Abstract

Plant peptide protease inhibitors are important molecules in seed storage metabolism and to fight insect pests. Commonly they contain multiple disulfide bonds and are exceptionally stable molecules. In this study, a novel peptide protease inhibitor from beetroot (Beta vulgaris) termed bevuTI-I was isolated, and its primary structure was determined via mass spectrometry-based amino acid sequencing. By sequence homology analysis a few peptides with high similarity to bevuTI-I, also known as the Mirabilis jalapa trypsin inhibitor subfamily of knottin-type protease inhibitors, were discovered. Hence, we assessed bevuTI-I for inhibitory activity toward trypsin (IC50 = 471 nM) and human prolyl oligopeptidase (IC50 = 11 μM), which is an emerging drug target for neurodegenerative and inflammatory disorders. Interestingly, using a customized bioinformatics approach, bevuTI-I was found to be the missing link to annotate 243 novel sequences of M. jalapa trypsin inhibitor-like peptides. According to their phylogenetic distribution they appear to be common in several plant families. Therefore, the presented approach and our results may help to discover and classify other plant-derived cystine knot peptides, a class of plant molecules that play important functions in plant physiology and are currently being explored as lead molecules and scaffolds in drug development.
Plants have evolved a sophisticated chemical arsenal to defend themselves against exogenous threats such as microbiota and herbivores. One successful strategy is the production of plant defense peptides.1 A variety of different classes of plant defense peptides have been described in the literature, such as defensins, lipid transfer proteins, snakins, and cyclotides.2 Among this diverse family of peptides, members forming three disulfide bonds in a knotted arrangement3,4 have gained special interest as potential lead candidates for the discovery and design of peptide-based therapeutics.5,6 They are characterized by high stability in acidic conditions and are resistant to heat-induced and proteolytic degradation.7 Presumably, this stability confers protection against degradation in the gastrointestinal tract when administered orally as drugs.
The concept of utilizing nature-derived peptides in drug discovery has successfully been initiated several decades ago with the discovery of the angiotensin converting enzyme inhibitor captopril, which was developed from the venom of the pit viper Bothrops jararaca in the 1970s.8 Over the years, this discovery has led to several venom-based and nature-derived peptide drugs available as therapeutics and in preclinical/clinical development.9,10
Plants are another rich source of peptide protease inhibitors. They protect plants against pests by inactivating digestive enzymes of, for example, plant-feeding insects, and they regulate breakdown/metabolism of storage proteins.11 These peptides are widely distributed in the plant kingdom, and at least nine plant peptide protease inhibitor families have been reported including Bowman–Birk, bifunctional trypsin/α-amylase, Mirabilis jalapa trypsin, mustard-type, potato type-I and -II, metallocarboxypeptidase, squash, and cyclotide inhibitors.12 Peptide protease inhibitors often contain cysteine-stabilizing motifs (for example a cystine knot), and hence, they are considered as ideal scaffolds for designing novel peptide-based drug candidates.12 Despite many efforts in the discovery of plant protease inhibitors,12 there is still a limited understanding of how certain protease inhibitors are distributed within the plant kingdom. Because it is difficult to define a generalized protocol for characterization and nomenclature of novel peptides, there is inconsistency in their classification and trivial names. The first step in utilizing the full pharmaceutical potential of these peptides requires a discovery process that involves chemical extraction and isolation, structural characterization combined with bioinformatics analysis, as well as peptide synthesis, and enzymatic bioassays. This systematic approach will aid cataloging these interesting peptides more precisely accounting for unique classification based on sequence homology, cysteine spacing patterns, and biological activity. Here we report the isolation and characterization of a novel cysteine-rich peptide protease inhibitor containing three disulfide bonds from beetroot (Beta vulgaris subsp. vulgaris; Amaranthaceae), termed Beta vulgaris trypsin inhibitor I (bevuTI-I). Utilizing sequence homology analysis and genome mining, bevuTI-I appeared to be a missing link for the annotation and description of at least 243 putative novel peptides from 42 plant families. Since bevuTI-I inhibited the enzymatic activity of prolyl oligopeptidase (POP), our results open the door for the discovery and development of peptide-based protease inhibitors.
Results and Discussion
In this study, cysteine-rich peptides from the well-known vegetable plant beetroot were analyzed. While studying the chemical extracts of leaves and pulp for the presence of three disulfide-containing peptides, a new serine protease inhibitor from B. vulgaris, bevuTI-I, was isolated, and its amino acid structure was determined by mass spectrometry. In-depth sequence homology analysis and genome mining allowed the classification of bevuTI-I and led to the annotation of at least 243 novel putative members of the Mirabilis jalapa trypsin inhibitor family. To confirm the endogenous peptide structure, bevuTI-I was produced by solid phase peptide synthesis and oxidative folding, and its pharmacological properties as an enzyme inhibitor of trypsin and human POP were assessed.
Screening for Cysteine-Rich Peptides in Beetroot
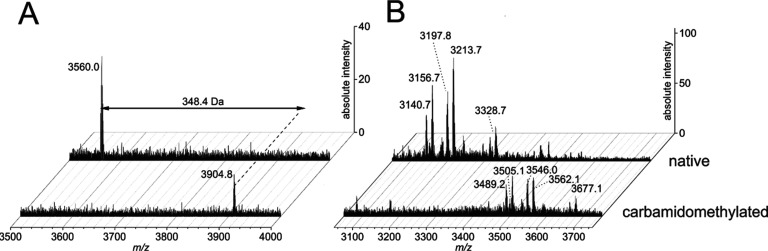
Plant-derived protease inhibitors often have high expression levels in plant seeds, fruit bodies, or kernels. Especially, the Bowman–Birk-, the Kunitz-, or the squash-type inhibitors contribute to the defense repertoire of plants against herbivores.12 Peptide-based protease inhibitors have secondary structure motifs for stabilization and to maintain their function. Often, they contain post-translational modifications such as a disulfide bond framework. For example, prototypic molecules of the Bowman–Birk family comprise seven disulfide bonds, the mustard-type TI four, the α-amylase-trypsin bifunctional inhibitors three to four, and the squash-TI, cyclotides, and potato metallocarboxypeptidase-type inhibitors three disulfide bonds.12 Knowing that beetroot and related species express peptides potentially involved in seed storage metabolism and insect defense,13,14 beetroot samples were analyzed for the presence of cysteine-containing peptides. For chemical extraction of beetroot plant material, 570 g of dried pulp and 270 g of dried leaves were used, and large-scale extraction of beetroot juice was performed with 5 L. Approximately 1–5 mg of the extracted and lyophilized powder was used for peptide screening experiments. Disulfide bonds were chemically reduced to cysteines using DTT (dithiothreitol) and S-carbamidomethylated with iodoacetamide to characterize the cysteine content by mass spectrometry. The chemical derivatization approach revealed the presence of one peptide containing three disulfide bonds in the pulp (Figure 1A) and at least five peptides containing three disulfide bonds in leaf samples (Figure 1B) based on an observed mass shift of +348.4 ± 0.4 Da, corresponding to six carbamidomethylated cysteines. Owing to its role as an important vegetable and food product ingredient, the peptide identified in pulp was isolated.
