomato fruit extracts. Integrals were normalized to Na-TSP-d4 as an internal reference and to the weight of the sample used for extraction. Mean centering and unit variance scaling were used in PCA. Correspondence of bucket numbers to spectral regions is given in Figure S3.
To identify the key metabolites that differentiate the ripening stages, the PCA scores plot (Figure 2a) was compared to the loadings plot (Figure 2b), which scores relative contributions of individual spectral buckets to PC1 and PC2 constructions. Key metabolites were identified thanks to previously acquired spectra of individual commercial metabolites.
Resulting assignment and red/green fruit association of metabolites are given in Table S1. This succeeds in revealing expected patterns, such as higher content of some amino acids (aspartic acid, asparagine, glutamine, leucine, and threonine) and malic acid in tomato green fruit extracts, whereas sugars (sucrose and fructose), citric acid, and glutamic acid have higher contents in tomato red fruit extracts. These are in global agreement with known features of tomato metabolism36 and for the first time shows the ability of hyperpolarized 13C metabolomics to provide reliable biological discrimination. Furthermore, univariate analysis of the discriminant buckets (box plots in Figure S4, Kruskal–Wallis, p < 0.05) highlights the high quality of the discrimination. Overall, our approach establishes a complete, robust workflow for hyperpolarized, 13C NMR-based metabolomic studies. Although the many assigned signals offer special benefits for metabolite identification, unidentified buckets also highlight a new promise of the hyperpolarized approach. Corresponding signals would not be observable by a thermally polarized approach, even with hours of signal-averaged acquisition.18 Thus, d-DNP-enhanced NMR opens a window to previously inaccessible metabolites. Future avenues to assign such signals may utilize the combination of d-DNP with heteronuclear ultrafast 2D NMR.37
In contrast to prior hyperpolarized 13C mixture analysis proof-of-concept papers,28,29 our workflow utilized a more streamlined d-DNP system, where a single person can perform the key dissolution steps. This, along with high robustness and repeatable usage (for the present 16 samples, plus several tens of experiments performed before and since), is in favor of a broader, general use of our method. The robustness and repeatability are impressive given the variety of (reusable) sample-handling components required for a d-DNP process involving steps at less than 1.5 K and room temperature and a superheated solvent to drive conversion and transfer between these conditions.
Key experimental optimizations enabled such performance. The first consisted of optimizing the line shape of hyperpolarized 13C signals to ensure well-resolved signals from 13C metabolites. We found that microbubbles from rapid (1–2 s) injection of dissolution output to a 5 mm NMR tube were responsible for initial broad, asymmetric line shapes. In order to avoid bubbles, we decreased the surface tension on the walls of the NMR tubes by precleaning with an alkaline solution (Hellmanex). Figure S5 shows the improvement of line shape and spectral resolution after this treatment. This also translates to 3 times better 13C sensitivity (Figure S6), since it enables a much shorter postdissolution delay to reach settled conditions and optimal S/N that balances lost 13C hyperpolarization during the transfer against initial intensity build-up as spectral lines narrow with homogenization (e.g., bubble dissipation) in the NMR sample.
Analytical metabolomics demands that variability is dominated by biological inputs, with insignificant (or small) contributions from the experimental method. Using the above treatment and timing optimizations, we demonstrate the needed precision in Figure 3. Progressive improvements in repeatability are shown for a series in which postdissolution timing of NMR observation was separately optimized for three protocols: A (ethanol-cleaned NMR tube, with 22.8 s delay to acquisition), B (Hellmanex treatment, with 12.8 s delay), and C (case B plus normalization of integrals according to the Na-TSP-d4 internal reference). Hellmanex alone provided 3%–5% drops in CV among metabolites in the sample (pyruvate, acetate, and alanine), and the internal reference yielded further 3%–9% drops for ultimate CV values of 2.0%, 4.8%, and 4.1% for the respective compounds. The improvements reflect both more reliable attainment of suitable conditions for solution-state NMR and the natural benefits of noted 3 times higher S/N.