Figure 1.
Mass spectrometry analysis of beetroot extracts. MALDI-TOF spectra of the crude extract (upper chromatograms) obtained after overnight extraction of beetroot pulp (A) and leaves (B) indicated the presence of one or several peptides containing six cysteine moieties, respectively, based on characteristic mass shifts of +348.4 ± 0.4 Da observed after reductive carbamidomethylation (lower chromatograms). All labeled m/z ions refer to monoisotopic [M + H]+ ions. Postacquisition processing of spectra was performed using baseline asymmetric least-squares filter (asymmetric factor: 0.001, threshold: 0.05, smoothing factor: 4, number of iterations: 10).
We initially applied an extraction protocol that used overnight extraction in CH2Cl2 and MeOH followed by liquid–liquid phase separation after the addition of water. This extraction protocol is widely used for the isolation of cyclotides.15 Hydrophobic molecules such as chlorophyll are partitioned into the lipophilic CH2Cl2 phase, whereas more hydrophilic peptides are enriched in the aqueous phase. As beetroots contain a relatively high amount of sugar (∼6.8 g/100 g according to the USDA food database16), which will also partition into the aqueous phase, the standard extraction protocol was changed to acetone precipitation of concentrated beetroot juice. This did not affect the peptide content of the extract, as demonstrated by comparative mass spectra of beetroot crude extract prepared with the standard protocol and with the modified extraction protocol (Figure S1, Supporting Information). Owing to the importance of beetroot, e.g., as a commercially available juice, the aim was to isolate the major peptide, termed bevuTI-I, with a molecular weight of 3560.0 Da.
Purification and de Novo Sequencing of BevuTI-I
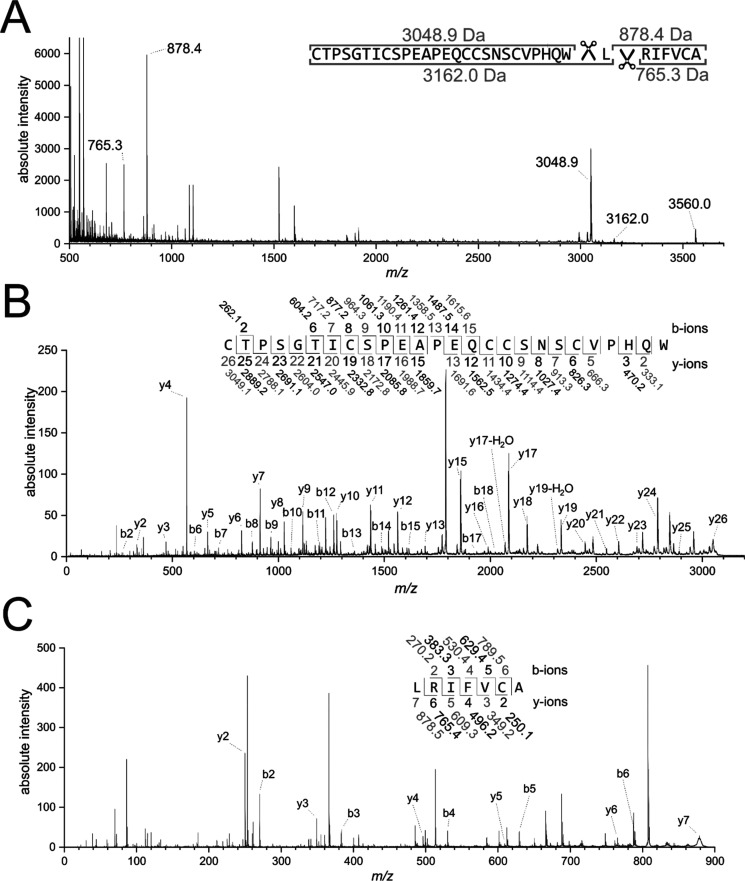
RP-HPLC was used for purification of bevuTI-I in order to determine its amino acid sequence. The pulp extract was refined by C18 solid-phase extraction and was purified with one run of preparative HPLC followed by one run of semipreparative HPLC. After derivatization of cysteines (as described above) a site-specific proteolytic cleavage of the full-length peptide was performed. To obtain the full amino acid sequence, samples were enzymatically processed with chymotrypsin (Figure 2), trypsin (Figure S2, Supporting Information), and endoproteinase GluC (Figure S3, Supporting Information), which were subsequently analyzed by MALDI-TOF/TOF MS. The chymotryptic digest yielded two cleavage product ions, m/z 3048 and 878 (Figure 2A), which in sum added up to the molecular weight of the precursor (plus addition of 18 Da corresponding to hydrolysis), indicating that bevuTI-I is a linear peptide. The proteolytic peptide products as well as the full-length peptide were subjected to postsource decay fragmentation experiments in MALDI-TOF/TOF. The obtained fragmentation spectra were manually annotated for characteristic b- and y-ion series as well as for diagnostic neutral loss ions (loss of amino groups, dehydration, etc.). Annotation of peptide fragments obtained by chymotrypsin (Figure 2B), trypsin (Figure S2, Supporting Information), and endoproteinase GluC (Figure S3, Supporting Information) digestion and of the full-length peptide (Figure S4, Supporting Information) allowed determination of the amino acid sequence of the novel peptide bevuTI-I as NH2-CTPSGTICSPEAPEQCCSNSCVPHQWLRIFVCA-COOH. A comparison of computed and measured b- and y-ions was performed as quality control (Tables S1–S4, Supporting Information). The isobaric residue Ile/Leu at position 27 was assigned as Leu based on the cleavage specificity of chymotrypsin at this residue.17 Chymotrypsin hydrolyses amide bonds in peptides containing, among others, leucine at the P1 position, but not isoleucine. Therefore, the analysis of chymotrypsin peptide fragments allows interpretation of the positions of Leu vs Ile. Assignment of Ile/Leu at positions 7 and 29 of the peptide is based on homology analysis and functional characterization described in the following paragraphs.
Figure 2.
De novo sequencing of bevuTI-I. (A) MALDI-TOF spectrum of the carbamidomethylated full-length bevuTI-I peptide after chymotryptic proteolysis. In total, four fragments were detected indicating cleavage of the peptide at two distinct positions. (B) Annotated MS/MS spectrum of the m/z 3162.0 and (C) the m/z 878.4 precursor ions. The b- and y-ions are indicated by sequence number and m/z (monoisotopic [M + H]+). Annotated MS/MS spectra of bevuTI-I as well as of fragments obtained by digestion with trypsin and endoproteinase GluC are shown in Figures S2–4 (Supporting Information).
Mining, Sequence Homology Analysis, and Phylogenetic Distribution of BevuTI-I-like Peptides
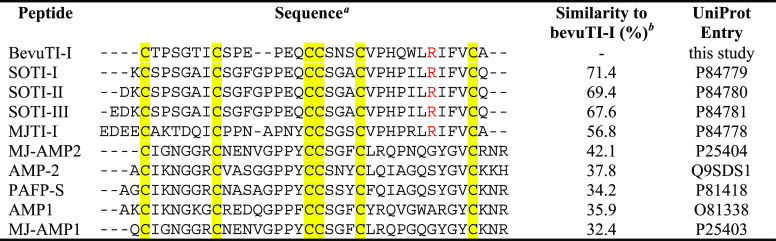
Elucidation of the amino acid sequence via mass spectrometry analysis led to performing database searches for the identification of peptides with sequence homology to bevuTI-I. The mature sequence of bevuTI-I was used as the query in a blastp search via UniProt’s blast interface. The search revealed considerable sequence homologies between the novel beetroot peptide and trypsin inhibitors from spinach (Spinacia oleracea trypsin inhibitor, SOTI-I, -II, and -III) as well as the four o’clock plant (M. jalapa) (Table 1). M. jalapa trypsin inhibitors have hitherto been characterized in two studies. Kowalska et al. have initially described S. oleracea trypsin inhibitor as a subfamily of knottin trypsin inhibitors with a unique cystine knot configuration and considerable sequence variation to many other peptide-based TIs of this class.18 In another study, crystallographic characterization of SOTI-III in complex with trypsin revealed that 12 hydrogen bonds participate in the interaction between the peptide and trypsin.19 Interestingly, eight interacting hydrogen bonds are formed by the arginine at position 32, and the importance of arginine in loop five was also noted earlier.18 The trypsin inhibitory activity was lost after modification of this arginine moiety by 1,2-cyclohexanedione.18 Peptides with a similar cysteine pattern, but without the arginine residue in loop five, have been isolated from M. jalapa,20Phytolaccaamericana,21,22 and Mesembryanthemum crystallinum. The high similarity of bevuTI-I to S. oleracea and M. jalapa trypsin inhibitors enabled the assignment of the amino acids at position 7 and 27 to isoleucine and at position 29 to leucine (Table 1). This agrees with similarity of bevuTI-I and other Beta spp. genome/transcriptome sequences (Figure S5, Supporting Information) and amino acid analysis of bevuTI-I (Table S5, Supporting Information).
Table 1. Alignment of BevuTI-I with Homologous Trypsin Inhibitors and Other Similar Plant Peptides.
Alignment of bevuTI-I with trypsin inhibitors from S. oleracea trypsin inhibitor I–III (SOTI I–III),18M. jalapa trypsin inhibitor I (MJTI-I)18 and other identified peptides with similarity, i.e., M. jalapa antimicrobial peptide 1–2 (MJ-AMP 1–2),20 , antimicrobial peptide 2 (AMP-2) from Phytolacca americana,21 antimicrobial peptide 1 (AMP1) from M. crystallinum, and antifungal protein 1 from P. americana seeds (PAFP-S).22 The arginine residue in loop five (responsible for trypsin inhibition) is colored in red. Cysteines are highlighted in yellow. Gaps were introduced to maximize the quality of the alignment. The cysteine connectivity CI–IV, CII–V, and CIII–VI is based on the structure of SOTI-III (PDB: 4AOR).18,19
Similarity was calculated based on a global alignment using the Needleman–Wunsch algorithm (matrix: BLOSUM62, gap open penalty: 10, gap extend penalty: 0.5, end gap open penalty: 10, end gap extend penalty: 0.5).
Based on the availability of few published homologous bevuTI-I peptides (Table 1), it was determined whether there are novel peptide sequences to be discovered based on the bevuTI-I sequence. Mining of transcriptome databases such as the 1-kp project or publicly available databases provided by NCBI is an emerging field for discovery and analysis of the phylogenetic distribution, sequence diversity, and precursor structure of peptides and proteins.23 The three most widely used technologies for this task are BLAST-based algorithms, hidden Markov model-based algorithms, and regular expression-based algorithms. As noted by Silverstein et al., cysteine-rich host-defense peptides are often underpredicted by automated annotation programs.24 Hence it is necessary to design custom computer scripts to obtain better information about a specific peptide family. In this study the programming language Python was used, which is well suited for this task, as it is widely used in the bioinformatics community. Other programs such as the blast+ package or HMMER can easily be called within a Python script.25 Triggered by the discovery and sequence determination of the beetroot peptide bevuTI-I, the analysis of 2433 transcriptomes and 664 genomes (provided by the 1-kp project26 and found on NCBI’s ftp-server on April 30, 2020) provided new insights into the sequence diversity, phylogenetic distribution, and precursor structure of the M. jalapa trypsin inhibitor peptide family.
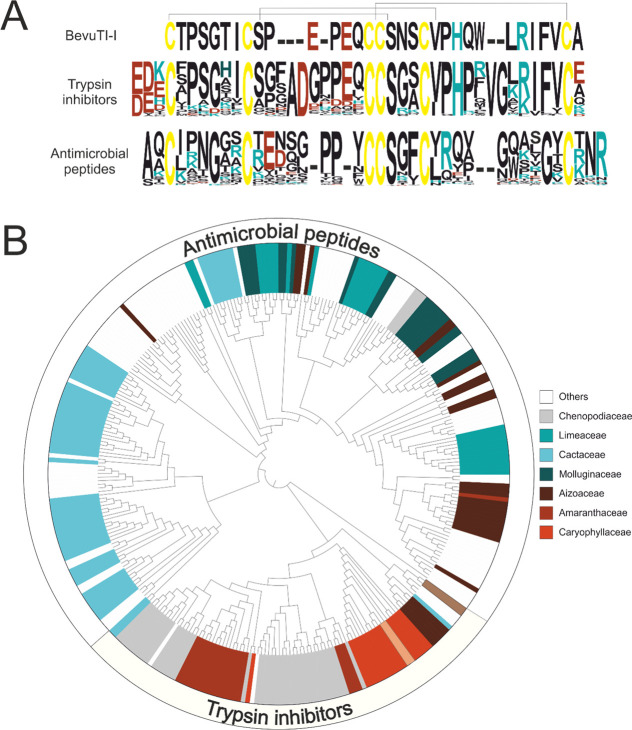
The sequences shown in Table 1 were selected as queries in blastp searches against six-frame translated genomes/transcriptomes. The search yielded 1147 hits with E-values below or equal to 10–4. Many of the hits were redundant, as they were found by several query sequences. A meta-analysis of the hits revealed that hits that were found when a trypsin inhibitor sequence was used as a query (SOTI-I, SOTI-II, SOTI-III, MJTI-I, bevuTI-I) were not found when an antimicrobial peptide (AMP-2, MJ-AMP1, PAFP-S, MJ-AMP2, AMP1) was used and vice versa (Data S1, Supporting Information). Therefore, it was hypothesized that M. jalapa cystine knot peptides can be divided into two branches: a trypsin inhibitor and an antimicrobial peptide branch. In total, 99 unique contigs belonging to the trypsin inhibitor branch and 221 unique contigs belonging to the antimicrobial peptide branch were identified. Nine contigs of the trypsin inhibitor branch and nine contigs of the antimicrobial peptide branch contained at least one ambiguous amino acid residue and were therefore not considered for further analysis. Of the identified trypsin inhibitor-like sequences, 60 (67%) contain an arginine residue in the second half of loop five and have therefore presumably trypsin inhibition activity (Data S2, Supporting Information). In total these 320 unique contigs yielded the annotation of 243 novel putative mature peptides. Only 40 sequences (18%) from the antimicrobial peptide branch contain an arginine residue in the second half of loop five. This makes sense since antimicrobial activity is less dependent on a single residue, but appears to be mainly affected by the ratio of hydrophobic vs charged residues.27 From both M. jalapa cystine knot peptide branches a sequence logo was prepared and aligned to bevuTI-I (Figure 3A). To examine whether bevuTI-I is a member of the trypsin inhibitor branch, every sequence of both branches was pairwise aligned to bevuTI-I. The comparison of bevuTI-I to the trypsin inhibitor branch yielded a score of 112.2 ± 14.7 and a score of 39.5 ± 6.8 when compared to the antimicrobial branch. Peptides belonging to the trypsin inhibitor branch and antimicrobial peptide branch have a calculated isoelectric point of 6.7 ± 1.4 and 8.0 ± 1.3, respectively. The hit with highest similarity to bevuTI-I was found in the Beta patula genome28 (GenBank ID: VASJ01001109.1). The only difference from bevuTI-I is an insertion of an alanine residue at position 11.