Figure 3.
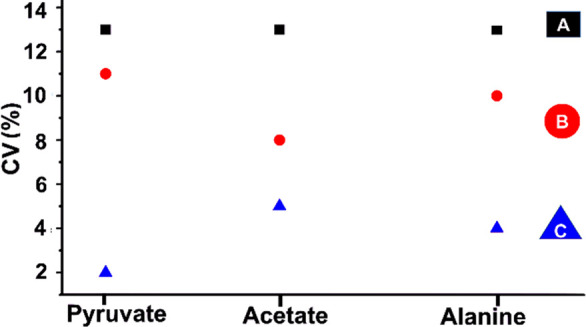
Coefficient of variation (CV (%)) of d-DNP enhanced 13C signal integrals (based on five successive experiments) for a standard metabolite mixture (pyruvate, acetate, alanine) at natural abundance using three experimental protocols (A, B, C). Solution-state spectra were detected with a 90° flip angle, after a postdissolution delay that ensured an optimum signal-to-noise ratio (S/N) for each case A, B, and C. CV at “A” was obtained using ethanol-washed NMR tubes, yielding optimum S/N at 22.8 s after dissolution. CV at “B” was obtained using NMR tubes additionally treated with Hellmanex (procedure in SI), yielding S/N optimum at 12.8 s after dissolution. CV at “C” was achieved under similar conditions as “B” but normalizing all signal integrals using the Na-TSP-d4 internal reference as described in the text.
In summary, we demonstrate that hyperpolarized, natural abundance 13C NMR is suitable for analytical metabolomics on relevant biological samples. This greatly expands the impact of NMR in the metabolomic field. This hyperpolarized 13C approach is complementary to valuable and more established 1H NMR methods for metabolomics. Indeed, 1H methods can be limited by spectral resolution and sensitivity issues, sometimes failing to clarify biochemical fingerprints or to pinpoint metabolic markers of functional states. For 13C NMR, only hyperpolarization can practically open a new window to the field. We have proven the needed precision, accuracy, and throughput using the sophisticated, but ultimately straightforward, approach of d-DNP. This success motivates both the broader adoption of natural abundance 13C metabolomics with application to challenging biological problems and the development of d-DNP instrumentation for the full community. Our work highlights development areas to further extend the value of NMR in metabolomics, including reduction of the time from dissolution to NMR observation to less than 5 s. Correspondingly lower hyperpolarization losses should enable analytical 1H metabolomics, as well as access to greater chemical functionality with natural abundance 13C. On the technical side, progresses are expected to make the experiment much less expensive, in particular, through the design of cryogen consumption-free systems that will undoubtedly broaden the application scope of hyperpolarized metabolomics.38,39
Acknowledgments
The authors acknowledge funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC Grant Agreements n° 814747/SUMMIT, DINAMIX n° 801774, n° 714519/HP4all, HYPROTIN n° 801936), support from Bruker BioSpin, the Corsaire metabolomics platform, and from MetaboHUB (ANR-11-INBS-0010). The authors acknowledge Isabelle Atienza for taking care of the tomato plants and kind assistance during d-DNP experiments from Corentin Jacquemmoz, Virginie Silvestre, Achille Marchand, and Ritu Raj Mishra.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c03510.
Experimental details, procedure of Hellmanex treatment, and figures and tables (PDF)
The authors declare the following competing financial interest(s): D.E., M.S., R.M., and J.G.K. are employees of Bruker Biospin, which supplied the d-DNP polarizer. It is not a commercial instrument but a step in ongoing Bruker R&D.