Figure 3.
Genome mining of bevuTI-I-like peptides. (A) Pairwise alignment of the bevuTI-I sequence with a frequency plot of the predicted mature sequence of trypsin inhibitor-like and antimicrobial-like peptides found by a blastp search. Cysteines are colored in yellow, amino acids with positive charged side chains (at pH 7) are colored in red (H, K, R), and those with negative charged side chains (at pH 7) are colored in cyan (D, E). Gaps were introduced to maximize the alignment. The cysteine connectivity is based on PDB 4AOR(19) and Kowalska et al.18 (B) Phylogenetic maximum likelihood tree of unique hits found by a blastp search.
To underline the power of this methodology, it was analyzed how many of the predicted major peptide domains occur in sequences of the UniProt database. Ten UniProt entries were identified, which are classified either as trypsin inhibitor domain containing protein (A0A0K9R0K7, A0A0K9R4D4, A0A0K9QYI6, A0A0K9QXT1, A0A0K9R2C8, A0A0K9R400) or as trypsin inhibitor (P84779, P84780, P84781), or antimicrobial seed protein (Q9SDS1). To obtain further confidence in the expression of the genome-derived sequences, the structure of the putative precursor peptides was analyzed with SignalP. It was possible to identify 145 signal peptides of the antimicrobial peptide branch and 39 signal peptides of the trypsin inhibitor branch (Data S2, Supporting Information). Interestingly, peptides belonging to the antimicrobial inhibitor branch have a signal sequence with an average length of 26 ± 5 amino acids followed by the mature domain without an N-terminal prodomain (except six of the sequences analyzed), whereas peptides belonging to the trypsin inhibitor branch have a signal sequence with an average length of 22 ± 3 amino acids followed by an N-terminal prodomain with an average length of 44 ± 16 amino acids followed by the mature domain.
To provide a better understanding on how the M. jalapa trypsin inhibitor family of cystine knot peptides is distributed within the plant kingdom, we constructed a maximum-likelihood phylogenetic tree (Figure 3B). Prior to the construction of the tree the precursor sequences, the mature peptide domains, and the C-terminal tail domains were separately aligned, and the alignments were merged. Peptides of the trypsin inhibitor branch were found in eight different plant families, of which seven (Amaranthaceae, Nyctaginaceae, Caryophyllaceae, Phytolaccaceae, Cactineae, Chenopodiaceae, Aizoaceae) belong to the order Caryophyllales. One contig was found in the species Arachis hypogaea (GenBank ID: GAER01017494.1) belonging to the family Fabaceae (Fabales), but since this contig contains no signal sequence, the expression of this peptide remains doubtful. A different picture arises from the analysis of the antimicrobial peptide branch: they were identified in 39 different plant families belonging to 17 different orders. Novel trypsin inhibitor/antimicrobial peptides were identified in 42 plant families. Although most peptides (184; 82%) were identified in the order Caryophyllales, one has to bear in mind that the genomes/transcriptomes of 118 species belonging to this order are available, but, for instance, only two genomes/transcriptomes of the order Canellales— where also one hit was identified—are available. Therefore, it is reasonable to hypothesize that the distribution of antimicrobial-type M. jalapa cystine knot peptides is much greater than for the trypsin inhibitor type, and possibly many yet-unknown sequences will be found when data of large sequencing programs like the 10-kp project become publicly available.29
As noted in at least two earlier studies19,30 similar plant cystine knot peptides can also be found outside the plant kingdom. Therefore, a blast search using bevuTI-I as query against a transcriptome database of species belonging to the phylum Arthropoda was performed, and indeed a novel peptide with homology to M. jalapa trypsin inhibitor peptides (data not shown) was identified, which should provide motivation for future studies to explore the evolutionary link between plant and animal “knottins”.
Synthesis and Protease Inhibition Activity of BevuTI-I
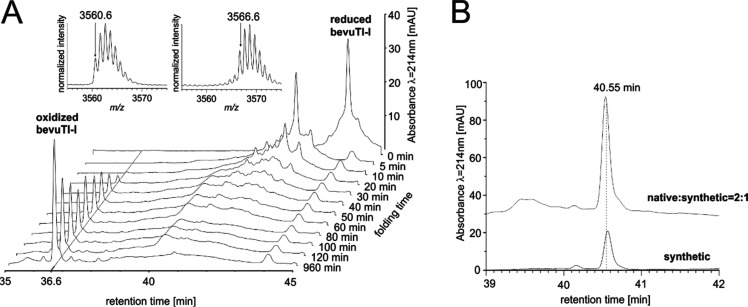
Following its detailed bioinformatics analysis, we aimed at chemical synthesis of bevuTI-I to confirm its endogenous structure and obtain enough material for pharmacological assays. The peptide was synthesized in a 0.1 mmol scale using fluorenylmethoxycarbonyl solid phase peptide synthesis (Fmoc-SPPS)31 and oxidative folding according to previously published protocols.32 Based on analytical HPLC data (Figure 4A) the folding half-time was determined to be 38 min (Figure S6, Supporting Information). During the folding reaction, linear bevuTI-I adopted its native tertiary structure within 24 h. For confirmation, analytical HPLC analysis was performed that demonstrated that the peptide peak coeluted with bevuTI-I isolated from beetroot juice (Figure 4B).
Figure 4.
Oxidative folding of bevuTI-I. (A) The folding was initiated by addition of oxidation buffer and thiol shuffling reagents at a peptide concentration of 0.5 mg/mL. The reaction was stopped with 10% TFA at different time points, and analytical HPLC spectra (bottom) and MS spectra (top) were recorded. A loss of 6 Da for the monoisotopic mass signal was observed after 16 h of folding. (B) In a coelution experiment of native and synthetic bevuTI-I (2:1 ratio; upper chromatogram; offset: 35 mAU) a single peak was observed at 40.6 min indicating the identity of native and synthetic bevuTI-I (bottom chromatogram).
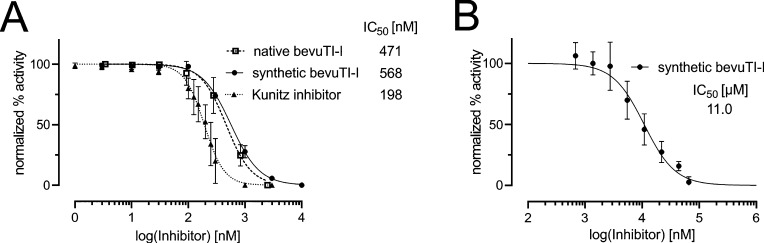
Based on the classification of bevuTI-I as putative trypsin inhibitor of the M. jalapa trypsin inhibitor family, enzymatic inhibition assays were performed.33 Bovine β-trypsin and bevuTI-I were used in a colorimetric trypsin inhibition assay to measure concentration-dependent inhibition. The half-maximal inhibition concentration (IC50) values of bevuTI-I isolated from beetroot juice and synthesized bevuTI-I were both within the range of error similar to each other, thus confirming the native fold of synthetic bevuTI-I. BevuTI-I exhibited moderate trypsin inhibition activity with an IC50 of 471 ± 160 nM (native) and 568 ± 36 nM (synthetic) (Figure 5). In comparison, the equilibrium association constants for S. oleracea-type inhibitors were reported to be 536 and 185 nM for M. jalapa TI-I and SOTI-I, respectively.18 The IC50 of bevuTI-I was slightly higher than that of the well-characterized Kunitz-type soybean trypsin inhibitor (IC50 = 198 ± 72 nM).
Figure 5.
Protease inhibition activity of bevuTI-I. (A) Concentration–response curves of bevuTI-I isolated from beetroots (native) and chemically synthesized (synthetic bevuTI-I) and Glycine max Kunitz-type trypsin inhibitor (as control) in a trypsin inhibitory assay. (B) Concentration–response data were generated by incubating various concentrations of synthetic bevuTI-I with human POP (50 ng). Data were fitted with a three-parameter logistic regression model. Data of the POP inhibition assay are presented as mean ± SD of three biological replicates.
As it was previously reported, plant cystine knot peptides belonging to the cyclotide family have the ability to inhibit human prolyl endopeptidase.33 Thus, we were interested whether the bevuTI-I was also able to inhibit this enzyme. Like psysol-2, bevuTI-I has three proline residues and presumably the same cysteine connectivity. The IC50 of bevuTI-I (IC50 = 11 ± 5.3 μM) is slightly lower than the reported value of psysol-2 (IC50 ≈ 25 μM).33 Hence, the most striking result to emerge from our data is that bevuTI-I is the first M. jalapa TI-type peptide with inhibitory activity toward human POP reported to date.
Cysteine-rich plant peptides, especially those comprising a cystine knot motif, are interesting scaffolds for the design of peptide-based therapeutics and have been successfully used in an ongoing clinical trial against the autoimmune disease multiple sclerosis.6 Beside their immune modulatory activity,34 intrinsic enzyme inhibitory activity (e.g., α-amylase inhibition, trypsin or chymotrypsin inhibition) might be another beneficial property for future drug development. The discovery of plant protease inhibitors in the field of drug development was hindered in the last decades since many identified molecules were only characterized toward a limited set of proteases.12 POP is an enzyme that cleaves peptides at the C-terminal side of proline residues. POP inhibitors have potential as drug candidates for the treatment of neurodegenerative disorders and inflammatory conditions.35,36 Although cystine knot peptides generally do not cross the blood brain barrier, attachment or incorporation of shuttling molecules such as mini-apamin might be a solution to overcome this obstacle.37 In this study, a peptide, namely, bevuTI-I, presumably containing a cystine knot motif (which will have to be confirmed by future NMR studies), was identified as a POP inhibitor. This discovery may spark the discovery of bevuTI-I or other M. jalapa trypsin inhibitor-based POP inhibitors.
In conclusion, we identified a novel trypsin inhibitor peptide, bevuTI-I, from beetroot. Genome mining combined with sequence analysis indicated that this beetroot peptide belongs to the M. jalapa trypsin inhibitor family. BevuTI-I not only inhibits the prototypic serine protease trypsin but is the first reported POP inhibitor from this family of peptides. In fact, the presented study provides a detailed description of this knottin peptide family, which comprises at least 243 unique sequences. Therefore, the new sequences may be the starting point to isolate and characterize further POP inhibitors. Overall, our study highlights great potential of plant protease inhibitors isolated from beetroot and other related plant species as a valuable source for peptide-based drug discovery for targets involved in diseases such as cancer, neurodegenerative disorders, and immune system related diseases.12
Experimental Section
General Experimental Procedures
MALDI-TOF MS analysis was performed on an ABSciex 4800 MALDI-TOF/TOF system (Applied Biosystems, Framingham, MA, USA). MS and MS/MS spectra were recorded in positive ion reflector mode, and ∼5000–10 000 laser shots per spectrum were acquired. The mass spectrometer was calibrated using the PepMix 4 from Laser Biolabs (Valbonne, France) before data acquisition. MS data were converted from the .t2d to the .txt format by a Python script and analyzed with the software “mmass”, version 5.5.0,38 or OriginPro, version 2018b (OriginLab Corporation, Northampton, MA, USA), or DataExplorer Software, version 4.9 (Applied Biosystems). For sample preparation 3 μL of α-cyano-4-hydroxycinnamic acid matrix (Sigma-Aldrich, Vienna, Austria), dissolved in 50% MeCN and 0.1% TFA (v/v) in ddH2O, was mixed with 0.5 μL of peptide solution. The mixed samples (0.5 μL) were spotted on a standard 384 target plate and incubated in the dark to air-dry. RP-HPLC was performed on a Dionex Ultimate 3000 system (Dionex, Sunnyvale, CA, USA) equipped with an HPG-3200SD standard binary pump, a TCC-3000SD column department, a WPS-3000TSL analytical autosampler, an AFC-3000 fraction collector, and a UV detector at a variable wavelength of 214, 254, and 280 nm. Kromasil C18 columns with a pore size of 300 Å, particle size of 10/5 μm, and dimensions of 250 × 21.2 mm, 250 × 10 mm, and 250 × 4.6 mm were used for preparative, semipreparative, and analytical RP-HPLC, respectively. For analytical runs the column department temperature was set to 30 °C. The polar solvent (here referred to as solvent A) consisted of ddH2O with 0.1% TFA (v/v), and the composition of the nonpolar solvent (here referred to as solvent B) was 90% MeCN, 10% ddH2O, and 0.1% TFA (v/v/v). For preparative/semipreparative/analytical HPLC runs the flow rate was set to 8 mL/4 mL/1 mL and a 60 min 1%/min gradient was applied.