Supplementary Material
References
- Puchades-Carrasco L.; Pineda-Lucena A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17 (24), 2740–2751. 10.2174/1568026617666170707120034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakis E. A.; Cagliani L. R.; Polissiou M. G.; Consonni R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by 1H NMR metabolite fingerprinting. Food Chem. 2015, 173, 890–896. 10.1016/j.foodchem.2014.10.107. [DOI] [PubMed] [Google Scholar]
- Freitas D. d. S.; Carlos E. F.; Gil M. C. S. d. S.; Vieira L. G. E.; Alcantara G. B. NMR-Based Metabolomic Analysis of Huanglongbing-Asymptomatic and -Symptomatic Citrus Trees. J. Agric. Food Chem. 2015, 63, 7582–7588. 10.1021/acs.jafc.5b03598. [DOI] [PubMed] [Google Scholar]
- Allwood J. W.; De Vos R. C.; Moing A.; Deborde C.; Erban A.; Kopka J.; Goodacre R.; Hall R. D. Plant Metabolomics And Its Potential for Systems Biology Research Background Concepts, Technology, and Methodology. Methods Enzymol. 2011, 500, 299–336. 10.1016/B978-0-12-385118-5.00016-5. [DOI] [PubMed] [Google Scholar]
- Viant M. R. Applications of Metabolomics to the Environmental Sciences. Metabolomics 2009, 5, 1–2. 10.1007/s11306-009-0157-3. [DOI] [Google Scholar]
- Gertsman I.; Barshop B. A. Promises and Pitfalls of Untargeted Metabolomics. J. Inherited Metab. Dis. 2018, 41, 355–366. 10.1007/s10545-017-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.-L.; Zhang A.-H.; Kong L.; Wang X.-J. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018, 8, 22335–22350. 10.1039/C8RA01574K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol K.; Brüschweiler R. Two Elephants in the Room: New Hybrid Nuclear Magnetic Resonance and Mass Spectrometry Approaches for Metabolomics. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 471–477. 10.1097/MCO.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. R.; Kwon H. N.; Nam H.; Kim J. J.; Park S.; Kim Y.-H. Urine-NMR Metabolomics for Screening of Advanced Colorectal Adenoma and Early Stage Colorectal Cancer. Sci. Rep. 2019, 9, 1–10. 10.1038/s41598-019-41216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Calabrò A.; De Carli V.; Luchinat C.; Nepi S.; Porfirio B.; Renzi D.; Saccenti E.; Tenori L. The Metabonomic Signature of Celiac Disease. Journal of Proteome Research. 2009, 8, 170–177. 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- Palama T. L.; Canard I.; Rautureau G. J. P.; Mirande C.; Chatellier S.; Elena-Herrmann B. Identification of Bacterial Species by Untargeted NMR Spectroscopy of the Exo-metabolome. Analyst 2016, 141, 4558–4561. 10.1039/C6AN00393A. [DOI] [PubMed] [Google Scholar]
- Castillo-Peinado L. S.; Luque de Castro M. D. Present and Foreseeable Future of Metabolomics in Forensic Analysis. Anal. Chim. Acta 2016, 925, 1–15. 10.1016/j.aca.2016.04.040. [DOI] [PubMed] [Google Scholar]
- Clendinen C. S.; Lee-McMullen B.; Williams C. M.; Stupp G. S.; Vandenborne K.; Hahn D. A.; Walter G. A.; Edison A. S. 13C NMR Metabolomics: Applications at Natural Abundance. Anal. Chem. 2014, 86, 9242–9250. 10.1021/ac502346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayrit F. M.; Dios A. C. d. 1H and 13C NMR for the Profiling of Natural Product Extracts: Theory and Applications. Intech 2017, 81–103. 10.5772/intechopen.71040. [DOI] [Google Scholar]