Plant Material
For peptide identification as well as de novo sequencing experiments Beta vulgaris (pulp and leaves) was collected at the organic garden “Gärtnerhof Gin” located in the 22nd district (Hänischgasse 12) of Vienna at the end of August 2017. Plants were grown from a breeding line named “Rote Kugel” purchased from the organic seed company “ReinSaat” (ReinSaat, Leonhard am Hornerwald, Austria). For large-scale extraction beetroot juice produced by “Naturfrucht GmbH”, A-2120 Wolkersdorf for the brand “Ja Natürlich”, was purchased in a local Austrian supermarket.
Chemical Extraction of Beetroot Plant Material
After the field collection of B. vulgaris, beets were washed with water, peeled, cut into slices of about 10 mm in diameter, and dried at 35 °C for 2 weeks. Leaves were dried at 35 °C for 2 weeks. The semidry plant material was chopped with a kitchen grinder and extracted with a 1:1 (v/v) mixture of CH2Cl2 and MeOH (about 200 g of plant material per L of solvent) for 12 h under continuous agitation at 25 °C. The suspension was filtered, 0.5 volume of ddH2O was added, and the peptide-enriched aqueous MeOH phase was obtained by liquid/liquid phase separation. After evaporation of the organic solvent the remaining aqueous phase, referred to as crude extract, was lyophilized.
Chemical Extraction of Beetroot Juice
Beetroot juice was lyophilized and dissolved in cold water. To precipitate proteins and polysaccharides, an equal amount (v/v) of cold acetone (−20 °C) was added. After stirring for 30 min with an overhead stirrer, the peptide of interest (bevuTI-I) was found in the supernatant, and the dark red viscous bottom layer was discarded. The acetone was evaporated, and the remaining water was removed by lyophilization, yielding a crude extract.
Screening for Cysteine-Rich Peptides in Crude Extracts
To explore the peptidome of B. vulgaris with respect to disulfide-rich peptides, a chemical derivatization approach to observe characteristic mass shifts was used. The crude extract was dissolved in 0.1 M NH4HCO3 buffer (pH 8.5), and cysteine-rich peptides were reduced in 10 mM DTT (Sigma-Aldrich) for 30 min at 60 °C and moderate agitation. Carbamidomethylation was conducted with a 50 mM final concentration of iodoacetamide (Sigma-Aldrich) for 10 min at 25 °C while shaking at 600 rpm. The reaction was terminated by addition of 1 μL of concentrated formic acid and directly afterward the sample was analyzed by MALDI-MS.
Preparation of Peptide-Enriched Fractions
The crude extracts were dissolved in 10% MeOH/90% ddH2O/0.1% TFA (v/v/v) and loaded onto 40–63 μm ZEOprep C18 material (Zeochem AG, Rüti, Switzerland) that had been preactivated with MeOH and equilibrated with 100% ddH2O/0.1% TFA (v/v). After washing with solvent A, a 10% stepwise elution gradient from 10% to 80% solvent B was applied. All fractions were examined by MALDI-MS, and those containing bevuTI-I were used for isolation by preparative and semipreparative HPLC.
Isolation of BevuTI-I for de Novo Sequencing
To isolate bevuTI-I from peptide-enriched pulp fractions, an automatic preparative HPLC separation was performed (linear 60 min gradient segment from 20% to 80% solvent B, flow rate 8 mL/min, collection period: 1 fraction/10 min). Fractions containing bevuTI-I (retention time between 35 and 30 min) were pooled and lyophilized. A subsequent semipreparative HPLC was carried out to achieve higher chromatographic peak resolution (linear 120 min gradient segment from 20% to 80% solvent B, flow rate 4 mL/min, manual collection). To isolate bevuTI-I from peptide-enriched fractions obtained by acetone precipitation, a preparative HPLC followed by two runs of semipreparative HPLC was performed. Fractions were collected manually, and the gradient was adjusted during the HPLC run based on the chromatogram. The extinction coefficient at 280 nm of bevuTI-I was calculated as 6050 M–1 cm–1 based on the sum of the molar extinction coefficients of tryptophan (5690 M–1 cm–1) and three cystines (120 M–1 cm–1 each).
De Novo Sequencing and Synthesis of BevuTI-I
The purified peptide was reduced, carbamidomethylated, and proteolytically cleaved by the enzymes (Sigma-Aldrich) trypsin, chymotrypsin, and endoproteinase GluC in separate experiments. The sequence of bevuTI-I was elucidated by assigning y- and b-ions in the fragmentation spectra of the proteolytic fragments and the full-length peptide. Afterward, the linear peptide (NH2-CTPSGTICSPEAPEQCCSNSCVPHQWLRIFVCA-COOH) was automatically synthesized using fluorenylmethoxycarbonyl (Fmoc)-based solid phase peptide synthesis on an Fmoc-Ala-Tenta-Gel PHB resin (90 μm, 0.19 mmol/g (Rapp Polymere, Tübingen, Germany)) and a CEM Liberty Blue synthesizer (CEM, NC, USA) in a 0.1 mmol scale. The peptide chain was elongated by double coupling amino acids (10-fold excess) using 4 min microwave cycles. Couplings were performed with dicyclohexylcarbodiimide and ethyl cyanohoydroxyiminoacetate (Oxyma), respectively. The protecting group was cleaved off with 20% piperidine in DMF. The full-length peptide was cleaved off the resin and fully deprotected with 150 μL of cleavage cocktail composed of TFA/triisopropylsilane/water/1,2-ethanedithiol (92.5:2.5:2.5:2.5% (v/v/v/v)) per mg empty resin at room temperature on a rotating wheel for 2 h. TFA was removed under a flow of nitrogen, and the peptide was precipitated in ice-cold Et2O. The solid peptide was dissolved in 50% MeCN/50% ddH2O/0.01% TFA (v/v/v) and immediately treated with 20 mM tris(2-carboxyethyl)phosphine at room temperature for 2 h. The freeze-dried peptide material was analyzed with HPLC and MS, confirming the mass of expected linear reduced peptide with an m/z of 3566.5. This crude peptide (200 mg) was purified by HPLC, yielding approximately 40 mg of linear bevuTI-I.
Oxidative Folding of BevuTI-I
To form the native cystine knot, bevuTI-I was dissolved in diluted HOAc (pH 4) and was mixed with folding buffer consisting of 0.1 M NH4HCO3 (pH 8.4)/isopropanol, 1:1 (v/v), supplemented with the thiol shuffling reagents reduced (2 mM) and oxidized (0.5 mM) glutathione at a concentration of 0.5 mg/mL. To monitor the folding process, aliquots were collected at several time points, acidified with 10% TFA, and analyzed by MALDI-MS and HPLC to ensure cysteine oxidation and formation of native bevuTI-I and to determine the folding kinetics.
Trypsin Inhibition Assays
The trypsin inhibition assay was performed in 96-well microtiter plates. To 140 μL of a 20 mM Tris-HCl buffer (pH 8.0) was added 20 μL of inhibitor (dissolved in buffer)/buffer (negative control). Afterward, 20 μL of bovine trypsin (0.125 mg/mL, Sigma-Aldrich) was added, and the plate was incubated for 10 min at 22 °C. A volume of 20 μL of benzoylarginine-4-nitroanilide hydroxychloride (Bachem, Bubendorf, Switzerland) dissolved in 20% DMSO at a concentration of 900 μM was added automatically with the dispenser of the Synergy H4 Hybrid reader (BioTek, VM, USA). The rate of enzymatic reaction with different concentrations of inhibitor was measured for 1 h every 5 min at a wavelength of 410 nm. For calculations, the linear portion of data, corresponding to the initial rate of reaction, was used. The highest rate was set to 100% activity. A nonlinear regression (log inhibitor vs normalized response) was performed with GraphPad Prism version 8.0.0 for Windows (GraphPad Software, CA, USA). The top and bottom parameters were set to 100% and 0%, respectively.
Prolyl Oligopeptidase Inhibition Assays
Recombinant human POP (NM_002726.4) was produced in E. coli BL21 (New England Biolabs, MA, USA). The full-length cDNA was cloned into the BamHI and EcoRI cloning sites of the commercially available expression plasmid pGEX-6P1 by a customized manufacturer service from GenScript (Leiden, The Netherlands). Cell culture was prepared in Lennox broth, and protein expression was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside at 23 °C for 6 h. The cells were pelleted by centrifugation and dissolved in 50 mM phosphate buffer at pH 8.2, including 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1-fold protease inhibitor complete cocktail (Roche, Basel, Switzerland). Cell lysis was performed with freeze–thaw cycles and with three-times pulsed ultrasonic bar treatments. The soluble protein fraction was incubated with GSH-sepharose (Serva GmbH, Heidelberg, Germany), and nonspecific protein was washed off twice with washing buffer (lysis buffer containing 20 and 50 mM imidazole). The free POP protein was obtained by site-specific cleavage using PreScission protease, fusion protein of human rhinovirus 3C protease, and glutathione-S-transferase. The protein concentration of the eluate was determined by the bicinchoninic acid assay (ThermoFisher Scientific), and aliquots of stock protease in storage buffer (including 20% glycerol) were kept at −80 °C. POP inhibition assays were performed similarly to those described previously.33 The substrate was dissolved as a 10-fold stock solution in an assay buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 0.5 mM DTT) including 20% (v/v) isopropanol and final concentration of 45 μM. The inhibition experiments were performed in 96 black wells in a total volume of 200 μL. Enzyme activity was measured by fluorescence signal as a result of free fluorophore from the Z-Gly-Pro-AMC substrate with an excitation of 360 ± 20 nm and emission of 450 ± 20 nm. Kinetic inhibition data were recorded in 5 min intervals, and the amount of inhibition was calculated from the steepness of the linear initial velocity. A substrate control was used for background correction, and the maximum protease activity was defined as POP activity in assay buffer without inhibitors. Concentration–response experiments were evaluated with a fitted nonlinear regression function in GraphPad Prism.
Mining and Analysis of Genomic Data
Viridiplantae genomes and transcriptomes were retrieved from the NCBI’s FTP-server, either the representative or latest version available (Data S3–S4, Supporting Information). Additional Viridiplantae transcriptomes were retrieved from the 1-kp project39 (Data S5, Supporting Information). All genetic data were six-frame translated using Biopython25 before further analysis was conducted. BLAST databases were constructed and searches were done with NCBI’s blast+ package version 2.9.0.40 Sequence logos were made with the online tool WebLogo 3,41 alignments were made with MUSCLE v3.8.31,42 and a phylogenetic maximum-likelihood tree was computed with the software MEGAX version 10.1.743 and visualized with iTOL version 5.5.44 For pairwise alignment the following settings were used: global alignment, gap open penalty 10, gap extend penalty −0.5, substitution matrix blosum62. The cleavage position of the signal peptide and the N-terminal pro-peptide was predicted with the software SignalP version 5.0.45 At several stages of data processing and filtering, customized self-written Python (Python 3.6) scripts were used to facilitate automatic analysis of data in the described working steps.
Acknowledgments
We thank M. Felkl, M. Muttenthaler, and C. Becker (Institute for Biological Chemistry, University of Vienna) for help and access to the peptide synthesis facility. In addition, we thank D. Kümmel (WWU Münster, Germany) for the gift of the PreScission protease expression plasmid.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jnatprod.0c00648.
Author Contributions
C.W.G. designed the research. B.R., R.H., E.M., and M.E.F.P. performed experiments. C.W.G., B.R., R.H., E.M., M.E.F.P., and V.S.B. analyzed the data. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This project has been supported by the Austrian Science Fund (FWF) through project P32109. M.E.F.P. was supported by São Paulo Research Foundation (FAPESP) grant #2019/04381-5 and CAPES grant BEX #9875/11-5.
The authors declare no competing financial interest.
Supplementary Material
References
- Campos M. L.; de Souza C. M.; de Oliveira K. B. S.; Dias S. C.; Franco O. L. J. J. Exp. Bot. 2018, 69, 4997–5011. 10.1093/jxb/ery294. [DOI] [PubMed] [Google Scholar]
- Tam J. P.; Wang S.; Wong K. H.; Tan W. L. Pharmaceuticals 2015, 8, 711–57. 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald N. Q.; Hendrickson W. A. Cell 1993, 73, 421–4. 10.1016/0092-8674(93)90127-C. [DOI] [PubMed] [Google Scholar]
- Pallaghy P. K.; Nielsen K. J.; Craik D. J.; Norton R. S. Protein Sci. 1994, 3, 1833–9. 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J.; Daly N. L.; Waine C. Toxicon 2001, 39, 43–60. 10.1016/S0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Thell K.; Hellinger R.; Sahin E.; Michenthaler P.; Gold-Binder M.; Haider T.; Kuttke M.; Liutkeviciute Z.; Goransson U.; Grundemann C.; Schabbauer G.; Gruber C. W. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 3960–5. 10.1073/pnas.1519960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgrave M. L.; Craik D. J. Biochemistry 2004, 43, 5965–75. 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- Bakhle Y. S. Br. J. Pharmacol. 2020, 177, 2657–65. 10.1111/bph.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J. L.; Dunn M. K. Bioorg. Med. Chem. 2018, 26, 2700–7. 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Pennington M. W.; Czerwinski A.; Norton R. S. Bioorg. Med. Chem. 2018, 26, 2738–58. 10.1016/j.bmc.2017.09.029. [DOI] [PubMed] [Google Scholar]
- Clemente M.; Corigliano M. G.; Pariani S. A.; Sanchez-Lopez E. F.; Sander V. A.; Ramos-Duarte V. A.. Int. J. Mol. Sci. 2019, 20, 1345. 10.3390/ijms20061345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinger R.; Gruber C. W. Drug Discovery Today 2019, 24, 1877–89. 10.1016/j.drudis.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. K.; Nielsen J. E.; Madrid S. M.; Mikkelsen J. D. Plant Physiol. 1997, 113, 83–91. 10.1104/pp.113.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catusse J.; Strub J.-M.; Job C.; Dorsselaer A.; Job D. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10262–7. 10.1073/pnas.0800585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehbach J.; Attah A. F.; Berger A.; Hellinger R.; Kutchan T. M.; Carpenter E. J.; Rolf M.; Sonibare M. A.; Moody J. O.; Wong G. K.; Dessein S.; Greger H.; Gruber C. W. Biopolymers 2013, 100, 438–52. 10.1002/bip.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Agriculture Survey (FNDDS) FDC ID: 787777. https://fdc.nal.usda.gov/fdc-app.html#/food-details/169145/nutrients.