- Clendinen C. S.; Stupp G. S.; Ajredini R.; Lee-McMullen B.; Beecher C.; Edison A. S. An overview of methods using (13)C for improved compound identification in metabolomics and natural products. Front. Plant Sci. 2015, 6, 1–13. 10.3389/fpls.2015.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtunov K. V.; Pokochueva E. V.; Salnikov O. G.; Cousin S. F.; Kurzbach D.; Vuichoud B.; Jannin S.; Chekmenev E. Y.; Goodson B. M.; Barskiy D. A.; Koptyug I. V. Hyperpolarized NMR Spectroscopy: d-DNP, PHIP, and SABRE Techniques. Chem. - Asian J. 2018, 13, 1857–1871. 10.1002/asia.201800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou P.; Goodson B. M.; Chekmenev E. Y. NMR Hyperpolarization Techniques for Biomedicine. Chem. - Eur. J. 2015, 21, 3156–3166. 10.1002/chem.201405253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardenkjær-Larsen J. H.; Fridlund B.; Gram A.; Hansson G.; Hansson L.; Lerche M. H.; Servin R.; Thaning M.; Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (18), 10158–10163. 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plainchont B.; Berruyer P.; Dumez J.-N.; Jannin S.; Giraudeau P. Dynamic Nuclear Polarization Opens New Perspectives for NMR Spectroscopy in Analytical Chemistry. Anal. Chem. 2018, 90, 3639–3650. 10.1021/acs.analchem.7b05236. [DOI] [PubMed] [Google Scholar]
- Jannin S.; Dumez J.-N.; Giraudeau P.; Kurzbach D. Application and methodology of dissolution dynamic nuclear polarization in physical, chemical and biological contexts. J. Magn. Reson. 2019, 305, 41–50. 10.1016/j.jmr.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardenkjær-Larsen J. H. On the present and future of dissolution-DNP. J. Magn. Reson. 2016, 264, 3–12. 10.1016/j.jmr.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Bornet A.; Melzi R.; Perez Linde A. J.; Hautle P.; van den Brandt B.; Jannin S.; Bodenhausen G. Boosting Dissolution Dynamic Nuclear Polarization by Cross Polarization. J. Phys. Chem. Lett. 2013, 4, 111–114. 10.1021/jz301781t. [DOI] [PubMed] [Google Scholar]
- Jannin S.; Bornet A.; Melzi R.; Bodenhausen G. High field dynamic nuclear polarization at 6.7 T: Carbon-13 polarization above 70% within 20 min. Chem. Phys. Lett. 2012, 549, 99–102. 10.1016/j.cplett.2012.08.017. [DOI] [Google Scholar]
- Vuichoud B.; Milani J.; Bornet A.; Melzi R.; Jannin S.; Bodenhausen G. Hyperpolarization of Deuterated Metabolites via Remote Cross-Polarization and Dissolution Dynamic Nuclear Polarization. J. Phys. Chem. B 2014, 118, 1411–1415. 10.1021/jp4118776. [DOI] [PubMed] [Google Scholar]
- Lerche M. H.; Yigit D.; Frahm A. B.; Ardenkjær-Larsen J. H.; Malinowski R. M.; Jensen P. R. Stable Isotope-Resolved Analysis with Quantitative Dissolution Dynamic Nuclear Polarization. Anal. Chem. 2018, 90, 674–678. 10.1021/acs.analchem.7b02779. [DOI] [PubMed] [Google Scholar]
- Frahm A. B.; Jensen P. R.; Ardenkjær-Larsen J. H.; Yigit D.; Lerche M. H. Stable isotope resolved metabolomics classification of prostate cancer cells using hyperpolarized NMR data. J. Magn. Reson. 2020, 316, 106750. 10.1016/j.jmr.2020.106750. [DOI] [PubMed] [Google Scholar]