- Hashempour H.; Koehbach J.; Daly N. L.; Ghassempour A.; Gruber C. W. Amino Acids 2013, 44, 581–95. 10.1007/s00726-012-1376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska J.; Pszczola K.; Wilimowska-Pelc A.; Lorenc-Kubis I.; Zuziak E.; Lugowski M.; Legowska A.; Kwiatkowska A.; Sleszynska M.; Lesner A.; Walewska A.; Zablotna E.; Rolka K.; Wilusz T. Phytochemistry 2007, 68, 1487–1496. 10.1016/j.phytochem.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Glotzbach B.; Schmelz S.; Reinwarth M.; Christmann A.; Heinz D. W.; Kolmar H. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 114–20. 10.1107/S0907444912043880. [DOI] [PubMed] [Google Scholar]
- Cammue B. P.; De Bolle M. F.; Terras F. R.; Proost P.; Van Damme J.; Rees S. B.; Vanderleyden J.; Broekaert W. F. J. Biol. Chem. 1992, 267, 2228–2233. [PubMed] [Google Scholar]
- Liu Y.; Luo J.; Xu C.; Ren F.; Peng C.; Wu G.; Zhao J. Plant Physiol. 2000, 122, 1015–24. 10.1104/pp.122.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G. H.; Liu W.; Dai J. X.; Wang J. F.; Hu Z.; Zhang Y.; Wang D. C. Biochemistry 2001, 40, 10973–8. 10.1021/bi010167k. [DOI] [PubMed] [Google Scholar]
- Shelenkov A.; Slavokhotova A.; Odintsova T. Antibiotics 2020, 9, 9. 10.3390/antibiotics9020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein K. A.; Moskal W. A. Jr.; Wu H. C.; Underwood B. A.; Graham M. A.; Town C. D.; VandenBosch K. A. Plant J. 2007, 51, 262–80. 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Cock P. J.; Antao T.; Chang J. T.; Chapman B. A.; Cox C. J.; Dalke A.; Friedberg I.; Hamelryck T.; Kauff F.; Wilczynski B.; de Hoon M. J. Bioinformatics 2009, 25, 1422–3. 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter E. J.; Matasci N.; Ayyampalayam S.; Wu S.; Sun J.; Yu J.; Jimenez Vieira F. R.; Bowler C.; Dorrell R. G.; Gitzendanner M. A.; Li L.; Du W.; K K. U.; Wickett N. J.; Barkmann T. J.; Barker M. S.; Leebens-Mack J. H.; Wong G. K. GigaScience 2019, 8, 1–7. 10.1093/gigascience/giz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehbach J.; Craik D. J. Trends Pharmacol. Sci. 2019, 40, 517–28. 10.1016/j.tips.2019.04.012. [DOI] [PubMed] [Google Scholar]
- Rodriguez Del Rio A.; Minoche A. E.; Zwickl N. F.; Friedrich A.; Liedtke S.; Schmidt T.; Himmelbauer H.; Dohm J. C. Plant J. 2019, 99, 1242–53. 10.1111/tpj.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.; Melkonian M.; Smith S. A.; Brockington S.; Archibald J. M.; Delaux P.-M.; Li F.-W.; Melkonian B.; Mavrodiev E. V.; Sun W.; Fu Y.; Yang H.; Soltis D. E.; Graham S. W.; Soltis P. S.; Liu X.; Xu X.; Wong G. K.-S.. GigaScience 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboye T. L.; Strömstedt A. A.; Gunasekera S.; Bruhn J. G.; El-Seedi H.; Rosengren K. J.; Göransson U. ChemBioChem 2015, 16, 1068–77. 10.1002/cbic.201402704. [DOI] [PubMed] [Google Scholar]
- Hansen P. R.; Oddo A. Methods Mol. Biol. 2015, 1348, 33–50. 10.1007/978-1-4939-2999-3_5. [DOI] [PubMed] [Google Scholar]
- Wong C. T.; Taichi M.; Nishio H.; Nishiuchi Y.; Tam J. P. Biochemistry 2011, 50, 7275–83. 10.1021/bi2007004. [DOI] [PubMed] [Google Scholar]
- Hellinger R.; Koehbach J.; Puigpinos A.; Clark R. J.; Tarrago T.; Giralt E.; Gruber C. W. J. J. Nat. Prod. 2015, 78, 1073–82. 10.1021/np501061t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thell K.; Hellinger R.; Schabbauer G.; Gruber C. W. Drug Discovery Today 2014, 19, 645–53. 10.1016/j.drudis.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarcbahs R.; Julku U.; Kilpelainen T.; Kyyro M.; Jantti M.; Myohanen T. T. Biochem. Pharmacol. 2019, 161, 113–20. 10.1016/j.bcp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Waumans Y.; Baerts L.; Kehoe K.; Lambeir A. M.; De Meester I. Front. Immunol. 2015, 6, 387. 10.3389/fimmu.2015.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster C.; Varese M.; Garcia J.; Giralt E.; Sanchez-Navarro M.; Teixido M. J. Pept. Sci. 2019, 25, e3172. 10.1002/psc.3172. [DOI] [PubMed] [Google Scholar]
- Niedermeyer T. H.; Strohalm M. PLoS One 2012, 7, e44913. 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J. H.; Barker M. S.; Carpenter E. J.; et al. Nature 2019, 574, 679–685. 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C.; Coulouris G.; Avagyan V.; Ma N.; Papadopoulos J.; Bealer K.; Madden T. L. BMC Bioinf. 2009, 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E.; Hon G.; Chandonia J. M.; Brenner S. E. Genome Res. 2004, 14, 1188–90. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Nucleic Acids Res. 2004, 32, 1792–7. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Li M.; Knyaz C.; Tamura K. Mol. Biol. Evol. 2018, 35, 1547–9. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Nucleic Acids Res. 2011, 39, W475–8. 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros J. J.; Tsirigos K. D.; Sonderby C. K.; Petersen T. N.; Winther O.; Brunak S.; von Heijne G.; Nielsen H. Nat. Biotechnol. 2019, 37, 420–3. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.