- Zhang G.; Ahola S.; Lerche M. H.; Telkki V.-V.; Hilty C. Identification of Intracellular and Extracellular Metabolites in Cancer Cells Using 13C Hyperpolarized Ultrafast Laplace NMR. Anal. Chem. 2018, 90, 11131–11137. 10.1021/acs.analchem.8b03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumez J.-N.; Milani J.; Vuichoud B.; Bornet A.; Lalande-Martin J.; Tea I.; Yon M.; Maucourt M.; Deborde C.; Moing A.; Frydman L.; Bodenhausen G.; Jannin S.; Giraudeau P. Hyperpolarized NMR of plant and cancer cell extracts at natural abundance. Analyst 2015, 140, 5860–5863. 10.1039/C5AN01203A. [DOI] [PubMed] [Google Scholar]
- Bornet A.; Maucourt M.; Deborde C.; Jacob D.; Milani J.; Vuichoud B.; Ji X.; Dumez J. N.; Moing A.; Bodenhausen G.; Jannin S.; Giraudeau P. Highly Repeatable Dissolution Dynamic Nuclear Polarization for Heteronuclear NMR Metabolomics. Anal. Chem. 2016, 88, 6179–6183. 10.1021/acs.analchem.6b01094. [DOI] [PubMed] [Google Scholar]
- Bornet A.; Milani J.; Vuichoud B.; Perez Linde A. J.; Bodenhausen G.; Jannin S. Microwave frequency modulation to enhance Dissolution Dynamic Nuclear Polarization. Chem. Phys. Lett. 2014, 602, 63–67. 10.1016/j.cplett.2014.04.013. [DOI] [Google Scholar]
- Bornet A.; Pinon A.; Jhajharia A.; Baudin M.; Ji X.; Emsley L.; Bodenhausen G.; Ardenkjær-Larsen J. H.; Jannin S. Microwave-gated dynamic nuclear polarization. Phys. Chem. Chem. Phys. 2016, 18, 30530–30535. 10.1039/C6CP05587G. [DOI] [PubMed] [Google Scholar]
- Bowen S.; Hilty C. Rapid sample injection for hyperpolarized NMR spectroscopy. Rapid sample injection for hyperpolarized NMR spectroscopy. Phys. Chem. Chem. Phys. 2010, 12, 5766–5770. 10.1039/c002316g. [DOI] [PubMed] [Google Scholar]
- Chen H. Y.; Hilty C. Implementation and characterization of flow injection in dissolution dynamic nuclear polarization NMR spectroscopy. ChemPhysChem 2015, 16, 2646–2652. 10.1002/cphc.201500292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova R.; Brown T. R. NMR spectral quantitation by principal component analysis. NMR Biomed. 2001, 14, 271–277. 10.1002/nbm.700. [DOI] [PubMed] [Google Scholar]
- Stoyanova R.; Brown T. R. NMR spectral quantitation by principal component analysis. III. A Generalized Procedure for Determination of Lineshape Variations. J. Magn. Reson. 2002, 154, 163–75. 10.1006/jmre.2001.2486. [DOI] [PubMed] [Google Scholar]
- Lemaire-Chamley M.; Mounet F.; Deborde C.; Maucourt M.; Jacob D.; Moing A. NMR-Based Tissular and Developmental Metabolomics of Tomato Fruit. Metabolites 2019, 9, 93. 10.3390/metabo9050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudeau P.; Shrot Y.; Frydman L. Multiple Ultrafast, Broadband 2D NMR Spectra of Hyperpolarized Natural Products. J. Am. Chem. Soc. 2009, 131, 13902–13903. 10.1021/ja905096f. [DOI] [PubMed] [Google Scholar]
- Baudin M.; Vuichoud B.; Bornet A.; Bodenhausen G.; Jannin S. A cryogen-consumption-free system for dynamic nuclear polarization at 9.4 T. J. Magn. Reson. 2018, 294, 115–121. 10.1016/j.jmr.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Ardenkjær-Larsen J. H.; Bowen S.; Petersen J. R.; Rybalko O.; Vinding M. S.; Ullisch M.; Nielsen N. C. Cryogen-free dissolution dynamic nuclear polarization polarizer operating at 3.35 T, 6.70 T, and 10.1 T. Magn. Reson. Med. 2019, 81, 2184–2194. 10.1002/mrm.27537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.